Abstract
The incidence of astrovirus infection was determined among infants and young children admitted to Alice Springs Hospital (central Australia) with gastroenteritis from 1995 to 1998. Astrovirus was detected by reverse transcription-PCR in 33 of 495 stool samples, and this represented 4.3% of a total of 774 stool samples tested for astrovirus, rotavirus, and Norwalk-like viruses. Astrovirus incidence was substantially lower than that of rotavirus but higher than that of Norwalk-like viruses both overall and in each of the 4 years individually. Over the period from 1981 to 1998, including the period from 1981 to 1994 during which astrovirus was identified only by electron microscopy, astrovirus serotypes (deduced from genotypes) 1, 2, 3, and 4 were identified. Deduced serotypes 1, 3, and 4 all appeared regularly over this 18-year period. Also over this period, nucleotide variation (in some cases substantial) in a section of the capsid protein precursor region of the virus genome was evident among strains of all four of the deduced central Australian serotypes. Consequent amino acid changes were, however, only evident among deduced serotype 3 strains. Geographic variation at both the genome and the resultant amino acid levels was evident among strains of all four of the deduced central Australian serotypes and their respective prototypes isolated in the United Kingdom.
Astroviruses, so called because of the characteristic five- or six-pointed star visible on some particles when viewed by electron micoscopy (1, 12), are small, nonenveloped, single capsid layered viruses with a positive-sense single-stranded RNA genome (14). Three open reading frames (ORFs)—ORF1a, ORF1b, and ORF2—have been identified in the genome. The first two encode the viral protease and polymerase, respectively, whereas ORF2, at the 3′ end of the genome, encodes the capsid protein precursor (9, 21).
Astroviruses have been classified into serotypes by using polyclonal sera or monoclonal antibodies (14). However, astroviruses have also been able to be classified into genotypes on the basis of the sequence of a 348-bp section of the capsid gene region of ORF2. This is conserved in virus strains of the same serotype but varies between strains of different serotypes that have been determined by serological methods (17). As good correlation was found between genotype and serotype (17), this proxy method of serotyping by reverse transcription-PCR (RT-PCR) and sequencing has been used in surveys for astrovirus types in recent years (15, 16, 18). Serotype-specific primers designed to identify all eight known astrovirus serotypes have also recently been used to type astroviruses in a survey in Japan (19).
Although astroviruses have been known to be associated with gastroenteritis since 1975 (1, 12), it has only in relatively recent years been found how widespread their role in particularly infantile gastroenteritis is (7). This has in part been due to the improvement in diagnostic and/or detection methods (7). In fact, some reports have shown that in certain settings astrovirus is second only to rotavirus as the most common cause of diarrhea in children (7).
Astroviruses have mainly been found to be associated with sporadic outbreaks of diarrhea in children, occurring in such settings as hospital children's wards, day care centers, kindergartens, and schools (7). The main symptoms of infection are watery diarrhea (often associated with vomiting), fever, and abdominal pain (14). However, diarrheal outbreaks among adults caused by astroviruses have also been described, having been found to occur in nursing homes for the elderly and among military recruits (2, 7). Asymptomatic infections also occur (5, 13).
The incidence of astrovirus-associated diarrhea in developed countries has, from different surveys, ranged from 2 to 6% (3, 4, 16, 18, 19). In developing countries the incidence has often been higher, ranging up to 26% of all children with diarrhea in a study of a Mayan community (5, 6, 8, 13). Surveys have also shown that serotype 1 is the most frequently detected diarrhea-causing type, with serotypes 2, 3, 4, and 5 being less common (11, 16, 18, 19). Serotypes 6, 7, and 8 have rarely been detected.
We determined the prevalence and monthly distribution of astrovirus infections in infants and young children with gastroenteritis admitted to Alice Springs Hospital in central Australia over the 4-year period from 1995 to 1998. Also, a number of similarly collected astrovirus-positive stool samples were available from the 14 years prior to this period, although these were most probably relatively restricted in number due to the method of virus detection that was used. However, enough were available to allow us to also report on variations in deduced astrovirus serotypes and relative genomic variation between different strains within serotypes over the 18-year period from 1981 to 1998.
MATERIALS AND METHODS
Stool samples.
Stool samples were obtained from infants and young children admitted to Alice Springs Hospital with gastroenteritis from 1981 to 1998. The hospital serves an area of central Australia within an ∼500-km radius of Alice Springs and a total population of ca. 25,000. For the survey of the incidence of astrovirus, carried out from January 1995 to December 1998, stool samples that were negative for rotavirus and Norwalk-like viruses were used. Rotavirus had been tested for by using commercial enzyme immunoassay and electron microscopy, and Norwalk-like viruses had been tested for by using RT-PCR (20). A total of 495 samples, 153 from 1995, 111 from 1996, 119 from 1997, and 112 from 1998 were tested for astrovirus. For the period from 1981 to 1994, only stool samples that had been shown to contain astrovirus by electron microscopy were available for testing. The ages of the children from whom stool samples were tested for astrovirus ranged from 3 weeks to 5 years. This study was approved by the Alice Springs Institutional Ethics Committee and the La Trobe University Human Ethics Committee.
Extraction and purification of viral RNA.
Viral RNA was extracted and purified from 400 μl of ca. 20% stool suspension according to the method of Jiang et al. (10) for Norwalk virus. Briefly, this involved genetron extraction and polyethylene glycol precipitation of virus, followed by proteinase K, sodium dodecyl sulfate, and cetyltrimethylammonium bromide (CTAB) treatment before extraction with phenol-chloroform and finally chloroform alone. The RNA was ethanol precipitated at −20°C and after pelleting and vacuum drying it was resuspended in 20μl of sterile distilled water and stored at −80°C until required.
Virus detection and RT-PCR.
Virus was detected by RT-PCR analysis of extracted viral RNA by using the previously published universal astrovirus-specific primer pair Mon269 and Mon270 of Noel et al. (17), which amplifies a 449-nucleotide (nt) region of ORF2 of the astrovirus genome. The conditions employed were the same as those of Noel et al. except that the gelatin and dimethyl sulfoxide were omitted. Bands at ca. 449 nt relative to known size markers on 2.0% agarose gels were considered indicative of the presence of astrovirus. These bands were then confirmed as being of astrovirus origin by Southern blot analysis.
Southern blot analysis.
Probes were prepared by extraction of RNA from prototype astrovirus serotypes 1 to 7 (obtained from J. Kurtz and T. Lee, John Radcliffe Hospital, Oxford, United Kingdom), followed by RT-PCR as outlined above. The astrovirus cDNA was then labeled with digoxigenin according to the manufacturer's instructions (Roche Biochemicals, Mannheim, Germany). For hybridization, bands were transferred from agarose gels to nylon membranes by alkaline transfer and then probed with an equal mix of the seven prototype astrovirus serotype probes. The mix had been tested to detect each of the seven astrovirus serotypes. Formamide was not used, the hybridization temperature was 55°C, and the two final washes were carried out with 0.1% citrate saline plus 0.1% sodium dodecyl sulfate at 55°C. Although only probes from astrovirus serotypes 1 to 7 were used, it was considered that astrovirus serotype 8 would also have been detected as a result of the cross-reactions that are obtained between probes from the different serotypes.
Nucleotide sequencing and astrovirus genotyping.
RT-PCR products (cDNA bands at 449 nt) that gave a positive reaction with the astrovirus cDNA probes were excised from agarose gels and further purified by using the QIAEX II DNA purification kit (Qiagen) according to the manufacturer's instructions. Manual sequencing was carried out by using the Thermo-Sequanase radiolabeled terminator cycle sequencing kit (Amersham Pharmacia Biotech) after a pretreatment of the purified cDNA samples with the enzymes from the Amersham Pharmacia Biotech PCR product presequencing reagent kit according to the manufacturer's instructions. Sequences were generated in both directions with the primers Mon269 and Mon270 used for the RT-PCR, and a consensus sequence for each of the isolates was deduced for the resulting 348-nt product (17). The genotypes, and hence the proxy serotypes, of the isolates were then determined by comparison of these sequences with those of the prototype astrovirus serotypes (18). The equivalent sequence for astrovirus serotype 8 was obtained from the GenBank database. A strain was defined as such when the sequence differed from that of another of the central Australian isolates by at least 1 nt.
Nucleotide sequence accession numbers.
The nucleotide sequences determined in the present study have been deposited in the GenBank database and have been assigned accession numbers AF440292 to AF440309 and AF440791 to AF440797.
RESULTS
Epidemiology.
Over the 4-year period from January 1995 to December 1998 the incidence of astrovirus was found to be 4.3%, having been identified in 33 of an overall total of 774 stool samples from children in Alice Springs Hospital with gastroenteritis during this period. The yearly incidences were 4.3% in 1995, 4.3% in 1996, 1.8% in 1997, and 6.6% in 1998 (Table 1). Overall, and in each of the 4 years the incidence of astrovirus was substantially lower than that of rotavirus but was higher than that of Norwalk-like viruses (Table 1). The overall incidence of rotavirus was 34.5%, and that of Norwalk-like viruses was 1.6%. The ages of the children from whom the astrovirus-positive stools came ranged from 6 weeks to 2.5 years. Of these, approximately half were under 12 months of age.
TABLE 1.
Incidence of astrovirus (compared to that of rotavirus and Norwalk-like virus) in children with gastroenteritis in Alice Springs Hospital (1995 to 1998)
| Yr | Total no. of stool samples tested | No. (%) of astrovirus-positive samples | No. (%) of rotavirus-positive samples | No. (%) of Norwalk-like virus-positive samples |
|---|---|---|---|---|
| 1995 | 235 | 10 (4.3) | 79 (33.6) | 3 (1.3) |
| 1996 | 185 | 8 (4.3) | 69 (37.3) | 5 (2.7) |
| 1997 | 171 | 3 (1.8) | 51 (29.8) | 1 (0.6) |
| 1998 | 183 | 12 (6.6) | 68 (37.2) | 3 (1.6) |
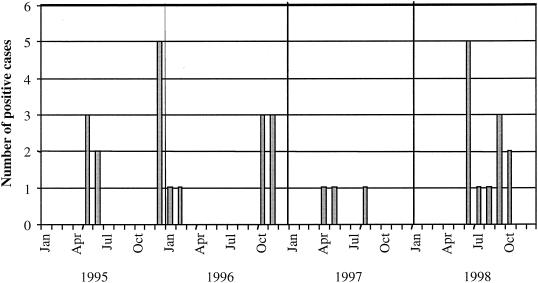
The monthly distribution of astrovirus identification is shown in Fig. 1. There was no consistent seasonal incidence over the 4 years of the survey, but the positive cases were detected in the colder months in 3 of the 4 years, in the winter months in 1995, and exclusively in the colder months in 1997 and 1998. However, no astrovirus-positive cases were detected in these months in 1996. Positive cases were also identified in the very hot summer months at the end of 1995 and the beginning of 1996. It was also of interest that the 11 cases of astrovirus infection detected in 1993 were all in the four coldest months of the year.
FIG. 1.
Monthly distribution of astrovirus-positive cases among children with gastroenteritis in Alice Springs Hospital from 1995 to 1998.
Astrovirus serotypes (deduced from genotypes) detected.
Serotypes 1, 2, 3, and 4 were identified in central Australia over the period 1981 to 1998. In the survey period (1995 to 1998), serotypes 1 and 4 were each detected in 3 of the 4 years and serotypes 2 and 3 were each detected in 2 of the 4 years (Table 2). No one serotype was therefore detected in each of the 4 years. More than one serotype was, however, detected in each of the 4 years (Table 2).
TABLE 2.
Astrovirus serotypes (deduced from genotypes) from children with gastroenteritis in Alice Springs Hospital (1981 to 1998)a
| Yr | No. of astrovirus isolates or no. of samples testedb | Serotype deduced (n) | Strain(s) |
|---|---|---|---|
| 1981 | 2 | 2 (2) | CAV2A |
| 1982 | 3 | 1 (1), 3 (1), 4 (1) | CAV1A, CAV3A, CAV4A |
| 1984 | 1 | 1 (1) | CAV1A |
| 1985 | 2 | 1 (1), 4 (1) | CAV1A, CAV4B |
| 1990 | 1 | 3 (1) | CAV3B |
| 1991 | 1 | 3 (1) | CAV3C |
| 1992 | 1 | 3 (1) | CAV3C |
| 1993 | 11 | 1 (11) | CAV1B, CAV1C |
| 1994 | 2 | 3 (1), 4 (1) | CAV3D, CAV4C |
| 1995 | 153 (10) | 1 (5), 2 (1), 4 (4) | CAV1D, CAV1E, CAV2B, CAV4D, CAV4E |
| 1996 | 111 (8) | 1 (5), 2 (3) | CAV1F, CAV2B, CAV2C |
| 1997 | 119 (3) | 3 (2), 4 (1) | CAV3E, CAV3F, CAV4F |
| 1998 | 112 (12) | 1 (7), 3 (2), 4 (3) | CAV1G, CAV3G, CAV3H, CAV4G |
In the years prior to 1995, astrovirus was identified only by electron microscopy, and those astrovirus isolates were typed. In the years not listed in the table, astrovirus was not identified. In the years 1995 to 1998, astrovirus was detected by RT-PCR. See the text for further details.
For the years 1981 to 1994, the number of astrovirus isolates identified and typed is given. For the years 1995 to 1998, the number of samples tested is given, with the number of astrovirus-positive samples detected and typed provided in parentheses.
In the period prior to 1995 serotype 3 was identified in 5 of the 9 years in which astrovirus was detected, serotype 1 in 4, serotype 4 in 3, and serotype 2 only in 1981 (Table 2). The incidence of astrovirus in this period cannot be compared directly to that in the period from 1995 to 1998 because of the lower sensitivity of electron microscopy compared to RT-PCR for the detection of astrovirus. However, overall, it can still be concluded that serotypes 1, 3, and 4 appeared consistently and at relatively regular intervals throughout the 18-year period from 1981 to 1998. It can also be noted that a relatively major outbreak of gastroenteritis attributable to astrovirus serotype 1 appears to have occurred in 1993 (Table 2).
Genomic variation among astrovirus isolates. (i) Deduced serotype 1.
Over the period of 17 years (1982 to 1998) in which serotype 1 (deduced from genotype) astrovirus strains were identified in central Australia, the differences in nucleotides between strains in the sequenced 348-nt region of the genome varied by up to a maximum of 7.2% (25 nt) (Table 3). This difference was between strain CAV1C (year 1993) and strain CAV1F (year 1996). The difference between strains detected in 1982 to 1985 (CAV1A) and 1998 (CAV1G), the extreme ends of the period under study, was not as large but of the same order at 5.7% (Table 3). Interestingly, strain CAV1A was identified over a period of 4 years (1982 to 1985) without any detected change in the 348-nt sequence. Of more significance, however, was the fact that over the 17-year period none of these nucleotide changes among the central Australian deduced serotype 1 strains translated into amino acid changes (Table 3). As well as the observed genome variation among strains, the determined sequences of a number of the deduced serotype 1 isolates in addition to those of strain CAV1A were identical. This included 10 isolates of strain CAV1B, 4 isolates of strain CAV1D, 5 isolates of strain CAV1F, and 7 isolates of strain CAV1G.
TABLE 3.
Sequence identity of a 348-nt region of ORF2 among serotype 1 (deduced from genotype) central Australian astrovirus strains and prototype serotype 1 astrovirus
| Strain | % Sequence identity in (yr of isolation)a:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Prototype | 1982, 1984, and 1985 | 1993
|
1995
|
1996 (CAV1F) | 1998 (CAV1G) | |||
| CAV1A | CAV1B | CAV1C | CAV1D | CAV1E | ||||
| Prototype | 96.6 | 97.1 | 96.6 | 96.8 | 96.6 | 91.1 | 91.1 | |
| CAV1A | 96.7 | 98.9 | 98.3 | 98.9 | 98.6 | 94.0 | 94.3 | |
| CAV1B | 96.7 | 100 | 99.4 | 100 | 99.7 | 93.4 | 93.7 | |
| CAV1C | 96.7 | 100 | 100 | 99.4 | 99.1 | 92.8 | 93.1 | |
| CAV1D | 96.7 | 100 | 100 | 100 | 99.7 | 93.7 | 94.0 | |
| CAV1E | 96.7 | 100 | 100 | 100 | 100 | 94.0 | 94.3 | |
| CAV1F | 96.7 | 100 | 100 | 100 | 100 | 100 | 98.9 | |
| CAV1G | 96.7 | 100 | 100 | 100 | 100 | 100 | 100 | |
Numbers at the top right indicate results of a pairwise comparison of the 348-nt regions, and numbers at the bottom left indicate results of pairwise comparison of the deduced amino acid sequences of the regions.
Nucleotide sequence differences between the central Australian deduced serotype 1 strains and the prototype strain isolated in the United Kingdom varied between 2.9 and 8.9% (Table 3). In contrast to the situation among the central Australian strains, four of these nucleotide differences resulted in amino acid changes. All of the central Australian strains differed from the United Kingdom prototype by the same four amino acids.
(ii) Deduced serotype 2.
Although serotype 2 (deduced from genotype) was less frequently identified than any of the other astrovirus serotypes detected in central Australia, the highest degree of nucleotide sequence variation was found among these strains. Deduced serotype 2 strains were identified over a 16-year period (from 1981 to 1996), but with the greatest difference of 14.1% (49 nt) being between strains CAV2B (December 1995 and early 1996) and CAV2C (November 1996), detected less than a year apart (Table 4). However, a similar difference (13.8%) was evident between strains CAV2A (year 1981) and CAV2B (Table 4). Interestingly, the difference in nucleotides between strains CAV2A and CAV2C was only 1.1% despite this being over a 16-year period. In spite of the substantial nucleotide variations between some of the strains, all were silent with respect to amino acid coding (Table 4). In addition to the genome variation, two strain CAV2A and two strain CAV2B isolates were identical in the determined genome sequence.
TABLE 4.
Sequence identity of a 348-nt region of ORF2 among serotype 2 (deduced from genotype) central Australian astrovirus strains and prototype serotype 2 astrovirus
| Strain | % Sequence identity in (yr of isolation)a:
|
|||
|---|---|---|---|---|
| Prototype | 1981 (CAV2A) | 1995 and 1996 (CAV2B) | 1996 (CAV2C) | |
| Prototype | 87.1 | 94.3 | 87.1 | |
| CAV2A | 99.1 | 86.2 | 98.9 | |
| CAV2B | 99.1 | 100 | 85.9 | |
| CAV2C | 99.1 | 100 | 100 | |
Numbers at the top right indicate results of a pairwise comparison of the 348-nt regions, and numbers at the bottom left indicate results of pairwise comparison of the deduced amino acid sequences of the regions.
The nucleotide differences between the central Australian deduced serotype 2 strains, and the prototype strain isolated in the United Kingdom ranged from 5.7 to 13.9%, and one of these differences resulted in an amino acid change. The amino acid was the same for all of the central Australian deduced serotype 2 strains identified.
(iii) Deduced serotype 3.
Serotype 3 (deduced from genotype) was identified in central Australia over the period of 17 years (1982 to 1998), and the maximum difference in nucleotides between strains was similar to that for the deduced serotyps 1 strains, varying by up to a maximum of 7.5% (26 nt). This was between strains CAV3A (year 1982) and CAV3H (year 1998) (Table 5). In contrast to the deduced serotype 1 strains, the maximum difference among the deduced serotype 3 strains was between strains identified at the two extremes of the time period under study. Also, in the case of the deduced serotype 3 strains, the nucleotide differences were not all silent. All strains identified after CAV3A differed by one amino acid from this strain, and strain CAV3H varied from the other deduced serotype 3 strains by a different amino acid. Strains CAV3A and CAV3H therefore differed by two amino acids (Table 5). Relative to the frequency of appearance, fewer isolates among the deduced serotype 3 strains were identical in the determined genome sequence, with only two strain CAV3C isolates identical.
TABLE 5.
Sequence identity of a 348-nt region of ORF2 among serotype 3 (deduced from genotype) central Australian astrovirus strains and prototype serotype 3 astrovirus
| Strain | % Sequence identity in (yr of isolation)a:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Prototype | 1982 (CAV3A) | 1990 (CAV3B) | 1991 and 1992 (CAV3C) | 1994 (CAV3D) | 1997
|
1998
|
|||
| CAV3E | CAV3F | CAV3G | CAV3H | ||||||
| Prototype | 95.1 | 99.1 | 98.9 | 98.3 | 98.9 | 98.6 | 96.8 | 96.3 | |
| CAV3A | 99.1 | 95.1 | 94.8 | 94.0 | 94.3 | 94.0 | 93.1 | 92.5 | |
| CAV3B | 100 | 99.1 | 99.7 | 98.0 | 98.0 | 97.7 | 96.6 | 96.0 | |
| CAV3C | 100 | 99.1 | 100 | 97.7 | 97.7 | 97.4 | 96.3 | 95.7 | |
| CAV3D | 100 | 99.1 | 100 | 100 | 98.0 | 97.7 | 98.3 | 97.7 | |
| CAV3E | 100 | 99.1 | 100 | 100 | 100 | 99.7 | 96.3 | 95.7 | |
| CAV3F | 100 | 99.1 | 100 | 100 | 100 | 100 | 96.0 | 95.4 | |
| CAV3G | 100 | 99.1 | 100 | 100 | 100 | 100 | 100 | 98.9 | |
| CAV3H | 99.1 | 98.3 | 99.1 | 99.1 | 99.1 | 99.1 | 99.1 | 99.1 | |
Numbers at the top right indicate results of a pairwise comparison of the 348-nt regions, and numbers at the bottom left indicate results of pairwise comparison of the deduced amino acid sequences of the regions.
Central Australian deduced serotype 3 strains varied in the determined nucleotide sequence from the prototype serotype 3 strain isolated in the United Kingdom by up to 4.9% (strain CAV3A). Strains CAV3A and CAV3H each differed from the prototype strain by one amino acid, but the amino acid sequences of all of the other central Australian deduced serotype 3 strains were identical to that of the prototype strain after translation of this region of the genome (Table 5).
(iv) Deduced serotype 4.
Serotype 4 (deduced from genotype) was also identified in central Australia over the 17-year period (1982 to 1998), with the maximum difference in nucleotides between strains in the sequenced region of the genome being 9.8% (34 nt) between strains CAV4F (year 1997) and CAV4G (year 1998) detected 10 months apart (Table 6). The difference between the strains identified in 1982 (CAV4A) and 1998 (CAV4G) was 6.6% (Table 6). As in the case of the deduced serotype 1 and serotype 2 strains, none of the nucleotide differences translated into amino acid changes (Table 6). It was also noted that three strain CAV4D and three strain CAV4G isolates were identical in the determined genome sequence.
TABLE 6.
Sequence identity of a 348-nt region of ORF2 among serotype 4 (deduced from genotype) central Australian astrovirus strains and prototype serotype 4 astrovirus
| Strain | % Sequence identity in (yr of isolation)a:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Prototype | 1982 (CAV4A) | 1985 (CAV4B) | 1994 (CAV4C) | 1995
|
1997 (CAV4F) | 1998 (CAV4G) | ||
| CAV4D | CAV4E | |||||||
| Prototype | 96.3 | 94.3 | 96.6 | 96.0 | 96.0 | 90.8 | 93.7 | |
| CAV4A | 99.1 | 94.3 | 98.9 | 98.6 | 98.6 | 91.7 | 93.4 | |
| CAV4B | 99.1 | 100 | 94.5 | 94.3 | 94.3 | 90.8 | 97.4 | |
| CAV4C | 99.1 | 100 | 100 | 99.1 | 99.1 | 91.4 | 93.7 | |
| CAV4D | 99.1 | 100 | 100 | 100 | 99.4 | 91.1 | 92.8 | |
| CAV4E | 99.1 | 100 | 100 | 100 | 100 | 91.7 | 92.8 | |
| CAV4F | 99.1 | 100 | 100 | 100 | 100 | 100 | 90.2 | |
| CAV4G | 99.1 | 100 | 100 | 100 | 100 | 100 | 100 | |
Numbers at the top right indicate results of a pairwise comparison of the 348-nt regions, and numbers at the bottom left indicate results of pairwise comparison of the deduced amino acid sequences of the regions.
In common with the other central Australian deduced astrovirus serotype strains relative to their respective prototype strains, all of the central Australian deduced serotype 4 strains differed in nucleotide sequence from the prototype serotype 4 strain isolated in the United Kingdom. In this case by from 3.4% to up to 9.2%, with the maximum difference being for strain CAV4F (year 1997) (Table 6). All of the central Australian deduced serotype 4 strains differed from the prototype strain by one amino acid (Table 6), the same one in each case.
DISCUSSION
The results of this survey suggest that astrovirus is a consistent enteric pathogen among infants and young children in central Australia, ranking well behind rotavirus but ahead of Norwalk-like viruses. The incidence, which may be a slight underestimate since rotavirus-positive and Norwalk-like virus-positive stools were not tested and simultaneous infection with astrovirus and rotavirus is possible, was comparable to that found in surveys carried out in developed countries. This appeared to be irrespective of whether the virus had been detected via the genome or by enzyme immunoassay (3, 16, 18, 19). The incidence was lower than that generally found in studies in developing countries (5, 6, 8, 13).
Astrovirus infection usually shows annual winter peaks in studies carried out in temperate climates (14) and, based on the results of the 1995 to 1998 survey period, this is possibly also the case in central Australia, with 1996 perhaps having been an unusual year. The apparent winter astrovirus outbreak of 1993 adds weight to this argument. However, a longer survey will be necessary to establish this more conclusively. The summer peak of 1995 to 1996 was possibly exceptional not only for its time of occurrence but also in that it appeared to be associated only with the generally less commonly detected serotypes 2 and 4.
Melbourne, a large metropolitan city with a population of ca. 3.5 million, is the only location in Australia for which detailed data on the incidence of astrovirus serotypes has so far been published. In common with the results of surveys carried out in other parts of the world serotype 1 was the most frequently detected type in surveys carried out in that city (16, 18). Alice Springs, a very small city, and rural central Australia in which the present study was carried out, are very different compared to Melbourne, and in central Australia the situation with respect to predominant astrovirus serotypes is not as clearcut. The frequency of appearance of serotype 4 was the same as that of serotype 1 in the survey years from 1995 to 1998, although the actual number of cases of serotype 1-associated infection was higher. Despite the fact that we were not able to make a direct comparison with the 1995 to 1998 period because of the more-sensitive method of astrovirus detection used then, it was of interest that the frequency of appearance of astrovirus serotypes 1, 3, and 4 over the 14-year period prior to 1995 was comparable to that observed over the 1995 to 1998 survey period. Serotype 3 was actually slightly more frequently detected in the earlier period. Overall, therefore, it would appear that in central Australia astrovirus serotypes 1, 3, and 4 are in constant circulation, with possibly one of these serotypes occasionally causing more-prominent outbreaks. It was also of interest that, whereas serotype 3 was very regularly detected over the 18 years of the central Australian study, including the last 4 survey years of 1995 to 1998, serotype 3 was not detected at all in the Melbourne studies (16, 18). This suggests that there may be a regional incidence for some astrovirus serotypes.
None of the rarely detected astrovirus serotypes were identified in central Australia, nor was the less commonly detected serotype 5, which had been identified in Melbourne (18). One case of serotype 8 infection had also been detected in Melbourne (16). It would not have been surprising to see the occasional introduction of other serotypes of astrovirus into Alice Springs and central Australia since, although relatively isolated geographically, they are major national and international tourist destinations.
The present study involved only the incidence of astrovirus in acute cases of gastroenteritis, cases severe enough for the children to have been admitted to hospital, and therefore it may be that other astrovirus serotypes causing clinically less severe symptoms and not resulting in hospitalization of the children may have remained undetected in central Australia. A more community-based study will be required to determine this. Asymptomatic cases of astrovirus infection have been detected in such surveys (5, 13). Such a study could also determine whether serotype 2 infection occurs but is relatively rare in central Australia, as it appears from the present study, or whether serotype 2 is just more likely to be associated with infections causing less-severe symptoms.
Genome sequence variation was found among central Australian deduced serotype 1 astrovirus strains, but the fact that none of the nucleotide changes translated into amino acid changes over the long period of 17 years indicates significant stabilty. The results are comparable with the findings of Mustafa et al. (16) and Palombo et al. (18) in their analysis of the same 348-nt region of ORF2 of such strains in their Melbourne astrovirus surveys, although Mustafa et al. did find one amino acid change occurring among the serotype 1 isolates they investigated over the 4 years of their survey. The lack of any amino acid changes among the central Australian deduced serotype 1 strains, despite the nucleotide changes, could be related to the fact that this 348-nt region of ORF2 is in a relatively conserved part of the astrovirus capsid protein precursor gene (22). Analysis of the more variable 3′ end of ORF2 may give a better indication of genomic and potential antigenic diversity, as also suggested by Mustafa et al. (16). Geographic variation between the central Australian deduced serotype 1 strains and the United Kingdom prototype serotype 1 strain was more pronounced, with differences between the central Australian and United Kingdom strains evident at not only the nucleotide level but more particularly also at the amino acid level. Medina et al. (15) and Mustafa et al. (16) also reported significant variation among astrovirus isolates from different countries.
Genomic variation, particularly among some of the central Australian deduced serotype 2 strains and also among some of the deduced serotype 4 strains, was relatively high, and it is tempting to suggest that strain CAV2B may be a recent introduction into the region or the result of recombination in a dual infection. However, at the amino acid level variation among the deduced serotype 2, 3, and 4 strains was in general similar to that among the deduced serotype 1 strains, underlining a general long-term stability. That the relatively large nucleotide differences between particularly some of the central Australian deduced serotype 2 strains did not lead to amino acid differences is surprising, but perhaps just underlines the stability in amino acid structure required in this region of the capsid protein. Whether the amino acid differences specifically between the central Australian deduced serotype 3 strains and generally between the central Australian strains and their respective United Kingdom prototype strains is indicative of any antigenic difference has yet to be determined.
Astrovirus has been shown to be an enteric pathogen of some importance in central Australia, with serotypes 1, 3, and 4 and, to a lesser extent, serotype 2 circulating relatively stably in the region. It is long-term or continuing surveys such as this one, in combination with other such studies, thereby allowing comparisons to be made on a broader national and international level, that are required in the determination of whether the development of an astrovirus vaccine would be worthwhile. Such studies will also be required in the prospective formulation of any such vaccine.
Acknowledgments
Support for this project was provided, in part, by a La Trobe University Central Large Research Grant.
REFERENCES
- 1.Appleton, H., and P. G. Higgins. 1975. Viruses and gastroenteritis in infants. Lancet i:1297. [DOI] [PubMed] [Google Scholar]
- 2.Belliot, G., H. Laveran, and S. S. Monroe. 1997. Outbreak of gastroenteritis in military recruits associated with serotype 3 astrovirus infection. J. Med. Virol. 51:101-106. [DOI] [PubMed] [Google Scholar]
- 3.Bon, F., P. Fascia, M. Dauvergne, D. Tenenbaum, H. Planson, A. M. Petion, P. Pothier, and E. Kohli. 1999. Prevalence of group A rotavirus, human calicivirus, astrovirus, and adenovirus type 40 and 41 infections among children with acute gastroenteritis in Dijon, France. J. Clin. Microbiol. 37:3055-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carter, M. J., and M. M. Willcocks. 1996. The molecular biology of astroviruses. Arch. Virol. Suppl. 12:277-285. [DOI] [PubMed] [Google Scholar]
- 5.Cruz, J. R., A. V. Bartlett, J. E. Herrmann, P. Caceres, N. R. Blacklow, and F. Cano. 1992. Astrovirus-associated diarrhea among Guatemalan ambulatory rural children. J. Clin. Microbiol. 30:1140-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaggero, A., M. O'Ryan, J. S. Noel, R. I. Glass, S. S. Monroe, M. Mamani, V. Prado, and L. F. Avendano. 1998. Prevalence of astrovirus infection among Chilean children with acute gastroenteritis. J. Clin. Microbiol. 36:3691-3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glass, R. I., J. Noel, D. Mitchell, J. E. Herrmann, N. R. Blacklow, L. K. Pickering, P. Dennehy, G. Ruiz-Palacios, M. L. de Guerrero, and S. S. Monroe. 1996. The changing epidemiology of astrovirus-associated gastroenteritis: a review. Arch. Virol. Suppl. 12:287-300. [DOI] [PubMed] [Google Scholar]
- 8.Guerrero, L. M., J. S. Noel, K. Mitchell, J. J. Calva, A. Morrow, J. Martinez, G. Rosales, R. Velazquez, S. S. Monroe, R. I. Glass, L. K. Pickering, and G. M. Ruiz-Palacios. 1998. A prospective study of astrovirus diarrhea of infancy in Mexico City. Pediatr. Infect. Dis. J. 17:723-727. [DOI] [PubMed] [Google Scholar]
- 9.Jiang, B., S. S. Monroe, E. V. Koonin, S. E. Stine, and R. I. Glass. 1993. RNA sequence of astrovirus: distinctive genomic organization and a putative retrovirus-like ribosomal frameshifting signal that directs the viral replicase synthesis. Proc. Natl. Acad. Sci. USA 90:10539-10543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang, X., J. Wang, D. Y. Graham, and M. K. Estes. 1992. Detection of Norwalk virus in stool by polymerase chain reaction. J. Clin. Microbiol. 30:2529-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee, T. W., and J. B. Kurtz. 1994. Prevalence of human astrovirus serotypes in the Oxford region 1976-92, with evidence for two new serotypes. Epidemiol. Infect. 112:187-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madeley, C. R., and B. P. Cosgrove. 1975. 28 nm particles in faeces in infantile gastroenteritis. Lancet ii:451-452. [DOI] [PubMed] [Google Scholar]
- 13.Maldonado, Y., M. Cantwell, M. Old, D. Hill, M. L. Sanchez, L. Logan, F. Millan-Velazco, J. L. Valdespino, J. Sepulveda, and S. M. Matsui. 1998. Population-based prevalence of symptomatic and asymptomatic astrovirus infection in rural Guatemala. J. Infect. Dis. 178:334-339. [DOI] [PubMed] [Google Scholar]
- 14.Matsui, S. M., and H. B. Greenberg. 1996. Astroviruses, p. 811-824. In B. N. Fields, D. M. Knipe, P. M. Howley, R. M. Chanock, J. L. Melnick, T. P. Monath, B. Roizman, and S. E. Straus (ed.), Fields virology, 2nd ed. Lippincott-Raven, Philadelphia, Pa.
- 15.Medina, S. M., M. F. Gutierrez, F. Liprandi, and J. E. Ludert. 2000. Identification and type distribution of astroviruses among children with gastroenteritis in Columbia and Venezuela. J. Clin. Microbiol. 38:3481-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mustafa, H., E. A. Palombo, and R. F. Bishop. 2000. Epidemiology of astrovirus infection in young children hospitalized with acute gastroenteritis in Melbourne, Australia, over a period of four consecutive years, 1995 to 1998. J. Clin. Microbiol. 38:1058-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noel, J. S., T. W. Lee, J. B. Kurtz, R. I. Glass, and S. S. Monroe. 1995. Typing of human astroviruses from clinical isolates by enzyme immunoassay and nucleotide sequencing. J. Clin. Microbiol. 33:797-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palombo, E. A., and R. F. Bishop. 1996. Annual incidence, serotype distribution, and genetic diversity of human astrovirus isolates from hospitalized children in Melbourne, Australia. J. Clin. Microbiol. 34:1750-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakamoto, T., H. Negishi, Q.-H. Wang, S. Akihara, B. Kim, S. Nishimura K. Kaneshi, S. Nakaya, Y. Ueda, K. Sugita, T. Motobiro, T. Nishimura, and H. Ushijima. 2000. Molecular epidemiology of astroviruses in Japan from 1995 to 1998 by reverse transcription-polymerase chain reaction with serotype-specific primers (1 to 8). J. Med. Virol. 61:326-331. [PubMed] [Google Scholar]
- 20.Schnagl, R. D., N. Barton, M. Patrikis, J. Tizzard, J. Erlich, and F. Morey. 2000. Prevalence and genomic variation of Norwalk-like viruses in central Australia in 1995-1997. Acta Virol. 44:265-271. [PubMed] [Google Scholar]
- 21.Willcocks, M. M., T. D. K. Brown, C. R. Madeley, and M. J. Carter. 1994. The complete sequence of a human astrovirus. J. Gen. Virol. 75:1785-1788. [DOI] [PubMed] [Google Scholar]
- 22.Willcocks, M. M., J. B. Kurtz, T. W. Lee, and M. J. Carter. 1995. Prevalence of human astrovirus serotype 4: capsid protein sequence and comparison with other strains. Epidemiol. Infect. 114:385-391. [DOI] [PMC free article] [PubMed] [Google Scholar]