Abstract
The expression of Staphylococcus aureus virulence proteins is under the control of RNA III, a central pleiotropic regulator transcribed from the agr locus. RNA III is activated by at least two two-component systems, one encoded by the agr locus (AgrC-AgrA) and another encoded outside of this locus (TRAP-RAP). In this work, we developed new typing methods based on genes encoding these two systems, which we used to characterize a nonclonal population of S. aureus bovine mastitis isolates. Twelve agr restriction types were identified in this population, but the majority of strains (56.3%) were grouped in the R III-A1 type. No strain isolated from humans, whose agr sequence is available from GenBank, was found to belong to this major type. Restriction maps constructed for all of those agr variants allowed the linking of all types in an evolution scheme and their grouping in one of the four agr interference groups. This analysis indicates that groups 2, 3, and 4 probably evolved from the more frequently encountered type, which belongs to group 1. agr group 1 was also found to be the most prevalent (69.0% of the strains) and the most polymorphic interference group. By developing an agr group-specific multiplex PCR, we confirmed the above classification of strains in the agr interference groups. Four allelic variants of trap were also identified, indicating that this two-component system is also polymorphic. The majority of strains was grouped in the trap 1 type (71.8%). Whereas no relationships between agr group and trap types were found, strains of similar agr restriction type were also of similar trap type (with the exception of strains belonging to the agr R IV-A5 and R VI-A8 types). Our analysis indicates that S. aureus isolated from cows has predominantly a clonal structure and that the highly prevalent agr R III-A1, trap 1 type (56.3% of the strains) probably possesses a genetic background which endows it with superior ability to infect the bovine mammary gland.
Staphylococcus aureus is a gram-positive bacterium responsible for various major diseases in both humans and domestic animals. In dairy animals, S. aureus is one of the major causes of intramammary infections (mastitis) of lactating females, from whose milk it is frequently isolated. The presence of this bacterium in raw milk represents a risk for human health and causes serious economic losses to milk producers around the world (17, 27).
The pathogenesis of S. aureus is complex and involves both surface-associated proteins implicated in the adhesion of the bacterium to host tissues and the secretion of toxins that not only cause disease but also contribute to the bacterial spread (22). The expression of the virulence proteins is under the control of RNA III, a central pleiotropic regulator transcribed from the accessory gene regulator (agr) locus. RNA III is activated by at least two two-component systems, one (AgrC-AgrA) encoded by agr and another (target of RNA III-activating protein [TRAP]-RNA III-activating protein [RAP]) encoded outside of this locus (3, 22, 23).
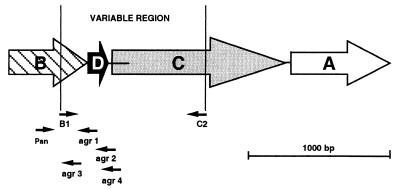
The agr system is a quorum-sensing system that, during the transition from the exponential to the stationary phase of growth, down-regulates the transcription of genes encoding some surface proteins and up-regulates the transcription of certain extracellular toxins (14, 22). The agr locus comprises two divergent transcriptional units, under the control of promoters P2 (RNA II) and P3 (RNA III). The P2 operon encodes a two-component signal transduction system (AgrC, transmembrane receptor-histidine kinase; AgrA, cytoplasmic regulator), a propeptide (AgrD), and an integral membrane protein (AgrB) that is probably involved in the processing and/or secretion of the peptide (Fig. 1). The resulting mature autoinducing peptide (AIP) accumulates in the extracellular environment during bacterial growth, reaches a threshold concentration (quorum sensing), and activates the two-component system by phosphorylation. The phosphorylated AgrA sensor then up-regulates the transcription from promoter P2, amplifying the response, and initiates transcription from promoter P3. The P3 transcript, RNA III, mediates up-regulation of secreted virulence factors as well as down-regulation of surface proteins (22). Interestingly, Ji et al. also described that AIP produced by a given strain of S. aureus activates its own agr locus but may inhibit the expression of agr in other strains. This phenomenon—due to polymorphism in a variable region of the agr locus comprising nucleotide sequences encoding AgrD, the C-terminal two-thirds of AgrB, and a portion of the N-terminal half of AgrC—actually led to the classification of S. aureus isolates in four different interference groups (12, 15). This type of bacterial interference could be implicated in the struggle for the colonization of infected sites (15).
FIG. 1.
Schematic map of the S. aureus agr operon (P2). The four members of the operon are represented with the position of the primers used in this work. agrA and agrC encode the response regulator and receptor-histidine protein kinase components of a two-component signal transduction pathway, whereas agrB and agrD combine to generate the activating ligand for the receptor. Large open arrows indicate the direction of transcription for each gene. Horizontal lines indicate intergenic regions (the usual length of the agrD-agrC intergenic region is 25 bp, but lengths up to 160 bp are also described). Thin arrows indicate the position of the primers used to amplify part of the operon. Primer lengths are not shown to scale, but their 5′ ends are correctly positioned.
On the other hand, RAP-TRAP, a second two-component system, was also shown to be able to activate transcription of RNA III (3). The proposed model taking into account this second system suggests that autoinduction of virulence occurs in a two-step process. At the beginning of the bacterial growth, the autoinducer RAP accumulates and induces the phosphorylation of the surface-associated protein TRAP. This results in up-regulation of the agr locus to produce RNA II. Once agr is activated (in the mid-exponential phase of growth), AIP and AgrC are produced. AIP accumulates in the environment and induces phosphorylation of AgrC, leading to phosphorylation of AgrA, up-regulation of RNA III synthesis, and down-regulation of TRAP phosphorylation (3, 32). Actually, it is not known if trap interstrain variation, similar to what was found for agr, does exist.
In this work, we developed new typing methods based on genes encoding the above-cited two-component systems, which we used to characterize the still poorly studied population of S. aureus strains isolated from cows with mastitis.
MATERIALS AND METHODS
Bacterial strains.
A total of 71 S. aureus strains isolated from milk of cows with mastitis were analyzed. All strains were identified as S. aureus using standard microbiological techniques (10). These strains were isolated from different locations and at different times and were chosen to be epidemiologically unrelated. Most of the strains (n = 65) were isolated from different regions of France by the Laboratory of Infectious Pathology and Immunology at the National Institute for Agronomic Research, but six strains were also isolated from different parts of the world: three strains were isolated in the United States (comprising strain 305 of Prasad and Newbould [ATCC 29470] [26]), one strain was isolated in the United Kingdom (the m strain of Neave and Oliver [ATCC 27543] [21]), and two strains were isolated in Japan (strains 125 and 130 of Takeuchi et al. [28]).
One strain was isolated in the 1950s, 15 strains were isolated in the 1960s, 16 strains were isolated in the 1970s, 12 strains were isolated in the 1980s, and 27 strains were isolated in the 1990s.
S. aureus agr reference strains RN6390 (agr group 1), RN6923 (agr group 2), RN8462 (agr group 3), and A880740 (agr group 4) (12, 15) and strains whose genomes are sequenced—N315 and Mu50 (18), COL (http://www.tigr.org/tdb/mdb/mbdinprogress.html), NCTC 8325 (http://www.genome.ou.edu/staph.html), and MRSA-252 and MSSA-476 (http://www.sanger.ac.uk/Projects/S_aureus/)—all from human origin, were used as controls.
Nucleic acid purification.
For nucleic acid purification, strains were grown overnight on brain heart infusion agar plates at 37°C for 24 h. Ten to 15 colonies were then scraped from plates, and spheroplasts were prepared as described by van Leeuwen et al. (29). Nucleic acids were then purified from the spheroplasts as described by Boom et al. (4). Briefly, guanidine thiocyanate (catalog no. 50990; Fluka Biochemika) was added for cell lysis and the nucleic acids were purified by affinity chromatography with diatoms (high-purity, analytical grade Celite [catalog no. 16,743-6]; Aldrich). The nucleic acids were finally eluted from the diatom particles with 100 μl of a 10−4 M EDTA-10−2 M Tris · Cl (pH 8.0) solution and stored at −20°C until use.
PCR amplification of the variable region of the agr operon.
PCR amplification of the 1,070-bp variable region of the agr operon was performed with primers B1 (5′-TAT GCT CCT GCA GCA ACT AA-3′) and C2 (5′-CTT GCG CAT TTC GTT GTT GA-3′) described by van Leeuwen et al. (Fig. 1) (29). The variable agr region was amplified from 2 μl of the purified nucleic acid solution in a 100-μl reaction mixture containing 2.5 U of Taq DNA polymerase (Taq DNA polymerase in storage buffer A [Promega]), 200 μM deoxynucleotide triphosphates (dNTPs) (Promega), 0.5 μM primer B1, 0.5 μM primer C2, 2 mM MgCl2, 50 mM KCl, 0.1% Triton X-100, and 10 mM Tris · Cl (pH 9.0). Amplifications were carried out in a Perkin-Elmer thermocycler (GeneAmp PCR system 9600) through the following temperature program: 1 cycle of 4 min at 94°C; 40 cycles of 1 min at 94°C, 1 min at 50°C, and 2 min at 74°C; and finally 1 cycle at 74°C for 3 min. For some samples that were not amplified with the above protocol, the concentration of MgCl2 in the PCR mixture was increased to 2.5 mM, and the annealing and elongation times were increased to 2 and 3 min, respectively. After precipitation with ethanol and centrifugation, the pellet was dissolved in 70 μl of a 10−4 M EDTA-10−2 M Tris · Cl (pH 9.0) solution. All samples were stored at −20°C before restriction.
Agr group-specific multiplex PCR.
The agr sequences were amplified from 2 μl of the purified nucleic acid solutions in a 25-μl reaction mixture containing 1.25 U of Taq DNA polymerase (Taq DNA polymerase in storage buffer A [Promega]), 200 μM dNTPs (Promega), 5 mM MgCl2, 50 mM KCl, 0.1% Triton X-100, 10 mM Tris · Cl (pH 9.0), and a 0.3 μM concentration of each of the following primers: Pan (5′-ATG CAC ATG GTG CAC ATG C-3′), agr1 (5′-GTC ACA AGT ACT ATA AGC TGC GAT-3′), agr2 (5′-TAT TAC TAA TTG AAA AGT GGC CAT AGC-3′), agr3 (5′-GTA ATG TAA TAG CTT GTA TAA TAA TAC CCA G-3′), and agr4 (5′-CGA TAA TGC CGT AAT ACC CG-3′). These primers allow the amplification of a 441-bp DNA fragment of the agr group 1 strains, of a 575-bp DNA fragment of the agr group 2 strains, of a 323-bp DNA fragment of the agr group 3 strains, and of a 659-bp DNA fragment of the agr group 4 strains. Amplifications were carried out in an MJ Research thermocycler (PTC-100) through the following temperature program: 1 cycle of 5 min at 94°C; 26 cycles of 30 s at 94°C, 30 s at 55°C, and 60 s at 72°C; and finally 1 cycle of 72°C for 10 min. Amplification products were electrophoresed in a 1.5% agarose gel containing ethidium bromide and visualized by transillumination under UV.
PCR amplification of the trap gene.
The entire 504-bp open reading frame of the trap gene was amplified with sense primer 5′-ACA TAA GGG GGA CCT TTC AT-3′ (ending 1 nucleotide before the start codon) and antisense primer 5′-ACC AAT GGA AGT TTT CTT CG-3′ (ending 4 nucleotides after the stop codon). The trap open reading frame was amplified from 2 μl of the purified nucleic acid solutions in a 100-μl reaction mixture containing 1.25 U of Taq DNA polymerase (Taq DNA polymerase in storage buffer A [Promega]), 200 μM dNTPs (Promega), 1 μM sense primer, 1 μM antisense primer, 1 mM MgCl2, 50 mM KCl, 0.1% Triton X-100, and 10 mM Tris · Cl (pH 9.0). Amplifications were carried out in a Perkin-Elmer thermocycler (GeneAmp PCR system 9600) through the following temperature program: 1 cycle of 45 s at 95°C; 35 cycles of 45 s at 95°C, 60 s at 52°C, and 60 s at 72°C; and finally 1 cycle at 72°C for 10 min. All samples were stored at −20°C before restriction.
Restrictions of the PCR products.
The trap amplicons were restricted with MseI (New England Biolabs), whereas the agr amplicons were restricted with RsaI (Roche Molecular Biochemicals) and AluI (Roche Molecular Biochemicals), according to manufacturer's instructions. The restriction fragments were then separated by electrophoresis on a 3% agarose gel (SeaKel HGT agarose; FMC, Rockland, Maine) containing ethidium bromide and visualized by transillumination under UV.
RESULTS
agr restriction fragment length polymorphism.
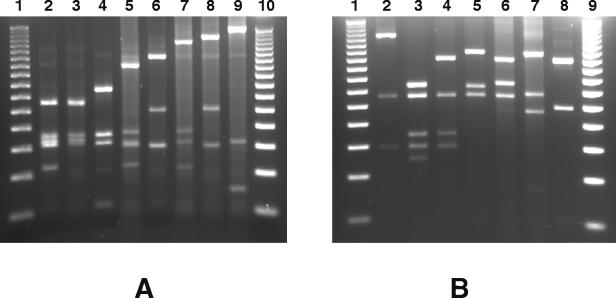
The polymorphism of the agr operon was analyzed by restriction endonuclease PCR in a population of 71 epidemiologically unrelated S. aureus bovine mastitis isolates. To this end, the 1,070-bp variable region of the agr operon was amplified by PCR with primers B1 and C2 (Fig. 1). Sixty-eight of the tested strains gave an amplicon of the expected molecular weight, and one strain gave an amplicon of around 740 bp, whereas no amplicon could be amplified from two strains (results not shown). The PCR products were then restricted with RsaI or AluI, giving 10 different profiles for each enzyme (Table 1; Fig. 2). The combination of these two restriction patterns allows the definition of 12 unique agr restriction types (Table 1). Most of the strains belong to the R III-A1 type (56.3%), the R IV-A5 type (12.7%), and the R IV-A7 type (8.4%). The other nine restriction types are shared by only one to three strains (Table 1). The numerical index of the discriminatory ability of the agr typing, calculated as described by Hunter and Gaston (11), indicates that if two strains were sampled randomly from the analyzed populations, then on 66.2% of occasions they would fall into different restriction types.
TABLE 1.
Characteristics of different restriction types of the agr operon identified among the analyzed S. aureus strains isolated from cows with mastitis
| agr type | agr groupa | trap type(s) | No. of occurrences (n = 71) | Lengthsb (bp) of agr restriction fragments
|
|
|---|---|---|---|---|---|
| RsaI | AluI | ||||
| R I-A6 | 4 | 2 | 1 | 642, 275, 155 | 626, 184, 155, 107 |
| R I′-A1 | 1 | 1 | 3 | 616, 276, 149, 32 | 274, 179, 168, 159, 155, 113, 25 |
| R II-A1 | 1 | 1 | 1 | 320, 276, 174, 149, 122, 32 | 274, 179, 168, 159, 155, 113, 25 |
| R III-A1 | 1 | 1 | 40 | 442, 276, 174, 149, 32 | 274, 179, 168, 159, 155, 113, 25 |
| R III-A2 | 1 | 2 | 1 | 442, 276, 174, 149, 32 | 274, 268, 179, 168, 159, 25 |
| R IV-A5 | 2 | 1, 3 | 9 | 484, 308, 280 | 502, 244, 149, 149, 28 |
| R IV-A7 | 2 | 2 | 6 | 484, 308, 280 | 679, 244, 149 |
| R V-A4 | 1 | 2 | 3 | 442, 323, 276, 32 | 442, 179, 159, 155, 113, 25 |
| R VI-A8 | 3 | 2, 3 | 2 | 478, 287, 230, 79 | 772, 155, 74, 73 |
| R VII-A3 | 1 | 1 | 1 | 442, 250, 50 | 325, 179, 159, 50, 25 |
| R VIII-A12 | 1→3 | 3 | 1 | 442, 230, 174, 149, 79 | 274, 179, 159, 155, 135, 74, 73, 25 |
| R IX-A11 | 1→2 | 2 | 1 | 442, 308, 174, 149 | 274, 244, 192, 179, 159, 25 |
| Not amplifiable | 2 | 4 | 2 | Not applicable | Not applicable |
Strains were classified in one of the four agr groups by agr group-specific PCR and agr restriction map analysis. The putative evolving stage between two groups is indicated by an arrow.
Lengths of restriction fragments were estimated by electrophoresis in the presence of molecular weight markers but, for some types, were also (R I-A6, GenBank accession no. AF288215; R I′-A1, GenBank accession no. X52543 [strain COL at The Institute for Genomic Research data bank and strain NCTC 8325 at the University of Oklahoma's ACGT data bank]; R III-A2, GenBank accession no. AF210055; R VI-A8, strain MSSA-476 at the Sanger data bank) or only (R VIII-A12, GenBank accession no. AB043555; R IX-A11, GenBank accession no. AB043554) calculated from computer-generated restrictions of agr nucleic acid sequences deposited in data banks.
FIG. 2.
Restriction polymorphism in the agr variable region of S. aureus strains isolated from cows with mastitis. The amplified variable region of the agr operon was digested with AluI (A) or RsaI (B) and electrophoresed on a 3% agarose gel. Examples of the different restriction types obtained are shown: type A1 (lane A2), type A2 (lane A3), type A3 (lane A4), type A4 (lane A5), type A5 (lane A6), type A6 (lane A7), type A7 (lane A8), type A8 (lane A9), type R I (lane B2), type R II (lane B3), type R III (lane B4), type R IV (lane B5), type R V (lane B6), type R VI (lane B7), and type R VII (lane B8). Type R I′, being difficult to differentiate from type R I in a 3% agarose gel, is not shown. Types R VIII, R IX, A11, and A12 were identified by computer analysis of sequences with GenBank accession no. AB043554 and AB043555. Molecular weight markers (50-bp DNA ladder; Promega) are shown in lanes A1, A10, B1, and B9.
Relationship between restriction types of the agr operon.
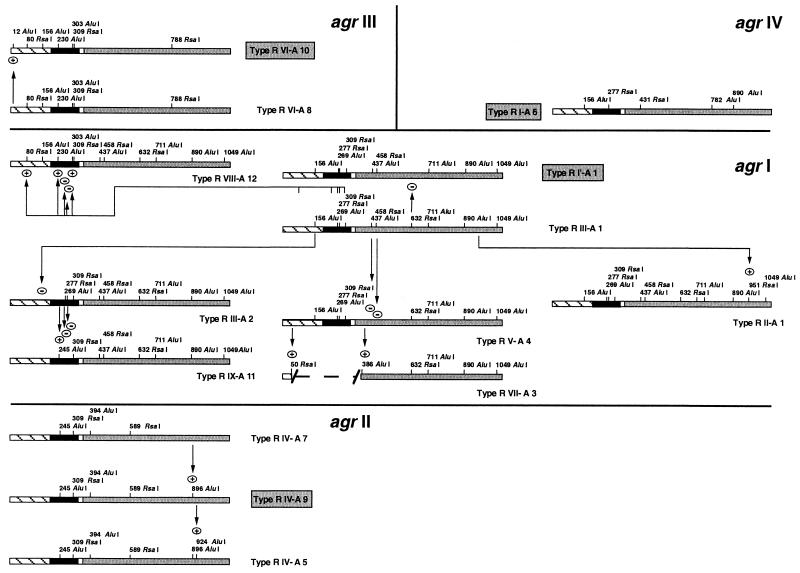
Restriction maps were constructed to know the relationship between the unique restriction types of the agr locus identified in the collection of strains isolated from cows with mastitis. To this end, partial hydrolysis was also conducted to correctly assign the position of some restriction fragments (results not shown). The maps were then compared with those corresponding to the same region of the agr operon sequenced from strains isolated from humans and available from nucleic acid databases (Fig. 3). The agr interference group reference strains isolated from humans were found to be type R I′-A1 for group 1 (GenBank accession no. X52543), type R IV-A9 for group 2 (GenBank accession no. AF001782), type R VI-A10 for group 3 (GenBank accession no. AF001783), and type R I-A6 for group 4 (GenBank accession no. AF288215). Strain CMRSA-1 (GenBank accession no. AF210055) classified by Papakyriacou et al. (25) as being a variant of the agr interference group 1 reference strain (Ia) was identified to be type R III-A2. It is worth noting that no type R VI-A10 and R IV-A9 were identified among the strains isolated from cows with mastitis. Similarly, after amplification and restriction of the agr operon, the restriction maps of six strains of human origin whose genomes are sequenced were also constructed. Strains N315 and Mu50 are type R IV-A9, strains NCTC 8325 and COL are type R I′-A1, strain MRSA-252 is type R VI-A10, and strain MSSA-476 is type R VI-A8.
FIG. 3.
Relationship between distinct restriction types of the agr operon identified among S. aureus strains isolated from cows with mastitis. The amplified variable region of the agr operon was digested with AluI and RsaI, and maps corresponding to the distinct restriction types identified in the population were constructed (3′ end of agrB [striped bars], agrD [solid bars], intergenic region [open bars], 5′ end of agrC [shaded bars]). These maps were compared with those corresponding to the same region of the agr group 1 to 4 reference sequences (type names in grey) obtained from GenBank (accession no. X52543 for group 1, accession no. AF001782 for group 2, accession no. AF001783 for group 3, and accession no. AF288215 for group 4). As the sequences of the agr operon deposited in GenBank do not always show any highly obvious translational start, the drafted length of the intergenic region represents the most frequently described length (25 bp). Based on the presence or absence of characteristic restriction sites, all restrictions types were classified in one of the four agr groups. The arrows indicate the appearance (+) or disappearance (−) of a restriction site with respect to the other types of the same agr group. The dashed line indicates the position of a deletion in the operon.
All restriction types were classified in one of the four agr groups on the basis of the presence or absence of a combination of restriction sites characteristic of each of the interference group reference sequences (Fig. 3 [types in grey]). All types were then linked one to the other inside each group, trying to produce the least gain or loss of restriction sites when evolving from one type to another (Fig. 3). This analysis allows the classification of all agr restriction types into the four previously known agr groups (Table 1). Nevertheless, it is of interest that the 3′ extremity of the variable region of the agr locus (plus or minus two-thirds of the molecules) of two strains isolated from cows in Japan, the strain 125 (type R IX-A11) and the strain 130 (type R VIII-A12), possess the characteristic restriction sites of the agr group 1 strains, whereas the 5′ extremity of this region (plus or minus one-third of the molecule) contains restriction sites characteristic of the agr group 2 and of the agr group 3 strains, respectively (Fig. 3). As the agr loci of both strains were sequenced (GenBank accession no. AB043555 and AB043554), this property was verified by aligning the nucleotide sequences of their variable agr region with those of the agr group 1 to 3 reference strains (results not shown).
The entire agr variable region of strains 125 and 130 being nevertheless more similar to the agr group 1 reference sequence (88.2 and 91.1% of nucleotides identical, respectively) than to the agr reference sequences of group 2 (69.7 and 59.0% of nucleotides identical, respectively) or group 3 (64.9 and 76.0% of nucleotides identical, respectively), we tentatively classified these two strains in agr group 1, at the junction between groups 2 and 3, respectively (Fig. 3). Our analysis also showed that the isolate giving an agr amplicon of reduced length is an agr group 1 strain (type R VII-A3 [Fig. 3]) with a deletion of a 330-bp region comprising the entire agrD gene.
The restriction map analysis indicates that group 1 is the most diverse agr group. This group contains 8 out of the 14 restriction types identified, whereas group 2, group 3, and group 4 contain only 3 types, 2 types, and 1 type, respectively. Finally, we found that the sequenced human strains COL and NCTC 8325 belong to agr group 1, that strains N315 and Mu50 belong to agr group 2, and that strains MRSA-252 and MSSA-476 belong to agr group 3 (Fig. 3).
Development of an agr group-specific multiplex PCR.
The analysis of the restriction maps of the variable region of the agr operon (Fig. 3) suggests that agr group-specific primers could be found in this region and used in an agr group-specific multiplex PCR. We thus analyzed in detail the nucleotide sequences of each of the four different agr operons obtained from GenBank (accession no. X52543, AF001782, AF001783, and AF288215) by using the Gene Jockey software (Biosoft, Cambridge, United Kingdom). One consensus forward primer (Pan) and four group-specific reverse primers (agr1, agr2, agr3, and agr4), which would allow the identification of the agr group on the basis of the molecular weight of its PCR product, were identified (Fig. 1). These primers were then tested experimentally in a multiplex PCR and shown to correctly identify the agr group of the agr reference strains RN6390 (group 1), RN6923 (group 2), RN8462 (group 3), and A880740 (group 4) (Fig. 4). We then used the above multiplex PCR to analyze the population of strains isolated from cows with mastitis. All of the 71 strains but 1 gave an amplifiable product. The latter strain is the one giving an agr amplicon of reduced length and was shown by restriction analysis to have a deletion of the sequence corresponding to the agr group 1- and group 3-specific primers (Fig. 3). As the 3′ extremity of this amplicon possesses the characteristic restriction sites of group 1 strains and not those of group 3 strains, we definitely classify this strain in the agr group 1.
FIG. 4.
agr group-specific multiplex PCR. S. aureus DNA from each of the four agr reference strains (RN6390 [group 1, lane 1], RN6923 [group 2, lane 2], RN8462 [group 3, lane 3], A880740 [group 4, lane 4]) were amplified with multiplex primers Pan, agr1, agr2, agr3, and agr4. PCR products were separated on a 1.5% agarose gel and visualized under UV.
The agr group-specific multiplex PCR confirms the above agr group classification of strains made by restriction analysis. Nevertheless, strains 125 and 130 isolated in Japan, tentatively classified by restriction analysis in group 1, are now classified by multiplex PCR in groups 2 and 3, respectively. This is due to the fact that primers agr2 and agr3 are not localized in the 3′ extremity of the agr variable region, which for these two strains is characteristic of group 1. These two strains are probably in a process of evolution from group 1 to groups 2 and 3, respectively. The classification of all six human strains used as controls into agr groups was also confirmed. The majority of strains isolated from cows with mastitis belong to agr group1 (69.0%) and to agr group 2 (23.9%). Groups 3 and 4 contain only 2.8 and 1.4% of the analyzed strains, respectively. As discussed above, the classification of two strains (strains 125 and 130) is uncertain.
trap restriction fragment length polymorphism.
As TRAP has been proved to be a component of a membrane-associated sensor able to induce the synthesis of RNA III via a signal transduction pathway other than agr, we analyzed whether trap interstrain variation, similar to that which was found for agr, does exist. To this end, we first aligned the trap sequences of six strains whose genomes are sequenced. Theoretical restrictions with MseI (a frequently cutting enzyme in trap) show that the agr group 1 or 2 strains COL, NCTC 8325, N315, and Mu50 have a similar MseI restriction profile and that this profile is different from those of the agr group 3 strains MRSA-252 and MSSA-476 (results not shown). These preliminary results indicate that trap is polymorphic and suggest that a relation between trap types and agr group could perhaps exist. This prompted us to analyze the entire S. aureus population isolated from cows with mastitis for trap polymorphism.
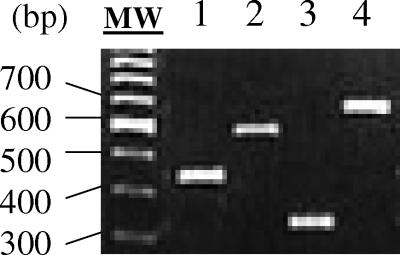
trap genes were amplified from all strains by PCR and restricted with MseI. This allowed the identification of four different restriction types among the analyzed population (Fig. 5). The vast majority of strains isolated from cows belong to trap type 1 (71.8%). Types 2, 3, and 4 account for 18.3, 7.0, and 2.8% of the analyzed strains, respectively (Table 1). It is worth noting that we were previously unable to amplify the variable region of the agr locus in the only two strains of trap type 4 identified (Table 1). The human sequenced strains COL, NCTC 8325, Mu50, and N315 belong to trap type 1, whereas strains MRSA-252 and MSSA-476 belong to trap types 2 and 3, respectively.
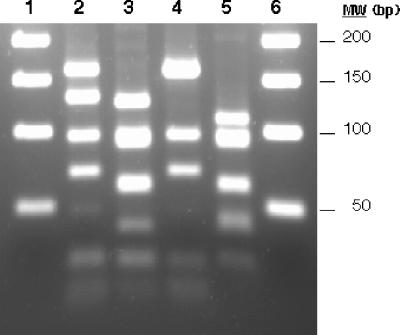
FIG. 5.
Restriction polymorphism in the trap gene. The trap gene was amplified by PCR, digested with MseI, and electrophoresed on a 3% agarose gel. Examples of the different restriction types obtained are shown: type 1 (lane 2), type 2 (lane 3), type 3 (lane 4), and type 4 (lane 5). Molecular weight markers (50-bp DNA ladder; Promega) are shown in lanes 1 and 6.
No particular relationship between unique agr group and trap type could be displayed. Indeed, our experiments showed that the agr groups 2, 1, and 3 contain strains of trap types 1 to 4, strains of trap types 1 to 3, and strains of trap types 2 and 3, respectively. The agr group 4 contains a unique strain of trap type 2 (Table 1). Whereas no relationship was found between agr groups and trap types, strains possessing a similar agr restriction type were also found to possess a similar trap type. Agr types R IV-A5 (containing strains of trap types 1 and 3) and R VI-A8 (containing strains of trap types 2 and 3) are nevertheless an exception to this rule (Table 1).
DISCUSSION
Polymorphism in the agr locus was first described by Ji et al. (15). This led to the classification of S. aureus isolates into four different interference groups (12, 15). Later, sequence variations within groups were also found (20, 25, 28, 29). In this work, we identified 14 different agr restriction types among the analyzed strains. Nevertheless, only 12 of them were present in our collection of bovine mastitis isolates. On the basis of the restriction maps of the agr variable region, we classified all agr types in one of the four interference groups (Fig. 3). This classification was confirmed by the agr group-specific PCR also developed in this work. Whereas strains belonging to each of the four agr groups were found, most of them (69.0%) were assigned to group 1. This repartition of strains among interference groups is quite similar to what was described by Moore and Lindsay for methicillin-sensitive hospital strains (19). The restriction sites used as markers to discriminate agr alleles indicate that the agrD sequences are stable within each interference group. Most mutations within groups appear to arise in agrC, the gene encoding the receptor of the AIP. Those mutations are probably not in sequences coding for amino acids interacting with the inducing peptide. The position of the AluI and RsaI sites in agrD seems to be sufficient to assign a strain to a particular agr group, whereas restriction sites characteristic of each group can also be found in other genes of this region (Fig. 3). This adds to previous reports showing that genes of the agr locus are submitted to a coevolutionary pressure, allowing the binding of a modified AIP to the receptor (15, 22). Our classification indicates that type R VII-A12 and type R IX-A11 are particular with respect to the coevolution of the propeptide and its receptor. Indeed, the receptor-encoding genes of type R VII-A12 and R IX-A11 are highly similar to those of the agr group 1 strains, whereas their propeptide-encoding genes are highly similar to those of groups 3 and 2, respectively. The fact that the propeptide and its receptor belong to different interference groups suggests that strains of type R VII-A12 and type R IX-A11 are impaired in the activation of RNA III by the agr system. We postulate that these two types are in a process of evolution from group 1 to groups 2 and 3, respectively. We also identified a strain (type R VII-A3) which has a deletion of the complete agrD gene and which should thus also be impaired in the activation of RNA III by the agr system. Strains of types R VII-A12, R IX-A11, and R VII-A3 are nevertheless virulent, because they were all isolated from the milk of cows with mastitis. In connection with this, Wesson et al. showed that a strain mutated in agr was internalized by cultured bovine mammary epithelial cells at a level greater than the wild-type strain but contrary to the wild type failed to induce apoptosis (31). Others have also isolated virulent S. aureus strains with an inactivated agr system (30). These strains show increased adherence and biofilm formation, and these properties were considered important for the development of chronic infection (24).
We were also interested to know if the RAP-TRAP system is polymorphic and ubiquitously associated with S. aureus strains. We identified the trap gene in all strains analyzed, and we proved that at least four different alleles exist. We also tried to learn if different alleles of the RAP-encoding gene exist. As the nucleotide sequence of RAP is unknown, we made BlastN searches with the available RAP NH2-terminal sequence (IKKYKPITN). Curiously, homologies were only found with the well-conserved L2 ribosomal protein of S. aureus (results not shown). The identification of the TRAP activator thus needs further clarification, as it is difficult to understand how a conserved ribosomal protein is able to activate the two-component system. RNA III-inhibiting peptide (RIP), a peptide of sequence YSPXTNF, isolated from culture supernatants of a coagulase-negative Staphylococcus species that is believe to be S. xylosus, was found to compete with RAP on inducing TRAP phosphorylation. This leads to inhibition of RNA III synthesis and to diminution of the virulence phenotype (2, 3, 9). These results need now to be extended to strains belonging to each of the four trap types identified in this work. With the exception of the Newbould 305 and the NCTC 8325 strains that we classified as trap type 1, nothing is known about the allelic variation of trap in the strains previously tested for inhibition by RIP. As we have shown that most of the S. aureus strains (71.8%) are trap type 1, it can be speculated that most, if not all, of the strains tested for inhibition by RIP are also trap type 1. It is thus still possible that RIP is not able to inhibit RNA synthesis in the other three trap types. Furthermore, it is also unknown if RAP purified from strains belonging to each of the four trap types are, as AIPs isolated from each of the four agr types, able to activate RNA III synthesis in strains of the same type and to inhibit this synthesis in strains of different types.
Most of the strains (56.3%) isolated from cows with mastitis belong to the agr R III-A1, trap 1 type. Our data indicate that strains belonging to this type have been able to infect cows from at least the end of the 1950s to date. The agr R III-A1, trap 1 type is also the type of the Newbould 305 strain (ATCC 29470), a strain isolated in the United States and widely used for experimental mastitis. This indicates that this type is not linked to a particular geographical location (under the circumstances of this work, France). The presence at a high prevalence of type agr R III-A1, trap 1 in the population of strains isolated from cows with mastitis suggests that this type has unique characteristics which, in contrast to the other rare types, endow it with superior ability to infect the bovine mammary gland. The agr R III-A1, trap 1 type is thus probably an S. aureus lineage that expands in the bovine population due to its possession of a unique combination of virulence genes. As no strain isolated from humans, whose agr sequence is available from GenBank, was found to belong to the R III-A1 type, it is tempting to think that on the contrary this S. aureus type is rarely isolated in the human population and that other agr and trap types are predominantly associated with human disease. This hypothesis will now be tested by analyzing the polymorphism of agr and trap in a population of S. aureus strains isolated from humans. Previous works using other methods also indicated that S. aureus isolated from humans and from cows has a predominantly clonal structure and that, whereas numerous types could be identified, only few of them are predominantly associated with a particular host and disease (1, 5-8, 13, 16, 33). The identification and characterization of a disease-dominant lineage(s) are very important for the development of vaccines and diagnostic tests. It could be expected that such works will lead in the future to the discovery of genetic determinants responsible for the tropism of different S. aureus lineages for specific hosts and tissues and to the development of new prophylactic and diagnostic tools.
ADDENDUM
The sequence of the whole genome of MW2, a strain of community-acquired methicillin-resistant S. aureus, became available after this paper was submitted (GenBank accession no. AP004822 to AP004832). This strain falls into the classifications agr group 3 (type R VI-A8) and trap type 3.
Acknowledgments
We thank Mark Enright (University of Bath, Bath, United Kingdom) for the gift of strains MRSA-252 and MSSA-476, John Iandolo (University of Oklahoma, Oklahoma City) for the gift of strain NCTC 8325, Keiichi Hiramatsu (Juntendo University, Tokyo, Japan) for the gift of strains N315 and Mu50, Philippe Moreillon (Centre Hospitalier Universitaire Vaudois, Lausanne, Switzerland) for the gift of strain COL, and Shotaro Takeuchi (Fukui Prefectural University, Fukui, Japan) for the gift of strains 125 and 130. The release of preliminary sequence data by the S. aureus NCTC 8325 Genome Sequencing Team at the University of Oklahoma Health Sciences Center, by the S. aureus MRSA-252 and MSSA-476 Sequencing Group at the Sanger Institute, and by the S. aureus COL Sequencing Team at The Institute for Genomic Research is acknowledged. We are grateful to Martine Braibant for careful reading of the manuscript.
This work was supported by a grant (AIP P00060, P00223) from the French association Bureau des Ressources Génétiques (BRG).
REFERENCES
- 1.Akineden, O., C. Annemüller, A. Hassan, C. Lämmler, W. Wolter, and M. Zschöck. 2001. Toxin genes and other characteristics of Staphylococcus aureus isolates from milk of cows with mastitis. Clin. Diagn. Lab. Immunol. 8:959-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balaban, N., L. V. Collins, J. S. Cullor, E. B. Hume, E. Medina-Acosta, O. Vieira da Motta, R. O'Callaghan, P. V. Rossitto, M. E. Shirtliff, L. Serafim da Silveira, A. Tarkowski, and J. V. Torres. 2000. Prevention of diseases caused by Staphylococcus aureus using the peptide RIP. Peptides 21:1301-1311. [DOI] [PubMed] [Google Scholar]
- 3.Balaban, N., T. Goldkorn, Y. Gov, M. Hirshberg, N. Koyfman, H. R. Matthews, R. T. Nhan, B. Singh, and O. Uziel. 2001. Regulation of Staphylococcus aureus pathogenesis via target of RNAIII-activating protein (TRAP). J. Biol. Chem. 276:2658-2667. [DOI] [PubMed] [Google Scholar]
- 4.Boom, R., C. J. Sol, M. M. Salimans, C. L. Jansen, P. M. Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Booth, M. C., L. M. Pence, P. Mahasreshti, M. C. Callegan, and M. S. Gilmore. 2001. Clonal associations among Staphylococcus aureus isolates from various sites of infection. Infect. Immun. 69:345-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Day, N. P., C. E. Moore, M. C. Enright, A. R. Berendt, J. M. Smith, M. F. Murphy, S. J. Peacock, B. G. Spratt, and E. J. Feil. 2001. A link between virulence and ecological abundance in natural populations of Staphylococcus aureus. Science 292:114-116. [DOI] [PubMed] [Google Scholar]
- 7.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fitzgerald, J., W. Meaney, P. Hartigan, C. Smyth, and V. Kapur. 1997. Fine-structure molecular epidemiological analysis of Staphylococcus aureus recovered from cows. Epidemiol. Infect. 119:261-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gov, Y., A. Bitler, G. Dell'Acqua, J. V. Torres, and N. Balaban. 2001. RNA III inhibiting peptide (RIP), a global inhibitor of Staphylococcus aureus pathogenesis: structure and function analysis. Peptides 22:1609-1620. [DOI] [PubMed] [Google Scholar]
- 10.Hogan, J., R. Gonzalez, R. Harmon, S. Nickerson, S. Oliver, J. Pankey, and K. Smith. 1999. Laboratory handbook on bovine mastitis. National Mastitis Council, Inc., Madison, Wis.
- 11.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jarraud, S., G. Lyon, A. Figueiredo, G. Lina, F. Vandenesch, J. Etienne, T. Muir, and R. Novick. 2000. Exfoliatin-producing strains define a fourth agr specificity group in Staphylococcus aureus. J. Bacteriol. 182:6517-6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarraud, S., C. Mougel, J. Thioulouse, G. Lina, H. Meugnier, F. Forey, X. Nesme, J. Etienne, and F. Vandenesch. 2002. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect. Immun. 70:631-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ji, G., R. Beavis, and R. Novick. 1995. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc. Natl. Acad. Sci. USA 92:12055-12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ji, G., R. Beavis, and R. P. Novick. 1997. Bacterial interference caused by autoinducing peptide variants. Science 276:2027-2030. [DOI] [PubMed] [Google Scholar]
- 16.Kapur, V., W. M. Sischo, R. S. Greer, T. S. Whittam, and J. M. Musser. 1995. Molecular population genetic analysis of Staphylococcus aureus recovered from cows. J. Clin. Microbiol. 33:376-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kluytmans, J., A. van Belkum, and H. Verbrugh. 1997. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin. Microbiol. Rev. 10:505-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuroda, M., T. Ohta, I. Uchiyama, et al. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 19.Moore, P. C., and J. A. Lindsay. 2001. Genetic variation among hospital isolates of methicillin-sensitive Staphylococcus aureus: evidence for horizontal transfer of virulence genes. J. Clin. Microbiol. 39:2760-2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mullarky, I. K., C. Su, N. Frieze, Y. H. Park, and L. M. Sordillo. 2001. Staphylococcus aureus agr genotypes with enterotoxin production capabilities can resist neutrophil bactericidal activity. Infect. Immun. 69:45-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neave, F., and J. Oliver. 1962. The relationship between the number of mastitis pathogens placed on the teats of dry cows, their survival, and the amount of intramammary infection caused. J. Dairy Res. 29:79-93. [Google Scholar]
- 22.Novick, R. 2000. Pathogenicity factors of Staphylococcus aureus and their regulation, p. 392-407. In V. Fischetti (ed.), Gram-positive pathogens. ASM Press, Washington, D.C.
- 23.Novick, R. P., H. F. Ross, S. J. Projan, J. Kornblum, B. Kreiswirth, and S. Moghazeh. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12:3967-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Otto, M. 2001. Staphylococcus aureus and Staphylococcus epidermidis peptide pheromones produced by the accessory gene regulator agr system. Peptides 22:1603-1608. [DOI] [PubMed] [Google Scholar]
- 25.Papakyriacou, H., D. Vaz, A. Simor, M. Louie, and M. McGavin. 2000. Molecular analysis of the accessory gene regulator (agr) locus and balance of virulence factor expression in epidemic methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 181:990-1000. [DOI] [PubMed] [Google Scholar]
- 26.Prasad, L., and F. Newbould. 1968. Inoculation of the bovine teat duct with Staphylococcus aureus: the relationship of teat duct length, milk yield and milking rate to development of intramammary infection. Can. Vet. J. 9:107-115. [PMC free article] [PubMed] [Google Scholar]
- 27.Sutra, L., and B. Poutrel. 1994. Virulence factors involved in the pathogenesis of bovine intramammary infections due to Staphylococcus aureus. J. Med. Microbiol. 40:79-89. [DOI] [PubMed] [Google Scholar]
- 28.Takeuchi, S., T. Maeda, N. Hashimoto, K. Imaizumi, T. Kaidoh, and Y. Hayakawa. 2001. Variation of the agr locus in Staphylococcus aureus isolates from cows with mastitis. Vet. Microbiol. 79:267-274. [DOI] [PubMed] [Google Scholar]
- 29.van Leeuwen, W., W. van Nieuwenhuizen, C. Gijzen, H. Verbrugh, and A. van Belkum. 2000. Population studies of methicillin-resistant and -sensitive Staphylococcus aureus strains reveal a lack of variability in the agrD gene, encoding a staphylococcal autoinducer peptide. J. Bacteriol. 182:5721-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vuong, C., H. L. Saenz, F. Gotz, and M. Otto. 2000. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J. Infect. Dis. 182:1688-1693. [DOI] [PubMed] [Google Scholar]
- 31.Wesson, C. A., L. E. Liou, K. M. Todd, G. A. Bohach, W. R. Trumble, and K. W. Bayles. 1998. Staphylococcus aureus Agr and Sar global regulators influence internalization and induction of apoptosis. Infect. Immun. 66:5238-5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.West, A., and A. Stock. 2001. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26:369-376. [DOI] [PubMed] [Google Scholar]
- 33.Zadoks, R., W. van Leeuwen, H. Barkema, O. Sampimon, H. Verbrugh, Y. H. Schukken, and A. van Belkum. 2000. Application of pulsed-field gel electrophoresis and binary typing as tools in veterinary clinical microbiology and molecular epidemiologic analysis of bovine and human Staphylococcus aureus isolates. J. Clin. Microbiol. 38:1931-1939. [DOI] [PMC free article] [PubMed] [Google Scholar]