Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) is one of the most important causes of hospital infections worldwide. High-level resistance to methicillin is caused by the mecA gene, which encodes an alternative penicillin-binding protein, PBP 2a. To determine the clonal relationships between methicillin-susceptible S. aureus (MSSA) and MRSA, we typed 1,069 S. aureus isolates (493 MSSA isolates and 576 MRSA isolates), collected mainly in North American and European hospitals between the 1960s and the year 2000, using pulsed-field gel electrophoresis and ribotyping. Of 10 widespread S. aureus lineages recognized, 8 had corresponding mecA-positive strains. Multiresistant MRSA strains are found in hospitals worldwide, while unrelated and more susceptible strains represent less than 1% of the MRSA population. This supports the hypothesis that horizontal transfer plays an important role in the dissemination of the mecA gene in the S. aureus population.
Staphylococcus aureus strains resistant to methicillin and many other antibiotics are major causes of nosocomial infections worldwide (8). Resistance to methicillin is determined by the mecA gene, which encodes the low-affinity penicillin-binding protein PBP 2A (3). The mecA gene is part of a 21- to 60-kb staphylococcal chromosome cassette mec (SCCmec), a mobile genetic element that may also contain genetic structures such as Tn554, pUB110, and pT181 which encode resistance to non-β-lactam antibiotics (15). Two hypotheses have been raised to explain the evolutionary origin of methicillin-resistant S. aureus (MRSA) strains. The single clone hypothesis, based on early analyses of the restriction fragment length polymorphisms obtained for MRSA isolates collected worldwide by using probes for mecA and Tn554, suggests that mecA entered the S. aureus population on one occasion and resulted in the formation of a single MRSA clone that has since spread around the world (15, 16). The second hypothesis, based on the detection of mecA in diverse S. aureus multilocus enzyme electrophoresis types, proposes that MRSA strains evolved a number of times by means of the horizontal transfer of mecA into phylogenetically distinct methicillin-susceptible S. aureus (MSSA) precursor strains (17). By using DNA microarray technology, mecA has been detected in at least five divergent lineages, implying that horizontal mecA transfer has played a fundamental role in the evolution of MRSA (9). The transfer of mecA from S. epidermidis to S. aureus was recently witnessed in vivo, suggesting that mecA may transfer more frequently to MSSA (21).
MRSA is also emerging in the community, particularly in the United States, where 28% of community-acquired S. aureus strains may be resistant to methicillin (4, 5, 8, 11). The prevalence of MRSA in the community is predicted to increase substantially due to the dissemination of a successful SCCmec type by horizontal transfer (5, 13). We present molecular typing data that support the theory of frequent mecA gene transfer into resident lineages of S. aureus, with the resulting formation of numerous MRSA clones. The population framework that we established can be exchanged between laboratories by use of automated riboprinting.
MATERIALS AND METHODS
Bacterial isolates.
The clonal relationships and antimicrobial susceptibilities of 1,069 S. aureus isolates, including 576 mecA-positive (mecA+) isolates (which are, by definition, MRSA) and 493 mecA-negative (mecA−) isolates (which are MSSA) were determined. These isolates were selected from different sources in order to study isolates from different temporal and geographic backgrounds. The origins of the isolates were as follows (Table 1): 397 MRSA and 260 MSSA isolates had been collected between April 1997 and December 1998 in 20 university hospitals in 12 European countries as part of the SENTRY Antimicrobial Surveillance Program (10). They included isolates from Athens, Greece (8 mecA− isolates, 19 mecA+ isolates); Düsseldorf (10 mecA−, 9 mecA+) and Freiburg (14 mecA−, 2 mecA+), Germany; Lausanne, Switzerland (56 mecA−, 1 mecA+); Linz, Austria (7 mecA−, 7 mecA+); Paris (group 1) (10 mecA−, 21 mecA+), Paris (group 2) (7 mecA−, 33 mecA+), Lille (7 mecA−, 22 mecA+), and Lyon (7 mecA−, 14 mecA+), France; Coimbra, Portugal (16 mecA−, 70 mecA+); Warsaw (7 mecA−, 17 mecA+) and Krakow (5 mecA−, 5 mecA+), Poland; Madrid (14 mecA−, 2 mecA+), Seville (18 mecA−, 29 mecA+), and Barcelona (7 mecA−, 6 mecA+), Spain; Rome (10 mecA−, 32 mecA+) and Genoa (9 mecA−, 34 mecA+), Italy; Brussels, Belgium (10 mecA−, 31 mecA+); London, United Kingdom (26 mecA−, 36 mecA+); and Istanbul, Turkey (12 mecA−, 7 mecA+). An additional 181 MSSA and 54 MRSA isolates had been collected between 1996 and 1999 at the University Medical Center (UMC), Utrecht, The Netherlands, where patients and staff coming from foreign hospitals are screened for MRSA carriage. These MRSA isolates, detected during 12 MRSA outbreak episodes, had evaded the hospital's search-and-destroy procedure and could not be linked epidemiologically to any foreign hospitals. One hundred three more MRSA isolates were selected to represent the genetic diversity of the MRSA collections of Kreiswirth et al. (16), Roberts et al. (18), de Lencastre et al. (7, 19), and Witte et al. (22). They included the earliest isolates collected from Europe and Africa during the 1960s, North American isolates collected from the 1970s to the 1990s, and European reference strains like the Iberian clone (7), the Brazilian clone (7), the North German clone(22), the South German clone (22), the Berlin clone (22), the Hannover clone (22), the Portuguese clone (7), the Pediatric clone (19), EMRSA 15 (14), and EMRSA 16 (14). Another 12 MRSA isolates that had been collected in South Africa during 1998 were studied. For comparison, MSSA strains ATCC 29213 and ATCC 12600 were included. Finally, 10 MRSA and 50 MSSA isolates that had been taken from colonized patients who had no clinical signs of S. aureus infection within 2 h after admission to the Cook County Hospital (Chicago, Ill.) were included.
TABLE 1.
Origins of isolates
| Collection | Description (reference) | No. of isolates
|
|
|---|---|---|---|
| mecA− (n = 493) | mecA+(n = 576) | ||
| SENTRY | European antimicrobial surveillance program (1997-1998) (10) | 260 | 397 |
| UMC | Dutch hospital with MRSA prevalence < 1% (1996-1999) | 181 | 54 |
| Chicago | Samples from community in Chicago | 50 | 10 |
| ATCCa | Control strains ATCC 29213 and ATCC 12600 | 2 | |
| South Africa | Samples from different hospitals | 12 | |
| W. Witte | Selected European MRSA genotypes (22) | 20 | |
| H. de Lencastre | Selected European MRSA genotypes (7, 19) | 4 | |
| B. Kreiswirth | Selected European, African, and American genotypes 1960-1990 (16) | 45 | |
| R. B. Roberts | Selected MRSA genotypes from New York City hospitals (18) | 34 | |
ATCC, American Type Culture Collection.
The isolates were identified as S. aureus by routine microbiological methods. Only one isolate per patient was included.
Susceptibility testing.
Susceptibility to oxacillin, erythromycin, clindamycin, rifampin, chloramphenicol, ciprofloxacin, gentamicin, and tetracycline was determined by the broth microdilution method defined by the National Committee for Clinical Laboratory Standards. Isolates were considered multiresistant when they displayed a decreased susceptibility to at least four of the eight antimicrobial agents tested.
Detection and amplification of mecA by PCR.
PCR was used to detect mecA DNA in methicillin-resistant isolates. A few colonies were picked from blood agar; suspended in 200 μl of a lysis buffer containing 10 mM Tris-HCl buffer (pH 8.0), 50 mM NaCl, lysostaphin (100 μg/ml), achromopeptidase (100 μg/ml), and RNase (100 μg/ml); incubated at 30°C for 45 min; boiled for 5 min; and then diluted by the addition of 400 μl of TE (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). For the PCR, 1 μl of lysate was added as a template to 24 μl of a reaction mixture containing 10 mM Tris-HCl (pH 9.0), 1.5 mM MgCl2, 50 mM KCl, 0.1% Triton X-100, 0.2 mM each deoxynucleoside triphosphate, and 0.75 U of Supertaq DNA polymerase (HT Biotechnology, Cambridge, United Kingdom). MecA DNA was amplified with the primers 5′-GTT GTA GTT GTC GGG TTT GG-3′ and 5′CTT CCA CAT ACC ATC TTC TTT AAC-3′ (20 μM). These primers were designed on the basis of the mecA sequence (GenBank accession no. X52593) with Primer software (Educational Software, State Line, Pa.). To test the quality of the template used for PCR, we added primers for the amplification of 16S ribosomal DNA (5′-AGG CCC GGG AAC GTA TTC AC-3′ and 5′-GAG GAA GGT GGG GAT GAC GT-3′) (20 μM) to each PCR mixture and monitored the amplification of 16S ribosomal DNA. Samples were subjected to 30 cycles consisting of 1 min at 95°C, 1 min at an annealing temperature ramped from 65 to 55°C during the first 10 cycles, and 1 min at 72°C in a thermocycler. The PCR product was visualized on a 1.5% agarose gel by using ethidium bromide and a UV transilluminator.
Pulsed-field gel electrophoresis (PFGE) analysis.
Genomic DNA was digested with SmaI and resolved with the CHEF-DRII system (Bio-Rad Laboratories, Hercules, Calif.), as described by the manufacturer.
Ribotyping.
Ribotypes were determined with an automated riboprinter system (Qualicon, Wilmington, Del.) and EcoRI, as described by the manufacturer.
Analysis of restriction patterns.
The restriction patterns were compared by calculating a similarity index by using the unweighted pair group method with arithmetic averages cluster algorithm and the Dice coefficient provided by the Bionumerics software (Applied Mathematics, Kortrijk, Belgium).
RESULTS
Clonal relationships among S. aureus isolates.
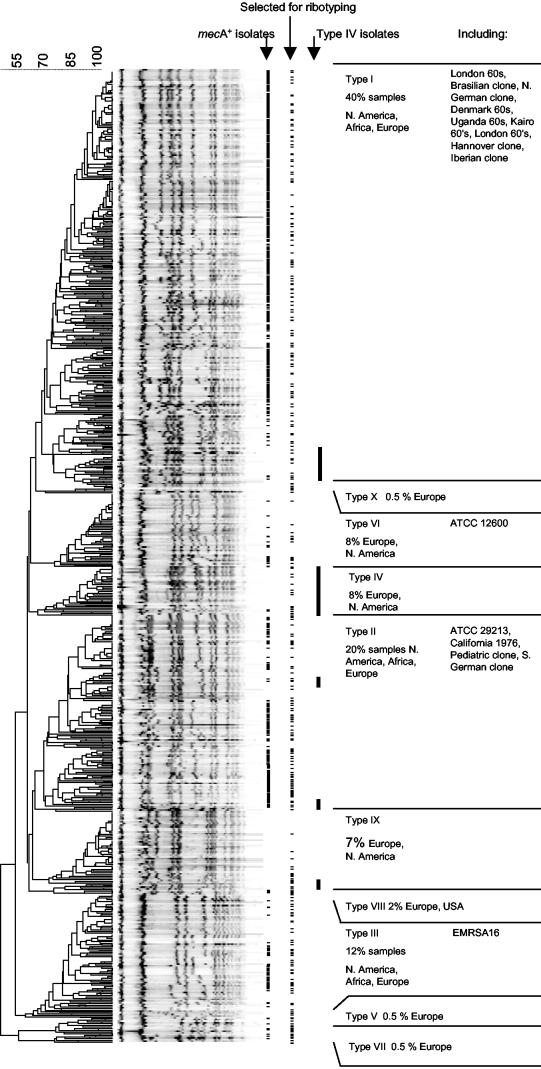
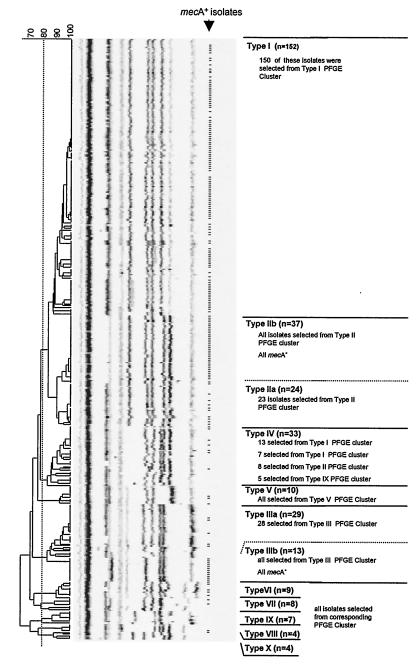
When the PFGE patterns of 576 MRSA isolates and 493 MSSA isolates were compared in a dendrogram, 10 major clusters of various sizes were discerned. To confirm the clonal relatedness of the isolates within these clusters, 330 isolates were selected for ribotyping, which covered the chromosomal diversity of the isolates (Fig. 1). Compared to the diversity of the PFGE patterns, the riboprints were much more conserved during evolution. Ten clusters were distinguished at the 80% similarity level, and these 10 clusters defined clonal lineages called S. aureus types I to X (Fig. 2). A very good correlation between PFGE typing and ribotyping was observed: except for the type IV isolates, 99% of the isolates typed by both methods clustered in corresponding branches of both the riboprint and the PFGE dendrograms. The PFGE patterns obtained for the type IV isolates were found not only in a cluster of their own but also in the PFGE clusters of other types as well (Fig. 1). Furthermore, both type II and type III isolates could be divided into two subtypes (subtypes a and b) that formed subclusters in the ribotyping dendrogram and that had different susceptibility patterns.
FIG. 1.
PFGE patterns from European and North American MSSA (n = 493) and MRSA (n = 576) isolates form 10 clusters. Isolates containing mecA are indicated by hyphens. In order to confirm the clonal relatedness of the isolates within the 10 clusters, a total of 330 MSSA and MRSA isolates representing the various PFGE types encountered were selected for ribotyping (indicated by hyphens). The positions of the type IV isolates in the PFGE dendrogram are indicated by black lines.
FIG. 2.
Ten clusters were obtained by ribotyping of 330 isolates, which represented the variability of PFGE types encountered. These clonal S. aureus lineages were called types I to X. There was an excellent correlation between PFGE and ribotyping. Except for the type IV isolates, 99% of the isolates typed by both methods were found in the corresponding PFGE and ribotype clusters. Isolates containing mecA are indicated by hyphens. Automated riboprinting allows standardized exchange of data between laboratories.
The more successful S. aureus types consisted of isolates which displayed numerous band-shift variations on widely disseminated and frequently isolated dominant PFGE patterns (Fig. 1). Pandemic clones of isolates yielding identical PFGE patterns were present in many hospitals on both continents. Of the type I (40% of all samples), type II (20%), and type III (12%) lineages, indistinguishable isolates were present in nearly all hospitals studied. Type IV (8%), type VI (8%), type IX (7%), and type VIII (2%) clones were also detected in Europe and North America, although less frequently. Isolates of the remaining lineages (type V [2%], type VII [0.5%], and type X [0.5%]) were referred only from European countries.
Dissemination of mecA in the different S. aureus lineages.
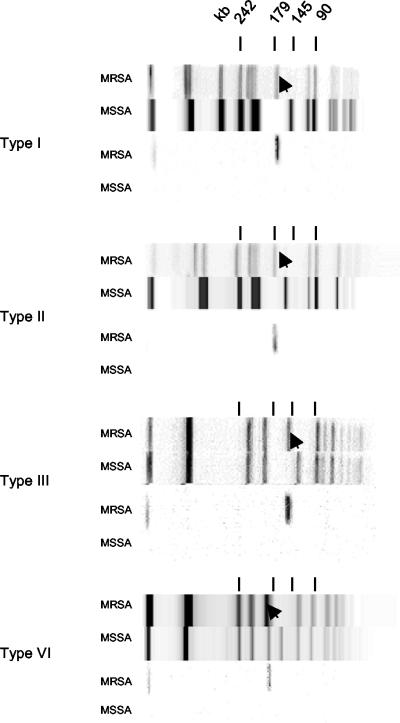
All isolates was assessed for the presence of mecA by PCR. MecA was detected in representatives of all but the type IX and X lineages. Some pandemic mecA− MSSA clones came with mecA+ MRSA counterparts that shared the identical ribotype, while their PFGE patterns differed by a single band shift due to insertion of a fragment containing mecA (Fig. 3). However, we found no mecA− counterparts among type IIb and type IIIb isolates, which, with no exception, all contained mecA.
FIG. 3.
PFGE patterns of mecA+ and mecA− counterparts (upper panels) that differ by a single band shift (arrowhead) due to the insertion of a fragment that hybridizes with a mecA probe (lower panels). The type II and type VI MSSA PFGE patterns are for control strains ATCC 29213 and ATCC 12600, respectively.
More than 60% of the mecA+ isolates belonged to the type I lineage. These isolates predominated in North America, Africa, and Europe. The type I MRSA isolates included all MRSA isolates from the 1960s, the Brazilian clone (7), the North German clone (22), the Hannover clone (22), the Iberian clone (7), and the Portuguese clone (7). One-quarter of the mecA+ isolates belonged to the type II lineage. Isolates of the type II lineage were dominant among the North American isolates from the 1980s and were later isolated in Europe and Africa. Type IIa MRSA isolates (14% of the mecA+ isolates) included the Pediatric clone (19). Type IIb MRSA isolates (11% of mecA+ isolates) included the South German clone (22).
Type III MRSA isolates were found among isolates recovered since the 1980s in Africa, Europe, and North America. Type IIIb MRSA isolates (6% of mecA+ isolates) included EMRSA 16 (14). In contrast, type IIIa, type IV (the Berlin clone [22]), type V (EMRSA 15 [14]), type VI, type VII, and type VIII MRSA strains, which appeared during the 1990s, were isolated only sporadically (<1% of mecA+ isolates). Interestingly, these sporadic MRSA types were relatively abundant among the different UMC genotypes (33% type IIa, type IV, and type VI isolates), which are epidemiologically unlinked to the hospital epidemic MRSA isolates, and among the mecA+ isolates from the community in Chicago (20% type IV isolates).
Multiresistance and dissemination of MRSA in Europe.
To correlate the resistance profiles of MRSA strains with their current dissemination, the susceptibilities of the recent European S. aureus isolates were compared. The mecA gene was present in all isolates resistant to four or more antibiotics. Moreover, this multiresistance was displayed by the most prevalent and geographically widespread MRSA types (types I, IIa, IIb, and IIIb), which together represented 99% of the mecA+ population in Europe. Of the European isolates that appeared to be susceptible to methicillin in the phenotypic test, 10% nevertheless contained mecA, and some of these were multiresistant. In contrast, 5% of the phenotypically methicillin-resistant isolates did not carry mecA and displayed low-level resistance (>8 μg/ml).
Most of the type I MRSA isolates, representing 68% of the recent European MRSA population and being present in 17 of 20 hospitals participating in the SENTRY program, were resistant to erythromycin (97%), gentamicin (98%), and clindamycin (89%) and showed decreased susceptibility to ciprofloxacin (98%), tetracycline (98%), and rifampin (98%). Although most of the type IIb MRSA isolates, which represented 16% of the recent European isolates and which were found in 5 of 20 hospitals participating in the SENTRY program, were resistant to erythromycin (98%), clindamycin (88%), ciprofloxacin (98%), and gentamicin (100%), they remained susceptible to tetracycline (98%) and rifampin (100%). Type IIIb MRSA isolates, which represented 8% of the recent European MRSA isolate population and which were found in 2 of 21 hospitals participating in the SENTRY program, were resistant to erythromycin (100%), ciprofloxacin (100%), and clindamycin (84%) but remained susceptible to rifampin (100%), tetracycline (100%), and gentamicin (84%). Also, the type IIa MRSA isolates, which represented 7% of the MRSA isolate population and which were found in 6 of 20 hospitals participating in the SENTRY program, were mostly resistant to erythromycin (75%), clindamycin (69%), and ciprofloxacin (90%) but remained susceptible to rifampin (100%), tetracycline (100%), and gentamicin (100%). In contrast, type IIIa, type IV, type V, type VI, type VII, and type VIII MRSA isolates, which were isolated only sporadically (<1% of recent European mecA+ isolates), mostly remained susceptible to all but the β-lactam antibiotics (clindamycin, 100%; tetracycline, 100%; gentamicin, 100%; rifampin, 100%; erythromycin, 60%; ciprofloxacin, 60%).
DISCUSSION
The mecA gene, which lies in the SCCmec resistance island (13), is carried by 95% of the isolates that display a phenotype of methicillin resistance and was detected in all multiresistant S. aureus isolates. This study aimed to examine the dissemination of mecA in the S. aureus population. Two methods were applied to determine the clonal relationships between mecA+ MRSA and mecA− MSSA isolates collected between 1960 and 2000 from over 50 locations in the Western world. The overall chromosomal organizations of the isolates were first compared by using SmaI-generated PFGE patterns, which provide a relatively quickly evolving genotypic marker. Because there is little evolutionary pressure to conserve the SmaI restriction sites per se, this technique has high discriminatory power and highlights some of the differences between the strains. Ribotyping was then used to combine evolutionarily closely related PFGE types into clonal lineages. The complete ribotyping procedure has been automated and is coupled to a database management system, allowing electronic data exchange between different centers. The genes that encode ribosomal DNA are more conserved during evolution and provide a relatively slowly evolving marker. Therefore, automated ribotyping results in fewer ribotypes compared to the diversity of PFGE types. Combination of PFGE and ribotyping allowed grouping of all geographically widespread isolates in distinct clonally related lineages except for some of the type IV isolates, which may be ancestrally related to many types.
S. aureus isolates of 10 different lineages, called types I to X, were present in the hospitals studied, and 8 of these have acquired mecA. Our data show the worldwide dissemination of both successful MSSA strains and successful MRSA strains. From the major lineages, both pandemic MSSA and MRSA clones yielding identical PFGE patterns were collected in many European and North American hospitals and the community. Several pandemic mecA− MSSA clones have mecA+ counterparts that share identical ribotypes, while their PFGE patterns differ by a single band shift due to acquisition of the element containing mecA. The worldwide appearance of specific MRSA clones has been shown before (7), but the existence of widespread MSSA counterparts was not described before in detail. In line with this observation, a comparison of MRSA and MSSA isolates isolated in the United Kingdom and Denmark in the early 1960s suggests that contemporary MSSA isolates served as an early recipient of the mecA gene in Europe (6).
The dissemination of particular MRSA lineages is correlated with their resistance profiles. The majority of the multiresistant type I MRSA isolates, predominant on both continents, lacked susceptibility to tetracycline, erythromycin, clindamycin, gentamicin, ciprofloxacin, and rifampin. A second multiresistant lineage (type IIb), found in North America and several European countries, was susceptible to tetracycline and rifampin. Smaller pandemics were caused by isolates susceptible to gentamicin, tetracycline, and rifampin (type IIa and type IIIb). Although antibiotic selection pressure by itself provides a reasonable explanation for the widespread dissemination of such multiresistant strains, additional factors, e.g., modifications in expression of virulence factors and binding capacities, may add to their high prevalence. In contrast, the sporadically isolated mecA+ MRSA types, type IIIa and types IV to VIII (1% of recent European isolates), generally remained susceptible to all except the β-lactam antibiotics. Sporadic MRSA types were overrepresented in the samples from the community in Chicago (20%) and UMC (30%), where the search-and-destroy procedure prevents epidemic MRSA strains from entering the hospital, keeping the prevalence of MRSA isolates below 1%. In both the community and the hospital, sporadic MRSA types, which are not descendants of the epidemic hospital clones, may be formed de novo by the horizontal transfer of mecA to resident MSSA lineages, as was witnessed recently in vivo when the gene was passed from S. epidermidis to S. aureus during antibiotic treatment (21).
Several investigators (2, 9, 17, 21) have questioned the hypothesis that the gene entered S. aureus on only one occasion. The data presented here show the repeated horizontal transfer of mecA to at least eight resident S. aureus lineages and the spread of more resistant clones favored most by antibiotic selection pressure. When DNA microarray technology was used to characterize the genetic diversity of 11 MRSA isolates, mecA was detected in at least five highly divergent chromosomal genetic groups (9). In addition, multilocus enzyme electrophoresis data showed that MRSA isolates comprise 15 electrophoretic types that form six clusters or lineages, indicating that multiple MRSA lineages arose by horizontal transfer to S. aureus (17). Analysis of the mecI and mecR1 region, which lies 5′ of mecA, revealed that older isolates lacked part of this region, whereas more recent isolates showed an organization at this position similar to that in coagulase-negative staphylococci (CoNS) (2, 12), and it has been proposed that CoNS serve as donors for the transfer of the mecA gene to S. aureus (1, 20, 21). In this context, it is noteworthy that 70 to 75% of all CoNS worldwide are now resistant to methicillin (8), thus representing a huge potential reservoir of resistance. The mechanism of transfer, however, remains unclear. There is evidence that mecA resides within a mobile genetic element, SSCmec, that encodes recombinases for its excision from and integration into the staphylococcal chromosome (15). This element may also contain other genetic elements, like Tn554, pUB110, and pT181, which encode resistance to non-β-lactam antibiotics, causing multiresistance (15). Thus, while new MRSA strains will continue to emerge by the horizontal transfer of the mecA gene, those strains disseminated widely possess additional resistance traits and are favored most by antibiotic selection pressure.
Acknowledgments
We acknowledge B. Kreiswirth, R. B. Roberts, H. de Lencastre, and W. Witte, who collected many of the isolates. In addition, we thank Adrienne Box, Mirjam Klootwijk, Karlijn Kusters, Stefan de Vaal, and Roland Geisel for proficient technical support and Max Heck for advice on PFGE.
REFERENCES
- 1.Archer, G. L., and D. M. Niemeyer. 1994. Origin and evolution of DNA associated with resistance to methicillin in staphylococci. Trends Microbiol. 2:343-347. [DOI] [PubMed] [Google Scholar]
- 2.Archer, G. L., D. M. Niemeyer, J. A. Thanassi, and M. J. Pucci. 1994. Dissemination among staphylococci of DNA sequences associated with methicillin resistance. Antimicrob. Agents Chemother. 38:447-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beck, W. D., B. Berger-Bachi, and F. H. Kayser. 1986. Additional DNA in methicillin-resistant Staphylococcus aureus and molecular cloning of mec-specific DNA. J. Bacteriol. 165:373-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 1999. Four pediatric deaths from community acquired methicillin-resistant S. aureus—Minnesota and North Dakota, 1997-1999. Morb. Mortal. Wkly. Rep. 48:707-710. [PubMed] [Google Scholar]
- 5.Chambers, H. F. 2001. The changing epidemiology of Staphylococcus aureus? Emerg. Infect. Dis. 7:178-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crisostomo, M. I., H. Westh, A. Tomasz, M. Chung, D. C. Oliveira, and H. de Lencastre. 2001. The evolution of methicillin resistance in Staphylococcus aureus: similarity of genetic backgrounds in historically early methicillin-susceptible and -resistant isolates and contemporary epidemic clones. Proc. Natl. Acad. Sci. USA 98:9865-9870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Sousa, M. A., I. S. Sanches, M. L. Ferro, M. J. Vaz, Z. Saraiva, T. Tendeiro, J. Serra, and H. de Lencastre. 1998. Intercontinental spread of a multidrug-resistant methicillin-resistant Staphylococcus aureus clone. J. Clin. Microbiol. 36:2590-2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diekema, D. J., M. A. Pfaller, F. J. Schmitz, J. Smayevsky, J. Bell, R. N. Jones, and M. Beach. 2001. Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY antimicrobial surveillance program, 1997-1999. Clin. Infect Dis. 32(Suppl. 2):S114-S132. [DOI] [PubMed] [Google Scholar]
- 9.Fitzgerald, J. R., D. E. Sturdevant, S. M. Mackie, S. R. Gill, and J. M. Musser. 2001. Evolutionary genomics of Staphylococcus aureus: insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. Proc. Natl. Acad. Sci. USA 98:8821-8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fluit, A. C., C. L. Wielders, J. Verhoef, and F. J. Schmitz. 2001. Epidemiology and susceptibility of 3,051 Staphylococcus aureus isolates from 25 university hospitals participating in the European SENTRY Study. J. Clin. Microbiol. 39:3727-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herold, B. C., L. C. Immergluck, M. C. Maranan, D. S. Lauderdale, R. E. Gaskin, S. Boyle-Vavra, C. D. Leitch, and R. S. Daum. 1998. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA 279:593-598. [DOI] [PubMed] [Google Scholar]
- 12.Hiramatsu, K. 1995. Molecular evolution of MRSA. Microbiol. Immunol. 39:531-543. [DOI] [PubMed] [Google Scholar]
- 13.Hiramatsu, K., L. Cui, M. Kuroda, and T. Ito. 2001. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol. 9:486-493. [DOI] [PubMed] [Google Scholar]
- 14.Hookey, J. V., J. F. Richardson, and B. D. Cookson. 1998. Molecular typing of Staphylococcus aureus based on PCR-restriction fragment length polymorphism and DNA sequence analysis of the coagulase gene. J. Clin. Microbiol. 36:1083-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito, T., and K. Hiramatsu. 1998. Acquisition of methicillin resistance and progression of multiantibiotic resistance in methicillin-resistant Staphylococcus aureus. Yonsei Med. J. 39:526-533. [DOI] [PubMed] [Google Scholar]
- 16.Kreiswirth, B., J. Kornblum, R. D. Arbeit, W. Eisner, J. N. Maslow, A. McGeer, D. E. Low, and R. P. Novick. 1993. Evidence for a clonal origin of methicillin resistance in Staphylococcus aureus. Science 259:227-230. [DOI] [PubMed] [Google Scholar]
- 17.Musser, J. M., and V. Kapur. 1992. Clonal analysis of methicillin-resistant Staphylococcus aureus strains from intercontinental sources: association of the mec gene with divergent phylogenetic lineages implies dissemination by horizontal transfer and recombination. J. Clin. Microbiol. 30:2058-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts, R. B., A. de Lencastre, W. Eisner, E. P. Severina, B. Shopsin, B. N. Kreiswirth, A. Tomasz, et al. 1998. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in 12 New York hospitals. J. Infect. Dis. 178:164-171. [DOI] [PubMed] [Google Scholar]
- 19.Sa-Leao, R., I. Santos Sanches, D. Dias, I. Peres, R. M. Barros, and H. de Lencastre. 1999. Detection of an archaic clone of Staphylococcus aureus with low-level resistance to methicillin in a pediatric hospital in Portugal and in international samples: relics of a formerly widely disseminated strain? J. Clin. Microbiol. 37:1913-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tesch, W., A. Strassle, B. Berger-Bachi, D. O'Hara, P. Reynolds, and F. H. Kayser. 1988. Cloning and expression of methicillin resistance from Staphylococcus epidermidis in Staphylococcus carnosus. Antimicrob. Agents Chemother. 32:1494-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wielders, C. L., M. R. Vriens, S. Brisse, L. A. de Graaf-Miltenburg, A. Troelstra, A. Fleer, F. J. Schmitz, J. Verhoef, and A. C. Fluit. 2001. Evidence for in-vivo transfer of mecA DNA between strains of Staphylococcus aureus. Lancet 357:1674-1675. [DOI] [PubMed] [Google Scholar]
- 22.Witte, W., M. Enright, F. J. Schmitz, C. Cuny, C. Braulke, and D. Heuck. 2001. Characteristics of a new epidemic MRSA in Germany ancestral to United Kingdom EMRSA 15. Int. J. Med. Microbiol. 290:677-682. [DOI] [PubMed] [Google Scholar]