Abstract
Cytomegalovirus (CMV) DNA amplification assays in plasma have shown limited sensitivity compared to the detection of pp65 antigen in leukocytes. Our goal was to increase the sensitivity of a commercial CMV DNA PCR quantitative assay. After modification, the new assay was able to reproducibly detect 20 CMV DNA copies/ml of plasma. We compared this new ultrasensitive PCR assay with the standard PCR and the pp65 test for CMV detection and quantification in 22 consecutive allogeneic hematopoietic stem cell recipients. CMV infection or reactivation was detected in 84 of 319 (26%) samples by the ultrasensitive PCR assay compared to 38 of 319 (12%) samples by the pp65 assay (P < 0.01). All samples positive by the pp65 assay were positive by the ultrasensitive PCR, and CMV episodes were detected on average 4 days earlier and 7 days later than the first and the last pp65-positive test, respectively. In addition, during CMV episodes, the ultrasensitive assay identified positive samples that were inconsistently detected by the pp65 assay. The ultrasensitive assay was also much more sensitive than the standard PCR, with 26 versus 12% of CMV DNA-positive samples (P < 0.01). This assay improved the monitoring of CMV infection or reactivation in hematopoietic allogeneic stem cell recipients.
Cytomegalovirus (CMV) is a leading cause of infection in allogeneic hematopoietic stem cell transplant patients. Preemptive therapy with ganciclovir or foscarnet is the most commonly used strategy to prevent CMV disease or complications, but it requires close quantification of the CMV load. Available methods for the detection and monitoring of the CMV load include the quantification of the pp65 antigen in blood leukocytes and CMV DNA amplification. The pp65 antigen assay remains a reference method and has been validated in several clinical trials (7, 9, 11, 15). The assay has several drawbacks, including its limited sensitivity for leukopenic hematopoeitic stem cell recipients and the labor-intensive procedures. CMV DNA amplification assays in the format of a quantitative plasma-based PCR have the potential for higher sensitivity, and they are not dependent on the leukocyte count. Another potential advantage is that plasma can easily be stored before being processed. However, in previous reports, plasma-based PCR assays were either not standardized or displayed limited sensitivity compared to whole-blood PCR or the pp65 assay (1, 13).
Our goal was to increase the sensitivity of a standardized plasma-based PCR assay, the Cobas Amplicor CMV Monitor test (Roche Diagnostic Systems, Inc., Branchburg, N.J.). The new ultrasensitive format showed ∼20-fold-increased sensitivity. After validation, we assessed whether this ultrasensitive PCR could improve the monitoring of the CMV load in allogeneic hematopoietic stem cell transplant patients. We compared the performance of the ultrasensitive PCR to that of the routinely used pp65 antigen assay and to that of the standard Amplicor CMV Monitor test.
MATERIALS AND METHODS
Consecutive and unselected allogeneic hematopoietic stem cell recipients who received the transplants in our institution were monitored for CMV infection by the routinely used pp65 antigen assay and by CMV DNA PCR of blood samples collected at the same time points. Measurements were performed as deemed necessary by the physicians in charge, generally two times a week in the early phase of transplantation. For the first 16 patients (214 samples), two PCR methods were compared: the standard Cobas Amplicor CMV Monitor test and the newly developed ultrasensitive format of the test system. After preliminary analysis of the first 214 samples, only the pp65 assay and the ultrasensitive Cobas Amplicor CMV Monitor test were performed on the subsequent specimens.
The routinely used pp65 assay was performed as described in the literature (3, 15, 16). Depending on the leukocyte count, 7 to 21 ml of EDTA blood was collected for leukocyte isolation by dextran sedimentation, and 250,000 leukocytes were applied on slides by cytospin centrifugation; then, the viral structural pp65 protein was stained by fluorescent antibodies (Argene, Varilhes, France) before the detection of positive cells by trained technicians. The Cobas Amplicor CMV Monitor test was performed according to the manufacturer's recommendations using 200 μl of plasma. The detection of the amplified product is performed by the hybridization of the products to a labeled oligonucleotide probe, which comprises a sequence of 365 nucleotides located in the amino terminus of the CMV DNA polymerase gene (4). A CMV quantitation standard amplicon (added to each specimen) and the final PCR products are measured by a colorimetric reaction, the lower detection limit of which is 400 CMV DNA copies/ml of plasma. Based on our previous experience, we modified this assay by increasing the DNA input through the addition of a plasma centrifugation step before extraction (10). Plasma (600 μl) in 1.5-ml sterile tubes (Sarstedt, Sevelen, Switzerland) was centrifuged at 27,000 rpm (50,000 × g) for 80 min using a refrigerated (4°C) centrifuge (Biofuge 28 RS; Heraeus AG, Osterode, Germany). All but 60 μl of the supernatant was discarded, and the pellet was resuspended in 600 μl of lysis buffer containing a proportion of quantitation standard similar to that introduced in the standard assay. The nucleic acid was precipitated with ethanol, the dried pellet was resuspended in 60 μl of the PCR buffer, and 50 μl of this mixture was used for the PCR. The remaining steps were similar to those of the standard Cobas Amplicor CMV Monitor test, and determination of the number of copies per milliliter was adjusted for plasma input. For each frozen (−75°C) plasma sample, the two PCR assays were conducted by one technician in the same run using the same reagents. The limit of detection of the ultrasensitive assay was determined using a pool of CMV DNA-positive plasma initially quantified by the standard PCR procedure. Serial dilutions with human plasma were then performed, aliquoted, and subsequently tested by both the standard and the ultrasensitive assays. Forty-six aliquots containing 20 CMV DNA copies/ml and 20 aliquots at 180 copies/ml were extracted and tested in further experiments. However, as it was a validation study and in order to show the consistency of the results, we also reported positive results that were below the currently validated limit of detection. We considered that a validation of the assay below 20 copies was not necessary and would not be clinically meaningful. The viral DNA quantifications measured by the two PCR methods were compared using linear regression. For comparison of the different assays, P values were calculated using the chi-square test.
RESULTS
Validation of the ultrasensitive PCR assay.
The limit of detection of the ultrasensitive PCR assay was assessed on 46 plasma samples containing 20 CMV DNA copies/ml; 44 of 46 (96%) were positive. The mean (± standard deviation) optical density (OD) produced by the hybridization reaction between the specific probe and the amplicon of the positive samples was 0.294 (±0.154). Samples from 33 unselected blood donors (21 of whom were positive for CMV immunoglobulin G) and 12 subjects on chronic dialysis and four lots of pooled plasma (each from 50 donors) were analyzed. The ultrasensitive Amplicor test was negative for CMV DNA in all these cases, with a mean OD of 0.020 (±0.003). Based on these experiments, the final OD detection threshold was set at 0.100 (25 standard deviations above the mean OD value from negative controls). In further experiments, 20 of 20 plasma samples containing 180 CMV DNA copies/ml were positive. Linearity of quantification of the ultrasensitive Amplicor was observed between 50 and 5,000 copies/ml, with an intra-assay coefficient of variation lower than 15% within these values and lower than 20% for values smaller than 50 copies/ml. The interassay coefficient of variation, tested with aliquots of frozen samples tested on different days by different technicians, was lower than 15%. We found a good correlation between the quantifications of the viral load measured by the standard and the ultrasensitive assays (r = 0.88).
Population studied.
Twenty-two consecutive allogeneic hematopoietic stem cell recipients were followed for a median time of 137 days (range, 29 to 275 days); the mean age was 37 years (range, 9 to 61 years), and 41% were male. The diagnosis leading to transplantation was chronic myeloid leukemia in six patients, acute myeloid leukemia in seven patients, acute lymphocytic leukemia in four patients, chronic lymphocytic leukemia in two patients, and other hematopoietic cancers in the other three patients. Conditioning therapies before transplantation were similar for all patients, including cyclophosphamide, a high dose of methylprednisone, and whole-body irradiation. All patients except one were treated with cyclosporine for graft-versus-host disease prevention in association with in vitro T-cell depletion using monoclonal anti-CD52 in 17 patients (methotrexate or mycophenolate mofetil was used in the remaining cases). During the follow-up period, all patients experienced severe neutropenia, defined as <500 neutrophils/mm3, for a median duration of 10 days (range, 2 to 19 days), and six presented a graft-versus-host disease. The donor (D) and recipient (R) CMV serostatus was D+ R+ in 15 patients, D+ R− in 5 patients, and D− R+ or D− R− in 1 patient each. During the follow-up period, all patients received at least one antiviral drug for prophylaxis or treatment of herpesvirus infections; intravenous acyclovir was given in 16 of the 22 cases for a median of 12 days (range, 1 to 39 days), and valacyclovir was given in 20 of the 22 cases for a median of 98 days (range, 5 to 273 days). Antivirals specifically directed against CMV were given in 21 of the 22 cases, including ganciclovir for a median of 12 days (range, 2 to 33 days) in 12 cases, foscarnet for a median of 6 days (range, 2 to 20 days) in 10 cases, and cidofovir for a median of 8 days (range, 2 to 18 days) in 3 cases.
PCR results in plasma.
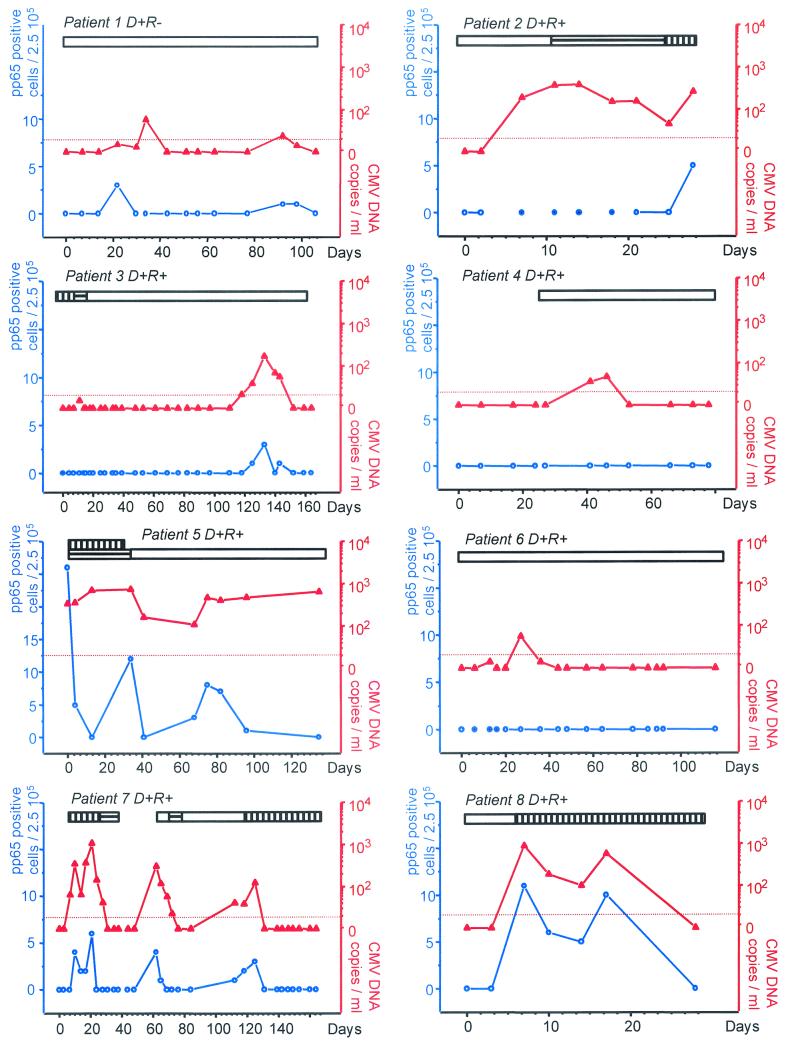
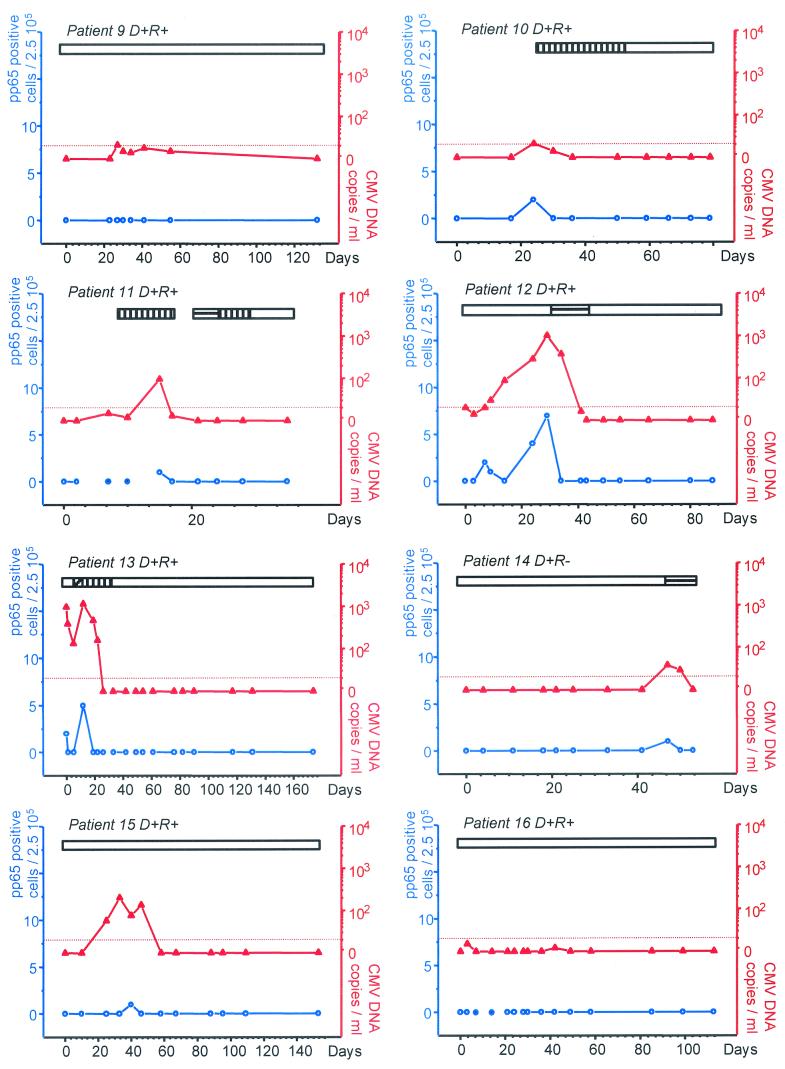
CMV infection was monitored in 319 leukocyte and plasma samples collected at the same time points. The CMV load was detected in 84 (26%) of the 319 samples by the ultrasensitive Amplicor test compared to 38 (12%) of the 319 samples by the pp65 assay (P < 0.01) (Table 1). The viral load detected by the ultrasensitive Amplicor in positive samples varied from 5 to 1,160 CMV DNA copies/ml (median, 106 copies/ml), whereas the dynamic range for the pp65 assay varied from 1 to 26 positive cells per 250,000 leukocytes (median, 3 positive cells). In six of the 22 patients, CMV remained undetectable by all the methods used. All samples positive by the pp65 assay were positive by the ultrasensitive PCR. On average, the ultrasensitive PCR detected CMV episodes 4 days earlier than the first pp65-positive test and 7 days later than the last pp65-positive test. In individual cases, the ultrasensitive PCR detected CMV episodes up to 21 days earlier (e.g., patient 2), and 12 to 39 days longer (patients 1, 5, and 12) than the pp65 assay. In addition, the pp65 assay was intermittently negative during several CMV episodes (patients 2, 5, 12, 13, and 15), whereas the ultrasensitive PCR remained consistently positive. In four cases, CMV episodes with limited viral load were detected by the ultrasensitive PCR and remained negative with the pp65 assay (patients 4, 6, 9, and 16). The results for all patients with a positive viral load are presented in Fig. 1 .
TABLE 1.
Numbers of CMV-positive samples according to three different assays in samples from hematopoietic stem cell recipients
| Method | No. (%) of CMV-positive samples
|
|
|---|---|---|
| All samples (n = 319) | Subgroup with comparison of 3 assays (n = 214) | |
| Ultrasensitive Amplicor CMV Monitor test | 84 (26)a | 56 (26)a |
| pp65 antigen assay | 38 (12)a | 28 (13) |
| Standard Amplicor CMV Monitor test | 25 (12)a | |
P < 0.01.
FIG. 1.
Monitoring of individual patients for CMV load by the pp65 assay or ultrasensitive PCR. On the abscissa are represented the days after transplantation. On the left y axis are reported the number of pp65 antigen-positive cells per 2.5 × 105 leukocytes (blue circles with white centers). On the right axis are reported the number of CMV DNA copies expressed per milliliter of plasma (red triangles). The blue circles with blue centers represent pp65 assays that were not possible because of severe leukopenia. The horizontal bars represent the durations of the following antiviral therapies; ganciclovir (vertical hatching), foscarnet (horizontal hatching), cidofovir (oblique hatching), and valaciclovir or intravenous acyclovir (open).
The ultrasensitive format, the pp65 assay, and the standard Amplicor CMV Monitor test were compared using 214 samples from the first 16 consecutive patients. CMV DNA was detected in 56 plasma samples (26%) by the ultrasensitive Amplicor compared to 25 samples (12%) by the standard Amplicor assay (P < 0.01) (Table 1). CMV DNA copy numbers quantified by the Amplicor CMV Monitor test ranged between 226 and 6,650 copies/ml (median, 671 copies/ml).
DISCUSSION
We demonstrated that a standardized and quantitative ultrasensitive plasma CMV PCR (limit of detection, 20 DNA copies/ml) improved the monitoring of CMV infection or reactivation in hematopoietic allogeneic stem cell recipients. The new ultrasensitive test format detected CMV DNA earlier and for a longer time than the pp65 assay and the standard Amplicor assay. Samples positive by the pp65 antigen assay were identified by the ultrasensitive PCR on average 4 days earlier and for 7 days longer. In addition, during CMV episodes, the ultrasensitive assay identified positive samples that were inconsistently detected by the pp65 assay, particularly in neutropenic patients. The test also proved to be much more sensitive for the detection of low levels of CMV DNA than the standard Amplicor Monitor test, with 26 versus 12% of samples, respectively, positive for CMV DNA. In previous reports, plasma PCR assays, including in-house assays detecting 30 to 50 DNA copies/ml, have showed limited sensitivity compared to the pp65 assay or PCR in other blood compartments (1, 2, 5, 6, 8, 13, 14). None of these studies has confirmed the superiority of plasma PCR to the pp65 assay. To our knowledge, the present study is the first to validate the clinical utility of a plasma-based ultrasensitive PCR for quantification of low CMV loads.
Preemptive therapy in high-risk patients needs to be introduced early, at the time of CMV reactivation or infection. In our population of stem cell recipients, the median viral load, as measured by the standard Amplicor CMV Monitor test, was low (mean, 671 copies/ml) and slightly above the detection threshold of 400 copies/ml. These low levels are possibly related to antiherpesvirus prophylaxis or treatment (i.e., intravenous acyclovir or oral valaciclovir), which are likely to impair and limit CMV replication. For this reason, tests that are able to detect and monitor low levels of CMV viremia in plasma are needed.
In solid-organ transplant recipients, the viral load before and at completion of treatment has been shown to predict the risk of relapsing CMV infection (12). Thus, tests that can quantify a low viral load at treatment completion could improve treatment management. Moreover, oral ganciclovir derivatives with improved, but still limited, bioavailability will soon be available. Patients on oral therapy will need careful monitoring of treatment response. Finally, an ultrasensitive assay is also of interest to rule out CMV replication. In case of a negative result, antiviral therapy can be safely withdrawn, avoiding unnecessary side effects or additional investigations.
We have shown that the ultrasensitive quantitative format of the Amplicor CMV Monitor test can reproducibly detect 20 CMV DNA copies/ml of plasma. This ultrasensitive PCR assay can quantify low levels of CMV DNA with high accuracy and is much more sensitive than the pp65 assay and the standard Cobas Amplicor CMV Monitor test. In high-risk patients, such as the neutropenic hematopoietic allogeneic stem cell recipients, this assay improved the monitoring of CMV infection or reactivation and thus can improve treatment management.
REFERENCES
- 1.Boivin, G., R. Belanger, R. Delage, C. Beliveau, C. Demers, N. Goyette, and J. Roy. 2000. Quantitative analysis of cytomegalovirus (CMV) viremia using the pp65 antigenemia assay and the COBAS AMPLICOR CMV MONITOR PCR test after blood and marrow allogeneic transplantation. J. Clin. Microbiol. 38:4356-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gault, E., Y. Michel, A. Dehee, C. Belabani, J. C. Nicolas, and A. Garbarg-Chenon. 2001. Quantification of human cytomegalovirus DNA by real-time PCR. J. Clin. Microbiol. 39:772-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerna, G., M. G. Revello, E. Percivalle, and F. Morini. 1992. Comparison of different immunostaining techniques and monoclonal antibodies to the lower matrix phosphoprotein (pp65) for optimal quantitation of human cytomegalovirus antigenemia. J. Clin. Microbiol. 30:1232-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kouzarides, T., A. T. Bankier, S. C. Satchwell, K. Weston, P. Tomlinson, and B. G. Barrell. 1987. Sequence and transcription analysis of the human cytomegalovirus DNA polymerase gene. J. Virol. 61:125-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Limaye, A. P., L. Corey, D. M. Koelle, C. L. Davis, and M. Boeckh. 2000. Emergence of ganciclovir-resistant cytomegalovirus disease among recipients of solid-organ transplants. Lancet 356:645-649. [DOI] [PubMed] [Google Scholar]
- 6.Limaye, A. P., M. L. Huang, W. Leisenring, L. Stensland, L. Corey, and M. Boeckh. 2001. Cytomegalovirus (CMV) DNA load in plasma for the diagnosis of CMV disease before engraftment in hematopoietic stem-cell transplant recipients. J. Infect. Dis. 183:377-382. [DOI] [PubMed] [Google Scholar]
- 7.Lo, C. Y., K. N. Ho, K. Y. Yuen, S. L. Lui, F. K. Li, T. M. Chan, W. K. Lo, and I. K. Cheng. 1997. Diagnosing cytomegalovirus disease in CMV seropositive renal allograft recipients: a comparison between the detection of CMV DNAemia by polymerase chain reaction and antigenemia by CMV pp65 assay. Clin. Transplant. 11:286-293. [PubMed] [Google Scholar]
- 8.Machida, U., M. Kami, T. Fukui, Y. Kazuyama, M. Kinoshita, Y. Tanaka, Y. Kanda, S. Ogawa, H. Honda, S. Chiba, K. Mitani, Y. Muto, K. Osumi, S. Kimura, and H. Hirai. 2000. Real-time automated PCR for early diagnosis and monitoring of cytomegalovirus infection after bone marrow transplantation. J. Clin. Microbiol. 38:2536-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niubo, J., J. L. Perez, J. T. Martinez-Lacasa, A. Garcia, J. Roca, J. Fabregat, S. Gil-Vernet, and R. Martin. 1996. Association of quantitative cytomegalovirus antigenemia with symptomatic infection in solid organ transplant patients. Diagn. Microbiol. Infect. Dis. 24:19-24. [DOI] [PubMed] [Google Scholar]
- 10.Schockmel, G. A., S. Yerly, and L. Perrin. 1997. Detection of low HIV-1 RNA levels in plasma. J. Acquir. Immune Defic. Syndr. 14:179-183. [DOI] [PubMed] [Google Scholar]
- 11.Sia, I. G., and R. Patel. 2000. New strategies for prevention and therapy of cytomegalovirus infection and disease in solid-organ transplant recipients. Clin. Microbiol. Rev. 13:83-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sia, I. G., J. A. Wilson, C. M. Groettum, M. J. Espy, T. F. Smith, and C. V. Paya. 2000. Cytomegalovirus (CMV) DNA load predicts relapsing CMV infection after solid organ transplantation. J. Infect. Dis. 181:717-720. [DOI] [PubMed] [Google Scholar]
- 13.Solano, C., I. Munoz, and A. Gutierrez. 2001. Qualitative plasma PCR assay (AMPLICOR CMV test) versus pp65 antigenemia assay for monitoring cytomegalovirus viremia and guiding preemptive ganciclovir therapy in allogeneic stem cell transplantation. J. Clin. Microbiol. 39:3938-3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka, N., H. Kimura, K. Iida, Y. Saito, I. Tsuge, A. Yoshimi, T. Matsuyama, and T. Morishima. 2000. Quantitative analysis of cytomegalovirus load using a real-time PCR assay. J. Med. Virol. 60:455-462. [DOI] [PubMed] [Google Scholar]
- 15.van den Berg, A. P., I. J. Klompmaker, E. B. Haagsma, A. Scholten-Sampson, C. M. Bijleveld, J. Schirm, M. van der Giessen, and M. J. Slooff. 1991. Antigenemia in the diagnosis and monitoring of active cytomegalovirus infection after liver transplantation. J. Infect. Dis. 164:265-270. [DOI] [PubMed] [Google Scholar]
- 16.van der Bij, W., R. Torensma, W. J. van Son, J. Anema, J. Schirm, and A. M. Tegzess. 1988. Rapid immunodiagnosis of active cytomegalovirus infection by monoclonal antibody staining of blood leukocytes. J. Med. Virol. 25:179-188. [DOI] [PubMed] [Google Scholar]