Abstract
Recent whole-genome analysis has demonstrated limited genetic variation in Chlamydia pneumoniae, with one strain (AR39) containing a 4,524 nucleotide single-stranded DNA bacteriophage, ΦCpn1. Using PCR, reverse transcription (RT)-PCR, and Western blotting, we confirmed the presence and functional expression of ΦCpn1 in C. pneumoniae strain AR39 and its absence in strain CWL029. Six additional epidemiologically distinct clinical isolates of C. pneumoniae also did not contain ΦCpn1. We generated recombinant viral protein 1 (Vp1) from ΦCpn1 in Escherichia coli and showed that Vp1 antigen is highly immunogenic in mice and that murine antisera readily recognize native Vp1 from C. pneumoniae strain AR39 elementary bodies (EB). We developed an enzyme-linked immunosorbent assay (ELISA) to measure antibodies to recombinant Vp1 in human sera collected from 32 patients with abdominal aortic aneurysm (AAA) and 40 controls. Among the 72 subjects, 61 had C. pneumoniae EB antibodies shown by ELISA. Antibodies to Vp1 were found in 39 of the 61 (64%) seropositive individuals and were significantly correlated with AAA (adjusted odds ratio, 13.9; 95% confidence interval, 1.1 to 175). Our studies indicate that phage-containing strains of C. pneumoniae are uncommonly found by isolation but may commonly infect individuals with vascular disease.
There is growing evidence linking infection with Chlamydia pneumoniae with vascular diseases, such as atherosclerosis and abdominal aortic aneurysm (AAA) (3, 5). However, the data remain inconclusive, and the clinical and public health importance of C. pneumoniae as a vasculopathic organism is unclear. Skeptics have suggested that C. pneumoniae is simply an “innocent bystander” that more readily infects diseased arteries (8) or that its association with vascular disease is confounded by its association with other atherosclerotic risk factors (13). Since seroepidemiological studies indicate that most people are infected by C. pneumoniae by the age at which the clinical manifestations of atherosclerosis usually appear (11), it is likely that if C. pneumoniae is a causal agent for vascular disease, there is variability in the characteristics of the organism, the host response to infection, or both. Variability in host characteristics could explain why some people might be more prone to develop vascular disease when infected with C. pneumoniae, and it is well known that family history is a strong risk factor for AAA. There may also be important variations in the vasculotrophisms and pathogenicities of different C. pneumoniae strains. In this regard, recent genomic studies have shown genetic variation in C. pneumoniae (16, 24, 29). One of the most prominent genetic differences among the three genomically characterized strains of C. pneumoniae was that one strain (AR39) contained a 4,524-nucleotide single-stranded DNA bacteriophage, ΦCpn1 (24). Though three phylogenetically related phages were identified in Chlamydia psittaci (14, 20, 26), ΦCpn1 was the first phage to be identified in C. pneumoniae. ΦCpn1 was predicted to contain up to eight open reading frames (ORFs), with ORFs 1 to 3, by analogy to the avian C. psittaci phage Chp1, encoding the viral structural proteins viral protein 1 (Vp1), Vp2, and Vp3, respectively (24). In a recent report of comparative analysis of chlamydia phages, the well-conserved Vp1 protein was predicted to contain a potential receptor binding site (25).
Phage-bearing strains of other bacteria are often more pathogenic than phage-free strains (6), and it may be that phage-containing strains of C. pneumoniae are more strongly correlated with vascular disease. However, the role played by the phage ΦCpn1 in the disease pathogenesis of C. pneumoniae is not obvious, since ΦCpn1 does not carry any known virulence genes, unlike other phages associated with pathogenic bacteria. The development of methodologies to detect this interesting phage is therefore necessary in order to carry out detailed epidemiologic studies of the association of the phage with C. pneumoniae-associated vascular diseases.
The purpose of the present study was to develop molecular methods to differentiate C. pneumoniae strains containing the phage ΦCpn1 from other strains that lack the phage based on the vp1 gene encoding Vp1 and to develop an enzyme-linked immunosorbent assay (ELISA) to detect Vp1 antibodies in order to determine whether exposure to C. pneumoniae strains containing the phage is associated with AAA.
MATERIALS AND METHODS
Strains of C. pneumoniae, growth, and purification.
The C. pneumoniae strains used in this study are described in Table 1. The human HL cell line was used for growth and propagation of the C. pneumoniae strains. Elementary bodies (EBs) were purified on discontinuous gradients of Renografin-76 (Squibb Canada, Montreal, Canada) as previously described (28). The purified EBs were resuspended in isotonic sucrose-phosphate-glutamate buffer and stored at −80°C.
TABLE 1.
C. pneumoniae strains evaluated for phage
| Strain | C. pneumoniae CP0543 sequence | ΦCpn1-specific sequences | Source and/or reference |
|---|---|---|---|
| AR39 | Yes | Yes | Laboratory strain (12) |
| CWL029 | Yes | No | Laboratory strain (17) |
| CM1 | Yes | No | Laboratory strain (2) |
| Kajaani7 | Yes | No | Kajaani, Finland (P. Saikku) |
| Bay 13 | Yes | No | Spartanburg, S.C. (M. Hammerschlag) |
| Bay 16 | Yes | No | Toledo, Ohio (M. Hammerschlag) |
| Bay 19 | Yes | No | Baltimore, Md. (M. Hammerschlag) |
| Bay 20 | Yes | No | Springfield, Mo. (M. Hammerschlag) |
| Bay 21 | Yes | No | Mesa, Az. (M. Hammerschlag) |
PCR analysis.
PCR was performed using a PTC-200 Peltier thermal cycler (MJ Research). The reaction mixture contained 1.5 mM MgCl2, 200 μM deoxyribonucleoside triphosphates, 5 U of Taq DNA polymerase enzyme, and 25 pmol of oligonucleotide primers (for the vp1 gene, forward [5′-CGCTCCTAGTGGGGGATTTACTGA-3′] and reverse [5′-CACAGCTTGCTCACCTAAATGGCT-3′]; for the 16S RNA gene, forward [5′-CGGTAATACGGAGGGTGCTAGC-3′] and reverse [5′-GAATTAAACCACATGCTCCACTGC-3′]; for the C. pneumoniae CP0543 gene, forward [5′-CTGTAAGGTGAAAAGTTTTTA-3′] and reverse [5′-CAGCTGTAAATGCAGCTTT-3′]) in a total volume of 50 μl. The PCR cycling conditions were as follows: one cycle of 95°C for 3 min and 35 cycles of 94°C for 15 s, 55°C for 30 s, and 72°C for 2 min. This was followed by strand elongation for 10 min at 72°C. The sizes of the amplicons were as follows: vp1, 925 bp; 16S RNA, 430 bp; CP0543, 266 bp.
RT-PCR.
Total RNA from HL cells infected with C. pneumoniae strains AR39 or CWL029 was isolated using Trizol (Life Technologies) and treated with RNase-free DNase (Roche). These RNA samples were used as templates for reverse transcription (RT) in a 20-μl reaction mixture containing 2 μg of random hexamers (Roche), 1 μl of 10 mM deoxyribonucleoside triphosphates, 2 μl of 0.1 M dithiothreitol, 1 μl of RNasin (Promega), and 200 U of Superscript II (Life Technologies). RT was carried out at 42°C for 50 min. PCR was performed with a PTC-200 Peltier Thermal Cycler. The reaction mixture contained 1.5 mM MgCl2, 200 μM deoxyribonucleoside triphosphates, 5 U of Taq DNA polymerase enzyme, 25 pmol each of oligonucleotide primers specific for the vp1 gene or the 16S RNA gene (see above), and 2 μl of RT product in a total volume of 50 μl. The PCR cycling conditions were as follows: one cycle of 95°C for 3 min and 35 cycles of 94°C for 15 s, 55°C for 30 s, and 72°C for 2 min. This was followed by strand elongation for 10 min at 72°C.
Molecular cloning, expression, and purification of recombinant Vp1.
The vp1 DNA fragments were generated by PCR using genomic DNA isolated from C. pneumoniae AR39 as the template. In order to subclone the PCR product as NcoI and XhoI fragments into the pET30b(+) vector (Novagen), forward (5′-AGTAGGGAAGCCATGGTTAGG-3′) and reverse (5′-ACTGACTCGAGATTAGAAATGATCAAT-3′) PCR primers were designed to contain NcoI and XhoI restriction sites, respectively. PCRs were carried out using Platinum Pfx DNA polymerase (Gibco BRL). The reaction mixture contained 1 mM MgSO4, 300 μM deoxyribonucleoside triphosphates, 2.5 U of Platinum Pfx DNA polymerase enzyme, and 25 pmol of each oligonucleotide primer in a total volume of 50 μl. The PCR cycling conditions were as follows: one cycle of 95°C for 2 min and 35 cycles of 94°C for 15 s, 58°C for 30 s, and 68°C for 2 min. This was followed by strand elongation for 10 min at 68°C. The PCR product was purified with the QIAquick PCR purification kit (Qiagen), and the purified DNA fragment was cloned into pET30b(+) after restriction enzyme digestion using standard molecular biology techniques. The sequence of the subcloned vp1 gene was confirmed by sequencing with dye-labeled terminators using the ABI PRISM kit (PE Biosystems). Plasmids carrying the vp1 gene (pET-vp1) were transformed into the Escherichia coli strain BL21(DE3), where Vp1 expression was carried out by inducing the lac promoter for expression of T7 RNA polymerase using isopropyl-β-d-thiogalactopyranoside. The expressed Vp1 protein with an N-terminal His tag was purified by QIAexpress protein purification system (Qiagen).
Antibody production against recombinant Vp1.
Antisera against Vp1 were raised in BALB/c mice (Charles River, Quebec, Canada) by intraperitoneal injection of the recombinant proteins (100 μg of protein in incomplete Freund adjuvant) followed by two booster injections (50 μg of protein in incomplete Freund adjuvant) at 2-week intervals. Sera were collected and pooled 4 weeks after the final boost.
Western blotting.
Samples for Western analysis were prepared by boiling purified EBs of C. pneumoniae strains AR39 and CWL029 for 5 min in the protein sample buffer. The samples were subjected to electrophoresis in sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis gels according to the method of Laemmli (18). The gels were blotted onto nitrocellulose membranes (Bio-Rad) at 70 V for 1 h in blotting buffer acccording to the method of Sambrook et al. (27). The filters were blocked overnight with Tris-buffered saline (TBS) containing 3% bovine serum albumin at 4°C prior to incubation with polyclonal anti-Vp1 antibodies and peroxidase-conjugated sheep anti-mouse immunoglobulin G secondary antibodies. The blots were processed for color detection using the substrate 5-bromo-4-chloro-3indolyl phosphate-nitroblue tetrazolium.
Selection of patients and controls.
The seroepidemiological analysis was based on stored frozen serum specimens from a convenient subset of participants in a previous retrospective hospital-based case control study of risk factors for AAA (4). The case control study was performed among ambulatory patients attending either of the ultrasound departments of the two tertiary-care hospitals in Winnipeg, Manitoba, Canada, between June 1992 and December 1995. In the original study, there were 98 AAA patients and 102 non-AAA controls. To minimize selection and misclassification biases, the controls were selected from among persons who were undergoing ultrasound examinations for indications similar to those of the patients. For this investigation, analysis was restricted to 32 patients and 40 controls from the original study for whom adequate leftover frozen serum specimens were available. This study was approved by the University of Manitoba Faculty of Medicine Research Ethics Board and by the Institutional Review Board of the Johns Hopkins University School of Hygiene and Public Health.
The definition of an AAA was based on the assessments of the ultrasound radiologists at the two ultrasound departments. Specific size criteria were not used. Instead, the definition was based on the shape of the infrarenal aorta such that any definite focal widening was classified as an AAA as suggested by Zwiebel (30). Nevertheless, all of the AAA patients and no control had an infrarenal aortic dilatation of ≥3.0 cm.
Data collection and risk factor definition.
An in-person interview was conducted with each study participant using a standardized questionnaire to collect data related to sociodemographics, symptomatology, indications for ultrasound examination, history of specific medical conditions, family history of AAA and other conditions, and cigarette smoking. Two blood pressure measurements were taken with the patient seated using a standard mercury sphygmomanometer. Fasting serum total-cholesterol levels were measured with the Hitachi-Boehringer-Mannheim 717 Autoanalyzer using established protocols.
The participants were classified as having hypertension if they had systolic blood pressure of ≥140 mm Hg or diastolic blood pressure of ≥90 mm Hg or they were currently taking antihypertensive medication prescribed by a physician. Hypercholesterolemia was defined as the presence of a fasting serum total-cholesterol level of >6.2 mmol/liter or current use of prescribed lipid-lowering medication. Lifelong pack years of cigarette smoking exposure were assessed by determining the average number of packs of cigarettes smoked per day for specific age periods, beginning with the age at initiation. These data were then cumulated to estimate the total pack years of exposure.
Serological assays for C. pneumoniae AR39 EB and bacteriophage ΦCpn1 Vp1.
Detection of anti-EB or anti-Vp1 antibodies in human immune sera was done by ELISA as described previously (9). Flat-bottom (96-well) polystyrene microtiter plates were coated with 100 μl of either C. pneumoniae AR39 EB or recombinant phage Vp1 protein (0.1 μg/well) or with coating buffer alone. The plates were incubated overnight at 4°C and then washed three times with TBS. Nonspecific binding sites were blocked by adding 150 μl of 3% bovine serum albumin in MTPBS (150 mM NaCl, 16 mM Na2HPO4, 4 mM NaH2PO4, pH 7.3)-Tween 20 detergent buffer to each well for 2 h at 37°C. After three washings with TBS, 100 μl of human sera diluted 1:500 was added in duplicate, and the plates were incubated for 1 h at 37°C. After three more washings with TBS, 100 μl of 1:2,000-diluted peroxidase-conjugated goat anti-human secondary antibody was added, and the plates were incubated for 1 h at 37°C. After five washings with TBS, 100 μl of 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) substrate was added and allowed to develop in the dark for 5 min. Readings of the optical density at 405 nm (OD405) were made on a microplate reader.
Statistical analyses.
For all analyses, the presence of antibodies to C. pneumoniae strain AR39 EB and ΦCpn1 Vp1 in sera was arbitrarily dichotomized such that those with measured levels of ≥0.5 OD units were classified as seropositive. This cutoff was chosen because visual inspection of the distribution of OD values to Vp1 antigen showed that an OD value of less than or greater than 0.5 separated the study subjects into two distinct populations. Odds ratios (OR), 95% confidence intervals (CI), and chi-square analyses were used to assess the relationship between these variables and AAA. Unconditional logistic regression was used to compute adjusted OR and 95% CI for the association between AAA and seropositivity to C. pneumoniae EB and Vp1. Other variables included in the logistic regression model were age, gender, pack years of smoking, the presence of hypertension, and the presence of hypercholesterolemia.
RESULTS
Differentiation of C. pneumoniae strains.
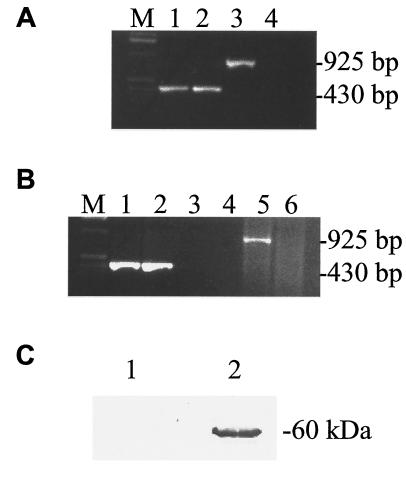
We developed molecular methods to identify C. pneumoniae strains that contain the phage ΦCpn1. Genome sequencing revealed the strain AR39 as a phage-containing strain and CWL029 as a phage-free strain (16, 24). The DNA sequence of the phage was originally found during the genome sequencing of strain AR39 (24), and no functional studies have been performed so far on this phage. We performed PCR, RT-PCR, and Western blot analysis to determine the presence and expression of the ΦCpn1 vp1 gene in C. pneumoniae strain AR39 in comparison to strain CWL029, which is known to lack the phage (16). As expected, results from PCR (Fig. 1A), RT-PCR (Fig. 1B), and Western blot (Fig. 1C) analyses confirm the presence and expression of the phage vp1 gene in C. pneumoniae strain AR39 and its absence in strain CWL029.
FIG. 1.
Differentiation of phage-containing (AR39) and phage-free (CWL029) strains of C. pneumoniae using PCR (A), RT-PCR (B), and Western blotting (C). (A) PCR analysis of DNA isolated from HL cells infected with C. pneumoniae strain AR39 (lanes 1 and 3) or CWL029 (lanes 2 and 4) using primers specific to chlamydial 16S RNA (lanes 1 and 2) or ΦCpn1 vp1 gene (lanes 3 and 4). Lane M, molecular size markers. (B) RT-PCR analysis of total RNA isolated from HL cells infected with C. pneumoniae strain AR39 (lanes 1, 3, and 5) or CWL029 (lanes 2, 4, and 6) using primers specific to chlamydial 16S RNA (lanes 1 to 4) or ΦCpn1 vp1 gene (lanes 5 and 6). Lane M, molecular size markers. Lanes 3 and 4 are similar to lanes 1 and 2 except that the template was total RNA instead of reverse-transcribed cDNA. (C) Western blot analysis of C. pneumoniae strains AR39 (lane 2) and CWL029 (lane 1). Boiled samples of purified AR39 and CWL029 EBs were subjected to electrophoresis in sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis gels, and the blotted nitrocellulose membranes were incubated with polyclonal anti-Vp1 antibodies followed by incubation with alkaline phosphatase-conjugated secondary antibodies and color detection.
Distribution of ΦCpn1 among C. pneumoniae strains.
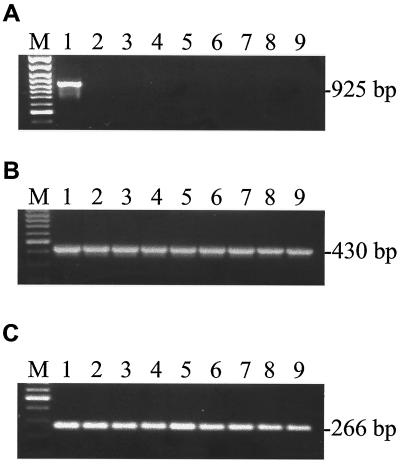
Among the three C. pneumoniae genomes sequenced so far, only one (AR39) contained the phage ΦCpn1 (16, 24, 29). It is of interest to know the distribution of ΦCpn1 among C. pneumoniae clinical isolates, and therefore we performed PCR analysis to determine the presence of the ΦCpn1 vp1 gene in six clinical isolates of C. pneumoniae in addition to the laboratory strains AR39 and CWL029. Our results demonstrate that none of the isolates other than AR39 contains phage ΦCpn1 (Table 1 and Fig. 2).
FIG. 2.
C. pneumoniae strain typing. DNA isolated from three laboratory strains (AR39 [lane 1], CWL029 [lane 2], and CM1 [lane 3]) and six clinical isolates (K7 [lane 4], Bay 13 [lane 5], Bay 16 [lane 6], Bay 19 [lane 7], Bay 20 [lane 8], and Bay 21 [lane 9]) were PCR analyzed using specific primers for the phage vp1 gene (A), C. pneumoniae 16S RNA (B), or C. pneumoniae ORF CP0543 (C). ORF CP0543 represents the truncated version of a ΦCpn1 ORF found in the C. pneumoniae genome (24) and used here as a control in addition to the 16S RNA. Lanes M, molecular size markers.
ELISA design and specificity.
In order to determine whether the ΦCpn1 Vp1 protein can be used as an antigen to detect the presence of the phage antibodies in sera collected from controls and patients with AAA, we raised antibodies in mice using recombinant Vp1 protein generated in E. coli and showed that Vp1 antigen is highly immunogenic in mice and that antibodies thus generated readily recognize the Vp1 protein in C. pneumoniae EBs (Fig. 1C). Antiserum from a nonimmunized mouse did not give any bands for C. pneumoniae EBs (data not shown).
We next used ELISA to measure human serum antibodies in 32 AAA patients and 40 controls. There were no significant differences in age, gender distribution, smoking history, or history of hypertension between the original study participants (98 patients and 102 controls) previously reported (4) and the subset analyzed for this study. Using a cutoff value of 0.5 OD units for both C. pneumoniae EB and ΦCpn1 Vp1 antibody levels, 61 (84.7%) of the 72 participants were seropositive for C. pneumoniae EB and 42 (58.3%) were seropositive for ΦCpn1 Vp1. Of the 61 who were seropositive for C. pneumoniae EB, 39 (63.9%) were also positive for ΦCpn1 Vp1. There were no consistent variations by age, gender, or smoking history for either C. pneumoniae EB or ΦCpn1 Vp1 seropositivity.
Correlation between C. pneumoniae EB and ΦCpn1 Vp1 antibodies and AAA.
Table 2 shows the relationship between AAA and C. pneumoniae EB and bacteriophage Vp1 seropositivity. Seropositivity (i.e., levels of >0.5 OD units) was more common among patients than controls for both C. pneumoniae EB (90.6 versus 80.0%) and ΦCpn1 Vp1 (68.8 versus 50.0%). There was a positive, but nonsignificant, association with AAA for both C. pneumoniae EB (crude OR, 2.4 [95% CI, 0.6 to 10.0]) and ΦCpn1 Vp1 (crude OR, 2.2 [95% CI, 0.8 to 5.8]) seropositivity. Multivariate analysis to control for age, gender, pack years of smoking, hypertension, and hypercholesterolemia showed a positive, but nonsignificant, association between AAA and C. pneumoniae EB seropositivity (adjusted OR, 2.3 [95% CI, 0.5 to 11.5]). Strikingly, multivariate adjustment resulted in a stronger and statistically significant association between AAA and ΦCpn1 Vp1 seropositivity (adjusted OR, 4.2 [95% CI, 1.2 to 14.4]) (P = 0.02). Seropositivity to both C. pneumoniae EB and ΦCpn1 Vp1 (versus seronegativity to both) was strongly associated with AAA (adjusted OR, 13.9 [95% CI, 1.1 to 175]). Furthermore, a significant association between AAA and ΦCpn1 Vp1 seropositivity persisted when adjusted for seropositivity to C. pneumoniae EB (adjusted OR, 3.9 [95% CI, 1.1 to 13.9]) (P = 0.04).
TABLE 2.
Association between AAA and seropositivity to C. pneumoniae EBs and ΦCpn1 Vp1
| Antibody | Seropositivity (%)
|
Crude OR (95% CI) | Adjusted ORa (95% CI) | |
|---|---|---|---|---|
| AAA patients (n = 32) | Controls (n = 40) | |||
| C. pneumoniae EBb | 90.6 | 80.0 | 2.4 (0.6-10.0) | 2.3 (0.5-11.5) |
| ΦCpn1 Vp1c | 68.8 | 50.0 | 2.2 (0.83-5.8) | 4.2 (1.2-14.4) |
| Both C. pneumoniae EB and ΦCpn1 Vp1 | 62.5 | 47.5 | 7.4 (0.83-65.7)d | 13.9 (1.1-175)d |
OR adjusted for age, sex, pack years of smoking, hypertension, and hypercholesterolemia using multiple logistic regression.
Antibodies to C. pneumoniae AR39 EB.
Antibodies to Vp1 of ΦCpn1.
Compared to a reference category of seronegativity to both C. pneumoniae EB and bacteriophage Vp1.
DISCUSSION
ΦCpn1 is the first bacteriophage to be identified as associated with a chlamydial strain that causes disease in humans. This observation immediately raises the question of whether the chlamydia phage contribute to C. pneumoniae virulence and disease pathogenesis. As a step to answer this question, we developed methodologies to detect the presence of the phage ΦCpn1 in clinical isolates of C. pneumoniae and of phage-related antibodies in sera from humans. We provide preliminary data supporting an association between AAA and infection with C. pneumoniae containing the phage ΦCpn1. We acknowledge that the results of this investigation cannot yet be used to draw firm conclusions with respect to a causal relationship between C. pneumoniae containing ΦCpn1 and AAA due to a modest sample size and statistical imprecision. However, the data are sufficiently intriguing to warrant further microbiological and seroepidemiological study.
Our data indicate that a subset (64.5%) of persons who are seropositive to C. pneumoniae have serological evidence of infection with ΦCpn1. Our data also suggest that serological evidence of exposure to strains of C. pneumoniae containing phage correlate better with the presence of AAA than does seropositivity to C. pneumoniae in general. This is demonstrated by the finding of a persistent positive association between Vp1 seropositivity and AAA among all subjects who were seropositive to C. pneumoniae and a much stronger association when seropositivity to both was compared to seronegativity to both.
It is striking that only one isolate of C. pneumoniae contained the phage among the three C. pneumoniae strains whose genomes were completely sequenced and the six clinical strains analyzed by PCR in this study. Thus, phage infection of C. pneumoniae isolates appears uncommon. Nonetheless, the ELISA results indicate that among C. pneumoniae EB-seropositive individuals, nearly two-thirds have antibodies against the phage. This difference between the infrequency of detection of C. pneumoniae containing phage in clinical isolates and the frequency of chlamydia phage antibodies in humans may be explainable in a number of ways. (i) C. pneumoniae strains containing the phage (such as AR39) may be more capable of causing a persistent infection. We are in the process of performing experiments using animal models to see whether a C. pneumoniae strain containing the phage (AR39) persists longer in a mouse lung infection model than does a phage-free strain. (ii) Since the phage is likely to be lytic based on its sequence homology to other lytic phages (25), phage-bearing strains may be relatively difficult to isolate and propagate in cell culture. (iii) The number of C. pneumoniae isolates analyzed for the phage may still be too small to reach a definite conclusion. Analysis of a larger collection of C. pneumoniae isolates for the phage is clearly needed, and analysis of strains directly isolated from artherosclerotic lesions for the presence of the phage will be particularly informative.
In summary, the molecular and serological methods developed in this study can be used to detect and analyze the ΦCpn1 phage in order to further understand its involvement in the pathogenesis of C. pneumoniae disease. The findings from ELISA analysis may have important implications regarding the association between C. pneumoniae and vascular disease. The data suggest that certain strains of C. pneumoniae, such as those carrying ΦCpn1, may be more vasculopathic than phage-free strains. There is some evidence in the literature to support this, in that the phage-containing C. pneumoniae strain AR39 (15, 21, 22), but apparently not the phage-negative Kajaani7 strain (1, 7), can exacerbate the development of atherosclerosis in LDLR−/− or ApoE−/− mice. Other factors must also contribute to disease outcome, since a proportion of New Zealand White rabbits infected with C. pneumoniae strain AR39 (10), CWL029 (23), or Kajaani7 (19) developed signs of atherosclerosis and not all AAA patients in the present study showed evidence of exposure to both C. pneumoniae and ΦCpn1. Nevertheless, a role for ΦCpn1 in the pathogenesis of C. pneumoniae disease may explain some of the inconsistencies in previous research on the association between C. pneumoniae seropositivity and vascular disease. More research to investigate the association between specific strains of C. pneumoniae, including those carrying ΦCpn1, and various vascular disease outcomes is warranted.
Acknowledgments
We thank M. Hammerschlag and P. Saikku for providing isolates of C. pneumoniae, as indicated in Table 1. S. Akbar and T. S. Lena are acknowledged for critical reading of the manuscript, and H. Lu is acknowledged for technical assistance.
This research was supported by a grant from the Canadian Institutes of Health Research and by the Manitoba Medical Services Foundation (grant 387-3143-66). K. P. Karunakaran was supported by a fellowship from the Michael Smith Foundation for Health Research.
REFERENCES
- 1.Aalto-Setala, K., K. Laitinen, L. Erkkila, M. Leinonen, M. Jauhiainen, C. Ehnholm, M. Tamminen, M. Puolakkainen, I. Penttia, and P. Saikku. 2001. Chlamydia pneumoniae does not increase atherosclerosis in the aortic root of apolipoprotein E-deficient mice. Arterioscler. Thromb. Vasc. Biol. 21:578-584. [DOI] [PubMed] [Google Scholar]
- 2.Black, C. M., J. E. Johnson, C. E. Farshy, T. M. Brown, and B. P. Berdal. 1991. Antigenic variation among strains of Chlamydia pneumoniae. J. Clin. Microbiol. 29:1312-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanchard, J. F., H. K. Armenian, R. Peeling, P. Poulter Friesen, C. Shen, and R. C. Brunham. 2000. The relation between Chlamydia pneumoniae infection and abdominal aortic aneurysm: a case-control study. Clin. Infect. Dis. 30:946-947. [DOI] [PubMed] [Google Scholar]
- 4.Blanchard, J. F., H. Armenian, and P. Poulter Friesen. 2000. Risk factors for abdominal aortic aneurysms: a case-control study. Am. J. Epidemiol. 151:575-583. [DOI] [PubMed] [Google Scholar]
- 5.Blasi, F., F. Denti, M. Erba, R. Cosentini, R. Raccanelli, A. Rinaldi, L. Fagetti, G. Esposito, U. Ruberti, and L. Allegra. 1996. Detection of Chlamydia pneumoniae but not Helicobacter pylori in atherosclerotic plaques of aortic aneurysms. J. Clin. Microbiol. 34:2766-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd, E. F., B. M. Davis, and B. Hochhut. 2001. Bacteriophage-bacteriophage interactions in the evolution of pathogenic bacteria. Trends Microbiol. 9:137-144. [DOI] [PubMed] [Google Scholar]
- 7.Caligiuri, G., M. Rottenberg, A. Nicoletti, H. Wigzell, and G. K. Hansson. 2001. Chlamydia pneumoniae infection does not induce or modify atherosclerosis in mice. Circulation 103:2834-2838. [DOI] [PubMed] [Google Scholar]
- 8.Capron, L. 1996. Chlamydia in coronary plaques—hidden culprit or harmless hobo? Nat. Med. 2:856-857. [DOI] [PubMed] [Google Scholar]
- 9.Cohen, C. R., R. Nguti, E. A. Bukusi, H. Lu, C. Shen, M. Luo, S. Sinei, F. Plummer, J. Bwayo, and R. C. Brunham. 2000. Human immunodeficiency virus type 1-infected women exhibit reduced interferon-gamma secretion after Chlamydia trachomatis stimulation of peripheral blood lymphocytes. J. Infect. Dis. 182:1672-1677. [DOI] [PubMed] [Google Scholar]
- 10.Fong, I. W., B. Chiu, E. Viira, D. Jang, and J. B. Mahony. 1999. De novo induction of atherosclerosis by Chlamydia pneumoniae in a rabbit model. Infect. Immun. 67:6048-6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grayston, J. T. 2000. Background and current knowledge of Chlamydia pneumoniae and atherosclerosis. J. Infect. Dis. 181(Suppl. 3):S402-S410. [DOI] [PubMed] [Google Scholar]
- 12.Grayston, J. T., C. C. Kuo, S. P. Wang, and J. Altman. 1986. A new Chlamydia psittaci strain, TWAR, isolated in acute respiratory tract infections. N. Engl. J. Med. 315:161-168. [DOI] [PubMed] [Google Scholar]
- 13.Haberbosch, W., and C. Jantos. 2000. Chlamydia pneumoniae infection is not an independent risk factor for arterial disease. Herz 25:79-83. [DOI] [PubMed] [Google Scholar]
- 14.Hsia, R. C., L. M. Ting, and P. M. Bavoil. 2000. Microvirus of Chlamydia psittaci strain guinea pig inclusion conjunctivitis: isolation and molecular characterization. Microbiology 146:1651-1660. [DOI] [PubMed] [Google Scholar]
- 15.Hu, H., G. N. Pierce, and G. Zhong. 1999. The atherogenic effects of chlamydia are dependent on serum cholesterol and specific to Chlamydia pneumoniae. J. Clin. Investig. 103:747-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalman, S., W. Mitchell, R. Marathe, C. Lammel, J. Fan, R. W. Hyman, L. Olinger, J. Grimwood, R. W. Davis, and R. S. Stephens. 1999. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat. Genet. 21:385-389. [DOI] [PubMed] [Google Scholar]
- 17.Kuo, C. C., and J. T. Grayston. 1988. Factors affecting viability and growth in HeLa 229 cells of Chlamydia sp. strain TWAR. J. Clin. Microbiol. 26:812-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 19.Laitinen, K., A. Laurila, L. Pyhala, M. Leinonen, and P. Saikku. 1997. Chlamydia pneumoniae infection induces inflammatory changes in the aortas of rabbits. Infect. Immun. 65:4832-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, B. L., J. S. Everson, B. Fane, P. Giannikopoulou, E. Vretou, P. R. Lambden, and I. N. Clarke. 2000. Molecular characterization of a bacteriophage (Chp2) from Chlamydia psittaci. J. Virol. 74:3464-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, L., H. Hu, H. Ji, A. D. Murdin, G. N. Pierce, and G. Zhong. 2000. Chlamydia pneumoniae infection significantly exacerbates aortic atherosclerosis in an LDLR−/− mouse model within six months. Mol. Cell Biochem. 215:123-128. [DOI] [PubMed] [Google Scholar]
- 22.Moazed, T. C., L. A. Campbell, M. E. Rosenfeld, J. T. Grayston, and C. C. Kuo. 1999. Chlamydia pneumoniae infection accelerates the progression of atherosclerosis in apolipoprotein E-deficient mice. J. Infect. Dis. 180:238-241. [DOI] [PubMed] [Google Scholar]
- 23.Muhlestein, J. B., J. L. Anderson, E. H. Hammond, L. Zhao, S. Trehan, E. P. Schwobe, and J. F. Carlquist. 1998. Infection with Chlamydia pneumoniae accelerates the development of atherosclerosis and treatment with azithromycin prevents it in a rabbit model. Circulation 97:633-636. [DOI] [PubMed] [Google Scholar]
- 24.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. DeBoy, J. Kolonay, G. McClarty, S. L. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28:1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Read, T. D., C. M. Fraser, R. C. Hsia, and P. M. Bavoil. 2000. Comparative analysis of Chlamydia bacteriophages reveals variation localized to a putative receptor binding domain. Microb. Comp. Genomics 5:223-231. [DOI] [PubMed] [Google Scholar]
- 26.Richmond, S. J., P. Stirling, and C. R. Ashley. 1982. Virus infecting the reticulate bodies of an avian strain of Chlamydia psittaci. FEMS Microbiol. Lett. 14:31-36. [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 28.Schachter, J., and P. B. Wyrick. 1994. Culture and isolation of Chlamydia trachomatis. Methods Enzymol. 236:377-390. [DOI] [PubMed] [Google Scholar]
- 29.Shirai, M., H. Hirakawa, M. Kimoto, M. Tabuchi, F. Kishi, K. Ouchi, T. Shiba, K. Ishii, M. Hattori, S. Kuhara, and T. Nakazawa. 2000. Comparison of whole genome sequences of Chlamydia pneumoniae J138 from Japan and CWL029 from USA. Nucleic Acids Res. 28:2311-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zwiebel, W. J. 1992. Aortic and iliac aneurysm. Semin. Ultrasound CT MR 13:69-80. [PubMed] [Google Scholar]