Abstract
The prevalence of microsporidiosis is likely underestimated due to the labor-intensive, insensitive, and nonspecific clinical laboratory methods used for the diagnosis of this disease. A real-time PCR assay was designed to assess DNA extraction methods and to detect three Encephalitozoon species in feces. Modifications of the MagNA Pure LC DNA isolation kit protocol (Roche Applied Sciences, Indianapolis, Ind.) were compared by using the automated MagNA Pure LC instrument (Roche) and fecal specimens spiked with Encephalitozoon intestinalis spores. Extracted DNA was amplified by the LightCycler (Roche) PCR assay. Assay sensitivity, reproducibility, and efficiency were assessed by comparing threshold crossover values achieved with different extraction and storage conditions (fresh, refrigerated, frozen, and preserved specimens). Optimal extraction conditions were achieved by using a commercial buffer, tissue lysis buffer (Roche), as the specimen diluent. LightCycler PCR results were compared to those obtained from routine stool microscopy with trichrome blue stain. The lower limit of detection for the LightCycler PCR assay varied by storage conditions from 102 to 104 spores/ml of feces, a value which represented a significant improvement over that achieved by staining (≥1.0 × 106 spores/ml). Melting temperature analysis of the amplicons allowed for the differentiation of three Encephalitozoon species (E. intestinalis, E. cuniculi, and E. hellem). The assay is readily adaptable to the clinical laboratory and represents the first real-time PCR assay designed to detect Encephalitozoon species.
Microsporidia can cause disease in human immunodeficiency virus-infected patients and other immunocompromised individuals (20, 29). Microsporidiosis has also been diagnosed in patients with chronic renal failure (4), in transplant patients (24, 25, 27, 35, 39, 47), and in children from and travelers to underdeveloped countries (11). In addition, serological studies have suggested that several species of microsporidia may commonly infect immunocompetent adults (51, 52). The organisms have been implicated in waterborne disease outbreaks and are ubiquitous in the environment (14, 33, 48). Species that are pathogenic for humans have been identified in cattle, pigs, other captive mammals, and chicken eggs (11, 34, 41, 43).
The diagnosis of microsporidiosis at present is difficult. Although a variety of methods are used to detect microsporidia in clinical laboratories, special stains and light microscopy are often used as a routine method of detection (3, 5-7, 13, 22, 32, 37, 46, 53). While the staining procedures are common, they can be time-consuming, nonspecific, and insensitive and require skilled, experienced technologists, factors contributing to what is probably a gross underreporting of this infection. Traditional PCR enhances the detection of microsporidia and has been used in some clinical laboratories. However, many of the assays developed thus far have been developed for research purposes and do not lend themselves to routine testing in clinical laboratories because of their cumbersome specimen processing and DNA extraction methods, false-positive results that are possibly caused by cross contamination, and false-negative results that occur because of low target concentration, inadequate specimen storage, or the presence of fecal inhibitors (1, 15, 21, 22, 36, 38, 42, 54).
Many issues exist regarding the reproducibility of any PCR assay that may have application for a clinical laboratory. Reproducibility depends on the entire system of extraction, amplification, detection, and contamination control. In one blinded, multicenter study, PCR diagnosis of microsporidian-spiked stool specimens was compared to diagnosis by light microscopy (42). Various primer sets were tested and shown to yield a typical PCR sensitivity of approximately 100 spores/g of stool. Differences in the ability to detect a true positive were noted between methods and between laboratories. In the multicenter study, the abilities of molecular tests to detect a true positive, with 102 to 106 spores/g, ranged from 36 to 96% detection. In addition, some laboratories reported a false-positive rate as high as 4.5% (42).
It is clear from the results of these studies that standardization of PCR protocols is needed and that PCR carryover prevention practices to prevent contamination are not always adequate. There are numerous reasons for this lack of consistency. A lack of standardized primers and PCR conditions makes interpretation of results and comparisons difficult. Little information is available on the specificity and sensitivity of various primer sets for PCR, and results can vary widely (21, 42, 54).
Inhibitors are commonly found in fecal specimens. These include heme compounds, complex acidic polysaccharides, fat, protein, DNases, proteinases, and interference from the DNA of other, normal enteric organisms or shed mucosal cells (2, 16, 26, 30, 36, 49, 55). Attempts have been made to remove microsporidia from fecal and environmental water matrix inhibitors (1, 38); however, these procedures are still relatively labor-intensive and do not easily lend themselves to routine incorporation into a clinical or environmental diagnostic laboratory. Any nucleic acid extraction requires the release of inhibitors, concentration of eluates into small volumes, and target elution into buffers compatible with amplification. Historical methods for the release of DNA from spores include boiling, freezing-thawing, glass bead disruption, and incubation with lyticase, chitinase, or proteinase K. DNA release is typically followed by various DNA extraction or purification methods, such as the use of chaotropic agents or phenol-chloroform extraction. Commercial kits have also been used with success (21, 54; D. M. Wolk, S. K. Schneider, L. M. Sloan, and J. E. Rosenblatt, Abstr. 101st Gen. Meet. Am. Soc. Microbiol., abstr. C9, 2001).
To address difficulties in the extraction and identification of microsporidia from stool specimens in the clinical diagnostic laboratory, we have developed an automated DNA extraction method designed for the MagNA Pure LC workstation (Roche Applied Sciences, Indianapolis, Ind.) and a rapid, real-time PCR assay designed for the LightCycler instrument (Roche). Experiments were designed to optimize spore lysis and DNA extraction from microsporidia in fecal matrices. The LightCycler PCR (LC PCR) assay sensitivity was compared to that of the reference method, a routine smear with trichrome blue stain. In addition, various storage conditions (specimens that were fresh, refrigerated at 4°C, frozen at −70°C, and preserved) were tested for use in the molecular detection of microsporidia. The designed LC PCR assay provides a rapid, real-time, robust, contamination-free method with improved detection of microsporidia compared with trichrome blue staining. In addition, the assay provides the ability to discriminate among three different Encephalitozoon species (Encephalitozoon intestinalis, E. cuniculi, and E. hellem) by using differences in amplicon melting temperatures (Tms).
MATERIALS AND METHODS
Preparation of spores of microsporidia.
Spores of microsporidia (E. intestinalis ATCC 50603 and ATCC 50506, E. hellem ATCC 50451, and E. cuniculi ATCC 50502) were cultivated and harvested from RK-13 cells (ATCC CCL-37) grown in RPMI medium (Invitrogen, Carlsbad, Calif.) with 7% fetal bovine serum (FBS; HyClone, Logan, Utah) (58, 59). Spores were purified with an isopycnic gradient of Percoll (Pharmacia, Piscatawy, N.J.) (58) and washed with sterile distilled cell-culture-grade water (Invitrogen) (58, 59). Hemocytometer counts of spore suspensions were obtained in duplicate, and spore concentrations were adjusted to the desired target concentrations in water (log dilutions ranging from 102 to 108 spores/ml). After dilution, selected spore counts were reanalyzed in duplicate.
Diluted spores were stored at 4°C and used within 3 months to seed normal human fecal specimens for subsequent extraction and PCR. Final target concentrations of spores in feces ranged from 101 to 107 spores/ml. Control spores were maintained for stability testing at 106 spores/ml of water. Spores for melting curve analysis were prepared from the appropriate Encephalitozoon strain and stored at 107 spores/ml of water.
Storage conditions for spore-spiked feces.
Fresh liquid human fecal specimens used as specimen matrices were randomly chosen from samples submitted to the Mayo Clinic Parasitology Laboratory for routine ovum and parasite testing, enteric bacterial culturing, or Clostridium difficile testing. These samples were found negative for ova and parasites by microscopy. Fecal samples were stored at 4°C and tested within 1 week of collection. E. intestinalis spore dilutions were used as positive controls in order to establish standard curves. Similarly, spores were used to seed fecal specimens and were stored under different conditions to evaluate their ability to maintain intact, amplifiable DNA. The following storage conditions were evaluated with eight different fecal samples each: (i) immediate processing after spiking, (ii) refrigeration at 4°C for 72 h, (iii) freezing at −70°C for 7 days, and (iv) preservation in Para-Pak ECOFIX (Meridian Diagnostics Inc., Cincinnati, Ohio) stool transport medium for 7 days at room temperature.
Traditional parasite examination with trichrome blue stain.
Two-milliliter portions of each spiked fecal sample were preserved in formalin and concentrated by using an FPC fecal parasite concentrator (Evergreen Scientific, Los Angeles, Calif.) according to the manufacturer's recommendations and standard recommendations for specimen processing (22). After processing, 10-μl aliquots of each sample were spread thinly over glass microscope slides, fixed with methanol, air dried, and stained with trichrome blue solution (Meridian Diagnostics) according to the manufacturer's instructions and standard practices for smear preparation (22, 46). Smears were examined by light microscopy according to standard procedures (×1,000, oil); microscopy was done by two independent and experienced laboratory personnel who were blinded to the identities of the specimens. Control slides of E. intestinalis were included with each staining batch for comparison.
MagNA Pure system.
The MagNA Pure LC workstation is an automated “walk-away” system for nucleic acid extraction which is based on the extraction technology described by Boom et al. (9). With a MagNA Pure LC DNA isolation kit, genomic DNA from organisms lysed in buffer containing guanidine isothiocyanate is bound to magnetic glass particles under chaotropic conditions. The magnetic particles are washed to remove unbound substances and impurities. The washed DNA is eluted from the magnetic particles under conditions of low salt concentration and elevated temperature.
With wide-bore micropipette tips, 0.3 ml of microsporidian-seeded fecal specimen was added to 0.3 ml of test diluent. The mixture was vortexed briefly and then heated to 95°C for 5 min in a dry-heat block. After heating, the specimen was centrifuged at 14,000 × g for 1 min, and 200 μl of the supernatant was transferred to the appropriate well of a MagNA Pure sample cartridge. The MagNA Pure software program DNA 1 Blood Cells High Performance, version 2.1, was used for extraction with settings for an initial sample volume of 200 μl and an elution volume of 100 μl.
Since it is known that PCR inhibition can occur when water is used as a specimen diluent (8), several buffers and diluents were tested for use with the MagNA Pure system. Two commercial extraction buffers, lysis binding buffer and tissue lysis buffer (both from Roche), were tested. FBS was chosen as a potential diluent, since the bovine alpha-casein found in serum is an enhancer of PCR (8). Also, it was hoped that FBS would act as a biological buffer for the extraction process.
LightCycler assay.
The LightCycler assay designed here uses target-specific hybridization probes to allow fluorescence resonance energy transfer-based detection of amplicons. Results, expressed as crossover points when the log-linear part of the amplification curve crosses a fluorescence background threshold, indicate the PCR cycle number at which the amplification enters the log-linear phase (56, 57). The crossover point is proportional to the initial DNA concentration of the sample and can be used as a measure of extraction efficiency (40). In addition, a melting curve for the PCR amplicon can be generated (44). In the assay for Encephalitozoon spp., the reporter probe was designed as a mutation probe so that sequence mismatches among the Encephalitozoon spp. examined resulted in amplicon Tm differences.
Encephalitozoon PCR.
Pan-Encephalitozoon primers and fluorescence hybridization probes were designed to amplify a 268-bp region of the 16S rRNA gene of three Encephalitozoon spp. (E. cuniculi [GenBank accession no. X98470], E. hellem [GenBank accession no. L19070], and E. intestinalis [GenBank accession no. U09929]) and to identify them by using amplicon melting curve analysis. Forward and reverse primers were synthesized at the Mayo Clinic Molecular Biology Core Facility with the following sequences: forward primer, 5′-GTC CGT TAT GCC CTG AGA T-3′ (the 5′ base corresponds to E. intestinalis nucleotide 1013), and reverse primer, 5′-ACA GCA GCC ATG TTA CGA CT-3′ (the 3′ base corresponds to nucleotide 1251). Hybridization probes were obtained from Qiagen Operon (Alameda, Calif.). The probes were labeled with fluorescein or red 640 reporter dye as follows: probe 1, 5′-GCC CGT CGC TAT CTA AGA TGA CGC A-fluorescein-3′ (the 5′ base corresponds to E. intestinalis nucleotide 1178), and probe 2, 5′-red 640-TGG ACG AAG ATT GGA AGG TCT GAG TC-phosphate-3′ (the 5′ base corresponds to nucleotide 1204).
The LC PCR assay was designed with dUTP and uracil-N-glycosylase (UNG) as a carryover prevention measure (12, 17, 31) and was performed in a closed system to minimize amplicon contamination events. The PCR hybridization mixture (minus target DNA) was as follows: 3.5 mM MgCl2 (Invitrogen), PCR buffer lacking Mg2+ (Invitrogen), 1 μM each primer, 0.03 U of Platinum Taq DNA polymerase (Invitrogen)/μl, 0.2 mM PCR Nucleotide MixPlus (Roche), 0.2% UNG (1 U/μl; Epicentre, Madison, Wis.), 0.025% bovine serum albumin (Sigma, St. Louis, Mo.), 0.2 μM probe 1, and 0.4 μM probe 2. Thermal conditions were as follows: amplicon inactivation (37°C, 5 min), DNA denaturation and UNG inactivation (95°C for 3 min), amplification (45 cycles of 95°C, 60°C for 10 s, and 72°C for 16 s at 20°C/s), amplicon melting (55 to 85°C at 0.2°C/s), and cooling (40°C, 30 s). Each run included negative controls (approximately 10 to 15% of the total specimens tested) consisting of unspiked specimens and diluent blanks. Positive results were determined based on mathematical algorithms included with the LightCycler system. Results were determined to be positive when the fluorescence signal crossed at the baseline at ≤43 cycles.
Assay verification.
The presence of extraction and/or PCR inhibitors in the fecal matrices was assessed by comparing the average amplification efficiencies for spiked fecal samples to those for spores in water. If inhibitors were present in the feces, then they would be expected to cause an increase in the crossover value relative to that for samples in water and a corresponding decrease in the assay efficiency. The slope of a standard curve of spore concentration versus crossover value was used to calculate amplification efficiency (E) with the equation E = 10−1/slope. A perfect amplification efficiency is 2.0, indicating a doubling of target DNA during each cycle.
Assay sensitivity and reproducibility were evaluated by testing log dilutions of E. intestinalis spores in water (n = 3) and in feces (n = 8) over a range of 0 to 107 spores/ml of feces. Samples were tested under three storage conditions (fresh, refrigerated at 4°C, and frozen at −70°C), and resulting crossover values for the LC PCR assay were used for the calculations. Tm reproducibility was analyzed for all E. intestinalis spore dilutions (102 to 107 spores/ml) and for all storage conditions.
For analytical specificity, LC PCR testing was performed with DNA extracts from organisms known to be pathogens or normal enteric flora (50) (Table 1). The presence of extracted DNA was confirmed for all potential cross-reacting bacterial organisms by performing routine PCR for the 16S rRNA gene with universal primers and assessing the presence of amplicons by gel electrophoresis (M. K. Hopkins, D. T. Lynch, P. S. Mitchell, et al., Abstr. 101st Gen. Meet. Am. Soc. Microbiol., abstr. C45, 2001). The presence of E. bieneusi DNA was confirmed by a PCR assay for E. bieneusi (L. Xiao, Centers for Disease Control and Prevention, personal communication). The presence of viable Cryptosporidium parvum DNA was confirmed by an excystation protocol (28) and reverse transcription-PCR (45). Nonspiked, liquid fecal matrices (n = 74) submitted to the Mayo Clinic for routine ovum and parasite examination, enteric bacterial culturing, or C. difficile testing were also assayed by LC PCR.
TABLE 1.
Specificity testinga
| Organism |
|---|
| Bacterial pathogens |
| Aeromonas hydrophila |
| Clostridium difficile |
| Escherichia coli O157:H7 |
| Plesiomonas shigelloides |
| Salmonella group B |
| Shigella flexneri |
| Yersinia enterocolitica |
| Toxin producers |
| Clostridium perfringens |
| Staphylococcus aureus |
| Mycobacteria and fungi |
| Aspergillus fumigatus |
| Aspergillus nidulans |
| Candida albicans |
| Mycobacterium avium |
| Mycobacterium kansasii |
| Saccharomyces cerevisiae |
| Parasites |
| Cryptosporidium parvum (purified Iowa strain from calf feces) |
| Enterocytozoon bieneusi (n = 5 in feces) |
| Giardia lamblia (in feces) |
| Enteric bacterial flora |
| Arcanobacterium pyogenes |
| Bacteroides distasonis |
| Bacteroides fragilis |
| Bacteroides thetaiotaomicron |
| Bacteroides vulgatus |
| Bifidobacterium odontolyticus |
| Citrobacter freundii |
| Clostridium ramosum |
| Enterobacter cloacae |
| Enterococcus faecalis |
| Escherichia coli |
| Escherichia hermanii |
| Escherichia vulneris |
| Eubacterium lentum |
| Fusobacterium nucleatum |
| Klebsiella pneumoniae |
| Lactobacillus sp. |
| Proteus mirabilis |
| Pseudomonas aeruginosa |
| Staphylococcus epidermidis |
| Viridans group streptococci |
Genomic DNA from the listed organisms was tested for cross-reactivity in the Encephalitozoon LC PCR assay (n = 1 unless otherwise specified). A total of 74 fecal specimens were negative for microsporidia.
RESULTS
Extraction optimization.
In preliminary testing with fecal specimens spiked with E. intestinalis at 106 spores/ml, the MagNAPure LC method, performed with various specimen diluents, such as water, FBS, and serum-free cell culture medium, was found to produce crossover point results that were equivalent (within 95% confidence intervals) to those for specimens processed by the QIAamp DNA stool minikit method (Qiagen, Valencia, Calif.), used according to the manufacturer's instructions with high-temperature pretreatment (n = 5) (Wolk et al., Abstr. 101st Gen. Meet. Am. Soc. Microbiol.). Since the QIAamp method was found to be more labor-intensive that the MagNAPure LC method, all further testing was performed with the MagNAPure LC method.
With crossover points as a measure of relative DNA concentration and therefore extraction yield, spore suspensions at concentrations of 107 spores/ml were tested with tissue lysis buffer, lysis binding buffer, and FBS as sample diluents. Tissue lysis buffer permitted more efficient DNA extraction, with a crossover point of 20.6 ± 0.6 cycles (here and below, means and standard deviations are given), than did FBS (26.6 ± 1.2 cycles) or lysis binding buffer (30.3 ± 2.8 cycles). Therefore, tissue lysis buffer was used as the sample diluent for all subsequent work.
MDL and LLOD.
The analytical accuracies of microsporidian detection by the LC PCR assay and the traditional trichrome blue staining method are reported in Table 2. LC PCR results for control and refrigerated spores were essentially equivalent, with a minimum detection limit (MDL) of ≥103 spores/ml of feces. Reduced accuracy was observed when spores were frozen or preserved in ECOFIX (MDL, 105 spores/ml of feces). Nevertheless, under all storage conditions, the LC PCR assay resulted in a 2- to 4-log-unit increase in sensitivity over that of the routine smear method with trichrome blue staining (MDL, ≥106 spores/ml). The lower limit of detection (LLOD) for trichrome blue staining was 106 spores/ml of feces. Again, under all conditions examined, the LC PCR assay showed a 2- to 4-log-unit improvement over the routine smear method, with an LLOD of 102 to 104 spores/ml of feces, a value which is equivalent to 0.5 to 50 spores/PCR.
TABLE 2.
Analytical accuracy of the Encephalitozoon LC PCR assay for the detection of E. intestinalis-spiked fecesa
| Assay and storage conditions | % Positive results at the following concn of spores/ml
|
|||||
|---|---|---|---|---|---|---|
| 102 | 103 | 104 | 105 | 106 | 107 | |
| LC PCR | ||||||
| Fresh | 50 | 88 | 100 | 100 | 100 | 100 |
| Refrigerated (4°C for 72 h) | 50 | 100 | 100 | 100 | 100 | 100 |
| Frozen (−70°C for 7 days) | 25 | 63 | 75 | 100 | 100 | 100 |
| ECOFIX (room temperature for 14 days) | 0 | 0 | 75 | 100 | 100 | 100 |
| Smear with trichrome blue stain (no PCR) | 0 | 0 | 0 | 0 | 75 | 100 |
Fecal samples spiked with microsporidian spores at the indicated levels were analyzed by using the indicated assays. MDLs are indicated by bold type. Eight spiked fecal samples were analyzed at each concentration.
Sensitivity and specificity.
The analytical sensitivity of the LC PCR assay for spiked specimens (n = 25) that were processed immediately was determined to be 96% at 104 spores/ml of feces. The sensitivity for specimens stored in ECOFIX for 7 days was determined to be 96% at 104 spores/ml of feces (n = 25). Despite equivalent sensitivity, the crossover values for specimens processed immediately were statistically lower than those for specimens in ECOFIX storage (P = 0.05), suggesting that storage in ECOFIX resulted in some loss of DNA integrity. Nonspecific amplification was absent for nonspiked fecal matrices (n = 74) and for typical enteric bacterial pathogens and nonpathogens as well as parasitic, mycobacterial, and fungal organisms that might be found in stool specimens (Table 1).
Intra- and interassay reproducibilities.
Intra-assay reproducibility was assessed by using samples spiked with 104 spores/ml of feces and processed immediately. The mean crossover point was 33.2 ± 0.9 cycles, with a coefficient of variation of 2.7, for eight samples. Interassay reproducibility was also assessed at 104 spores/ml of feces, with crossover points of 32.2 ± 1.6 (n = 8 assays) for fresh, 30.9 ± 1.5 (n = 8) for refrigerated, 34.4 ± 2.5 (n = 6) for frozen, and 38.3 ± 2.1 (n = 6) for ECOFIX-preserved samples.
Control spore stability.
The stability of the control spores in water at 4°C was assessed by monitoring the crossover point as a function of storage time. Trend analysis revealed that the crossover point was 25.4 ± 1.4 cycles over a 5-month storage period. A similar analysis of spore-spiked feces resulted in a crossover point of 25.9 ± 2.1 cycles, indicating slightly greater variation, as might be predicted given the complex nature of the fecal storage medium. Using Dunnett's test, no statistical differences in crossover points were observed when control spores in water were compared to control spores in fecal matrix for the 5-month storage period.
Linear range of the assay.
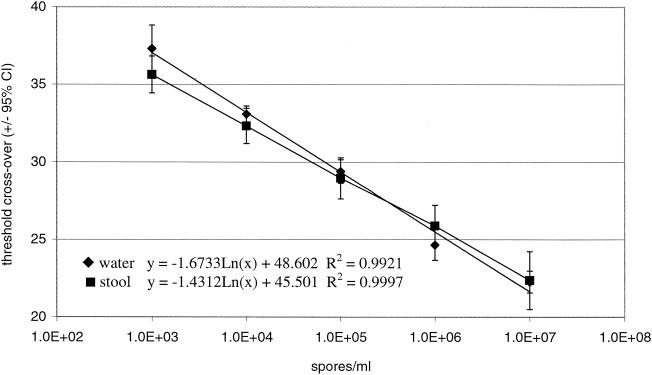
The LC PCR assay for Encephalitozoon spp. was linear from 103 to 107 spores/ml of water (Fig. 1). The same linear range was obtained for stool samples spiked with spores, regardless of whether they were assayed immediately, refrigerated, or frozen (Fig. 1). The regression lines for spores in water and for spores in stool samples (an average of all storage conditions) were equivalent (P = 0.05).
FIG. 1.
LC PCR assay linearity in water and stool samples. Spores were used to spike the specified matrix at the indicated concentrations, and then the samples were assayed immediately by the LC PCR method. The lines represent the linear regression analysis of each data set. The regression line for spores in stool samples is a combination of all data obtained for samples processed immediately, samples refrigerated at 4°C for 72 h, samples frozen at −70°C for 7 days, and samples preserved in ECOFIX for 14 days. CI, confidence interval. Spore concentrations are given as logarithmic dilutions of spores, i.e., 1.0 + E2 = 100 and 1.0 + E3 = 1,000.
PCR efficiency.
For serial dilutions of spores in water and feces, PCR efficiency proved to be statistically equivalent for all spore concentrations and storage conditions tested (P = 0.05). PCR efficiency for all assays ranged from 1.9 ± 0.1 to 2.1 ± 0.2, bracketing a perfect amplification doubling value of 2.0 in all instances.
Melting curve analysis.
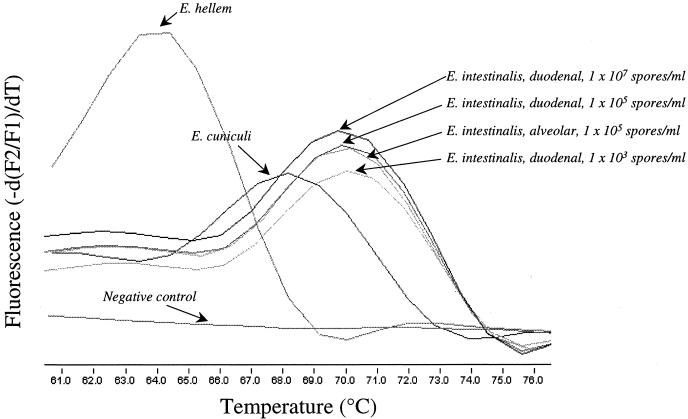
Another assessment of assay specificity was the repeated ability to differentiate among Encephalitozoon spp. by melting curve analysis (Fig. 2). The Tms for E. hellem, E. cuniculi, and E. intestinalis (alveolar and duodenal strains) were 63.7 ± 0.2, 67.9 ± 0.4, 69.7 ± 0.8, and 69.7 ± 0.7°C, respectively (n = 3). Although E. hellem and E. cuniculi are not expected to be found in fecal samples, the specificity testing demonstrates that E. intestinalis has a unique Tm even when the system is challenged by using organisms with very similar sequences. At 106 E. intestinalis spores/ml of feces, all Tms were significantly different, as judged by the 95% confidence interval, with the exception of those for the alveolar and duodenal strains of E. intestinalis, which one would expect to be alike. No statistical difference was observed in Tms for spore-spiked water or fecal matrix under any storage condition (n = 3) or as a function of spore dilution when spore concentrations ranging from 102 to 107 spores/ml of feces were tested.
FIG. 2.
Melting curve analysis of Enchephalitozoon species. Melting curves were obtained following the LC PCR assay for spiked stool samples (n = 3) containing E. hellem (63.7 ± 0.2°C), E. cuniculi (67.9 ± 0.4°C), and E. intestinalis (alveolar strain, 69.7 ± 0.8°C; duodenal strain, 69.7 ± 0.7°C) at 105 spores/ml of feces. Although E. hellem and E. cuniculi are not expected to be found in fecal samples, the specificity testing demonstrates that E. intestinalis has a unique Tm even when the system is challenged by using organisms with very similar sequences. The melting curves for E. intestinalis at various spore dilutions are provided to demonstrate Tm stability as a function of spore concentration. The melting curve for a negative control specimen is also provided. −d(F2/F1), change in fluorescence; dT, change in temperature.
DISCUSSION
The diagnosis of microsporidiosis is difficult because of the subjective nature of routine smears and their relative lack of sensitivity. Traditional molecular testing is promising (18, 19, 21, 22, 54), but routine molecular methods to identify microsporidia are resource intensive and not readily integrated into a clinical diagnostic laboratory setting. Furthermore, routine methods do not easily provide quantitative information, which may prove useful for correlation of organism burden with disease state.
In this study, an LC PCR assay was developed to detect three Encephalitozoon spp. and to assess DNA extraction and specimen storage methods. Of the extraction buffers tested, tissue lysis buffer afforded the best extraction efficiency in a fecal matrix. When MagNA Pure extraction with tissue lysis buffer was combined with LC PCR for the detection of E. intestinalis, assay sensitivity was greatly enhanced, as evidenced by a 1,000- to 10,000-fold decrease in the MDL compared to that obtained with traditional diagnosis by trichrome blue staining. The quantitative LC PCR assay can provide spore concentration information from fecal samples in a linear range of 103 to 107 spores/ml of feces.
Different storage conditions were examined in order to determine the best method for sample transport to and storage within the testing laboratory. Assay results for samples refrigerated for 72 h were shown to be equivalent to those for samples processed immediately and should provide adequate time for transport to a reference laboratory for testing. Alternatively, samples may be frozen at −70°C for 7 days or preserved in ECOFIX for 14 days with a minimal loss of sensitivity.
Inhibitor removal represents a major challenge to any DNA extraction process. One tactic to avoid assay inhibitors is to filter the spores away from the specimen. This approach has been successful for amplification from as little as 100 to 500 spores/ml of sample but is labor-intensive (38). While a number of PCR assays are available for microsporidia (18, 19, 21, 22, 54), none of these assays has a ready means by which to measure inhibition caused by the specimen matrix. The LC PCR assay is unique in that it allows an assessment of inhibition through monitoring of PCR efficiency. Since extraction efficiency and DNA product purity contribute to PCR efficiency and reproducibility, the LC PCR assay also provides an indirect measure of extraction efficiency and reproducibility. Finally, the LC PCR assay makes use of sequence-specific probe hybridization for distinguishing among three common Encephalitozoon spp. (E. intestinalis, E. hellem, and E. cuniculi) by using amplicon melting curve analysis. Although E. hellem and E. cuniculi are not expected to occur naturally in fecal samples, the Tm results demonstrate that E. intestinalis has a unique Tm even when the system is challenged by use of organisms with very similar sequences. Further analysis will be necessary to assess the discriminatory value of the Tm for other specimens, such as urine or respiratory specimens, where E. hellem and E. cuniculi are more likely to be detected.
In summary, the LC PCR assay greatly improves the sensitivity and accuracy of microsporidian detection over those of traditional staining methods and offers an improvement over previously described PCR assays in terms of ease of sample preparation, quantitation, species identification, and contamination control. The extraction method is a unique automated method that allows for processing of and inhibitor removal from 32 specimens in approximately 1.5 h with 15 to 20 min of hands-on preparation time. The PCR assay can be performed in a similar time frame. The system can be easily adapted for use in a high-throughput clinical laboratory.
The assay was validated by using liquid fecal specimens; thus, results may vary if formed specimens are tested. However, the symptoms of microsporidiosis have been consistently reported to be chronic diarrhea with liquid stools (10, 23). Therefore, only liquid fecal specimens should be tested at this time. Clearly, it will be important to adapt the assay for the detection of other microsporidian species, such as E. bieneusi, and for use with other specimen matrices, such as urine.
Acknowledgments
This work was supported by funds from the Division of Clinical Microbiology, Department of Laboratory Medicine and Pathology, Mayo Clinic.
We are grateful to Lihuo Xiao, Centers for Disease Control and Prevention (Atlanta, Ga.), for the contribution of five fecal specimens containing PCR-confirmed E. bieneusi for our assay specificity testing; Charles Sterling and Marilyn Marshall, University of Arizona (Tucson), for contributions of C. parvum; and Heather Swan, Parasitology Laboratory, Mayo Clinic, for laboratory assistance with this project. We also acknowledge the statistical consultation performed by the Center for Patient-Oriented Research, Mayo Clinic, and Mary Mays, Office of Clinical Research, University of Arizona.
REFERENCES
- 1.Accoceberry, I., M. Thellier, A. Datry, I. Desportes-Livage, S. Biligui, M. Danis, and X. Santarelli. 2001. One-step purification of Enterocytozoon bieneusi spores from human stools by immunoaffinity expanded-bed adsorption. J. Clin. Microbiol. 39:1947-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akane, A., K. Matsubara, H. Nakamura, S. Takahashi, and K. Kimura. 1994. Identification of the heme compound copurified with deoxyribonucleic acid (DNA) from bloodstains, a major inhibitor of polymerase chain reaction (PCR) amplification. J. Forensic Sci. 39:362-372. [PubMed] [Google Scholar]
- 3.Aldras, A. M., J. M. Orenstein, D. P. Kotler, J. A. Shadduck, and E. S. Didier. 1994. Detection of microsporidia by indirect immunofluorescence antibody test using polyclonal and monoclonal antibodies. J. Clin. Microbiol. 32:608-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ali, M. S., L. A. Mahmoud, B. E. Abaza, and M. A. Ramadan. 2000. Intestinal spore-forming protozoa among patients suffering from chronic renal failure. J. Egypt. Soc. Parasitol. 30:93-100. [PubMed] [Google Scholar]
- 5.Beckers, P. J., G. J. Derks, T. Gool, F. J. Rietveld, and R. W. Sauerwein. 1996. Encephalocytozoon intestinalis-specific monoclonal antibodies for laboratory diagnosis of microsporidiosis. J. Clin. Microbiol. 34:282-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berlin, O. G., L. R. Ash, C. N. Conteas, and J. B. Peter. 1999. Rapid, hot chromotrope stain for detecting microsporidia. Clin. Infect. Dis. 29:209. [DOI] [PubMed] [Google Scholar]
- 7.Berlin, O. G., C. N. Conteas, L. R. Ash, F. Sorvillo, C. V. Jacob, J. Yatabe, and J. B. Peter. 2000. Permanence of Fungi-Fluor epifluorescence stain to detect microsporidia. Am. J. Gastroenterol. 95:3147-3149. [DOI] [PubMed] [Google Scholar]
- 8.Boom, R., C. Sol, M. Beld, J. Weel, J. Goudsmit, and P. Wertheim-van Dillen. 1999. Improved silica-guanidinium thiocyanate DNA isolation procedure based on selective binding of bovine alpha-casein to silica particles. J. Clin. Microbiol. 37:615-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boom, R., C. J. Sol, M. M. Salimans, C. L. Jansen, P. M. Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brasil, P., D. B. de Lima, D. D. de Paiva, M. S. Lobo, F. C. Sodre, S. P. Silva, E. V. Villela, E. J. Silva, J. M. Peralta, M. Morgado, and H. Moura. 2000. Clinical and diagnostic aspects of intestinal microsporidiosis in HIV-infected patients with chronic diarrhea in Rio de Janeiro, Brazil. Rev. Inst. Med. Trop. Sao Paulo 42:299-304. [DOI] [PubMed] [Google Scholar]
- 11.Bryan, R. T., and D. A. Schwartz. 1999. Epidemiology of microsporidiosis, p. 502-516. In M. Wittner and L. M. Weiss (ed.), The microsporidia and microsporidiosis. American Society for Microbiology, Washington, D.C.
- 12.Burkardt, H. J. 2000. Standardization and quality control of PCR analyses. Clin. Chem. Lab. Med. 38:87-91. [DOI] [PubMed] [Google Scholar]
- 13.Chioralia, G., T. Trammer, H. Kampen, and H. M. Seitz. 1998. Relevant criteria for detecting microsporidia in stool specimens. J. Clin. Microbiol. 36:2279-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cotte, L., M. Rabodonirina, F. Chapuis, F. Bailly, F. Bissuel, C. Raynal, P. Gelas, F. Persat, M. A. Piens, and C. Trepo. 1999. Waterborne outbreak of intestinal microsporidiosis in persons with and without human immunodeficiency virus infection. J. Infect. Dis. 180:2003-2008. [DOI] [PubMed] [Google Scholar]
- 15.da Silva, A. J., F. J. Bornay-Llinares, I. N. Moura, S. B. Slemenda, J. L. Tuttle, and N. J. Pieniazek. 1999. Fast and reliable extraction of protozoan parasite DNA from fecal specimens. Mol. Diagn. 4:57-64. [DOI] [PubMed] [Google Scholar]
- 16.Demeke, T., and R. P. Adams. 1992. The effects of plant polysaccharides and buffer additives on PCR. BioTechniques 12:332-334. [PubMed] [Google Scholar]
- 17.Espy, M. J., T. F. Smith, and D. H. Persing. 1993. Dependence of polymerase chain reaction product inactivation protocols on amplicon length and sequence composition. J. Clin. Microbiol. 31:2361-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fedorko, D. P., N. A. Nelson, and C. P. Cartwright. 1995. Identification of microsporidia in stool specimens by using PCR and restriction endonucleases. J. Clin. Microbiol. 33:1739-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fedorko, D. P., N. A. Nelson, E. S. Didier, D. Bertucci, R. M. Delgado, and A. M. Hruszkewycz. 2001. Speciation of human microsporidia by polymerase chain reaction single-strand conformation polymorphism. Am. J. Trop. Med. Hyg. 65:397-401. [DOI] [PubMed] [Google Scholar]
- 20.Fournier, S., O. Liguory, C. Sarfati, F. David-Ouaknine, F. Derouin, J. M. Decazes, and J. M. Molina. 2000. Disseminated infection due to Encephalitozoon cuniculi in a patient with AIDS: case report and review. HIV Med. 1:155-161. [DOI] [PubMed] [Google Scholar]
- 21.Franzen, C., and A. Muller. 1999. Molecular techniques for detection, species differentiation, and phylogenetic analysis of microsporidia. Clin. Microbiol. Rev. 12:243-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia, L. S. 2002. Laboratory identification of the microsporidia. J. Clin. Microbiol. 40:1892-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goetz, M., S. Eichenlaub, G. R. Pape, and R. M. Hoffmann. 2001. Chronic diarrhea as a result of intestinal microsposidiosis in a liver transplant recipient. Transplantation 71:334-337. [DOI] [PubMed] [Google Scholar]
- 24.Guerard, A., M. Rabodonirina, L. Cotte, O. Liguory, M. A. Piens, S. Daoud, S. Picot, and J. L. Touraine. 1999. Intestinal microsporidiosis occurring in two renal transplant recipients treated with mycophenolate mofetil. Transplantation 68:699-707. [DOI] [PubMed] [Google Scholar]
- 25.Gumbo, T., R. E. Hobbs, C. Carlyn, G. Hall, and C. M. Isada. 1999. Microsporidia infection in transplant patients. Transplantation 67:482-484. [DOI] [PubMed] [Google Scholar]
- 26.Holland, J. L., L. Louie, A. E. Simor, and M. Louie. 2000. PCR detection of Escherichia coli O157:H7 directly from stools: evaluation of commercial extraction methods for purifying fecal DNA. J. Clin. Microbiol. 38:4108-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelkar, R., P. S. Sastry, S. S. Kulkarni, T. K. Saikia, P. M. Parikh, and S. H. Advani. 1997. Pulmonary microsporidial infection in a patient with CML undergoing allogeneic marrow transplant. Bone Marrow Transplant. 19:179-182. [DOI] [PubMed] [Google Scholar]
- 28.Korich, D. G., J. R. Mead, M. S. Madore, N. A. Sinclair, and C. R. Sterling. 1990. Effects of ozone, chlorine dioxide, chlorine, and monochloramine on Cryptosporidium parvum oocyst viability. Appl. Environ. Microbiol. 56:1423-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kotler, D. P., and J. M. Orenstein. 1998. Clinical syndromes associated with microsporidiosis. Adv. Parasitol. 40:321-349. [DOI] [PubMed] [Google Scholar]
- 30.Lantz, P. G., M. Matsson, T. Wadstrom, and P. Radstrom. 1997. Removal of PCR inhibitors from human fecal samples through the use of an aqueous two-phase system for sample preparation prior to PCR. J. Microbiol. Methods 28:159-167. [Google Scholar]
- 31.Longo, M. C., M. S. Berninger, and J. L. Hartley. 1990. Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene 93:125-128. [DOI] [PubMed] [Google Scholar]
- 32.Luna, V. A., B. K. Stewart, D. L. Bergeron, C. R. Clausen, J. J. Plorde, and T. R. Fritsche. 1995. Use of the fluorochrome calcofluor white in the screening of stool specimens for spores of microsporidia. Am. J. Clin. Pathol. 103:656-659. [DOI] [PubMed] [Google Scholar]
- 33.Marshall, M. M., D. Naumovitz, Y. Ortega, and C. R. Sterling. 1997. Waterborne protozoan pathogens. Clin. Microbiol. Rev. 10:67-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mathis, A., A. C. Breitenmoser, and P. Deplazes. 1999. Detection of new Enterocytozoon genotypes in faecal samples of farm dogs and a cat. Parasite 6:189-193. [DOI] [PubMed] [Google Scholar]
- 35.Metge, S., J. T. Van Nhieu, D. Dahmane, P. Grimbert, F. Foulet, C. Sarfati, and S. Bretagne. 2000. A case of Enterocytozoon bieneusi infection in an HIV-negative renal transplant recipient. Eur. J. Clin. Microbiol. Infect. Dis. 19:221-223. [DOI] [PubMed] [Google Scholar]
- 36.Monteiro, L., D. Bonnemaison, A. Vekris, K. G. Petry, J. Bonnet, R. Vidal, J. Cabrita, and F. Megraud. 1997. Complex polysaccharides as PCR inhibitors in feces: Helicobacter pylori model. J. Clin. Microbiol. 35:995-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moura, H., D. A. Schwartz, F. Bornay-Llinares, F. C. Sodre, S. Wallace, and G. S. Visvesvara. 1997. A new and improved “quick-hot Gram-chromotrope” technique that differentially stains microsporidian spores in clinical samples, including paraffin-embedded tissue sections. Arch. Pathol. Lab. Med. 121:888-893. [PubMed] [Google Scholar]
- 38.Orlandi, P. A., and K. A. Lampel. 2000. Extraction-free, filter-based template preparation for rapid and sensitive PCR detection of pathogenic parasitic protozoa. J. Clin. Microbiol. 38:2271-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rabodonirina, M., M. Bertocchi, I. Desportes-Livage, L. Cotte, H. Levrey, M. A. Piens, G. Monneret, M. Celard, J. F. Mornex, and M. Mojon. 1996. Enterocytozoon bieneusi as a cause of chronic diarrhea in a heart-lung transplant recipient who was seronegative for human immunodeficiency virus. Clin. Infect. Dis. 23:114-117. [DOI] [PubMed] [Google Scholar]
- 40.Read, S. J. 2001. Recovery efficiences on nucleic acid extraction kits as measured by quantitative LightCycler PCR. Mol. Pathol. 54:86-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reetz, J. 1994. Natural transmission of microsporidia (Encephalitozoon cuniculi) by way of the chicken egg. Tierarztl. Prax. Suppl. 22:147-150. [PubMed] [Google Scholar]
- 42.Rinder, H., K. Janitschke, H. Aspock, A. J. Da Silva, P. Deplazes, D. P. Fedorko, C. Franzen, U. Futh, F. Hunger, A. Lehmacher, C. G. Meyer, J. M. Molina, J. Sandfort, R. Weber, T. Loscher, et al. 1998. Blinded, externally controlled multicenter evaluation of light microscopy and PCR for detection of microsporidia in stool specimens. J. Clin. Microbiol. 36:1814-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rinder, H., A. Thomschke, B. Dengjel, R. Gothe, T. Loscher, and M. Zahler. 2000. Close genotypic relationship between Enterocytozoon bieneusi from humans and pigs and first detection in cattle. J. Parasitol. 86:185-188. [DOI] [PubMed] [Google Scholar]
- 44.Ririe, K. M., R. P. Rasmussen, and C. T. Wittwer. 1997. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal. Biochem. 245:154-160. [DOI] [PubMed] [Google Scholar]
- 45.Rochelle, P. A., D. M. Ferguson, T. J. Handojo, R. De Leon, M. H. Stewart, and R. L. Wolfe. 1997. An assay combining cell culture with reverse transcriptase PCR to detect and determine the infectivity of waterborne Cryptosporidium parvum. Appl. Environ. Microbiol. 63:2029-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ryan, N. J., G. Sutherland, K. Coughlan, M. Globan, J. Doultree, J. Marshall, R. W. Baird, J. Pedersen, and B. Dwyer. 1993. A new trichrome-blue stain for detection of microsporidial species in urine, stool, and nasopharyngeal specimens. J. Clin. Microbiol. 31:3264-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sax, P. E., J. D. Rich, W. S. Pieciak, and Y. M. Trnka. 1995. Intestinal microsporidiosis occurring in a liver transplant recipient. Transplantation 60:617-618. [DOI] [PubMed] [Google Scholar]
- 48.Sparfel, J. M., C. Sarfati, O. Liguory, B. Caroff, N. Dumoutier, B. Gueglio, E. Billaud, F. Raffi, J. M. Molina, M. Miegeville, and F. Derouin. 1997. Detection of microsporidia and identification of Enterocytozoon bieneusi in surface water by filtration followed by specific PCR. J. Eukaryot. Microbiol. 44:78S. [DOI] [PubMed] [Google Scholar]
- 49.Stacy-Phipps, S., J. J. Mecca, and J. B. Weiss. 1995. Multiplex PCR assay and simple preparation method for stool specimens detect enterotoxigenic Escherichia coli DNA during course of infection. J. Clin. Microbiol. 33:1054-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanock, G. W. 1999. The normal microflora: an introduction, p. 1-20. In C. W. Tannock (ed.), Medical importance of the normal microflora. Kluwer Academic Publishers, Boston, Mass.
- 51.van Gool, T., J. C. Vetter, B. Weinmayr, A. Van Dam, F. Derouin, and J. Dankert. 1997. High seroprevalence of Encephalitozoon species in immunocompetent subjects. J. Infect. Dis. 175:1020-1024. [DOI] [PubMed] [Google Scholar]
- 52.Weber, R., and R. T. Bryan. 1994. Microsporidial infections in immunodeficient and immunocompetent patients. Clin. Infect. Dis. 19:517-521. [DOI] [PubMed] [Google Scholar]
- 53.Weber, R., R. T. Bryan, R. L. Owen, C. M. Wilcox, L. Gorelkin, G. S. Visvesvara, et al. 1992. Improved light-microscopical detection of microsporidia spores in stool and duodenal aspirates. N. Engl. J. Med. 326:161-166. [DOI] [PubMed] [Google Scholar]
- 54.Weiss, L. M., and C. R. Vossbrinck (ed.). 1999. Molecular biology, molecular phylogeny, and molecular diganostic approaches to the microsporidia. American Society for Microbiology, Washington, D.C.
- 55.Widjojoatmodjo, M. N., A. C. Fluit, R. Torensma, G. P. Verdonk, and J. Verhoef. 1992. The magnetic immunopolymerase chain reaction assay for direct detection of salmonellae in fecal samples. J. Clin. Microbiol. 30:3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wittwer, C. T., M. G. Herrmann, A. A. Moss, and R. P. Rasmussen. 1997. Continuous fluorescence monitoring of rapid cycle DNA amplification. BioTechniques 22:130-131, 134-138. [DOI] [PubMed] [Google Scholar]
- 57.Wittwer, C. T., K. M. Ririe, R. V. Andrew, D. A. David, R. A. Gundry, and U. J. Balis. 1997. The LightCycler: a microvolume multisample fluorimeter with rapid temperature control. BioTechniques 22:176-181. [DOI] [PubMed] [Google Scholar]
- 58.Wolk, D. M. 1999. Development and application of cell culture and molecular techniques for the diagnosis, identification, and viability testing of human microsporidia. Ph.D. thesis. University of Arizona, Tucson.
- 59.Wolk, D. M., C. H. Johnson, E. W. Rice, M. M. Marshall, K. F. Grahn, C. B. Plummer, and C. R. Sterling. 2000. A spore counting method and cell culture model for chlorine disinfection studies of Encephalitozoon syn. Septata intestinalis. Appl. Environ. Microbiol. 66:1266-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]