Abstract
We report on a new Actinobaculum species, “Actinobaculum massiliae,” isolated from the urine of an elderly woman with recurrent cystitis. Its phenotypic pattern was similar to those of both of the other Actinobaculum species described to date. On 16S rRNA sequencing, the Marseille isolate shared 95% homology with Actinobaculum suis, 92 to 93% homology with Actinobaculum schaalii, 91 to 92% homology with Arcanobacterium spp., and 87 to 90% homology with Actinomyces species. A bootstrap value of 99% supports the node separating the Actinobaculum sp. from its closest neighbor (A. suis). In conclusion, on the basis of phenotypic, genotypic, and phylogenetic assessments, we show that the Marseille isolate is a previously unrecognized organism within the Actinobaculum genus, and we propose placement of the organism in the taxon “Actinobaculum massiliae.”
Actinobaculum suis, initially described as Eubacterium suis (7), was reclassified as Actinomyces suis in 1992 (4). Its taxonomy was further modified in 1997 by Lawson et al. (3), who proposed a new genus, Actinobaculum, that includes two species: Actinobaculum schaalii and A. suis. A. suis is pathogenic for sows, leading to cystitis (6) and abortion (9). The type strain of A. schaalii was isolated from the blood of a 64-year-old man with chronic pyelonephritis (3), suggesting that it may be an agent of urinary tract infection. Here we report on a new Actinobaculum species that we named “Actinobaculum massiliae.” The organism was isolated from the urine of an elderly woman with recurrent cystitis.
CASE REPORT
An 81-year-old woman presented with brachiofacial ischemic stroke in the summer of 1999, and a bladder catheter was placed. The catheter was associated with cystitis and was removed 1 month later. A gram-positive bacillus was the only bacterium isolated from her urine at that time. This bacterium, recovered by a private laboratory and thought to be a vaginal contaminant, was not identified, nor was it tested for its antibiotic susceptibility. Empirical therapy with amoxicillin-clavulanate was given for 2 weeks, with partial and temporary resolution of the symptoms. Repeated episodes of pollakiuria with urge urinary incontinence were reported starting in March 2000. In May 2001, a urine culture was done when symptoms of cystitis, i.e., dysuria and pollakiuria, reappeared. At that time, a gram-positive bacillus was again isolated from the patient's urine, but this time it was identified as an Actinobaculum sp. by 16S rRNA amplification and sequencing. It was found to be susceptible to all antibiotics used for the treatment of lower urinary tract infection. Despite successive therapy with amoxicillin and trimethoprim-sulfamethoxazole, in October 2001 the same organism was recovered from the patient's urine at a high concentration (106 bacteria/ml), and it was then resistant to trimethoprim-sulfamethoxazole. Rifampin therapy (900 mg/day) initiated in October 2001 failed, and it was later associated with in vitro resistance to rifampin (December 2001) and persistence of the pollakiuria. Cure was finally achieved with doxycycline.
MATERIALS AND METHODS
The strain isolated on sheep blood agar from the patient's urine in May 2001 was used for phenotypic, genotypic, and phylogenetic analyses. Growth requirements and colony morphology were studied with cultures set up on Columbia sheep blood agar (BioMérieux, Marcy l'Etoile, France) and chocolate agar (BioMérieux) and incubated at 37°C in a 5% CO2 atmosphere. In addition, we tested for the presence of growth under anaerobic conditions on sheep blood agar at 37°C (GENbag anaer; BioMérieux), and anaerobic conditions were confirmed by using the AnaerIndicator strip (Merck, Darmstadt, Germany). A mobility test was performed by the hanging-drop method. Oxidase and catalase activities were evaluated by routine procedures. Additional biochemical tests were performed by inoculation of the API 20Strep (BioMérieux) and API Coryne (BioMérieux) systems according to the instructions of the manufacturer, with the exception that the incubation time was increased to 72 h. The cell wall fatty acid composition was analyzed by gas chromatography (5).
DNA was extracted with the FastDNA kit (Bio 101, Carlsbad, Calif.) and a FastPrep120 grinder (Bio 101) according to the instructions of the manufacturer. PCR amplification of the 16S RNA gene was performed with primers fD1 and rP2 (8) and Taq DNA polymerase (Gibco BRL, Life Technologies), also as specified by the manufacturer. The success of the amplification was determined by electrophoresis of the PCR products in a 1% agarose gel and staining with ethidium bromide. The PCR products were purified by use of the QIAquick PCR purification kit (Qiagen, Courtaboeuf, France). Sequencing was performed by using the dRhodamine Terminator Cycle Sequencing Ready Reaction kit with one of nine different primers and AmpliTaq DNA (Perkin-Elmer Biosystems, Warrington, England) with a 3100 ABI Prism automated sequencer (Applied Biosystems, Courtaboeuf, France). Sequences derived from each primer were aligned, compared, and combined into a single 16S rRNA sequence by using Autoassembler software (version 2.1; Applied Biosystems). The validity of the sequence obtained was assessed by comparison with two additional sequences similarly obtained, but from two other PCRs of the same template DNA. The sequence was compared with all eubacterial 16S rRNA sequences available in the GenBank database by using the BLASTN 2.2.2 program available on the National Center for Biotechnology Information website (www.ncbi.nlm.nih.gov) (1). The 16S rRNA sequence of the Marseille isolate was aligned with those of other members of the family Actinomycetaceae, and the degree of 16S rRNA homology was determined by using the CLUSTWAL W program supported by the DDBJ website (www.ddbj.nig.ac.jp). Sequences were edited by removal of the longer 5′ and 3′ ends so that their lengths matched that of the shortest sequence. The edited sequences were then analyzed by the distance matrix program (Kimura's correction) supported by the DDBJ website. With these sequences, additional trees were constructed by using the distance matrix, parsimony, and maximum likelihood programs of the Phylip package (2). The isolates recovered in October and December were identified by 16S rRNA amplification and sequencing of a segment of about 500 bp with the following primers: 5′-CAGCAGCCGCGGTAATAC-3′ and 5′-CACGAGCTGACGACA-3′.
Nucleotide sequence accession number.
The 16S rRNA gene sequence of the Marseille “A. massiliae” isolate has been deposited in the GenBank database under accession number AF487679.
RESULTS
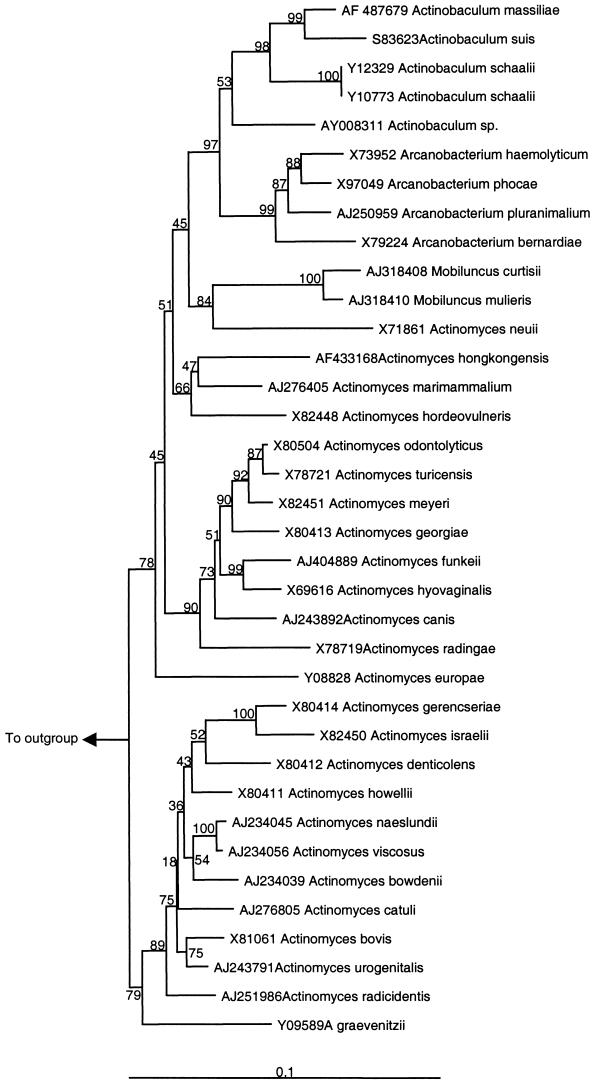
Primary isolation was obtained by inoculation of sheep blood agar (BioMérieux) with 10 μl of midstream urine and incubation at 37°C in a 5% CO2 atmosphere for 72 h. No growth was observed when urine was inoculated on bromocresol purple lactose agar (Becton Dickinson, Meylan, France). Nonhemolytic small greyish colonies could be seen on blood agar; the strain presented two colony types after 5 days of incubation. Microscopic examination showed nonmotile, non-acid-fast, non-spore-forming, gram-positive organisms. Some bacteria were poorly stained with the Gram stain, and a few exhibited branching. Growth was observed, although it was slower, under anaerobic conditions and on chocolate agar. Green hemolysis was observed with culture on chocolate agar. Bacteria grown on the latter medium were tested for catalase reactivity, which was absent. Acid was produced from glucose, maltose, ribose, xylose, and trehalose, but not from mannose, mannitol, sorbitol, or amidon. Slight acid production from raffinose was observed. Esculin and gelatin were not hydrolyzed. Nitrate was not reduced to nitrite. Hippurate was hydrolyzed. Pyrazinamidase and α-glucosidase activities were detected. Inversely, no pyrrolidonyl arylamidase, alkaline phosphatase, α-galactosidase, β-glucuronidase, β-galactosidase, N-acetyl-β-glucosaminidase, urease, leucine arylamidase, or arginine dihydrolase activity was detected (Table 1). The whole-cell fatty acid composition of the Marseille isolate demonstrated relatively large amounts of hexadecanoic acid (C16:0; 41.5%), cis-octadec-9-enoic-acid (C18:1ω9c; 24.2%), and octadecanoic acid (C18:0; 8.7%). The Marseille isolate shared 95% homology with A. suis, 93% homology with A. schaalii, 91 to 92% homology with Arcanobacterium spp., and 87 to 90% homology with Actinomyces species (Table 2). The shortest 16S rRNA sequence included in the phylogenetic analysis was that of Actinomyces turicensis (1310 bp); accordingly, alignment began at base 1 of the 16S rRNA gene of A. turicensis. The Marseille isolate clustered with the Actinobaculum spp. by phylogenetic analysis (matrix distance approach) of the 16S rRNA genes of species of the family Actinomycetaceae (Fig. 1). Thus, a bootstrap value of 97% in the neighbor-joining tree supported the fork separating the Actinobaculum spp. (including the Marseille isolate) from their closest relatives: members of the genus Arcanobacterium. More importantly, the isolate was clearly different from its closest neighbor (A. suis), with a bootstrap value of 99% (Fig. 1). Parsimony and maximum likelihood methods confirmed that the Marseille isolate clustered within the Actinobaculum spp. and that its lineage was clearly different from that of A. suis. The isolates recovered in October and December 2001 shared 100% sequence homology with the type strain, recovered in May 2001 from urine specimens from the same patient.
TABLE 1.
Comparison of the “A. massiliae” phenotype with those of other members of the family Actinomycetaceae, showing its relatedness to other Actinobaculum spp.a
| Organism | Nitrate reduction | Urease activity | Esculin hydrolysis | Acid produced from:
|
β-Galactosidase | α-Glucosidase | N-Acetyl-β- glucosaminidase | β-Hemolysis | |
|---|---|---|---|---|---|---|---|---|---|
| Mannitol | Xylose | ||||||||
| Actinobaculum schaalii | − | − | − | − | + | − | + | − | − |
| Actinobaculum suis | − | + | − | − | − | + | ND | ND | − |
| Actinomyces bovis | − | − | V | − | − | − | − | + | V |
| Arcanobacterium bernardiae | − | − | − | − | − | − | + | − | V |
| Arcanobacterium haemolyticum | − | − | − | − | − | + | ND | + | + |
| Arcanobacterium phocae | − | − | − | V | − | (+) | (+) | − | + |
| Arcanobacterium pyogenes | − | − | − | V | + | V | − | + | + |
| Clinical isolate from this study | − | − | − | − | + | − | + | − | − |
Adapted from Lawson et al. (3). Abbreviations and symbols: +, positive reaction; −, negative reaction; (+), weakly positive reaction; V, variable reaction; ND, not determined.
TABLE 2.
Comparison of 16S rRNA sequence homology of “A. massiliae” with those of other members of the family Actinomycetaceae and Corynebacterium amycolatum
| Species no. | Species | % Homologya
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | ||
| 1 | “Actinobaculum massiliae” | 100 | 93 | 95 | 92 | 91 | 91 | 91 | 89 | 89 | 88 | 80 | 88 |
| 2 | Actinobaculum schaalii | 100 | 93 | 91 | 89 | 90 | 91 | 88 | 89 | 88 | 80 | 87 | |
| 3 | Actinobaculum suis | 100 | 92 | 89 | 90 | 91 | 87 | 88 | 89 | 79 | 87 | ||
| 4 | Arcanobacterium bernardiae | 100 | 93 | 94 | 95 | 89 | 88 | 90 | 80 | 88 | |||
| 5 | Arcanobacterium haemolyticum | 100 | 97 | 95 | 89 | 88 | 88 | 80 | 88 | ||||
| 6 | Arcanobacterium phocae | 100 | 96 | 89 | 90 | 89 | 80 | 89 | |||||
| 7 | Arcanobacterium pluranimalium | 100 | 90 | 90 | 90 | 80 | 88 | ||||||
| 8 | Actinomyces bovis | 100 | 92 | 88 | 81 | 87 | |||||||
| 9 | Actinomyces meyeri | 100 | 90 | 80 | 87 | ||||||||
| 10 | Mobiluncus mulieris | 100 | 78 | 85 | |||||||||
| 11 | Denitrobacterium detoxifians | 100 | 80 | ||||||||||
| 12 | Corynebacterium amycolatum | 100 | |||||||||||
The number at the top of each column corresponds to the species as number defined for each row.
FIG. 1.
Phylogenetic tree showing relationship of “A. massiliae” sp. nov. with other members of the family Actinomycetaceae. The tree was constructed by using the neighbor-joining method, based on the nearly complete sequence (1,310 nucleotides) of the 16S rRNA gene. Bootstrap values resulting from 100 replications are presented at the branch points. Denitrobacterium detoxifians was used as the outgroup.
DISCUSSION
In this paper, we described a new species, “A. massiliae,” recovered from the urine of an 81-year-old woman with a chronic urinary tract infection. The same strain was recovered three times over a 7-month period, highlighting the persistence of the infection despite antibiotic treatment. Interestingly, this strain was not recovered from bromocresol purple lactose agar (Becton Dickinson), which is the medium routinely used in the Marseille laboratory, even after prolonged incubation. Our use of sheep blood agar and prolonged incubation, which permitted strain recovery, was motivated by the presence of 106 leukocytes/ml in the patient's urine. This is the second report of a human infection due to Actinobaculum species: the A. schaalii type strain was isolated from a 64-year-old man with chronic pyelonephritis (3). Of interest, A. suis has been isolated from sows with cystitis (6). It should be stressed that chronic urinary tract infection due to Actinobaculum species might be underdiagnosed because these organisms cannot be isolated by routine procedures. In addition, the phenotypic heterogeneity of the colonies after 5 days of incubation may lead to an erroneous interpretation of sample contamination with two different species of gram-positive bacilli. The infection described in the present study was cured only with doxycycline, which may be the treatment of choice for such infections. As recurrence occurred after therapy with amoxicillin and trimethoprim-sulfamethoxazole and as resistance to rifampin was observed after exposure to that drug, close follow-up with stringently controlled urine cultures seems mandatory to confirm a cure.
The phenotypic and biochemical patterns suggested that the Marseille isolate was related to A. schaalii. Indeed, tests for urease and β-galactosidase activities were found to be negative, and acid was produced from glucose, maltose, ribose, and xylose. However, as for A. suis, acid was produced from glycogen. In addition, the Marseille isolate showed only 95% 16S rRNA sequence homology with A. suis and showed even lower degrees of homology with all other known species of the family Actinomycetaceae. Our phylogenetic analysis demonstrated that the Marseille isolate shares some degree of evolutionary descent with members of the genus Actinobaculum. In conclusion, on the basis of phenotypic, genotypic, and phylogenetic assessments, we show that the Marseille isolate is a previously unrecognized organism within the genus Actinobaculum, and we propose placement of the organism in the taxon “Actinobaculum massiliae.”
Description of “A. massiliae” sp. nov.
Massilia is the old Greek and Roman name for Marseille, where the organism was isolated. The organism consists of straight to slightly curved rods, some of which exhibit branching. It has nonmotile, non-acid-fast, gram-positive (easily decolorized) cells. Colonies on sheep blood agar are 1 mm in diameter after 72 h of incubation at 37°C in a 5% CO2 atmosphere, and there is no hemolysis. It is facultatively aerobic. Esculin and gelatin were not hydrolyzed. Nitrate was not reduced to nitrite. Acid was produced from glucose, maltose, ribose, glycogen, and xylose. The type strain of “A. massiliae,” which was isolated from the urine of an 81-year-old woman with cystitis, has been deposited in the Collection de l'Institut Pasteur, Paris, France, as strain CIP 107404T.
Acknowledgments
We thank Christiane Bibard and Veronique Brice for help.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Felsenstein, J. 1989. PHYLIP—phylogeny inference package. Cladistics 5:164-166. [Google Scholar]
- 3.Lawson, P. A., E. Falsen, E. Akervall, P. Vandamme, and M. D. Collins. 1997. Characterization of some Actinomyces-like isolates from human clinical specimens: reclassification of Actinomyces suis (Soltys and Spratling) as Actinobaculum schaalii comb. nov. and description of Actinobaculum schaalii sp. nov. Int. J. Syst. Bacteriol. 47:899-903. [DOI] [PubMed] [Google Scholar]
- 4.Ludwig, W., G. Kirchhof, M. Weizenzgger, and N. Weiss. 1992. Phylogenetic evidence for transfer of Eubacterium suis to the genus Actinomyces as Actinomyces suis comb. nov. Int. J. Syst. Bacteriol. 42:161-165. [DOI] [PubMed] [Google Scholar]
- 5.Miller, L., and T. Berger. 1985. Bacterial identification by gas chromatography of whole cell fatty acid, p. 228-241. Hewlett-Packard Co., Palo Alto, Calif.
- 6.Walker, R. L., and N. J. MacLachlan. 1989. Isolation of Eubacterium suis from sows with cystitis. J. Am. Vet. Med. Assoc. 195:1104-1107. [PubMed] [Google Scholar]
- 7.Wegienek, J., and C. A. Reddy. 1982. Nutritional and metabolic features of Eubacterium suis. J. Clin. Microbiol. 15:895-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamini, B., and R. F. Slocombe. 1988. Porcine abortion caused by Actinomyces suis. Vet. Pathol. 25:323-324. [DOI] [PubMed] [Google Scholar]