Abstract
An enzyme immunoassay has recently been developed for the hepatitis C virus (HCV) core antigen. To evaluate the possible association between core antigen and HCV RNA levels with regards to the change in liver histology over time as well as study the effect of duration of storage on viral load results, sequential sera were analyzed from 45 patients with chronic HCV infection who had undergone two or more liver biopsies. A relatively strong association was found between the core antigen and HCV RNA concentrations (rs = 0.8), with a core antigen level of 1 pg/ml corresponding to approximately 1,000 IU/ml. All 42 sera with detectable HCV RNA at the time of the second biopsy had core antigen concentrations above 1 pg/ml, and the three sera without detectable HCV RNA had concentrations below 1 pg/ml. No association was found between HCV RNA or core antigen levels and the stage of fibrosis in biopsy samples, progression of fibrosis, necro-inflammatory grade, steatosis, genotype, alanine aminotransferase level, or alcohol consumption. A significant association was demonstrated between the storage time of the samples and both the HCV RNA and core antigen concentrations. The median log HCV RNA concentrations (international units/milliliter) were 3.92 for the sera obtained at the time of the first biopsy (median storage time, 13.0 years) and 4.41 for the sera obtained at the time of the second biopsy (median storage time, 6.6 years) compared to 5.96, the median for 102 different routine clinical patient samples.
Previous studies have demonstrated that age at the beginning of infection, alcohol consumption, male sex, and prolonged inflammatory response in the form of interface hepatitis are associated with the progression of fibrosis in patients with chronic hepatitis C virus (HCV) infection (11, 14). In addition, progression in fibrosis has been shown to be more pronounced earlier in the course of infection (11).
There have been conflicting reports regarding the possible association between the quantity of virus and histological changes in the liver. The majority of studies have not been able to find any relationship between HCV RNA concentrations and liver fibrosis (2, 4, 18). However, in a multivariate logistic analysis, Iijima et al. noted a significant association between deterioration of the histological stage and amount of HCV viremia though the odds ratio was extremely low (8).
The HCV core protein has potent effects on cellular and viral gene regulation (15, 16) and in vitro transforms primary rat embryo fibroblasts to the tumorigenic phenotype (15) as well as immortalizes primary human hepatocytes (17). Similarly, transgenic mice expressing the HCV core protein develop hepatic steatosis (13) as well as hepatocellular carcinoma (12). Given these diverse effects, quantification of the HCV core protein might potentially be a better correlate of fibrosis progression than HCV RNA levels. Recently, a new enzyme immunoassay (EIA) for HCV core antigen has been developed (1) and has been proposed as a complement to standard HCV viral load quantification based on reverse transcription (RT)-PCR and branched-DNA analysis (10, 23). In this study, our aim was to evaluate the possible association between core antigen and HCV RNA quantification with regards to the change in liver histology over time in untreated HCV-infected patients as well as to study the effect of duration of storage on sample integrity.
MATERIALS AND METHODS
Patients.
Between July 1971 and November 1996, 358 patients who had a known or retrospectively proven positive serology for HCV (second- or third-generation microparticle EIA [Abbott Axsym, Abbott Park, Ill.]) confirmed by RIBA strip immunoblot assay (Chiron Corporation, Emeryville, Calif.) and who had attended the Infectious Diseases Outpatient Clinic in Göteborg, Sweden (the major referral center in Göteborg for HCV-infected patients), underwent liver biopsy. Of these 358 patients, 119 underwent two or more liver biopsies prior to antiviral therapy. At least two biopsies could be retrieved for retrospective evaluation in 101 cases. Of these 101 cases, two patients were excluded from the study because of hemochromatosis and one because of coinfection with human immunodeficiency virus. All of the remaining 98 patients had negative serologies for human immunodeficiency virus and negative assays for hepatitis B surface antigen and had no other known liver disease. Of these 98 patients, 45 patients had serum samples stored from the time of both of the liver biopsies that could be retrieved. Of the 45 patients included in this study, 24 were women and 21 were men. Twenty had likely been infected through intravenous drug use, 11 through blood transfusion, and 2 through sexual transmission; 2 were healthcare workers; and 10 had an unknown route of infection. The mean age at the time of the first biopsy was 36.4 years (standard deviation [SD], 12.2 years), and the mean time between the two biopsies was 6.1 years (SD, 3.5). Twenty-three patients had genotype 1, 9 had genotype 2, 10 had genotype 3, and 3 could not be genotyped.
In addition, 102 sera from 102 different patients submitted for routine clinical diagnostic evaluation between 1 July and 31 December 2001 to the Department of Clinical Virology, Göteborg, Sweden, were also analyzed. No clinical information was available on these patients.
Serum samples.
For all patients, serum samples were obtained at the time of both liver biopsies and were frozen immediately in aliquots at −20°C. The mean storage time was 12.7 years (SD, 4.2) for the samples obtained at the time of the first liver biopsy and 6.7 years (SD, 2.3) for the samples obtained at the time of the second biopsy. Only one patient had sufficient serum stored from the first liver biopsy for HCV RNA analysis but not for core antigen testing.
The 102 routine clinical samples were immediately frozen after arrival at the Department of Clinical Virology, Göteborg, Sweden, and thereafter stored at −20°C for a median of 10 days before analysis.
Liver biopsy samples and their scoring.
All biopsies were performed as part of the routine medical follow-up and were obtained by the standard Menghini procedure (needle diameter, 1.6 mm) with a biopsy sample length of approximately 2 cm. For each biopsy sample, a hematoxylin-eosin stain and a reticulin stain were staged and graded according to the protocol established by Ishak et al. (9) by two independent observers in a blinded fashion. Equivocal issues were debated after the independent scores were noted, and a consensus score was obtained. The degree of interobserver variability between the observers in this study has previously been reported (24).
Alcohol consumption.
The patients' lifetime alcohol intake was evaluated by a posted questionnaire which was adapted from that of Skinner (20). The subjects were asked to report average drinking frequency, average quantity consumed on each occasion, and length of periods of abstinence, if any. From these data, we calculated the total cumulative alcohol intake (kilograms of 100% ethanol) at the first and second biopsy times for each subject. Of the 45 patients included in this study, 38 responded to the questionnaire.
HCV serology, genotype analysis, HCV RNA quantification, and HCV core antigen quantification.
All sera were tested with a second- or third-generation HCV EIA, and seropositivity was confirmed by a second- or third-generation RIBA HCV strip immunoblot assay (Chiron Corporation). Genotyping of HCV was done by using a multiplex PCR method with genotype-specific primers (25). The concentration of HCV RNA was determined by RT-PCR with the Cobas Amplicor HCV monitor test (Roche Diagnostics, Branchburg, N.J.). Total HCV nucleocapsid core antigen levels were determined by quantitative immunoassay using the Ortho trak-C assay (Ortho Clinical Diagnostics, Inc., Raritan, N.J.).
Statistical methods.
In the present study, a possible relationship between variables was evaluated using the Spearman rank order correlation coefficient (rs), possible differences in individual characteristics between groups were evaluated by means of the Wilcoxon-Mann-Whitney U test (where P values of <0.05 were considered significant), and a box plot displaying the 10th, 25th, 50th, 75th, and 90th percentiles was also used.
A multivariate logistic regression was performed on data from all 45 patients where the outcome variable was dichotomized as either an increase in the Ishak fibrosis score or an unchanged or decreased score between the biopsies. The potential explanatory variables analyzed were log HCV RNA concentrations, log core antigen levels, and the length of storage for the first and second biopsies.
The evaluation of change in liver fibrosis is based on a statistical approach designed for the evaluation of change in ordered categorical data (21, 22). This method allows for a comprehensive evaluation of the pattern of change in the fibrosis score, describes the level of change in common for the group separately from the level of individual variability within the group, and has previously been used to evaluate change in liver histology (11). The difference between the probabilities of systematic improvement and deterioration is called relative position (RP), ranging from −1 to 1, where RP = 0 means a lack of systematic change in the fibrosis score in common for the group. The 95% confidence interval (CI) of RP was estimated by means of the bootstrap technique (3).
When the HCV RNA and core antigen concentrations are analyzed, the log10 values are utilized for both analyses because this is the format in which the results are reported at the end of the assays.
Ethical committee.
This study has been approved by the Göteborg University Medical Faculty Ethical Committee, Göteborg, Sweden.
RESULTS
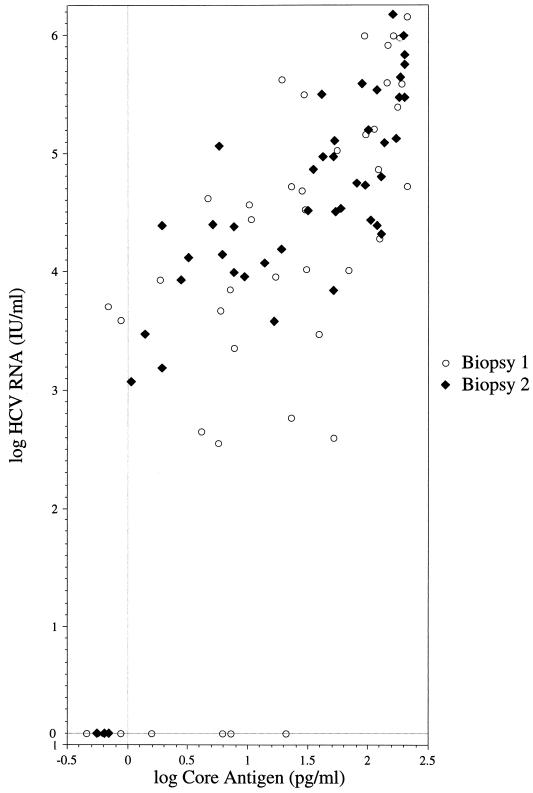
As seen in Fig. 1, a strong association between the log core antigen and log HCV RNA levels was found (rs = 0.8; P < 0.0001). All patients with detectable HCV RNA at the time of the second liver biopsy had core antigen concentrations above 1 pg/ml, corresponding to approximately 1,000 IU of HCV RNA/ml. All three samples with undetectable HCV RNA concentrations at the time of the second liver biopsy also had core antigen levels below 1 pg/ml.
FIG. 1.
Relationship between the log core antigen level (pg/ml) and the log HCV RNA level (international units/milliliter) for the 90 serum samples grouped according to the liver biopsy occasion (rs = 0.8; P < 0.0001 for both biopsies).
Among the samples obtained at the first biopsy, nine had undetectable HCV RNA of which four had core antigen levels greater than 1 pg/ml. Likewise, two serum samples had detectable HCV RNA concentrations but core antigen levels less than 1 pg/ml. These discrepancies may be attributable to the storage time, which was a mean of 16.1 years for the four samples that had undetectable HCV RNA and a core antigen level greater than 1 pg/ml, 12.8 years for the two samples that had detectable HCV RNA and a core antigen level less than 1 pg/ml, and 13.2 years for the four samples that had undetectable HCV RNA and core antigen concentrations less than 1 pg/ml compared to 12.0 years (SD, 3.9) for the remaining 34 samples that had detectable HCV RNA and core antigen levels greater than 1 pg/ml.
Of the four patients who had undetectable HCV RNA and core antigen levels less than 1 pg/ml in the samples obtained at the time of the first biopsy, three had detectable HCV RNA and core antigen concentrations greater than 1 pg/ml in the samples from the second biopsy. Only one patient continued to have undetectable HCV RNA and a core antigen level less than 1 pg/ml, although he had a slightly elevated alanine aminotransferase (ALT) level at the time of the second biopsy (1.1 times the upper limit of normal). This patient had Ishak fibrosis stage 1 in the first biopsy and Ishak fibrosis stage 2 in the second.
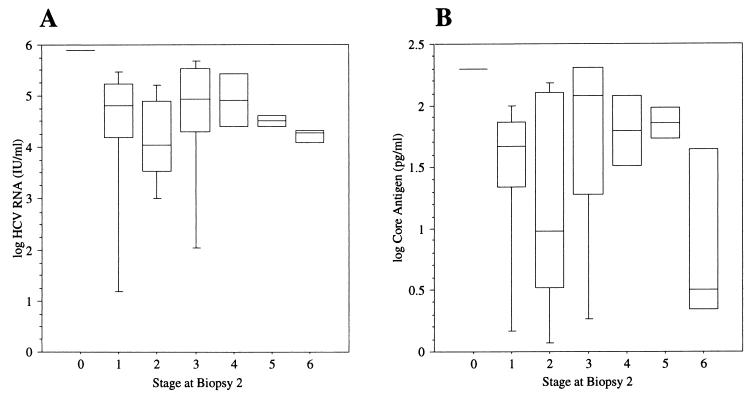
Figure 2 shows the log HCV RNA and log core antigen levels in the samples obtained at the time of the second biopsy grouped according to the Ishak fibrosis score. Possibly, lower concentrations of both were seen in the group with cirrhosis (i.e., Ishak fibrosis stage 6). However, there were only three patients in this group and thus caution must be used so as to not overinterpret this result.
FIG. 2.
Box plot displaying the 10th, 25th, 50th, 75th, and 90th percentiles of the log HCV RNA level (international units/milliliter) (A) and the log core antigen level (pg/ml) (B) grouped according to the Ishak fibrosis stage in the second biopsy.
No significant differences in the progression of fibrosis were seen whether the HCV RNA level was above (RP = 0.26, 95% CI = 0.07 to 0.47) or below (RP = 0.23, 95% CI = −0.04 to 0.48) the median concentration of 15,000 IU/ml or the core antigen level was above (RP = 0.25, 95% CI = 0.04 to 0.46) or below (RP = 0.24, 95% CI = 0 to 0.5) 20 pg/ml. Likewise, no statistically significant difference in the progression was seen if the HCV RNA or core antigen concentrations increased between the two samples compared to those for unchanged or decreased levels, though a trend was noted towards greater progression in the group that increased in core antigen concentrations (RP = 0.33 and 95% CI = 0.13 to 0.54 for increased levels versus RP = 0.12 and 95% CI = −0.15 to 0.37 for decreased or unchanged levels). When analyzing for the possible association between the necro-inflammatory activity (grade) in the biopsy samples and HCV RNA and core antigen levels, we were not been able to detect any such association with regards to interface hepatitis, focal necrosis, or portal inflammation (data not shown). Similarly, we found no association between steatosis, genotype, alcohol consumption, or ALT level and HCV RNA or core antigen concentrations (data not shown).
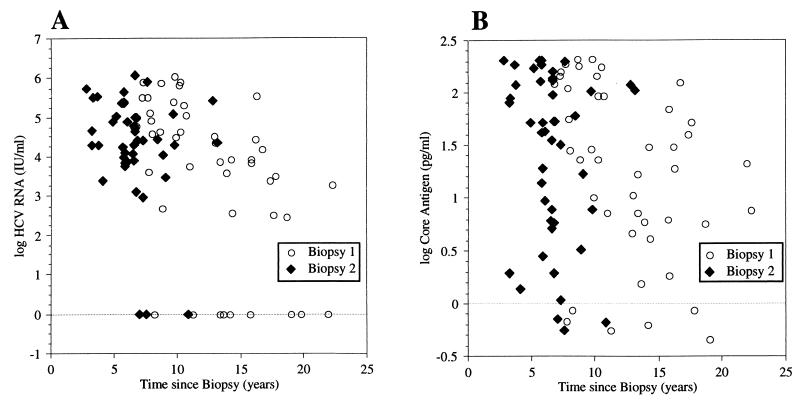
Figure 3 demonstrates the association between the duration of storage of the samples and the log HCV RNA (Fig. 3A) or log core antigen (Fig. 3B) concentrations for samples obtained in both biopsies. A significant association was found between the storage time of the samples obtained from the first biopsy with both log HCV RNA (rs = −0.6; P < 0.0001) and log core antigen (rs = −0.4; P = 0.0042) levels. When the HCV RNA concentrations from our 45 patients were compared with those from the 102 serum samples obtained from 102 different patients that were analyzed for routine diagnostic purposes between 1 July 2001 and 31 December 2001 at the Clinical Virology Department in Göteborg, Sweden, the levels obtained from the study patients were found to be significantly lower. For the 102 routine clinical samples, the median log HCV RNA concentration (international units/milliliter) was 5.96 (5.32 for the 1st quartile, 6.35 for the 3rd quartile) compared to the median concentration of 3.92 (2.55 for the 1st quartile, 5.1 for the 3rd quartile; P < 0.0001 based on the Wilcoxon-Mann-Whitney U test) for the 45 study sera obtained at the first biopsy (median storage time, 13.0 years) and the median concentration of 4.41 (3.97 for the 1st quartile, 5.09 for the 3rd quartile; P < 0.0001) for the sera obtained at the second biopsy (median storage time, 6.6 years). The core antigen assay is not presently a routine diagnostic test at the Clinical Virology Department in Göteborg, Sweden, and therefore was not used to analyze the 102 routine clinical patient samples. However, the median core antigen concentration was lower in the samples obtained at the time of the first biopsy (1.36 pg/ml) than that obtained at the time of the second biopsy (1.71 pg/ml).
FIG. 3.
Distribution of the log HCV RNA (international units/milliliter) (rs = −0.6; P < 0.0001 for biopsy 1) (A) and log core antigen (pg/ml) (rs = −0.4; P = 0.0042 for biopsy 1) (B) levels compared to the storage time grouped according to the first or second liver biopsy.
In the multivariate logistic regression, we attempted to compensate for the effect of storage time on the possible relationship between HCV RNA and core antigen levels and progression of fibrosis. However, in this model, no potential explanatory variable was statistically significant (data not shown).
DISCUSSION
The diagnosis of HCV infection is presently based on the detection of antibodies with confirmation by immunoblot assay or HCV RNA analysis. Our study showed a relatively good association between core antigen and HCV RNA concentrations, with a core antigen level of 1 pg/ml corresponding to approximately 1,000 IU/ml. This finding suggests that the assay might function as a confirmatory test when the presence of antibodies against HCV has been detected. When analyzing the sera from 145 HCV-negative blood donors, Kurtz et al. suggested that a cutoff of 200 pg/ml might be appropriate (10). However, this value was based on an earlier version of the core antigen assay that has since been replaced. Our data, based on a more recent version, would rather suggest a cutoff of 1 pg/ml when considering patients who are HCV antibody positive, which is in agreement with the manufacturer's suggested level of 0.8 to 1.5 pg/ml (personal communication).
The relatively strong relationship between the core antigen and HCV RNA levels may also warrant the use of the former assay in the monitoring of viral kinetics during ongoing therapy. Presently, quantitative assessments of HCV viral load are based on the amplification of target RNA by RT-PCR or signal amplification utilizing branched DNA. The use of both of these tests is limited by their relatively high cost. Closer monitoring of viral load during antiviral therapy is likely to occur if the cost can be lowered. In this setting, the core antigen test may prove useful.
In our study, we could not find any relationship between the HCV viral load as measured by the quantitative HCV RNA and core antigen assays and fibrosis stage, fibrosis progression, necro-inflammatory grade, genotype, steatosis, ALT level, or alcohol consumption. De Moliner et al. reported an association between the levels of viremia and the amount of virus in the liver but no relationship between viral load and ALT level, genotype, or histological diagnosis (2). Likewise, Rodriguez et al. demonstrated a significant association between the proportion of infected hepatocytes and viral load but no relationship with the histological activity index (18). Fanning et al., however, noted a weak association (rs = 0.26) between viral load and degree of inflammation but no relationship between viral load and degree of fibrosis or ALT level (4). Iijima et al., however, have reported a significant association between deterioration of the histological stage and amount of HCV viremia, though the odds ratio was 1.002, which is very low (8). Together, these findings suggest that the development of fibrosis in HCV may not be related principally to the amount of virus present. Instead, the vigor and duration over time of the immunological response are possibly of greater importance.
The potential degradation of HCV RNA and core antigen over time of course may have affected the above-mentioned analyses and thus distorted any relationship that was possibly present. HCV RNA has been demonstrated to be stable when stored at +4°C for 168 h (6). Similarly, Fong et al. reported that serum HCV RNA is resistant to degradation under routine laboratory handling and various storage conditions, including freezing at −20°C (5). Halfon et al., however, noted a 10% decline in HCV RNA as measured by branched-DNA assay after storage at −80°C for 6 months and a 23% decline at −20°C (7). Likewise, storage for up to 17 years at −70°C has been demonstrated to markedly lower HCV RNA concentrations as measured by the Superquant RT-PCR method (19). The sera used in our study were stored in aliquots at −20°C for a median time of 13.0 and 6.6 years for the samples obtained at the time of the first and second biopsies, respectively. There was a significant association between the storage time and HCV RNA, as well as core antigen, concentrations. Median HCV RNA levels were approximately 2 logs lower in the samples obtained at the time of the first biopsy and 1.5 logs lower in the samples obtained at the time of the second biopsy than those from the 102 routine patient samples.
In summary, we conclude that there is a strong association between the HCV RNA and core antigen concentrations which may warrant a possible role for the latter assay in confirming HCV infection when antibodies have been detected, in screening of patients, and in monitoring viral kinetics during therapeutic intervention. Additionally, we could not find any relationship between viral load and liver histology. However, this lack of association may have been due to the effect of storage time on both HCV RNA and core antigen levels.
ADDENDUM IN PROOF
Bouvier-Alias et al., using the same assay that we utilized in this study, recently suggested that 1 pg/ml of total HCV core antigen is equivalent to approximately 8,000 IU/ml (M. Bouvier-Alias, K. Patel, H. Dahari, S. Beaucourt, P. Larderie, L. Blatt, C. Hezode, G. Picchio, D. Dhumeaux, A. U. Neumann, J. G. McHutchinson, and J. Pawlotsky, Hepatology 36:211-218, 2002). The discrepancy between their results and ours may result from differences in storage conditions. Our samples were stored at −20°C. If the integrity of the HCV RNA is less than that of the core antigen under these conditions, our finding that 1 pg/ml corresponds to 1,000 IU/ml may be an underestimate. To resolve this issue, a prospective analysis of fresh unfrozen samples for both HCV RNA and core antigen simultaneously is required.
Acknowledgments
We thank Elisabeth Svensson and Nibia Aires for constructive criticism and statistical expertise, Nancy Nenonen for her technical assistance, and Kari Torgheim, Ingela Lindgren, and their staff at the pathology department, Sahlgrenska University Hospital, for helping in retrieving liver biopsy slides.
This study was financially supported by The Göteborg Medical Society.
REFERENCES
- 1.Aoyagi, K., C. Ohue, K. Iida, T. Kimura, E. Tanaka, K. Kiyosawa, and S. Yagi. 1999. Development of a simple and highly sensitive enzyme immunoassay for hepatitis C virus core antigen. J. Clin. Microbiol. 37:1802-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Moliner, L., P. Pontisso, G. L. De Salvo, L. Cavalletto, L. Chemello, and A. Alberti. 1998. Serum and liver HCV RNA levels in patients with chronic hepatitis C: correlation with clinical and histological features. Gut 42:856-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Efron, B., and R. J. Tibshirani. 1993. An introduction to the bootstrap. Chapman & Hall, London, United Kingdom.
- 4.Fanning, L., E. Kenny, M. Sheehan, B. Cannon, M. Whelton, J. O'Connell, J. K. Collins, and F. Shanahan. 1999. Viral load and clinicopathological features of chronic hepatitis C (1b) in a homogeneous patient population. Hepatology 29:904-907. [DOI] [PubMed] [Google Scholar]
- 5.Fong, T. L., F. Charboneau, B. Valinluck, and S. Govindarajan. 1993. The stability of serum hepatitis C viral RNA in various handling and storage conditions. Arch. Pathol. Lab. Med. 117:150-151. [PubMed] [Google Scholar]
- 6.Gessoni, G., P. Barin, A. Frigato, M. Fezzi, G. de Fusco, N. Arreghini, P. Galli, and G. Marchiori. 2000. The stability of hepatitis C virus RNA after storage at +4 degrees C. J. Viral Hepatol. 7:283-286. [DOI] [PubMed] [Google Scholar]
- 7.Halfon, P., H. Khiri, V. Gerolami, M. Bourliere, J. M. Feryn, P. Reynier, A. Gauthier, and G. Cartouzou. 1996. Impact of various handling and storage conditions on quantitative detection of hepatitis C virus RNA. J. Hepatol. 25:307-311. [DOI] [PubMed] [Google Scholar]
- 8.Iijima, A., E. Tanaka, M. Kobayashi, S. Yagi, M. Mizokami, and K. Kiyosawa. 2000. Relationship between histological prognosis of chronic hepatitis C and amount of hepatitis C virus core protein in serum. J. Gastroenterol. Hepatol. 15:311-319. [DOI] [PubMed] [Google Scholar]
- 9.Ishak, K., A. Baptista, L. Bianchi, F. Callea, J. De Groote, F. Gudat, H. Denk, V. Desmet, G. Korb, R. N. MacSween, et al. 1995. Histological grading and staging of chronic hepatitis. J. Hepatol. 22:696-699. [DOI] [PubMed] [Google Scholar]
- 10.Kurtz, J. B., E. Boxall, N. Qusir, J. Shirley, D. Coleman, and C. Chandler. 2001. The diagnostic significance of an assay for “total” hepatitis C core antigen. J. Virol. Methods 96:127-132. [DOI] [PubMed] [Google Scholar]
- 11.Lagging, L. M., J. Westin, E. Svensson, N. Aires, A. P. Dhillon, M. Lindh, R. Wejstal, and G. Norkrans. 2002. Progression of fibrosis in untreated patients with hepatitis C virus infection. Liver 22:136-144. [DOI] [PubMed] [Google Scholar]
- 12.Moriya, K., H. Fujie, Y. Shintani, H. Yotsuyanagi, T. Tsutsumi, K. Ishibashi, Y. Matsuura, S. Kimura, T. Miyamura, and K. Koike. 1998. The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat. Med. 4:1065-1067. [DOI] [PubMed] [Google Scholar]
- 13.Moriya, K., H. Yotsuyanagi, Y. Shintani, H. Fujie, K. Ishibashi, Y. Matsuura, T. Miyamura, and K. Koike. 1997. Hepatitis C virus core protein induces hepatic steatosis in transgenic mice. J. Gen. Virol. 78:1527-1531. [DOI] [PubMed] [Google Scholar]
- 14.Poynard, T., P. Bedossa, P. Opolon, et al. 1997. Natural history of liver fibrosis progression in patients with chronic hepatitis C. Lancet 349:825-832. [DOI] [PubMed] [Google Scholar]
- 15.Ray, R. B., L. M. Lagging, K. Meyer, and R. Ray. 1996. Hepatitis C virus core protein cooperates with ras and transforms primary rat embryo fibroblasts to tumorigenic phenotype. J. Virol. 70:4438-4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ray, R. B., L. M. Lagging, K. Meyer, R. Steele, and R. Ray. 1995. Transcriptional regulation of cellular and viral promoters by the hepatitis C virus core protein. Virus Res. 37:209-220. [DOI] [PubMed] [Google Scholar]
- 17.Ray, R. B., K. Meyer, and R. Ray. 2000. Hepatitis C virus core protein promotes immortalization of primary human hepatocytes. Virology 271:197-204. [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez-Inigo, E., J. Bartolome, S. de Lucas, F. Manzarbeitia, M. Pardo, C. Arocena, J. Gosalvez, H. Oliva, and V. Carreno. 1999. Histological damage in chronic hepatitis C is not related to the extent of infection in the liver. Am. J. Pathol. 154:1877-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmid, P., M. Tong, A. Conrad, J. McHutchison, and L. M. Blatt. 1999. Analysis of the viability of freezer stored serum samples for hepatitis C virus RNA analysis by the SUPERQUANT method: results of a 16 year retrospective study. J. Virol. Methods 82:201-206. [DOI] [PubMed] [Google Scholar]
- 20.Skinner, H. A. 1982. Development and validation of a lifetime alcohol consumption assessment procedure. Addiction Research Foundation, Toronto, Canada.
- 21.Svensson, E. 1993. Analysis of systematic and random differences between paired ordinal categorical data. Almqvist & Wiksell International, Stockholm, Sweden.
- 22.Svensson, E. 1998. Ordinal invariant measures for individual and group changes in ordered categorical data. Stat. Med. 17:2923-2936. [DOI] [PubMed] [Google Scholar]
- 23.Tanaka, E., C. Ohue, K. Aoyagi, K. Yamaguchi, S. Yagi, K. Kiyosawa, and H. J. Alter. 2000. Evaluation of a new enzyme immunoassay for hepatitis C virus (HCV) core antigen with clinical sensitivity approximating that of genomic amplification of HCV RNA. Hepatology 32:388-393. [DOI] [PubMed] [Google Scholar]
- 24.Westin, J., L. M. Lagging, R. Wejstal, G. Norkrans, and A. P. Dhillon. 1999. Interobserver study of liver histopathology using the Ishak score in patients with chronic hepatitis C virus infection. Liver 19:183-187. [DOI] [PubMed] [Google Scholar]
- 25.Widell, A., S. Shev, S. Mansson, Y. Y. Zhang, U. Foberg, G. Norkrans, A. Fryden, O. Weiland, J. Kurkus, and E. Nordenfelt. 1994. Genotyping of hepatitis C virus isolates by a modified polymerase chain reaction assay using type specific primers: epidemiological applications. J. Med. Virol. 44:272-279. [DOI] [PubMed] [Google Scholar]