Abstract
We evaluated the precision and accuracy of a procedure for detecting recent human immunodeficiency virus (HIV) infections, specifically, the avidity index (AI) calculated using a method based on an automated AxSYM HIV 1/2gO assay (Abbott). To evaluate precision, we performed multiple replicates on eight HIV-positive serum samples. To evaluate the accuracy in identifying recent infections (i.e., within 6 months of seroconversion), we used 216 serum samples from 47 persons whose dates of seroconversion were known. To evaluate the sensitivity and specificity of the procedure for different AI cutoff values, we performed receiver operating characteristic (ROC) analysis. To determine the effects of antiretroviral treatment, advanced stage of the disease (i.e., low CD4-cell count), and low HIV viral load on the AI, we analyzed 15 serum samples from 15 persons whose dates of seroconversion were unknown. The precision study showed that the procedure was robust (i.e., the total variance of the AI was lower than 10%). Regarding accuracy, the mean AI was significantly lower for samples collected within 6 months of seroconversion, compared to those collected afterwards (0.68 ± 0.16 versus 0.99 ± 0.10; P < 0.0001), with no overlap of the 95% confidence intervals. The ROC analysis revealed that an AI lower than 0.6 had a sensitivity of 33.3% and a specificity of 98.4%, compared to 87.9 and 86.3%, respectively, for an AI lower than 0.9. Antiretroviral treatment, low CD4-cell count, and low viral load had no apparent effect on the AI. In conclusion, this procedure is reproducible and accurate in identifying recent infections; it is automated, inexpensive, and easy to perform, and it provides a quantitative result with different levels of sensitivity and specificity depending on the selected cutoff.
For persons infected with the human immunodeficiency virus (HIV), knowing at what point in time infection occurred would be useful for a variety of purposes, including surveillance, the planning of vaccine trials, making decisions with regard to treatment, tailoring and evaluating preventive measures, and partner notification. Although for other infections the time of infection can be approximated based on the dynamics of the antibody response, these procedures have not been standardized for HIV infection. Moreover, although the classic sequence of the antibody response, in which a primary immunoglobulin M (IgM) response is followed by an increase in IgG and the IgG response increases with repeated exposures (20), is generally applicable to HIV infection, current screening assays are not able to discriminate among the immunoglobulin classes of anti-HIV antibodies.
In the attempt to discover a means of distinguishing recent HIV infections from established infections in single sampling, researchers have studied various antibody assays and testing strategies (4, 12). However, some of these assays have a number of drawbacks, specifically, high costs, difficulties in performing the assay, low reproducibility, qualitative rather than quantitative results, different performance depending on viral subtypes, and infections being misclassified as recent in persons who have developed end-stage AIDS or who are undergoing treatment with protease inhibitors (13, 15).
To diagnose recent infections, a procedure based on the avidity index (AI) of anti-HIV antibodies has been recently proposed (7, 14). The AI is a marker of recent primary infection and is routinely used for several infectious diseases, including toxoplasmosis, rubella, and cytomegalovirus infection (1, 5, 9). Moreover, it has been applied for a number of other infectious agents, such as hepatitis C virus, hepatitis B virus, human herpes viruses 6 and 7, and varicella-zoster virus (6, 17, 18, 19). The AI is based on the rationale that antibodies produced in the early phase of infection show a low avidity for the antigen. In fact, it is well known that antibody avidity increases progressively with time after exposure to an immunogen (2); thus, a low avidity is likely to indicate recent primary infection.
In the present study, we evaluated the AI for detecting recent HIV infections with an automated anti-HIV enzyme immunoassay (EIA). The objective of the evaluation was twofold; specifically, we attempted to evaluate both the precision of the entire procedure and the accuracy of the AI in discriminating between recent infections (within 6 months after seroconversion) and established infections (more than 1 year after seroconversion).
MATERIALS AND METHODS
Precision study.
To evaluate the reproducibility of the entire procedure, we tested multiple replicates of selected samples, following the recommendations of the U.S. National Committee for Clinical Laboratory Standards (11).
(i) Study samples.
We selected eight serum samples from eight different individuals that had previously tested positive for antibodies against HIV type 1 (HIV-1) by the AxSYM HIV 1/2gO test (Abbott Diagnostics Division, Delkenheim, Germany). To establish the consistency of the AI calculation across the dynamic range of the anti-HIV assay, we selected two samples with a weak positive signal (i.e., a sample/cutoff [S/CO] ratio of less than 10) and six with a strong signal (i.e., an S/CO ratio ranging from 20 to 30). These eight samples had been confirmed as positive by commercial Western blots and by PCR testing for HIV-1 RNA. All samples were stored frozen at −20°C before testing. We also analyzed the two positive controls (recalcified human plasma positive for anti-HIV-1 and anti-HIV-2) from the test kit, with a very low S/CO ratio (i.e., from 2 to 3).
(ii) Sample preparation.
For each sample (both the study samples and the two positive controls from the AxSYM assay), after thawing, two aliquots of 0.2 ml each were subjected to a pretest dilution: one aliquot was diluted 1:10 with phosphate-buffered saline (PBS), and the other was diluted 1:10 with 1 M guanidine (G). All samples were vortexed and incubated at room temperature for 10 min.
(iii) Anti-HIV-1-HIV-2 assay.
Both aliquots from each sample were subjected to the automated AxSYM HIV 1/2gO assay, following the manufacturer's procedures without any modifications. The AxSYM assay is a microparticle EIA with a third-generation sandwich format; it is thus able to recognize antibodies of all immunoglobulin classes. In this assay, the solid phase is represented by polystyrene microparticles coated with recombinant HIV-1 env and gag, HIV-1gO env and HIV-2 gag proteins. Anti-HIV antibodies forming an antigen-antibody complex with the solid phase are detected by incubation with recombinant biotinylated HIV-1 env and gag, HIV-1gO env, and HIV-2 gag proteins. The addition of alkaline phosphatase-conjugated antibiotin and of an appropriate substrate generates a fluorescent signal which is automatically read and transformed into a numeric signal which is compared to the assay cutoff, established by a calibration. The assay is fully automated and provides a qualitative result based on the S/CO ratio. The result is deemed positive when the S/CO ratio is greater than or equal to 1. The addition of a denaturing agent (G in our study) elutes low-avidity and -affinity antibody after antigen-antibody bonds have formed. In the present assay, this procedure results in a low S/CO ratio for the G aliquot compared to that for the PBS aliquot.
(iv) Replicates.
For each sample, five replicates of the assay were performed every day for 3 days, representing a total of 15 replicates for each sample.
(v) AI calculation.
After obtaining the S/CO ratios for the PBS and the G aliquots, the AI of HIV antibodies was calculated using the following formula: AI = (S/CO ratio of the G aliquot)/(S/CO ratio of the PBS aliquot).
(vi) Statistical methods.
For each daily session we calculated the mean and standard deviations of the AI obtained on each sample or control. The intraday, interday, and total imprecision of the procedure were evaluated by conventional analysis of variance and expressed as coefficients of variation (CV).
Validation study.
The validation study was conducted to evaluate the accuracy of the AI in discriminating between recent seroconversions (i.e., within 6 months of seroconversion) and established infections (more than 1 year after seroconversion).
(i) Serum samples.
A total of 216 serum samples from 47 HIV-positive individuals whose dates of seroconversion were known were collected. All participants had a documented negative HIV test followed by a positive test within 24 months. Specimens were considered positive when reactive by conventional HIV enzyme-linked immunosorbent assay and confirmed by Western blotting and PCR. The seroconversion date was estimated as the midpoint between the date of the last negative test and the date of the first positive test. None of the participants had AIDS upon enrollment. For all individuals, at least one serum sample collected within the first year after seroconversion was available, plus one or more serum samples collected at different times after seroconversion.
We also attempted to evaluate the effect of certain clinical and immunological factors on the AI, specifically, receiving antiretroviral treatment, being in an advanced stage of disease (revealed by a low CD4-cell count), and having a low HIV viral load (under 500 copies/ml or undetectable). To this end, we studied 15 serum samples from an additional 15 HIV-infected individuals who presented one or more of the above-mentioned factors (for these persons, it was not possible to estimate the date of seroconversion). Serum samples were collected at a median time of 8.2 years after the first positive HIV test result; nine persons were under antiretroviral treatment (eight included a protease inhibitor), five had a CD4-cell count under 200 cells/ml, and three had an HIV viral load under the cutoff when blood was drawn. All sera were frozen at −20°C.
(ii) Laboratory methods.
The procedures used for diluting and assaying the serum samples and for calculating the AI were identical to those used for the precision study (described above).
(iii) Statistical methods.
Ninety-five percent confidence intervals (95% CI) were calculated from the standard error of the mean. Positive predictive values (PPV) and negative predictive values (NPV) for recent seroconversion were calculated for different AI levels. The distribution of the AI at different times from seroconversion was graphically shown using box plots. The diagnostic accuracy of the AI in predicting recent seroconversions was evaluated by receiver operating characteristic (ROC) analysis.
RESULTS
Precision study.
The results of the precision study are reported in Table 1. Both the intraday and the interday CV were consistently lower than 8%. The whole procedure was robust, as indicated by the total variance of the AI, which was consistently lower than 10%; therefore, an AI variation of 10% or more can be considered to be representative of a true variation of the antibody avidity.
TABLE 1.
Precision of the AI for detecting recent HIV infections
| Specimen no. (signal) | Anti-HIV S/CÔ | Mean AIa | CV (%)
|
||
|---|---|---|---|---|---|
| Intraday | Interday | Total | |||
| 1 (weakly positive) | 5.08 | 0.62 | 7.03 | 6.89 | 9.84 |
| 2 (weakly positive) | 5.33 | 0.63 | 6.90 | 6.80 | 9.69 |
| 3 (strongly positive) | 22.53 | 1.02 | 5.08 | 1.82 | 5.39 |
| 4 (strongly positive) | 20.24 | 1.00 | 4.55 | 1.76 | 4.88 |
| 5 (strongly positive) | 21.07 | 0.99 | 3.63 | 2.02 | 4.15 |
| 6 (strongly positive) | 21.82 | 0.99 | 3.18 | 0.73 | 3.26 |
| 7 (strongly positive) | 21.11 | 0.93 | 3.98 | 1.21 | 4.16 |
| 8 (strongly positive) | 21.07 | 0.94 | 3.56 | 3.52 | 5.01 |
| 9 (positive control) | 2.24 | 0.95 | 6.32 | 1.50 | 6.49 |
| 10 (positive control) | 2.07 | 1.03 | 6.34 | 5.23 | 8.22 |
Mean of 15 replicates.
Validation study.
The 216 serum samples were collected from the 47 HIV-positive individuals whose dates of seroconversion were known from 4 days to 10.3 years after seroconversion (median of 836 days). More specifically, 17 (7.9%) samples were collected less than 3 months after seroconversion, 17 (7.9%) samples were collected 4 to 6 months after seroconversion, 10 (4.6%) samples were collected 7 to 9 months after seroconversion, 20 (9.2%) samples were collected 10 to 12 months after seroconversion, 42 (19.5%) samples were collected 13 to 24 months after seroconversion, 45 (20.8%) samples were collected 25 to 48 months after seroconversion, and 65 (30.1%) samples were collected more than 48 months after seroconversion. Samples collected within 6 months from seroconversion were considered to represent recent seroconversions. From two to nine samples were collected per individual.
A total of 158 serum samples were analyzed one time, whereas the remaining 58 samples were analyzed three times each for the PBS and the G aliquots. Since the variance among the results of the three replicates was lower than 5%, we used the results from the first replicate to calculate the AI.
A CD4-cell count was available for all serum samples. The HIV viral load was measured in 113 samples; in 24 of these samples, the viral load was undetectable. One hundred sixty-six samples were collected from persons undergoing antiretroviral treatment (n = 35 individuals). Twenty-seven persons had an interval of less than 6 months between the last negative test and the first positive test.
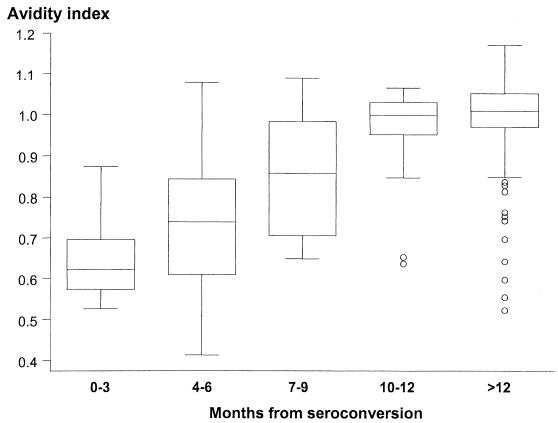
The distribution of the AI by time elapsed since seroconversion is shown in Fig. 1. In the first year, the AI clearly increased, reaching approximately 1 at the end of the first year after seroconversion; afterwards, the AI stabilized at around 1. The 13 outlying samples represent eight patients; the average CD4-cell count in these samples was 479.2 lymphocytes/ml; four of these patients had an interval of more than 300 days between the last negative test and the first positive test.
FIG. 1.
Box-and-whisker plots of the AI by time elapsed since seroconversion (in months). Boxes at each time unit extend from the 25th percentile (x[25]) to the 75th percentile (x[75]) (i.e., the interquartile range [IQR]); the lines inside the boxes represent the median values. The lines emerging from the boxes (i.e., the whiskers) extend to the upper and lower adjacent values. The upper adjacent value is defined as the largest data point that is ≤x[75] + 1.5 × IQR; the lower adjacent value is defined as the smallest data point that is ≥x[25] − 1.5 × IQR. Those values not within the range of the adjacent values are individually plotted (circles).
The AI was not influenced by CD4-cell count, undetectable viral load, or antiretroviral treatment, even when the analysis was restricted to samples obtained from individuals with an interval of less than 6 months between the last negative and the first positive test (data not shown).
For serum samples collected within 6 months of seroconversion, both the mean and the median AI were lower than 0.8. The mean AI for these samples was significantly lower than that of the samples collected after 6 months from seroconversion (0.68 ± 0.16 versus 0.99 ± 0.10, respectively; P < 0.0001), with no overlap of the 95% confidence intervals.
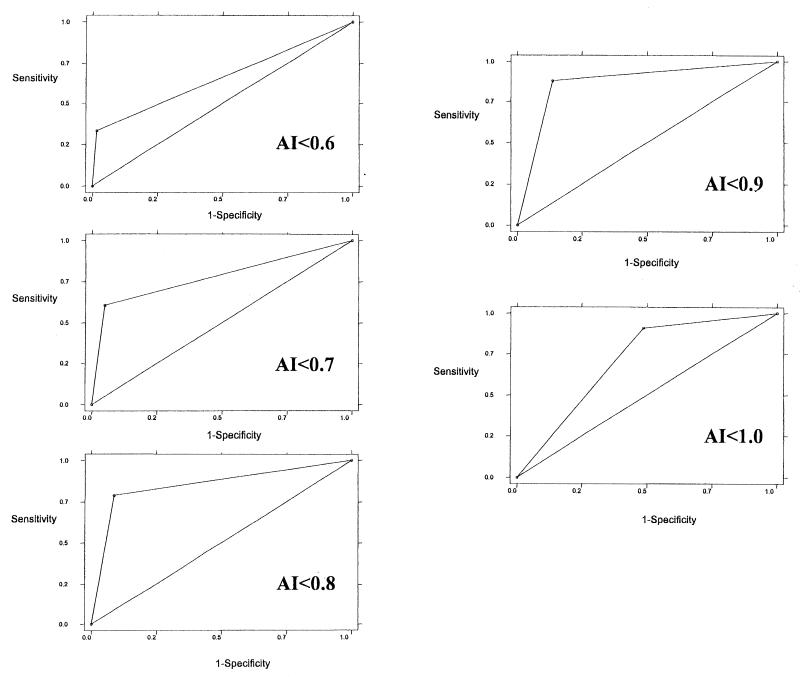
The diagnostic accuracy of the AI in predicting recent infections, determined by ROC analysis, is shown in Fig. 2. An AI lower than 0.6 had a sensitivity of 33.3% and a specificity of 98.4%, whereas an AI lower than 0.9 had a sensitivity of 87.9% and a specificity of 86.3%.
FIG. 2.
Use of different cutoffs of the AI to identify recent HIV infections (ROC curves).
Taking as reference the sensitivity and specificity levels found in our study, we calculated the PPV and NPV that would be observed if the AI were used to identify recent infections among HIV-positive populations with different proportions of recent seroconverters. These percentages are merely theoretical. As shown in Table 2, the PPV increased with lower AI cutoff values and with higher proportions of recent seroconversions, whereas the NPV increased with higher AI cutoffs and lower proportions of recent seroconversions.
TABLE 2.
Use of PPV and NPV in identification of recent HIV infectionsa
| AI cutoff | Sensitivity (%) | Specificity (%) | Result for indicated PRIb
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 5%
|
10%
|
20%
|
40%
|
|||||||
| % PPV | % NPV | % PPV | % NPV | % PPV | % NPV | % PPV | % NPV | |||
| <0.6 | 26.4 | 98.9 | 56.5 | 96.1 | 74.3 | 92.1 | 86.7 | 84.3 | 94.5 | 66.7 |
| <0.7 | 61.8 | 95.1 | 39.0 | 97.8 | 57.9 | 95.7 | 75.6 | 90.9 | 89.5 | 78.9 |
| <0.8 | 79.4 | 92.3 | 35.4 | 98.9 | 52.3 | 97.5 | 71.8 | 94.6 | 86.8 | 86.8 |
| <0.9 | 88.2 | 86.8 | 26.3 | 99.3 | 42.9 | 98.5 | 62.9 | 96.7 | 81.9 | 91.6 |
PPV and NPV were calculated for populations with different proportions of recent infections (PRI) and grouped according to AI (sensitivity and specificity are based on the results of the present study).
PRI (proportion of recent infections) = (no. of recent infections)/(no. of HIV-positive persons).
The results of the analysis to determine the effect of clinical and immunological factors on the AI showed that the AI was always higher than 0.90 (range 0.91 to 1.06) for the 15 samples analyzed and that there was no association with antiretroviral treatment, advanced stage of disease, or low HIV viral load (results not shown).
DISCUSSION
That the antibody response to an acute infection matures is a well established principle in the immunology of infectious diseases (20). Although there exists individual variability in the maturation process, some general patterns can be identified; in fact, in practice, the serological algorithms used for the diagnosis of several infections (e.g., rubella, cytomegalovirus infection, and toxoplasmosis) require that IgG avidity be estimated in order to rule out a recent infection.
For HIV infection, early studies evaluated antibody maturation and avidity to specific antigens (i.e., p24 and gp41) (16) as indicators of clinical progression of HIV disease. The first instance of the broad-range epidemiological application of HIV antibody maturation was the study of Janssen et al. (4), who used two tests in order to detect recent infections (i.e., a standard HIV-1 antibody assay approved by the Food and Drug Administration and a “detuned” version of the same assay in which both sample dilution and incubation times were modified). Specifically, according to these authors, in the period from 109 to 149 days after seroconversion, the result of the standard assay is positive and that of the detuned assay is negative; thus, these two assays can be combined to detect recent infections. Since then, the detuned assay has been used in several studies, mostly in the United States (3, 8, 10, 15). However, this strategy has several drawbacks. First, since the result is expressed qualitatively (positive or negative) and not quantitatively, interlot variability could influence the results. Second, the detuned assay is an indirect second-generation EIA and does not perform as well as current third-generation assays in persons with non-B-subtype HIV-1 infection. Third, the standard assay on which the detuned assay is based is no longer available in Europe, where third-generation immunoassays were introduced in 1995, and it will soon become obsolete in the United States. Fourth, the detuned assay may misclassify HIV-positive persons with an established infection as having a recent infection when they are at the end stage of AIDS or when they are undergoing antiretroviral treatment that includes a protease inhibitor (15).
For the method described in this paper, the preparation of the sample includes elution with G under conditions whereby bonds between antigens and antibodies that bind weakly with these antigens or that have a low avidity would be disrupted, whereas strongly binding or high-avidity antibodies would remain complexed with the antigens. Differently from the detuned assay, this method is quantitative and allows the level of antibody avidity to be determined as a continuous variable, which ranges from 0 to 1 depending primarily on the time elapsed from infection. Moreover, as for the other commonly used avidity assays (1, 5, 9), the AI is calculated by the ratio of two measures obtained at the same time using the same testing kit, thus eliminating interlot variation.
Being a third-generation EIA, the assay is more sensitive to seroconversion because it detects both IgG and IgM, differently from the second-generation EIA, which detects only IgG. Moreover, unlike the second-generation EIA, which only includes antigens and whole viral lysates of HIV-1, the present assay includes antigens of both HIV-1 and HIV-2, as well as antigens of the subtype O of HIV-1, making it more sensitive to different subtypes of the virus.
This method is simple to perform because it is not necessary to modify the manufacturer's procedures. Moreover, the cutoff does not need to be changed. For these reasons, additional approval for its use does not have to be obtained. Only a single additional step is required (i.e., sample preparation), and the agent used for dilution, namely, G, is easy to obtain and inexpensive. Moreover, the assay is an automated EIA that is commonly used in many countries, thus allowing results from different geographic areas to be compared.
The first authors to propose using this procedure for identifying recent HIV infections were Le Guillou et al. (7); however, their evaluation of the procedure had some limitations. Specifically, they did not perform a precision study, and the accuracy in predicting recent infections was evaluated on a small number of serum samples, which, moreover, were obtained from persons for whom the date of infection was estimated only on the basis of clinical symptoms or self-reported at-risk exposure. Furthermore, the published description of this evaluation does not include the results from all of the samples tested.
With specific regard to the present study, the results clearly show that this assay is quite sound. The precision study, which was based on a standardized protocol, confirmed the reproducibility of the assay, showing a consistently low intra-assay variability. As expected, the AI was low in the first 6 months after seroconversion, and afterwards it was significantly higher. The ROC curves and the PPV and NPV revealed that a low AI cutoff results in a low sensitivity and a high specificity for recent seroconversions, whereas higher cutoff values correspond to a higher sensitivity and a lower specificity. In practice, the choice of the cutoff value (and thus of the level of sensitivity and specificity of the assay) would depend on the specific objectives, for example, epidemiological surveys would benefit most from a higher sensitivity and thus a higher cutoff, whereas a lower cutoff would be preferable for clinical purposes, such as decision-making in antiretroviral treatment. Finally, since the AI is apparently not influenced by any type of antiretroviral treatment, by the CD4 level, or by the HIV viral load, its use does not result in established infections being misclassified as recent infections or in the incidence of HIV infection being overestimated, both of which have been observed when using the detuned assay.
Acknowledgments
We thank Francesca Farchi for secretarial support and Mark Kanieff for editorial assistance.
REFERENCES
- 1.Eggers, M., U. Baeder, and G. Enders. 2000. Combination of microneutralization and avidity assays: improved diagnosis of recent primary human cytomegalovirus infection in single serum sample of second trimester pregnancy. J. Med. Virol. 60:324-330. [DOI] [PubMed] [Google Scholar]
- 2.Eisen, H. N., and G. W. Siskind. 1964. Variations in affinities of antibodies during the immune response. Biochemistry 3:389-393. [DOI] [PubMed] [Google Scholar]
- 3.Gupta, S. B., O. N. Gill, C. Graham, A. D. Grant, P. A. Rogers, and G. Murphy. 2000. What a test for recent infection might reveal about HIV incidence in England and Wales. AIDS 14:2597-2601. [DOI] [PubMed] [Google Scholar]
- 4.Janssen, R. S., G. A. Satten, S. L. Stramer, B. D. Rawal, T. R. O'Brien, B. J. Weiblen, F. M. Hecht, N. Jack, F. R. Cleghorn, J. O. Kahn, M. A. Chesney, and M. P. Busch. 1998. New testing strategy to detect early HIV-1 infection for use in incidence estimates and for clinical and prevention purposes. JAMA 280:42-48. [DOI] [PubMed] [Google Scholar]
- 5.Jenum, P. A., B. Stray-Pedersen, and A.-G. Gundersen. 1997. Improved diagnosis of primary Toxoplasma gondii infection in early pregnancy by determination of antitoxoplasma immunoglobulin G avidity. J. Clin. Microbiol. 35:1972-1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kangro, H. O., S. Manzoor, and D. R. Harper. 1991. Antibody avidity following varicella zoster virus infections. J. Med. Virol. 33:100-105. [DOI] [PubMed] [Google Scholar]
- 7.Le Guillou, H., A. Le Meur, S. Bourdon, M. Riou, J. Loison, P. Flalaire, J.-M. Chennebault, S. Kouyoumdjian, and C. Payan. 2001. Avidité des anticorps: application au diagnostic d'infection récente à VIH. Ann. Biol. Clin. 59:41-47. [PubMed] [Google Scholar]
- 8.Machado, D. M., E. L. Delwart, R. S. Diaz, C. F. de Oliveira, K. Alves, B. D. Rawal, M. Sullivan, M. Gwinn, K. A. Clark, and M. P. Busch. 2002. Use of the sensitive/less-sensitive (detuned) EIA strategy for targeting genetic analysis of HIV-1 recently infected blood donors. AIDS 16:113-119. [DOI] [PubMed] [Google Scholar]
- 9.Matter, L., K. Kogelschatz, and D. Germann. 1997. Serum levels of rubella virus antibodies indicating immunity: response to vaccination of subjects with low or undetectable antibody concentration. J. Infect. Dis. 175:749-755. [DOI] [PubMed] [Google Scholar]
- 10.McFarland, W., M. P. Busch, T. A. Kellogg, B. D. Rawal, G. A. Satten, M. H. Katz, J. Dilley, and R. S. Janssen. 1999. Detection of early HIV infection and estimation of incidence using a sensitive/less sensitive enzyme immunoassay testing strategy at anonymous counseling and testing sites in San Francisco. J. Acquir. Immun. Defic. Syndr. 22:484-489. [DOI] [PubMed] [Google Scholar]
- 11.NCCLS. 1999. Evaluation of precision performances of clinical chemistry devices; approved guidelines. NCCLS document EP5-A. NCCLS, Wayne, Pa.
- 12.Parekh, B., C-P. Pau, S. Kennedy, T. L. Bobbs, and J. S. McDougal. 2001. Assessment of antibody assays for identifying and distinguishing recent from long-term HIV type 1 infection. AIDS Res. Hum. Retrovir. 17:137-146. [DOI] [PubMed] [Google Scholar]
- 13.Parekh, B. S., Hu, D. J., S. Vanichseni, G. A. Satten, D. Candal, N. L. Young, D. Kitayaporn, L. O. Srisuwanvilai, S. Rakhtam, R. Janssen, K. Choopanya, and T. D. Mastro. 2001. Evaluation of a sensitive/less-sensitive testing algorithm using the 3A11-LS assay for detecting recent HIV seroconversion among individuals with HIV-1 subtype B or E infection in Thailand. AIDS Res. Hum. Retrovir. 17:453-458. [DOI] [PubMed] [Google Scholar]
- 14.Riou, M., G. Renier, S. Mattman, P. Flalaire, J. Loison, J.-M. Chennebault, and C. Payan. 2000. Etude dynamique de l'avidité des anticorps anti-VIH lors de la renstauration de l'immunité cellulaire sous traitement antirétroviral. Ann. Biol. Clin. 58:715-720. [PubMed] [Google Scholar]
- 15.Schwarcz, S., T. Kellogg, W. McFarland, B. Louie, R. Kohn, M. Busch, M. Katz, G. Bolan, J. Klausner, and H. Weinstock. 2001. Differences in the temporal trends of HIV seroincidence and seroprevalence among sexually transmitted disease clinic patients, 1989-1998: application of the serologic testing algorithm for recent HIV seroconversion. Am. J. Epidemiol. 153:925-934. [DOI] [PubMed] [Google Scholar]
- 16.Thomas, H. I. J., S. Wilson, C. M. O'Toole, C. M. Lister, A. M. Saeed, R. P. F. Watkins, and P. Morgan-Capner. 1996. Differential maturation of avidity of IgG antibodies to gp41, p24 and p17 following infection with HIV-1. Clin. Exp. Immunol. 103:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomas, H. I. J. 1997. Relative functional affinity of specific anti-core IgG in different categories of hepatitis B virus infection. J. Med. Virol. 51:189-197. [DOI] [PubMed] [Google Scholar]
- 18.Ward, K. N., D. J. Turner, X. Couto Parada, and D. Thiruchelvam. 2001. Use of immunoglobulin G antibody avidity for differentiation of primary human herpesvirus 6 and 7 infections. J. Clin. Microbiol. 39:959-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ward, K. N., W. Dhaliwal, K. L. Ashworth, E. J. Clutterbuck, and C. G. Teo. 1994. Measurement of antibody avidity for hepatitis C virus distinguishes primary antibody responses from passively acquired antibody. J. Med. Virol. 43:367-372. [DOI] [PubMed] [Google Scholar]
- 20.Weinblen, B. J., R. T. Schumacher, and R. Hoff. 1990. Detection of IgM and IgA antibodies after removal of IgG with recombinant protein G. J. Immunol. Methods 126:199-204. [DOI] [PubMed] [Google Scholar]