Abstract
During the course of a molecular epidemiology study of mechanisms of antibiotic resistance in the area served by our hospital (516,000 inhabitants), we isolated the gene encoding CTX-M-14 β-lactamase. Thirty clinical strains (27 Escherichia coli and 3 Klebsiella pneumoniae isolates) with a phenotype of extended-spectrum β-lactamase were collected from January to October 2001 and studied for the presence of the CTX-M-14 β- lactamase gene. By isoelectric point determination, PCR, and nucleotide sequencing, we detected the presence of this gene in 17 E. coli strains belonging to 15 different genotypes (REP-PCR) causing infections in 17 different patients. Epidemiological studies based on medical records did not suggest any relationship between the patients infected with these E. coli strains and, interestingly, 7 of 30 patients harboring strains with extended-spectrum β-lactamases never had contact with the hospital environment before the clinical E. coli isolation. Conjugation experiments revealed that this gene was plasmid mediated in the 17 E. coli strains, and plasmid restriction fragment length polymorphisms showed 9 different patterns in the 17 E. coli strains. By PCR, the sequence of the tnpA transposase gene of the insert sequence ISEcp-1 was detected in all the plasmids harboring the CTX-M-14 gene. These results strongly suggest that plasmid dissemination between different E. coli strains in addition to a mobile element (transposon) around the β-lactamase gene may be involved in the spreading of the CTX-M-14 gene. This study reinforces the hypothesis that the epidemiology of the prevalence of the β-lactamase genes is changing and should alert the medical community to the increase in the emergence of the CTX-M β-lactamases worldwide.
The emergence of plasmid-mediated extended-spectrum β-lactamases (ESBLs) in members of the family Enterobacteriaceae has become a worldwide problem (6, 12, 21). ESBL molecular class A possesses an extended hydrolysis spectrum toward oxyimino-β-lactams and aztreonam but remains susceptible to cefoxitin and β-lactamase inhibitors (7). Most ESBLs are derivatives of TEM-1, TEM-2, or SHV-1 enzymes; however, reports describing the emergence of β-lactamases belonging to other families, such as CTX-M and/or OXA derivatives, are increasing worldwide (5, 8, 10, 11, 16).
CTX-M-related enzymes have been found in Europe, South America, and Mediterranean countries (3, 13, 15, 16, 18, 21). Unlike microorganisms harboring TEM and SHV β-lactamases, reports of outbreaks caused by organisms harboring non-TEM or non-SHV ESBL are very scarce. CTX-M-14 β-lactamase has recently been described in clinical isolates in Korea, China, and France (8, 11, 16, 19). In this communication we not only report for the first time the presence of the CTX-M-14 gene in Spain but we also show clear evidence of the dissemination of the gene encoding CTX-M-14 among 17 different patients, who had no apparent contact, living in the northwest part of Spain. This CTX-M-14 gene was detected in 15 different genotypes of Escherichia coli strains causing urinary tract infection in 17 patients in an area of 516,000 inhabitants. Plasmid dissemination and the presence of a putative transposon around the CTX-M-14 gene may account for the spreading of this gene. This finding emphasizes the increasing role of the CTX-M β-lactamases in antibiotic resistance worldwide and leads to consideration of empirical treatment for infections caused by E. coli strains, especially in patients compromised by underlying disease or immunological status.
MATERIALS AND METHODS
Selection of clinical isolates and patients.
Between January and October 2001, 27 E. coli and 3 Klebsiella pneumoniae clinical strains with an ESBL phenotype were collected in the Department of Microbiology, Juan Canalejo Hospital, a 1,200-bed hospital serving a population of 516,000 in the northwest area of Spain. These isolates were identified by DadeMicroScan and the API 20E system (bioMérieux, Marcy l'Etoile, France).
Susceptibility tests and confirmation of ESBL production.
The MICs of the β-lactams were determined by using E-test strips (AB Biodisk, Dalvägen, Sweden). The ESBL phenotype was detected by using ESBL detection E-test strips (AB Biodisk) as specified by the manufacturer. The double disk diffusion test was concomitantly used to confirm the E-test results.
IEF assay.
β-Lactamases were characterized by isoelectric focusing (IEF) of ultrasonicated bacterial extracts. Bacteria growing exponentially at 37°C in Luria-Bertani (LB) medium were harvested, and cell-free lysates were prepared by sonication. β-Lactamases were analyzed by isoelectric focusing of cell extracts on polyacrylamide gels containing ampholytes with a pH range of 3 to 9 (PhastGel; Amersham Pharmacia Biotech AB) in a PhastSystem apparatus (Amersham Pharmacia Biotech AB). The focused β-lactamases were detected by overlaying the gel with nitrocefin (0.5 mg/ml) in 100 mM phosphate buffer (pH 7.0). pIs were determined by comparison with those of β-lactamases with a known pI: TEM-1 (pI 5.4), SHV-1 (pI 7.6), IMP-2 (pI 8.2), and AmpC from A. baumannii (pI 9.4).
DNA extraction.
Bacterial chromosomal DNA was obtained with the High Pure PCR template preparation kit (Roche Diagnostics Corp., Indianapolis, Ind.). The chromosomal DNA was checked by electrophoresis in agarose gels, and the concentrations of the different extracts were standardized by spectrophotometric measurements.
Conjugation experiments, plasmid purification, cloning experiments, and DNA sequencing.
Transfer of resistance by conjugation was attempted using E. coli BM21 and E. coli XL-10-Gold Kan cells (Stratagene, La Jolla, Calif.) as recipients. Overnight mating experiments were performed at 37°C, and the transconjugants were selected on LB agar plates supplemented with ampicillin (50 μg/ml) and nalidixic acid (50 μg/ml) for E. coli BM21 and ampicillin (50 μg/ml) and kanamycin (25 μg/ml) for E. coli XL-10-Gold Kan cells. Afterward, transconjugants were selected for the cefotaxime resistance phenotype.
Plasmids from different E. coli strains were purified by the alkaline-lysis method with the High Pure plasmid isolation kit (Roche Diagnostics Corp.). Cloning procedures were performed as described by Sambrook et al. (20). Restriction enzymes were purchased from Boehringer (Mannheim, Germany) and were used as specified by the manufacturer.
For cloning the CTX-M-14 gene, plasmid DNA from transconjugant E. coli clinical strain 3 (see Table 2) was digested with BamHI. The resulting fragments were ligated into pBGS18 and digested with BamHI, and the mixture was transformed into E. coli TG1 made competent by the calcium chloride method (20). After transformation, a few clones grew on LB agar plates supplemented with kanamycin (10 μg/ml) and ampicillin (50 μg/ml). They harbored an identical plasmid (pC) with an insert of about 6 kb. These plasmids were used as templates to determine the nucleotide sequence. Templates were sequenced on both strands by the method of Sanger et al. Sequencing was carried out with the Taq DyeDeoxyTerminator cycle-sequencing kit using primers specific to the coding sequence, and the sequence was analyzed in an automatic DNA sequencer (377 ABI Prism; Perkin-Elmer).
TABLE 2.
MICs for the clinical strains included in this study
| Isolate no. | pI | MIC (μg/ml) ofa:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMX | AMC | FOX | AT | CTX | CTX-CL | CAZ | CAZ-CL | IMP | MRP | CEP | ||
| 1 | 5.4, 8.0 | >256 | 4 | 8 | 4 | 24 | 0.094 | 1.5 | 0.25 | 0.25 | 0.016 | 2 |
| 2 | 8.0 | >256 | 4 | 2 | 2 | 24 | 0.047 | 1.0 | 0.094 | 0.25 | 0.016 | 2 |
| 3 | 8.0 | >256 | 6 | 4 | 3 | 16 | 0.064 | 1.0 | 0.25 | 0.25 | 0.016 | 2 |
| 3 (Tc)b | 8.0 | >256 | 4 | 4 | 3 | 32 | 0.047 | 1.0 | 0.125 | 0.125 | 0.032 | 4 |
| 4 | 5.4, 8.0 | >256 | 4 | 2 | 0.75 | 16 | 0.023 | 0.5 | <0.064 | 0.094 | 0.012 | 1.5 |
| 5 | 5.4, 8.0 | >256 | 4 | 12 | 2 | 48 | 0.094 | 1.0 | 0.125 | 0.125 | 0.023 | 4 |
| 6 | 5.4, 8.0 | >256 | 4 | 4 | 2 | 24 | 0.047 | 0.75 | 0.125 | 0.19 | 0.016 | 2 |
| 7 | 8.0 | >256 | 4 | 2 | 2 | 24 | 0.032 | 1.0 | 0.064 | 0.19 | 0.012 | 1.5 |
| 8 | 8.0 | >256 | 4 | 4 | 2 | 16 | 0.064 | 1.0 | 0.125 | 0.125 | 0.016 | 4 |
| 9 | 5.4, 8.0 | >256 | 8 | 24 | 8 | >128 | >1.0 | 6.0 | 4.0 | 0.25 | 0.023 | 4 |
| 10 | 5.4, 8.0 | >256 | 6 | 12 | 2 | 64 | 0.094 | 1.0 | 0.125 | 0.125 | 0.016 | 4 |
| 11 | 5.4, 8.0 | >256 | 4 | 3 | 2 | 32 | 0.047 | 1.0 | 0.064 | 0.25 | 0.016 | 4 |
| 12 | 8.0 | >256 | 4 | 4 | 2 | 64 | 0.047 | 1.0 | 0.094 | 0.19 | 0.016 | 4 |
| 13 | 8.0 | >256 | 4 | 4 | 4 | >128 | 0.047 | 1.0 | 0.094 | 0.125 | 0.012 | 2 |
| 14 | 8.0 | >256 | 4 | 4 | 2 | 48 | 0.064 | 1.5 | 0.19 | 0.125 | 0.012 | 4 |
| 15 | 8.0 | >256 | 4 | 3 | 1.5 | >128 | 0.047 | 1.0 | 0.125 | 0.19 | 0.016 | 4 |
| 16 | 5.4, 8.0 | >256 | 4 | 4 | 2 | 16 | 0.064 | 1.0 | 0.19 | 0.19 | 0.023 | 2 |
| 17 | 8.0 | >256 | 4 | 4 | 2 | 24 | 0.064 | 1.0 | 0.125 | 0.125 | 0.012 | 2 |
| 18 | 5.4, 8.5 | >256 | 4 | 2 | 8 | 24 | >1 | >32 | >4 | 0.25 | 0.032 | 0.38 |
| 19 | 5.4 | >256 | 16 | 16 | 24 | 0.5 | 0.19 | >32 | 0.75 | 0.25 | 0.032 | 2 |
| 20 | 5.4 | >256 | 2 | 1 | 0.5 | <0.016 | <0.016 | 12 | 0.064 | 0.19 | 0.023 | 0.032 |
| 21 | 5.4 | >256 | 4 | 12 | 6 | 0.5 | <0.016 | >32 | 0.19 | 0.125 | 0.023 | 0.75 |
| 22 | 9.0 | >256 | 4 | 2 | 4 | >256 | 0.047 | 6 | 0.094 | 0.125 | 0.016 | 8 |
| 23 | 9.0 | >256 | 12 | >256 | >256 | >256 | >1.0 | >32 | >4 | 0.125 | 0.064 | >128 |
| 24 | 9.0 | >256 | 12 | >256 | >256 | >256 | >1.0 | >32 | >4 | 0.125 | 0.047 | 128 |
| 25 | 5.9 | >256 | 4 | 2 | 1 | 2 | 0.094 | 2 | 0.19 | 0.5 | 0.032 | 0.094 |
| 26 | 5.4, 5.9 | >256 | 4 | 8 | 6 | 0.19 | 0.047 | >32 | 0.125 | 0.125 | 0.016 | 0.5 |
| 27 | 7.6 | >256 | 8 | 4 | 0.125 | 0.064 | 0.032 | 4 | 0.19 | 0.19 | 0.032 | 0.5 |
| 28c | 7.4 | >256 | 4 | 1 | 6 | 24 | <0.016 | >32 | 0.19 | 0.19 | 0.032 | 3 |
| 29c | 7.9 | >256 | 4 | 12 | 6 | 0.5 | 0.064 | >32 | 0.19 | 0.125 | 0.023 | 0.75 |
| 30c | 7.4 | >256 | 6 | 2 | 1 | 1 | 0.047 | 1.5 | 0.125 | 0.19 | 0.032 | 0.094 |
| E. coli TG1 | 3 | 3 | 2 | 0.047 | 0.023 | 0.023 | 0.064 | 0.064 | 0.125 | 0.032 | 0.094 | |
| E. coli TG1 (pC)d | 8.0 | >256 | 6 | 2 | 48 | >256 | 0.064 | 4 | 0.125 | 0.19 | 0.023 | 64 |
AMX, amoxicillin; AMC, amoxicillin-clavulanic acid; FOX, cefoxitin; AT, aztreonam; CTX, cefotaxime; CTX-CL, cefotaxime-clavulanic acid; CAZ, ceftazidime; CAZ-CL, ceftazidime-clavulanic acid; IMP, imipenem; MRP, meropenem; CEP, cefepime.
This isolate corresponds to the transconjugant of clinical strain 3.
This isolate corresponds to K. pneumoniae.
E. coli transformed with plasmid pC, which carries the CTX-M-14 gene.
Detection of the CTX-M-14 gene.
Plasmid DNA from different E. coli strains was used as the template in a PCR amplification. Amplification reactions were performed in a final volume of 50 μl. Mg2+-free PCR buffer was purchased as a 10× concentrate consisting of 500 mM KCl, 100 mM Tris-HCl (pH 9.0), and 1% Triton X-100 (Perkin-Elmer, Roche Molecular Systems, Inc., Nutley, N.J.) with 200 μM (each) dATP, dCTP, dGTP, and dTTP (Perkin-Elmer, Roche Molecular Systems, Inc.). The Mg2+ concentration was 2.5 mM, and the primers were used at 0.5 μM each. The primer pair CTX-M-1 (5′-AACACGGATTGACCGTATTG-3′) and CTX-M-2 (5′-TTACAGCCCTTCGGCGAT-3′) was used to amplify the CTX-M-14 gene in the plasmid DNA. Amplification reactions were carried out in an Eppendorf thermal cycler (Eppendorf AG, Hamburg, Germany), with an initial denaturation (10 min at 94°C) followed by 30 cycles of denaturation (30 s at 94°C), annealing (30 s at 58°C), and extension (2 min at 72°C), with a single final extension of 10 min at 72°C. Aliquots (15 μl) of each sample were subjected to electrophoresis in 1.0% agarose gels. Amplified products were detected after staining with ethidium bromide (50 μg/ml) and photographed with Polaroid type 665 film.
REP-PCR.
Amplification reactions were performed in a final volume of 50 μl. Mg2+-free PCR buffer was purchased as a 10× concentrate consisting of 500 mM KCl, 100 mM Tris-HCl (pH 9.0), and 1% Triton X-100 (Perkin-Elmer, Roche Molecular Systems, Inc.), with 200 μM (each) dATP, dCTP, dGTP, and dTTP (Perkin-Elmer, Roche Molecular Systems, Inc.). The Mg2+ concentration was 3 mM, and the primers were used at 0.5 μM each. The primer pair REP1 (5′-IIIGCGCCGICATCAGGC-3′) and REP2 (5′-ACGTCTTATCAGGCCTAC-3′) was used to amplify putative REP-like elements in the genomic bacterial DNA (23). A total of 500 ng of chromosomal DNA was added to the reaction mixture. Amplification reactions were carried out in an Eppendorf thermal cycler, with an initial denaturation (10 min at 94°C) followed by 30 cycles of denaturation (1 min at 94°C), annealing (1 min at 45°C), and extension (2 min at 72°C), with a single final extension of 16 min at 72°C. Aliquots (20 μl) of each sample were subjected to electrophoresis in 1.0% agarose gels. Amplified products were detected after staining with ethidium bromide (50 μg/ml) and photographed with Polaroid type 665 film. Strains belonging to the same DNA group showed identical or highly similar profiles (up to two bands different).
Plasmid restriction fragment length polymorphisms (RFLPs).
Plasmid DNA from transconjugant E. coli strains 1 to 17 (see Table 2) was isolated as described above. The DNA was then digested with HindIII, and the resulting fragments were loaded on a 0.8% agarose gel. After electrophoresis, DNA fragments were detected by staining with ethidium bromide (50 μg/ml) and photographed with Polaroid type 665 film.
Detection of the gene encoding the tnpA transposase.
To detect the presence of the tnpA gene of the insert sequence ISEcp-1, we used the primer pair tnpa1 (5′-GAATTCATCAATTGTATT-3′) and tnpa2 (5′-CAAGAAATACATACTTCAA-3′). About 10 ng of plasmid DNA was added to the reaction mixture. Amplification reactions were carried out in an Eppendorf thermal cycler, with an initial denaturation (10 min at 94°C), 30 cycles of denaturation (1 min at 94°C), annealing (1 min at 48°C), and extension (2 min at 72°C), and a single final extension of 16 min at 72°C. Aliquots (20 μl) of each sample were subjected to electrophoresis in 1.0 % agarose gels. Amplified products were detected after staining with ethidium bromide (50 μg/ml) and photographed with Polaroid type 665 film.
RESULTS
Selection of clinical isolates and patients.
Between January and October 2001, 27 E. coli and 3 K. pneumoniae clinical strains with a phenotype suggestive of ESBL were selected. These strains were isolated from 30 different patients. One isolate per patient was selected (Table 1). The strains were associated with urinary tract infections (77%), wound infection (13.3%), catheter infection (3.3%), blood infection (3.3%), and conjuntival infection (3.3%). Most patients had underlying diseases. Moreover, although most of the patients were hospitalized before the E. coli and/or K. pneumoniae isolation, seven patients had no hospital admission noted in their medical records.
TABLE 1.
Patients from whom the E. coli and K. pneumoniae strains used in this study were isolated
| Patient data (no., sex/age [yr]) | Type of infectiona | Underlying disease | Specimen | Hospitalization before the E. coli isolation |
|---|---|---|---|---|
| 1. F/81 | UTI | Diabetes | Urine | No |
| 2. F/9 | UTI | Acute pyelonephritis | Urine | Yes |
| 3. M/66 | UTI | Prostate tumor | Urine | Yes |
| 4. F/1 | UTI | Respiratory tract infection | Urine | Yes |
| 5. F/Unknown | UTI | Unknown | Urine | Unknown |
| 6. F/77 | UTI | Blood infection, diabetes | Urine | Yes |
| 7. F/80 | UTI | No | Urine | No |
| 8. F/Unknown | UTI | Unknown | Urine | Unknown |
| 9. M/67 | UTI | Colon cancer | Urine | Yes |
| 10. M/58 | UTI | Prostate tumor | Urine | Yes |
| 11. F/73 | UTI | HBP,b arthrosis | Urine | Yes |
| 12. F/75 | UTI | HPB, urinary incontinence | Urine | No |
| 13. F/41 | UTI | Unknown | Urine | No |
| 14. F/57 | WI | Stomach ulceration | Exudate | Yes |
| 15. F/76 | UTI | Type II diabetes, chronic hepatitis | Urine | Yes |
| 16. M/86 | WI | Unknown | Exudate | Unknown |
| 17. F/37 | UTI | No | Urine | No |
| 18. F/73 | UTI | HBP, subarachnoid hemorrhagia | Urine | Yes |
| 19. F/32 | UTI | Unknown | Urine | Yes |
| 20. F/51 | UTI | Unknown | Urine | Yes |
| 21. F/73 | UTI | HBP | Urine | Yes |
| 22. M/25 | WI | Unknown | Exudate | No |
| 23. F/14 | CI | Unknown | Catheter | Unknown |
| 24. M/79 | WI | Pneumonia | Exudate | Yes |
| 25. F/66 | UTI | Unknown | Urine | No |
| 26. M/73 | UTI | Prostate tumor | Urine | Yes |
| 27. M/5 | UTI | Several UTIs | Urine | Yes |
| 28. M/53 | UTI | Leukemia | Urine | Yes |
| 29. F/1 | BI | Hepatitis | Blood | Yes |
| 30. F/1 | CoI | Premature birth | Conjunctival swab | Yes |
UTI, urinary tract infection; WI, wound infection; CI, catheter infection; BI, blood infection; CoI, conjunctival infection.
HBP, high blood pressure.
Determination of antibiotic susceptibility and ESBL production.
MICs for the 30 clinical strains were determined by the DadeMicroScan. In addition, all MICs were confirmed by the E test (AB Biodisk) according to the NCCLS criteria (14). The overall results are shown in Table 2. For all strains, the amoxicillin MICs were higher than 256 μg/ml; moreover, this concentration decreased drastically in the presence of clavulanic acid, suggesting the presence of a class A β-lactamase. Regarding broad-spectrum cephalosporins, strains 1 to 17 and 22 had a phenotype suggesting cefotaximase β-lactamase activity as shown by the cefotaxime MICs (the cefotaxime MICs were clearly higher than the ceftazidime MICs).
Interestingly, the K. pneumoniae isolates included in this study with an ESBL phenotype (strains 28 to 30) showed unique SHV-like β-lactamase activity band by IEF. This band probably corresponds to the SHV chromosomal β-lactamase that is consitutively present in K. pneumoniae strains; alternatively, it is an SHV ESBL indistinguishable from the chromosomal SHV-1. The presence of a second antibiotic resistance mechanism in these K. pneumoniae strains cannot be ruled out.
IEF assay.
Protein extracts of the 30 clinical strains and their transconjugants obtained by sonication were resolved by IEF (3-9). Different pIs corresponding to different β-lactamases were obtained. Interestingly, strains 1 to 17 yielded an identical pI of 8.0; in addition, some of them yielded a pI of 5.4, suggestive of a TEM-1-like β-lactamase. Strains 18 to 30 showed different pIs such as 5.4, 5.9, 7.4, 7.6, 7.9, 8.5, and 9.0 (Table 2).
Cloning of the CTX-M-14 gene.
The cefotaxime MICs and pIs obtained by IEF suggested the presence of an identical or similar β-lactamase gene associated with resistance to cefotaxime in strains 1 to 17 (Table 2). We attempted to clone the β-lactamase gene responsible for this selective resistance to cefotaxime.
For this purpose, strain 3 (Table 2) was chosen for the cloning experiments. First, we tried to show whether the β-lactamase gene was plasmid or chromosomally mediated. A conjugation experiment was performed using E. coli XL-1-blue as a host, as described in Materials and Methods. Transconjugants were selected on LB agar plates supplemented with kanamycin and ampicillin. Several transconjugants were obtained, and IEF of the sonicated extracts of the clinical strain and the transconjugants showed a pI 8.0 activity band in all cases. Plasmid analysis of the clinical strain (strain 3) and one transconjugant (strain 3Tc) showed that the β-lactamase gene was associated with a plasmid of ca. 40 kb (data not shown). Moreover, MIC experiments performed with both parental (strain 3) and transconjugant (strain 3Tc) isolates showed almost identical antibiotic susceptibilities (Table 2), clearly suggesting that the β-lactamase gene was plasmid mediated.
Cloning of the CTX-M-14 gene was performed as described in Materials and Methods. The nucleotide sequence of the fragment revealed only one open reading frame with homology with β-lactamase genes. The CTX-M-14 open reading frame was 876 bp long and encoded a protein of 291 amino acid residues. This gene has previously been reported (GenBank accession numbers AJ416341 and AF252622). In the upstream region of the CTX-M-14 gene, the nucleotide sequence of the tnpA gene, the transposase gene of insertion sequence ISEcp1, was also detected (24).
Spread of the CTX-M-14 gene in different E. coli strains.
To elucidate whether the CTX-M-14 gene was plasmid mediated in all E. coli strains, conjugation experiments were performed. Regarding the kanamycin and/or nalidixic acid susceptibility of the E. coli clinical strains, two different E. coli isolates were used as hosts in the conjugation experiments, E. coli BM21 (resistant to nalidixic acid) when the E. coli clinical strains were susceptible to nalidixic acid and E. coli XL-10-Gold Kan (resistant to kanamycin) when the E. coli clinical strains were susceptible to kanamycin. Transconjugants were obtained in all E. coli strains (isolates 1 to 17 in Table 2) that yielded a β-lactamase activity band of pI 8.0 with cefotaximase activity, strongly suggesting that in all cases the β-lactamase with cefotaximase activity was plasmid mediated.
To assess the possibility of the spread of this CTX-M-14 gene in the 16 remaining E. coli strains, a PCR assay was performed. Plasmid DNA obtained from the 27 E. coli and 3 K. pneumoniae strains was used as a template in a PCR amplification with CTX-M-1 (5′-AACACGGATTGACCGTATTG-3′) and CTX-M-2 (5′-TTACAGCCCTTCGGCGAT-3′) primers, which annealed at the promoter and the 3′ part of the CTX-M-14 gene, respectively. Interestingly, a positive PCR result, indicated by a band of 905 bp, was detected in 17 out of 30 clinical strains, including strain 3 (data not shown). The overall results showed that the CTX-M-14 gene was disseminated among 17 E. coli strains in the northwest area of Spain. Moreover, nucleotide sequence analysis of these 17 amplicons showed total identity to the CTX-M-14 nucleotide sequence.
It is worth mentioning that strain 18, although possessing a pI 8.5 band of β-lactamase activity, did not yield an amplification product with the CTX-M-1 and CTX-M-2 primers. Also, the antibiotic susceptibility pattern of this strain did not show the putative cefotaximase profile detected in the other 17 E. coli strains.
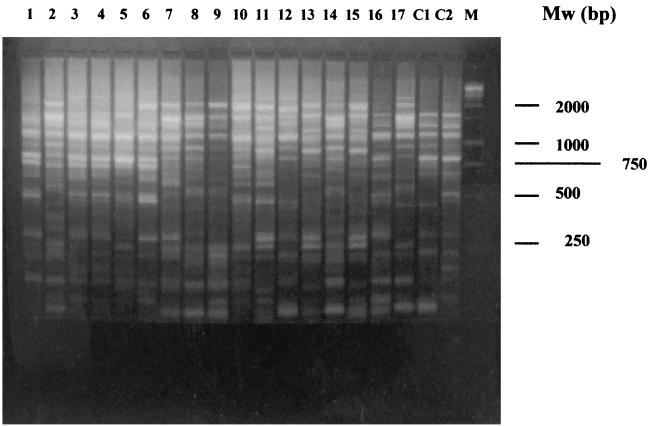
The next step was to try to determine whether these 17 E. coli strains were genetically related. This experiment was necessary to distinguish between two putative hypotheses: (i) one epidemic E. coli strain has been disseminated among all the patients, and so one ancestor strain may be responsible for the spreading; it implies a link between all the patients, and epidemiological studies must confirm this hypothesis; and (ii) the CTX-M-14 gene has disseminated among different E. coli strains. Therefore, the possibility that a plasmid harboring the CTX-M-14 gene or a mobile genetic element carrying this gene had been disseminated in different E. coli strains cannot be ruled out. A REP-PCR assay was performed as described in Materials and Methods. This assay showed that most of the E. coli strains were genetically unrelated (Fig. 1). Fifteen different genotypes may be assigned to the 17 E. coli clinical strains (isolates 3 and 4 and isolates 13 and 15 yielded the same band patterns). This result therefore eliminated the possibility of clonal dissemination of one epidemic E. coli strain.
FIG. 1.
REP-PCR of the 17 E. coli strains and 2 genetically unrelated E. coli controls (C1 and C2). The strain numbers are shown above the gel. Mw (molecular size markers) corresponds to the 1-kb DNA ladder (Promega, Madison, Wis.).
Plasmid RFLP.
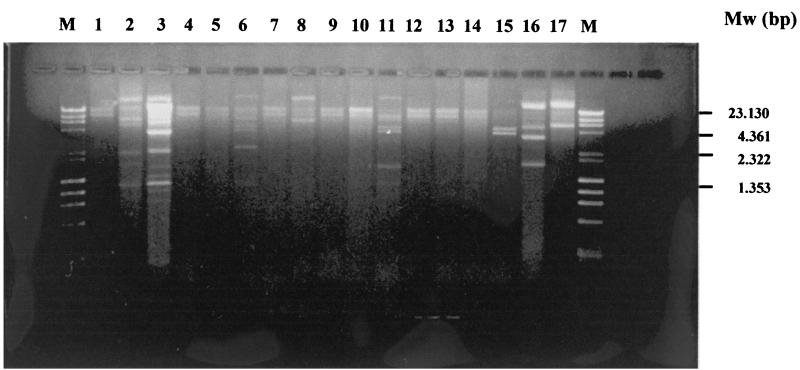
Purified plasmids from transconjugants of strains 1 to 17 (Table 2) were digested with HindIII, and the resulting fragments were separated on 0.8% agarose gels (Fig. 2). The overall results showed a similarity between plasmids in strains 1, 4, 5, 7, 9, 10, 12, 13, and 14 (RFLP pattern I in Table 3) obtained from the 17 E. coli strains, thus suggesting identity in the plasmids harboring CTX-M-14 on these strains. However, strains 2, 3, 6, 8, 11, 15, 16, and 17 did not show a similarity in their RFLP patterns, and their patterns were different from RFLP pattern I. Therefore, the CTX- M-14 gene was harbored in different plasmids on these strains, and so the possibility that a mobile genetic element might be involved in the dissemination of the CTX-M-14 gene in these plasmids cannot be ruled out.
FIG. 2.
Plasmid RFLP obtained by HindIII digestion with plasmid purifications of the transconjugants of the 17 E. coli strains. The strain numbers are shown above the gel. Mw (molecular size markers) corresponds to the lambda DNA HindIII digest and bacteriophage Φ174 HaeIII digest (Fynnzymes OY).
TABLE 3.
REP-PCR pattern, detection of the CTX-M-14 gene and RFLP of the plasmids of the transconjugants of the clinical strains used in this study
| Isolate no. | pI | REP-PCR pattern | PCR CTX-M-14 | Transconjugant plasmid RFLP pattern |
|---|---|---|---|---|
| 1 | 5.4, 8.0 | 1 | + | I |
| 2 | 8.0 | 2 | + | II |
| 3 | 8.0 | 3 | + | III |
| 4 | 5.4, 8.0 | 3 | + | I |
| 5 | 5.4, 8.0 | 4 | + | I |
| 6 | 5.4, 8.0 | 5 | + | IV |
| 7 | 8.0 | 6 | + | I |
| 8 | 8.0 | 7 | + | V |
| 9 | 5.4, 8.0 | 8 | + | I |
| 10 | 5.4, 8.0 | 9 | + | I |
| 11 | 5.4, 8.0 | 10 | + | VI |
| 12 | 8.0 | 11 | + | I |
| 13 | 8.0 | 12 | + | I |
| 14 | 8.0 | 13 | + | I |
| 15 | 8.0 | 12 | + | VII |
| 16 | 5.4, 8.0 | 14 | + | VIII |
| 17 | 8.0 | 15 | + | IX |
| 18 | 5.4, 8.5 | NDa | − | ND |
| 19 | 5.4 | ND | − | ND |
| 20 | 5.4 | ND | − | ND |
| 21 | 5.4 | ND | − | ND |
| 22 | 9.0 | ND | − | ND |
| 23 | 9.0 | ND | − | ND |
| 24 | 9.0 | ND | − | ND |
| 25 | 5.9 | ND | − | ND |
| 26 | 5.4, 5.9 | ND | − | ND |
| 27 | 7.6 | ND | − | ND |
| 28 | 7.4 | ND | − | ND |
| 29 | 7.9 | ND | − | ND |
| 30 | 7.4 | ND | − | ND |
ND, not done.
Detection of the tnpA transposase gene.
Plasmid DNAs from the conjugants of the 17 E. coli clinical strains (strains 1 to 17 in Table 2) were subjected to PCR amplification with primers which annealed to the tnpA gene, as described in Materials and Methods. A positive band of 405 bp was obtained in all the plasmid purifications (data not shown), suggesting that a putative functional transposon was present in all E. coli strains and that this transposon may be involved in the mobilization of the CTX-M-14 gene.
DISCUSSION
In 1992 (1, 2) a novel type of ESBL that conferred a high level of resistance to cefotaxime but not to ceftazidime was identified in some members of Enterobacteriaceae. This new family of plasmid-mediated ESBLs of Ambler class A was assigned to the cefotaximase (CTX-M) family. Cefotaximases (CTX-M) have been detected mainly in South America, Eastern Europe, Japan, Spain, Kenya, and France (3, 13, 15, 17, 22).
The family of the CTX-M-type ESBLs comprises several members belonging to four major phylogenetic trees on the basis of their amino acid sequence similarities (4): the CTX-M-1 group (CTX-M-1, CTX-M-3, and CTX-M-10), the CTX-M-2 group (CTX-M-2, Toho-1, CTX-M-4, CTX-M-5, and CTX-M-6), the CTX-M-9 group (CTX-M-9 and Toho-2), and the CTX-M-8 group. The CTX-M-14 β-lactamase is an Ala231-Val mutant of CTX-M-9; therefore, it may be assigned to the CTX-M-9 group.
We have detected the CTX-M-14 β-lactamase for the first time in Spain. This β-lactamase has previously been described in China, Korea, and France (8, 11, 16, 19).
As described above, the CTX-M-14 gene is plasmid mediated; indeed, we detected it in the plasmids of the 15 different genotypes of E. coli strains. The plasmid fingerprints yielded 9 different patterns; therefore, 9 of the 17 profiles were identical. This result suggests that the spreading of this plasmid among different E. coli strains was important in the dissemination of the CTX-M-14 gene. However, in the other eight E. coli strains, different plasmids were detected, as shown by restriction analysis. This result implies some kind of mobilization of the CTX-M-14 gene in the different E. coli strains in addition to the horizontal transfer. The analysis of the CTX-M-14 upstream region showed the presence of the transposase gene (tnpA) of the insertion sequence named ISEcp-1 in all plasmids detected in the E. coli strains. It has previously been suggested that the ISEcp-1 element could be associated with the mobility of the CTX-M genes (11). Therefore, the possibility that the ISEcp-1 insertion sequence can be associated with the spreading of the CTX-M-14 genes in our E. coli strains cannot be ruled out. In fact, this is the only genetic element we have found associated with the high dissemination of the CTX-M-14 gene.
These 17 E. coli strains (15 genotypes) caused infections in 17 patients. In 15 patients the E. coli strain harboring CTX-M-14 caused urinary tract infections whereas in the other two they caused wound infections. Furthermore, most patients had underlying diseases and, interestingly, no hospital admission was noted in the medical records of 7 patients. Thus, on the basis of these data, we may assume the possible existence of outpatients with bacterial strains causing infections harboring antibiotic resistance mechanisms. Therefore, the data presented here raise the hypothesis that microorganisms with antibiotic resistance mechanisms can be present in patients in the community. Another interesting issue is that no apparent contacts between the 17 patients occurred. Indeed, most inpatients were admitted to different hospital wards of the hospital complex. The outpatients live in different areas far away from each other. With the current data, how can we explain the presence of up to 62% of the ESBL-producing strains as harboring the CTX-M-14 gene? Several reports have been published describing the detection of CTX-M-type enzymes in different areas worldwide. Here we report a practical study showing that CTX-M-type β-lactamases are highly disseminated; indeed, they are replacing the TEM and SHV derivative β-lactamases (9). The reasons why this is happening remain to be elucidated.
In a hospital environment, spreading between patients can be easy when a bacterium with an antibiotic resistance mechanism has been established, especially if this mechanism is associated with mobile genetic elements. However, we report here that seven patients from whom bacterial strains harboring ESBL were isolated never had contact with the hospital environment before the clinical E. coli isolation. Further epidemiological studies need to be performed to explain these data.
In summary, we have found a high prevalence of the CTX-M-14 β-lactamase in the northwest area of Spain. On the basis of these data, the empirical administration of cefotaxime in serious infections should be avoided in order to prevent clinical complications. In addition, we alert the medical community of the increase of these β-lactamases worldwide.
Acknowledgments
We express our gratitude to Luis de Rafael for English corrections.
REFERENCES
- 1.Barthélémy, M., J. Péduzzi, H. Bernard, C. Tancrede, and R. Labia. 1992. Close amino acid sequence relationship between the new plasmid-mediated extended-spectrum β-lactamase MEN-1 and chromosomally encoded enzymes of Klebsiella oxytoca. Biochim. Biophys. Acta 1122:15-22. [DOI] [PubMed] [Google Scholar]
- 2.Bauernfeind, A., J. M. Casellas, M. Goldberg, M. Holley, R. Jungwirth, P. Mangold, T. Rohnisch, S. Schweighart, and R. Wilhem. 1992. A new plasmidic cefotaximase from patients infected with Salmonella typhimurium. Infection 20:158-163. [DOI] [PubMed] [Google Scholar]
- 3.Bonnet, R., C. Dutour, J. L. Samapaio, C. Chanal, D. Sirot, R. Labia, C. D. Champs, and J. Sirot. 2001. Novel cefotaximase (CTX-M-16) with increased catalytic efficiency due to substitution Asp-240-Gly. Antimicrob. Agents Chemother. 45:2269-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonnet, R., J. L. Sampaio, R. Labia, C. De Champs, D. Sirot, C. Chanal, and J. Sirot. 2000. A novel CTX-M β-lactamase (CTX-M-8) in cefotaxime-resistant Enterobacteriaceae isolated in Brazil. Antimicrob. Agents Chemother. 44:1936-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bou, G., A. Oliver, and J. Martínez-Beltrán. 2000. OXA-24, a novel class D β-lactamase with carbapenemase activity in an Acinetobacter baumannii clinical strain. Antimicrob. Agents Chemother. 44:1556-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brun-Bruisson, C., P. Legrand, A. Philippon, F. Montravers, M. Ansquer, and J. Duval. 1987. Transferable enzymatic resistance to third-generation cephalosporins during nosocomial outbreak of multiresistant Klebsiella pneumoniae. Lancet ii:302-306. [DOI] [PubMed]
- 7.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chanawong, A., F. H. M'Zali, J. Heritage, J. H. Xiong, and P. M. Hawkey. 2002. Three cefotaximases, CTX-M-9, CTX-M-13, and CTX-M-14, among Enterobacteriaceae in the People's Republic of China. Antimicrob. Agents Chemother. 46:630-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coque, T. M., A. Oliver, J. C. Pérez-Díaz, F. Baquero, and R. Cantón. 2002. Genes encoding TEM-4, SHV-2, and CTX-M-10 extended-spectrum β-lactamases are carried by multiple Klebsiella pneumoniae clones in a single hospital (Madrid, 1989 to 2000). Antimicrob. Agents Chemother. 46:500-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donald, H. M., W. Scaife, S. G. B. Amyes, and H.-K. Young. 2000. Sequence analysis of ARI-1, a novel OXA β-lactamase, responsible for imipenem resistance in Acinetobacter baumannii 6B92. Antimicrob. Agents Chemother. 44:196-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dutour, C., R. Bonnet, H. Marchandin, M. Boye, C. Chanal, D. Sirot, and J. Sirot. 2002. CTX-M-1, CTX-M-3, and CTX-M-14 β-lactamases from Enterobacteriaceae isolated in France. Antimicrob. Agents Chemother. 46:534-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacoby, J. A., and P. Han. 1996. Detection of extended-spectrum β-lactamases in clinical isolates of Klebsiella pneumoniae and Escherichia coli. J. Clin. Microbiol. 34:908-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kariruki, S., J. E. Corkill, G. Revathi, R. Musoke, and C. A. Hart. 2001. Molecular characterization of a novel plasmid-encoded cefotaximase (CTX-M-12) found in clinical Klebsiella pneumoniae isolates from Kenya. Antimicrob. Agents Chemother. 45:2141-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.National Committee for Clinical Laboratory Standards. 1997. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A4. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 15.Oliver, A., J. C. Pérez-Díaz, T. M. Coque, F. Baquero, and R. Cantón. 2001. Nucleotide sequence and characterization of a novel cefotaxime-hydrolyzing β-lactamase (CTX-M-10) isolated in Spain. Antimicrob. Agents Chemother. 45:616-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pai, H., E. H. Choi, H. J. Lee, J. Y. Hong, and G. A. Jacoby. 2001. Identification of CTX-M-14 extended-spectrum β-lactamase in clinical isolates of Shigella sonnei, Escherichia coli, and Klebsiella pneumoniae in Korea. J. Clin. Microbiol. 39:3747-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palucha, A., B. Mikiewicz, W. Hryniewicz, and M. Gniadkowski. 1999. Concurrent outbreaks of extended spectrum β-lactamase-producing organism of the family Enterobacteriaceae in a Warsaw hospital. J. Antimicrob. Chemother. 44:489-499. [DOI] [PubMed] [Google Scholar]
- 18.Sabate, M., R. Tarrago, F. Navarro, E. Miro, C. Verges, J. Barbe, and G. Prats. 2000. Cloning and sequence of the gene encoding a novel cefotaxime-hydrolyzing beta-lactamase (CTX-M-9) from Escherichia coli in Spain. Antimicrob. Agents Chemother. 44:1970-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saladin, M., V. Bao Cao, T. Lambert, J. L. Donay, J. L. Herrmann, Z. Ould- Hocine, C. Verdet, F. Deleisle, A. Philippon, and G. Arlet. 2002. Diversity of CTX-M β-lactamases and their promoter regions from Enterobacteriaceae isolated in three Parisian hospitals. FEMS Microbiol. Lett. 209:161-168. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Siu, L. K., P. Lu, P. R. Hsueh, F. M. Lin, S. C. Chang, K. T. Luh, et al. 1999. Bacteremia due to extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumonia in a pediatric oncology ward: clinical features and identification of different plasmids carrying both SHV-5 and TEM-1 genes. J. Clin. Microbiol. 37:4020-4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tzouvelekis, L. S., E. Tzelepi, P. T. Tassios, and N. J. Legakis. 2000. CTX-M-type β-lactamases: an emerging group of extended-spectrum enzymes. Int. J. Antimicrob. Agents 14:137-142. [DOI] [PubMed] [Google Scholar]
- 23.Vila, J., M. A. Marcos, and M. T. Jiménez de Anta. 1996. A comparative study of different PCR-based DNA fingerprinting techniques for typing of the Acinetobacter calcoaceticus-A. baumannii complex. J. Med. Microbiol. 44:482-489. [DOI] [PubMed] [Google Scholar]
- 24.Wu, S. W., K. Dornbusch, G. Kronvall, and M. Norgren. 1999. Characterization and nucleotide sequence of a Klebsiella oxytoca cryptic plasmid encoding a CMY-type β-lactamase: confirmation that the plasmid-mediated cephamycinase originated from the Citrobacter freundii AmpC β-lactamase. Antimicrob. Agents Chemother. 43:1350-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]