Abstract
Broad-range amplification of bacterial DNA from clinical specimens has proved useful for the diagnosis of various bacterial infections, especially during antimicrobial treatment of the patient. Optimal sample processing protocols for diagnostic broad-range bacterial PCR should release DNA from an array of target organisms with equal efficiencies and wash out inhibitory factors from various sample types without introducing bacterial DNA contamination to the amplification reaction. In the present study, two physical cell wall disintegration methods, bead beating and sonication, for enhanced detection of organisms with difficult-to-lyse cell walls were studied. The analytical sensitivities of several commercially available DNA purification kits, which were used with and without additional cell disintegration steps, were compared by using dilution series of model bacteria. Selected purification methods were used to process routine clinical specimens in parallel with the standard phenol-ether DNA extraction, and the results obtained by bacterial PCR and sequencing with the two template preparations were compared. The method with the DNA isolation kit with the lowest detection limits from the bacterial suspensions (Masterpure) did not prove to be superior to the standard method when the two methods were applied to 69 clinical specimens. For another set of 68 clinical specimens, DNA purified with a glass fiber filter column (High Pure) with an additional sonication step yielded results well in accord with those obtained by the standard method. Furthermore, bacterial DNA was detected in four samples that remained PCR negative by the standard method, and three of these contained DNA from gram-positive pathogens. Three samples were positive by the standard method only, indicating the limitations of applying any single method to all samples.
Direct amplification of bacterial DNA from clinical specimens with broad-range primers provides an alternative approach to the recognition of pathogens infecting normally sterile body compartments. In comparing the molecular diagnosis obtained by broad-range bacterial PCR and partial sequencing of the amplicon to the results of routine bacterial cultures, we found 83% overall agreement for a set of 536 clinical specimens from hospitalized patients (7). The molecular approach proved superior to culture during antimicrobial treatment of the patient and in detection of bacteria with unusual growth requirements. A major drawback of the molecular method in comparison to bacterial culture was the difficulty in detecting species with gram-positive cell walls and mycobacteria. This was associated with problems in breaking bacterial cell walls and releasing bacterial DNA for amplification when standard phenol-ether DNA extraction was used. This finding led us to search for better methods to prepare the clinical specimens for broad-range bacterial PCR assay.
In general, an optimal sample processing method should concentrate the DNA, especially that derived from the target organism, and wash out inhibitory factors commonly present in biological fluids. To be applicable for routine diagnostic use, the process should be suitable for use with an array of clinical specimens, simple or preferably at least semiautomatic, reproducible, and safe for the staff handling the specimens. Furthermore, design of the process should prevent cross-contamination between the samples; i.e., samples with large amounts of target microbial cells or their nucleic acids should not contaminate other specimens in the batch.
In contrast to species-specific PCR assays, the possible target organisms of a clinical broad-range bacterial PCR test can be highly variable in their capability to resist chemical and physical treatments. In other words, an optimal sample preparation procedure should efficiently break very resistant bacterial cell walls, like those of streptococci and mycobacteria, without being too harsh for the DNA released from cells that are easily lysed. Another aspect specific for broad-range bacterial PCR is the danger of introducing bacterial DNA from the reagents used in various phases of sample processing and amplification (9), which may result in false-positive PCR results. Elimination of this background bacterial DNA from the reactions has proved very difficult (1). Thus, aiming at maximal analytical sensitivity in the broad-range bacterial PCR assay may eventually result in impaired clinical performance of the test in diagnosing true infectious conditions.
In the study described here we studied the use of bead beating and sonication to enhance the lysis of bacterial cell walls prior to standard DNA extraction and 23S ribosomal DNA (rDNA)-targeted PCR. These lysis methods were then combined with some DNA purification protocols available in kit format, and the analytical sensitivities were compared by using dilution series of some model bacteria. Finally, selected processing methods were applied to clinical specimens in parallel with the previously used sample preparation procedures, and the results obtained by bacterial PCR and sequencing were evaluated.
MATERIALS AND METHODS
Bacteria.
Streptococcus pyogenes ATCC 8184 and Mycobacterium avium ATCC 35712 were used as model organisms with difficult-to-lyse cell walls, and Haemophilus influenzae ATCC 49247 was used as an example of a more fragile bacterial cell. S. pyogenes and H. influenzae were grown on blood and chocolate agar plates, respectively, and several colonies were suspended in sterile phosphate-buffered saline (PBS). The numbers of CFU in the suspensions were calculated after cultivation of 50 μl of appropriate dilutions on agar plates. Suspensions of M. avium cells were prepared and enumerated similarly by use of Middlebrook 7 H10 agar plates. Tenfold dilution series containing 2,000 to 0.02 CFU per μl were prepared in sterile PBS, aliquoted, and stored at −20°C until use. Each dilution series also included a negative control, i.e., PBS without added bacteria.
Lysis of bacterial cells.
A total of 100 μl of S. pyogenes or H. influenzae cell suspensions (containing a total of 200,000 to 2 CFU) was transferred into sterile screw-cap Eppendorf tubes with 0.3 g of zirconia-silica beads (diameter, 0.1 mm; Biospec Products, Bartlesville, Okla.), and 100 μl of sterile UV-irradiated water (two times for 90 s each time with UV Stratalinker [Stratagene, La Jolla, Calif.]) was added. Before addition of the samples the tubes with the beads had been UV irradiated as described above for the water. The dilution series were homogenized with a Mini-beadbeater (Biospec Products) for 30 s or 1, 2, 3, or 5 min, after which the DNA was extracted with two phenol-chloroform-isoamyl alcohol extractions followed by one ether wash (3). Similarly, dilution series of bacterial cells were subjected to sonication (51 kHz; Branson 5200; Branson Cleaning Equipment Company, Shelton, Conn.), with or without glass beads, for 1, 3, 5, or 10 min prior to standard DNA isolation as described above.
DNA isolation kits and instruments.
A mixture of bacterial dilution series (100 μl containing 200,000 to 2 CFU) and human monocytes (106 human monocytes in 100 μl of PBS to simulate the excess of human cells in clinical specimens) was used as a sample in the testing of various DNA isolation methods. The commercial DNA isolation kits used in this study were the Masterpure DNA purification kit (Epicentre Technologies, Madison, Wis.), DNA-Pure Yeast Genomic kit (CPG Inc., Lincoln Park, N.J.), Qiamp DNA Mini kit (Qiagen GmbH, Hilden, Germany), High Pure PCR template preparation kit (Roche Diagnostics GmbH, Mannheim, Germany), and Magna Pure LC DNA isolation kit III (bacteria, fungi) with the Magna Pure instrument (Roche Diagnostics). In the first two kits, lysis of cells and precipitation of proteins are followed by precipitation of DNA with isopropanol, drying, and resuspension of the DNA pellet. In the Qiamp and High Pure kits, the released DNA is bound on a silica gel membrane and a glass fiber filter, respectively, and washed before elution. In the Magna Pure application, the DNA released by use of lysis buffer is bound on glass-covered magnetic beads, which the instrument transfers through several washing steps. Finally, the DNA is eluted and the beads are discarded. All the kits were used according to the manufacturers' instructions, with the exception that the final volume of the DNA preparation was always adjusted to 200 μl with the elution or resuspension buffer provided with the kit. According to the manufacturers, the capacities of the Qiagen and High Pure kits are at least 60 and 10 μg of DNA, respectively. A total of 200,000 genomes of our model bacteria, S. pyogenes and H. influenzae, are estimated to contain about 0.0004 μg of DNA, and a million human cells are estimated to contain about 3.2 μg of DNA. When used in combination with the kits, the bead-beating and sonication steps were performed after incubation with the lysis buffer provided with the kit. A total of 5 μl of purified template DNA was used in the PCR.
Bacterial PCR.
An 850-bp sequence of bacterial 23S rDNA was amplified with primers MS37 and MS38, which have been described previously (4). The 30-cycle amplification was performed in a DNA Thermal Cycler 480 (Applied Biosystems, Foster City, Calif.) or a PTC-200 Thermal Cycler (MJ Research, Watertown, Mass.) in 50-μl reaction mixtures with the reagents, cycling times, and temperatures described earlier (4). For detection of PCR products, 10 μl of the amplified DNA was run on a 1.5% SeaKem agarose gel (FMC BioProducts, Rockland, Maine), stained with ethidium bromide, and visualized under UV light.
Before addition of the template DNA, all reaction mixtures were UV irradiated for 3 min to degrade endogenous bacterial DNA. A positive control (50 ng of DNA from Bacillus subtilis ATCC 6051), a negative isolation control (PBS without added bacteria for bacterial dilution series and sterile UV-irradiated distilled water for patient samples), and negative reagent controls (for which sterile UV-irradiated distilled water instead of DNA template was added to the reaction mixture) were included in all runs. The precautions taken to avoid carryover contamination have been described earlier (7). In the case of clinical specimens, inhibition of PCR was assessed by amplification of a fragment of the human growth hormone gene from all template DNA preparations, as described previously (7).
Clinical specimens.
A total of 137 clinical specimens sent to our laboratory for routine bacterial PCR were divided in two. One half of each specimen was processed by the routine proteinase K-phenol-ether protocol (7), and the other half was processed by one of the selected test protocols, i.e., with the Masterpure kit (69 samples) or the High Pure kit with an additional 5-min sonication step after the lysis buffer-proteinase K treatment (68 samples). In the case of the liquid samples, two 1-ml aliquots were concentrated by centrifugation (8,000 × g, 5 min) and 800 μl of the supernatant was removed from each tube. The remaining 200 μl was used for DNA isolation by the standard method (aliquot 1) or one of the test methods (aliquot 2). Apart from the additional sonication step in the High Pure protocol, the kits were used according to the manufacturers' instructions.
The 23S rDNA-targeted PCR described above was used to screen for the presence of bacteria in the samples. The bacterial DNA present in a 23S rDNA PCR-positive sample was identified by sequencing the 16S and/or 23S rDNA by our routine laboratory procedure (7). The 16S rDNA was preferably used for sequencing, and 23S rDNA was sequenced if sequencing of 16S rDNA failed. The 16S rDNA PCR primers and conditions have been described previously (4).
The PCR product was purified by use of a GFX PCR and Gel Band Purification kit (Amersham Pharmacia Biotech Inc., Piscataway, N.J.). Eleven microliters of the purified product was sequenced by use of the ABI Prism DNA sequencing kit, Big Dye Terminator Cycle Sequencing (version 2.0 or 3.0), and ABI Prism 310 genetic analyzer (Applied Biosystems). Sequencing primer 533f targets bases 515 to 533 in the bacterial 16S rDNA sequence (Escherichia coli numbering), and primer JJ04 targets bases 1601 to 1629 in the bacterial 23S rDNA sequence (4). Additional sequencing primers (3) were used if they were considered necessary for clinical purposes. Sequence comparisons were done by using an in-house algorithm in a local database (4) and the FastA program (5) in the European Molecular Biology Laboratory (EMBL) prokaryote database (8).
RESULTS
More than 200,000 CFU of S. pyogenes in a sample was required to yield a visible PCR product after standard phenol DNA extraction and 23S rDNA-targeted PCR. By using H. influenzae cell suspensions, the limit of detection was determined to be 20,000 CFU per sample (Table 1).
TABLE 1.
Comparison of different DNA isolation methods
| Isolation method | Detection limit (no. of CFU/sample) of PCRa
|
Hands-on + hands-off times for seven samplesb | Price ($) per samplec | ||
|---|---|---|---|---|---|
| S. pyogenes ATCC 8184 | M. avium ATCC 33712 | H. influenzae ATCC 49247 | |||
| Proteinase K, phenol-ether | >200,000 | >200,000 | 20,000 | 1 h 30 min + 30 min | 1.33 |
| Bead beating for 1 min, proteinase K, phenol-ether | 20,000 | 2,000 | 20,000 | ||
| Sonication for 5 min, proteinase K, phenol-ether | 20,000 | 20,000 | 20,000 | ||
| High Pure PCR template preparation kit, tissue protocol | 200,000 | >200,000 | 20,000 | 2 h | 2.70 |
| Bead beating for 1 min, High Pure kit | 20,000 | 20,000 | 20,000 | ||
| Sonication for 5 min, High Pure kit | 20,000 | 20,000 | 2,000 | ||
| Masterpure DNA purification kit | 20,000 | 20,000 | 2,000 | 1 h 30 min | 1.72 |
| Sonication for 5 min., Masterpure Kit | 20,000 | 20,000 | 2,000 | ||
| Qiamp DNA mini kit, buffer ATL | 200,000 | 200,000 | 2,000 | 1 h | 2.76 |
| Sonication for 5 min, Qiamp DNA Mini kit | 200,000 | 200,000 | Not done | ||
| CPG DNA-pure yeast genomic kit | 20,000 | 200,000 | 20,000 | 1 h 15 min + 30 min | 4.26 |
| Sonication for 5 min, DNA-pure yeast | 20,000 | Not done | Not done | ||
| MagNA Pure LC DNA isolation kit III (bacteria, fungi) | 20,000 | >200,000 | Not done | 45 min + 1 h | 6.23 |
Samples were 10-fold dilution series of bacteria in PBS containing 200,000 to 2 CFU and 1 million human mononuclear cells in a volume of 200 μl. After extraction, DNA was eluted in 200 μl of sterile water or the elution buffer included in the kit, and 5 μl was amplified with broad-range bacterial primers. The number of CFU in the most dilute sample producing a clear-cut band on a 1.5% agarose gel in at least two independent extractions was considered the detection limit.
Single times apply to the total time for the methods that do not include longer hands-off steps.
Prices are for reagents and plastic ware. Plastic ware includes Eppendorf tubes not provided in the kit, aerosol-resistant tips, and the plastic ware used by the MagNA Pure instrument.
Lysis by bead beating and sonication.
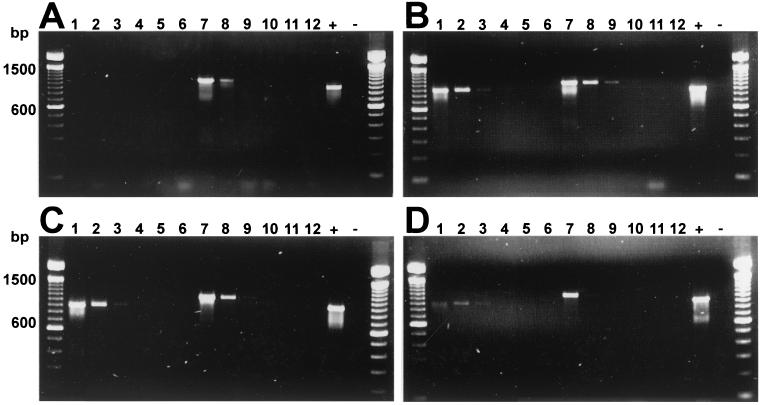
Two hundred-microliter suspensions of S. pyogenes and H. influenzae containing 200,000 to 2 CFU in PBS were beaten with glass beads for 30 s or 1, 2, 3, or 5 min prior to standard DNA isolation and were subjected to 23S rDNA PCR. Bead beating for 1 to 2 min yielded the lowest detection limit for S. pyogenes (a clear-cut band with 20,000 CFU per sample and weak bands in two successive dilutions). A total of 2,000 CFU of H. influenzae yielded a clear-cut band after bead beating for 30 s and a weak one after bead beating for 1 min. Continued bead beating resulted in impaired detection of both bacteria, probably reflecting the progressive degradation of DNA, which seemed to occur more readily for H. influenzae than for S. pyogenes (Fig. 1).
FIG. 1.
Effect of bead beating on detection of bacteria by 23S rDNA-targeted PCR. Bacterial cell suspensions were treated by the standard proteinase K-phenol-ether DNA extraction protocol, and 5 μl of the DNA was used in PCR (A). The lysis of bacteria was enhanced by bead beating for 30 s (B), 1 min (C), or 2 min (D). The samples on each gel were as follows: lanes 1 to 6, 200,000, 20,000, 2,000, 200, and 20 CFU of S. pyogenes in 200 μl of PBS and a negative isolation control with PBS only, respectively; lanes 7 to 12, 200,000, 20,000, 2,000, 200, and 20 CFU of H. influenzae in 200 μl of PBS and a negative isolation control with PBS only, respectively; lanes +, positive control (50 ng of DNA from B. subtilis ATCC 6051); lanes −, negative reagent control (5 μl of sterile UV-irradiated distilled water as a template). The lanes on both sides of each gel contain a molecular size marker (100-bp DNA ladder; Life Technologies).
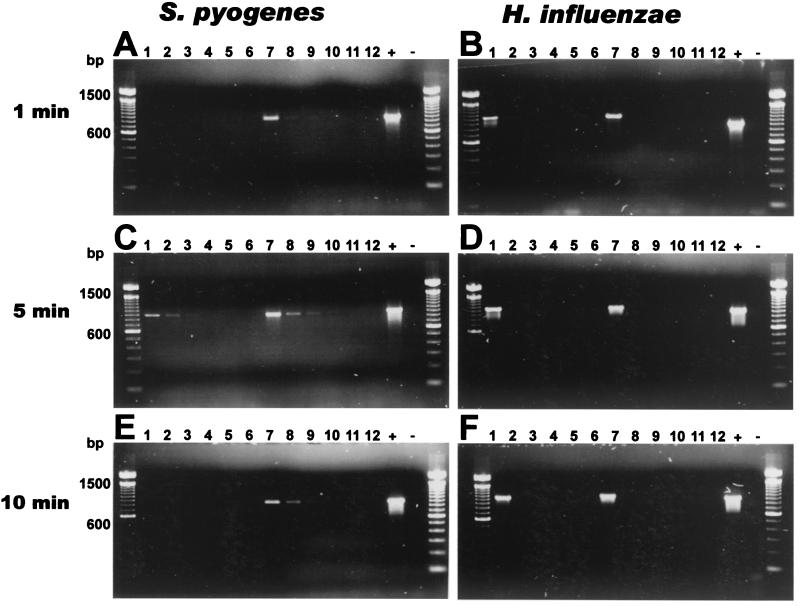
As for sonication, the optimum sonication time for the detection of S. pyogenes was determined to be 5 min both in the presence and in the absence of glass beads, the lowest limit of detection being 20,000 CFU per sample. Short sonication without glass beads yielded the same detection limit for H. influenzae as phenol-ether extraction without sonication, but the detection limit rose if the sonication period exceeded 3 min. In the presence of glass beads the release of S. pyogenes DNA was further enhanced, but lower numbers (below 200,000 CFU) of H. influenzae cells were not detected at any time point (Fig. 2).
FIG. 2.
Effect of sonication with and without glass beads on detection of bacteria by 23S rDNA-targeted PCR. Bacterial cell suspensions were treated by the standard proteinase K-phenol-ether DNA-extraction protocol, and 5 μl of the DNA was used in PCR. The lysis of bacteria was enhanced by sonication for 1 min (A and B), 5 min (C and D) or 10 min (E and F). The samples on the gels in panels A, C, and E were as follows: lanes 1 to 6, 200,000, 20,000, 2,000, 200, and 20 CFU of S. pyogenes in 200 μl of PBS and a negative isolation control with PBS only, respectively; lanes 7 to 12, replicates of the samples in lanes 1 to 6, respectively, with 0.3 g of zirconia beads in each sample. The samples on the gels in panels B, D, and F were as follows: lanes 1 to 6, 200,000, 20,000, 2,000, 200, and 20 CFU of H. influenzae in 200 μl of PBS and a negative control with PBS only, respectively; lanes 7 to 12, replicates of the samples in lanes 1 to 6, respectively, with 0.3 g of zirconia beads in each sample. Lanes +, positive control (50 ng of DNA from B. subtilis ATCC 6051); lanes −, negative reagent control (5 μl of a sterile UV-irradiated distilled water as a template). The lanes on both sides of each gel contain a molecular size marker (100-bp DNA ladder; Life Technologies).
Comparison of DNA isolation kits.
Table 1 shows the detection limits obtained for S. pyogenes, M. avium, and H. influenzae by use of standard phenol-ether extraction and various commercial DNA isolation kits alone and in combination with bead beating and sonication. Also, the prices of the kit reagents and plastic ware per reaction and the hands-on and hands-off times used for the extractions are shown. Among the commercial kits tested, the High Pure kit showed improved detection of S. pyogenes and M. avium after inclusion of the additional bead-beating or sonication steps in the protocol. The effect of bead beating was not tested in combination with the Masterpure, DNA-Pure Yeast, and Qiamp kits.
Clinical specimens.
On the basis of the results presented above and the ease of integration in the routine work flow, the Masterpure DNA purification kit without additional lysis steps and the High Pure PCR template purification kit with an additional sonication step were selected for comparison with the standard phenol-ether extraction method (which included no additional lysis steps). Sixty-nine consecutive specimens (12 tissue biopsy specimens [including 3 cardiac valve specimens], 15 synovial fluid specimens, 11 pleural fluid specimens, 10 cerebrospinal fluid specimens, 7 amniotic fluid specimens, 3 pericardial fluid specimens, 1 ascitic fluid specimen, and 10 pus samples) were treated by standard phenol-ether extraction and purified with the Masterpure kit. Of these, 48 samples were negative by the broad-range bacterial PCR after both DNA isolation protocols. Eleven samples were positive for bacterial DNA by both methods, two were positive by the standard method only, and one was positive only by the Masterpure kit (Table 2). A biopsy specimen from an aortic valve was inhibitory to the PCR after purification with the Masterpure kit, whereas streptococcal DNA was amplified from the phenol-purified half (Table 2, sample 12). A cerebrospinal fluid sample was inhibitory to the PCR after routine purification, as judged by the failure to detect human DNA also after 90 ng was spiked into the sample (7); the aliquot purified with the Masterpure kit was negative by bacterial PCR. Four samples purified with the Masterpure kit yielded weak bands in the PCR and were classified as contaminated on the basis of a weak band from the negative isolation control of the batch. All remained negative by PCR with the phenol-purified aliquot.
TABLE 2.
Comparison of sequencing results obtained with aliquots of clinical samples processed by the phenol-ether and Masterpure protocols with positive 23S rDNA PCR results
| Specimen no. | Specimen type | 23S rDNA PCR result for aliquot processed by:
|
Sequencing result for PCR product amplified from the aliquot processed by:
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Phenol-ether protocol
|
Masterpure protocol
|
||||||||
| Phenol-ether protocol | Masterpure protocol | Speciesa | % Similarityb | Overlap (bp) | Species | % Similarity | Overlap (bp) | ||
| 1 | Synovial fluid | + | + | Neisseria gonorrhoeae | 98.96 | 289 | N. gonorrhoeae | 98.76 | 159 |
| 2 | Synovial fluid | + | + | Staphylococcus sp. (S. aureus) | 98.21 | 493 | Staphylococcus sp. (S. aureus) | 99.07 | 212 |
| 3 | Pleural fluid | + | + | Streptococcus sp. (S. parauberis) | 99.26 | 268 | Streptococcus sp. (S. parauberis) | 98.50 | 263 |
| 4 | Pleural fluid | + | + | Streptococcus sp. (S. infantis) | 98.91 | 455 | Streptococcus sp. (S. pneumoniae) | 99.80 | 506c |
| 5d | Pleural fluid | + | + | Sequencing unsuccessfule | Listeria (S. monocytogenes/ L. innocua) | 99.28 | 274 | ||
| 6d | Pleural fluid | + | + | Listeria (L. innocua/ L. monocytogenes) | 99.45 | 181 | Sequencing unsuccessful | ||
| 7 | Amniotic fluid | + | + | Ureaplasma urealyticum | 98.88 | 177 | Sequencing unsuccessful | ||
| 8 | Abscess | + | + | Klebsiella pneumoniae | 95.82 | 275 | K. pneumoniae | 95.53 | 278 |
| 9 | Mitral valve | + | + | Staphylococcus sp. | 99.75 | 395 | Staphylococcus sp. | 93.02 | 172 |
| 10 | Synovial fluid | + | + | Staphylococcus sp. | 100.0 | 174 | Sequencing unsuccessful | ||
| 11 | Pus | + | + | Sequencing unsuccessful | Sequencing unsuccessful | ||||
| 12 | Aortic valve | + | Inhibitory | Streptococcus sp. (S. mitis/ S. oralis) | 99.30 | 857 | |||
| 13 | Biopsy | + | − | Sequencing unsuccessful | |||||
| 14 | Pericardial fluid | − | + | Sequencing unsuccessful | |||||
The probable species names are reported in parentheses.
The similarity comparisons are based on the bacterial 16S rDNA sequences available in the EMBL prokaryote database or the 23S rDNA sequences for those samples for which only 23S rDNA sequencing was successful (phenol-ether-purified aliquot of sample 6, and Masterpure protocol-purified aliquots of samples 2 and 9).
All samples except the Masterpure protocol-purified aliquot of sample 4 were sequenced by use of version 2.0 of the Big Dye Terminator Cycle Sequencing kit; the Masterpure protocol-purified aliquot of sample 4 was amplified by use of version 3.0 of the kit.
Samples 5 and 6 were from the same patient.
Sequencing failure was reported if the sequencing signals reported by the instrument were weak (average signal intensities, <100 relative light units for two or more dyes).
A total of 68 specimens (15 biopsy specimens, 9 synovial fluid specimens, 8 pleural fluid specimens, 9 cerebrospinal fluid specimens, 8 amniotic fluid specimens, 3 ascitic fluid specimens, and 16 pus samples from abscesses) were processed by the routine protocol and the High Pure protocol with the additional sonication step. Forty-eight samples remained negative for bacterial DNA after both DNA isolation procedures. Both aliquots of 11 samples were PCR positive with consistent sequencing results, 3 samples were PCR positive by the phenol purification protocol only, and 4 samples were positive by the High Pure purification protocol only (Table 3). A cerebrospinal fluid sample was inhibitory to the PCR after routine purification, and a pus sample was inhibitory after purification with the High Pure kit, with both samples being negative for bacterial DNA by PCR with the aliquot prepared by the other method.
TABLE 3.
Comparison of sequencing results obtained with aliquots of clinical samples processed by the phenol-ether and High Pure sonication protocols with positive 23S rDNA PCR results
| Specimen no. | Specimen type | 23S rDNA PCR result for aliquot processed by:
|
The sequencing result of the PCR product amplified from the aliquot processed by:
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Phenol-ether protocol
|
High Pure Protocol
|
||||||||
| Phenol-ether protocol | High Pure protocol | Speciesa | % Similarityb | Overlap (bp) | Species | % Similarity | Overlap (bp) | ||
| 1 | Pleural fluid | + | + | Streptococcus sp. (S. intermedius) | 100 | 480c | Streptococcus sp. (S. intermedius) | 100.0 | 500c |
| 2 | Cyst fluid | + | + | Streptococcus sp. (S. anginosus/ S. millen) | 99.41 | 338 | Streptococcus sp. (S. anginosus) | 100.0 | 515 |
| 3 | Amniotic fluid | + | + | Staphylococcus sp. (S. aureus) | 100 | 451 | Staphylococcus sp. (S. aureus) | 99.8 | 497c |
| 4 | Amniotic fluid | + | + | Campylobacter (C. jejuni) | 99.56 | 453 | Campylobacter (C. jejuni) | 100 | 534 |
| 5 | Liver biopsy | + | + | Mycobacterium (M. tuberculosis) | 98.4 | 307 | Mycobacterium (M. tuberculosis) | 100 | 530 |
| 6 | Abscess | + | + | Staphylococcus sp. (S. aureus) | 100 | 510 | Staphylococcus sp. (S. aureus) | 100 | 533 |
| 7 | Pus from valve | + | + | Staphylococcus sp. | 99.64 | 275 | Staphylococcus sp. | 99.81 | 532 |
| 8 | Pus | + | + | Staphylococcus sp. (S. aureus) | 100 | 533 | Staphylococcus sp. (S. aureus) | 100.0 | 574c |
| 9 | Pus | + | + | Staphylococcus sp. (S. caprae/ S. capitis/S. epidermidis) | 100 | 450c | Staphylococcus sp. (S. caprae/ S. capitis/S. epidermidis) | 99.8 | 497c |
| 10 | Brain abscess | + | + | Fusobacterium nucleatum | 98.67 | 444 | Fusobacterium nucleatum | 98.6 | 494 |
| 11 | Biopsy | + | + | Porphyromonas (P. levii), and othersd | 95 | 424 | Porphyromonas (P. levii), and othersd | 92.20 | 423 |
| 12 | Pleural fluid | + | − | Streptococcus sp. (S. peroris) | 100 | 520 | |||
| 13 | Synovial fluid | + | − | Staphylococcus sp. (S. warneri) | 100 | 878 | |||
| 14 | Mandibular bone | + | − | Sequencing unsuccessfule | |||||
| 15 | Pleural fluid | − | + | Streptococcus sp. (S. constellatus/ S. intermedius) | 98.14 | 370 | |||
| 16 | Biopsy | − | + | Haemophilus (H. paraprohemolyticus) | 99.81 | 527 | |||
| 17 | Vesicle (skin) | − | + | Streptococcus sp. (S. pyogenes) | 100.0 | 495c | |||
| 18 | Biopsy | − | + | Staphylococcus sp. (S. aureus) | 100.0 | 498c | |||
The probable species names are reported in parentheses.
The similarity comparisons are based on bacterial 16S rDNA sequences available in the EMBL prokaryote database, except for the High-Pure protocol-purified aliquot of sample 11, for which only 23S rDNA sequencing was successful.
The sample was sequenced by use of version 3.0 of the Big Dye Terminator Cycle Sequencing kit. The previous version (version 2.0) was used for the other samples.
The sample was interpreted to contain several bacteria, as the electropherogram showed strong signals but multiple overlapping peaks in some locations.
Sequencing failure was reported if the sequencing signals reported by the instrument were weak (average signal intensities, <100 relative light units for two or more dyes).
DISCUSSION
In the present work we demonstrate that molecular detection of bacteria with resistant cell walls in clinical samples can be enhanced by use of physical cell wall disintegration methods. Furthermore, the amplification of DNA derived from bacteria that are easily lysed is not compromised if the treatments are carefully optimized.
In the early diagnostic applications of PCR, classic phenol extraction was successfully used to prepare DNA for amplification. A major advantage of phenol is that it inactivates microbes very efficiently, including, e.g., spores of Bacillus anthracis, which are very resistant to inactivation by other methods (unpublished observations). Unfortunately, phenol is corrosive and toxic; and ether, which is used to remove it from the samples, is explosive. Although the reagents are inexpensive, the classic organic extraction is relatively laborious and unpractical for the processing of large numbers of samples that arrive in the laboratory at different times during the day. Residual phenol may also inhibit amplification of the extracted DNA. An array of commercially available DNA isolation systems has been developed to circumvent these drawbacks. Ideally, use of DNA isolation kits offers standardized, quality-controlled reagents with optimized compositions for all steps of the process. However, the kits are often designed for isolation of human DNA from human tissue or microbial DNA from cultivated cells rather than for detection of a minor amount of microbial DNA among an abundance of human DNA. Preliminary tests by use of several commercial DNA isolation kits failed to detect the relatively low numbers of bacterial cells in our experimental setting.
Hendolin et al. (2) demonstrated the problem of finding a DNA isolation procedure for clinical specimens that would produce DNA from both gram-positive and gram-negative bacteria with equal efficiencies. They analyzed middle ear effusions for the presence of H. influenzae, Streptococcus pneumoniae, Alloiococcus otitidis, and Moraxella catarrhalis by multiplex PCR after processing the samples either by the classic phenol-ether method or a modified Qiamp DNA extraction protocol with an additional boiling step in sodium dodecyl sulfate-NaOH-chaotropic salt. The phenol method yielded more positive PCR results (48 of 49 specimens; 98%) than the Qiamp method (20 of 24 specimens; 83%). Interestingly, the proportion of specimens positive for gram-negative bacteria (H. influenzae, M. catarrhalis) was significantly higher (P < 0.001) among phenol-extracted effusions, while the Qiagen protocol produced a higher proportion of samples positive for gram-positive organisms (S. pneumoniae and A. otitidis) with an equal statistical significance (P < 0.001).
The cell walls of gram-positive bacteria can be efficiently broken by use of the peptidoglycan-degrading enzymes lysozyme and mutanolysin. However, to minimize the number of reagents (and possible sources of bacterial DNA) in the PCR and to find a method equally efficient for cell walls of streptococci, staphylococci, and mycobacteria, we preferred the use of physical disintegration methods, i.e., bead beating and sonication. We aimed at determining a treatment time window that would enhance the release of DNA from difficult-to-lyse bacterial cells without severely compromising the detection of gram-negative organisms. A short bead beating was the most efficient lysis method for this purpose, but use of glass beads in combination with the membrane-formatted DNA purification methods proved difficult, as a small number of beads tended to end up in the purification column with the lysate and block the column. A sonication step was also integrated into the routine work flow more easily than bead beating. The sonicator used in these experiments is originally a washing sonicator; i.e., the sample tubes are placed in a water bath, which mediates the oscillation produced by the ultrasound crystal. As the tubes remain closed during and immediately after the sonication, there is no risk of cross-contamination.
The variability of clinical specimen types adds to the complexity of diagnostic bacterial PCR. This is reflected by the difficulty of extrapolating the experimentally determined detection limits to the true sensitivity in finding bacterial DNA in the clinical samples. The only DNA purification kit (Masterpure) which detected bacteria in lower numbers than the standard method without additional lysis treatments (Table 1) did not prove to be superior to the standard method when it was applied to clinical specimens. In comparison to the results obtained by the phenol-ether extraction procedure, the gram-positive bacteria in two samples remained unidentified, and those in one sample were not detected from the aliquot purified by the kit. Identifiable bacterial DNA was not detected after Masterpure purification in any sample that remained negative by the standard method. Modification of the downstream steps, i.e., amplification and sequencing reactions, might have improved the performance of this and some other kits, but these steps were not optimized for analysis of the template DNA produced by each purification method. According to our experience, amplification of the same template DNA by use of different enzymes and amplification systems may result in highly variable detection limits (6; unpublished observations). The Masterpure kit is inexpensive in comparison to the other kits tested, rapid, and applicable to an array of adequate sample types, including biological fluids, cells, and fresh and paraffin-embedded tissues. The contamination observed for four samples was probably cross-contamination. As the isolation control of this run was positive and the reagent control was negative, the contamination is likely to have occurred in the DNA isolation process. The run included a sample (Table 2, sample 7) with a very large amount of bacteria (as judged from the bands on the agarose gel), which could have been a source of cross-contamination. Unfortunately, sequencing was unsuccessful and insufficient quantities of the original samples were left for reextraction to verify this hypothesis. The isolation controls of the other runs remained negative.
The High Pure kit produced DNA suitable for our downstream applications, as the PCR and sequencing results were well in accord with those obtained by the routine method (Table 3). Four additional samples were found to be PCR positive by the High Pure method, three of which contained DNA of gram-positive organisms; among those were the important pathogens S. pyogenes and Staphylococcus aureus. Three samples were positive by the standard method but negative after purification by the High Pure method; Streptococcus peroris in a pleural fluid specimen and Staphylococcus warneri in a synovial fluid specimen would have remained undetected if only purification by the High Pure method had been used. By use of the standard method, bacterial DNA was also detected in a mandibular bone biopsy specimen, but sequencing of the PCR product failed. These findings indicate that no single method results in optimal recovery of all bacteria possibly present in the samples and that at least two different methods should be used, whenever possible.
The frequent detection of sequences related to the anginosus group of streptococci among the clinical specimens tested in the present study raised some concern about the validity of the findings. The results of bacterial cultures for these samples were clarified in order to rule out the possibility of PCR artifacts. Except for the pleural fluid specimen positive by the phenol purification method only (Table 3, sample 12), microaerophilic or viridans group streptococci had been isolated from the same specimens (Table 3, samples 1, 2, and 15). Thus, these streptococcal DNAs were most likely derived from the samples rather than reagents. Use of kit-form DNA isolation systems does not necessarily diminish the problem of reagent-derived bacterial DNA, as contamination of commercial purification columns with Legionella DNA was recently reported (10).
Regarding the removal of inhibitors, the three methods tested seemed equally efficient for the samples analyzed here. In a previous study we found the High Pure purification procedure to be superior to phenol-ether purification in preparing simulated sputum and bronchoalveolar lavage specimens for detection of Legionella pneumophila by PCR (6). These findings were associated with the possibility of efficient washing of DNA bound to the glass fiber filter. An additional buffer that removes inhibitors has been included in the new versions of the High Pure kit but was not used in this study.
In recent years, several instruments have been introduced for automated DNA extraction. We had the possibility to perform some isolations with the Magna Pure instrument. The preliminary results by use of the kit intended for bacteria were rather promising, and the process is applicable to an array of clinical specimens. However, the need for manual pretreatment steps, the relatively long time required to load the reagents on the instrument, and the high cost of the plastic ware that it uses make isolation with this instrument uneconomical if less than several tens of samples are processed daily.
The preanalytical phases are crucial for the success of any diagnostic laboratory method, and this is no less the case for tests applying molecular biology methods; but often, little attention is paid to their optimization in relation to the efforts made to polish other stages of the assays. Here we evaluated the use of physical lysis methods and commercial DNA purification kits to prepare DNA from clinical specimens for a broad-range bacterial PCR assay, which has previously proved to be a valid diagnostic test for the detection of bacterial DNA in normally sterile anatomical sites (7). We succeeded in finding a DNA isolation method which released and purified bacterial DNA, especially that from streptococci and staphylococci, at least as efficiently as the standard phenol-ether method and avoided the use of toxic and explosive components. Furthermore, sequencing of the PCR products revealed that the amplicons were derived from clinically relevant bacteria, and the results were well concordant with the results obtained by the validated standard method. However, our results also indicate that no single method is optimal for the detection of all the bacteria that might be present in tissues and biological fluids, and the use of more than one method is recommended, at least in research settings.
Acknowledgments
This work was supported by the University of Turku Foundation.
Matti Viljanen is acknowledged for useful comments on the manuscript. Kirsi Sundholm, Tiina Haarala, Kaisa Arvonen, Tarja Laine, and Merja Mikkola are thanked for excellent technical assistance; and Tuula Närä is thanked for processing of the figures.
REFERENCES
- 1.Corless, C. E., M. Guiver, R. Borrow, V. Edwards-Jones, E. B. Kaczmarski, and A. J. Fox. 2000. Contamination and sensitivity issues with a real-time universal 16S rRNA PCR. J. Clin. Microbiol. 38:1747-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hendolin, P. H., L. Paulin, and J. Ylikoski. 2000. Clinically applicable multiplex PCR for four middle ear pathogens. J. Clin. Microbiol. 38:125-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jalava, J., P. Kotilainen, S. Nikkari, M. Skurnik, E. Vanttinen, O. P. Lehtonen, E. Eerola, and P. Toivanen. 1995. Use of the polymerase chain reaction and DNA sequencing for detection of Bartonella quintana in the aortic valve of a patient with culture-negative infective endocarditis. Clin. Infect. Dis. 21:891-896. [DOI] [PubMed] [Google Scholar]
- 4.Kotilainen, P., J. Jalava, O. Meurman, O. P. Lehtonen, E. Rintala, O. P. Seppala, E. Eerola, and S. Nikkari. 1998. Diagnosis of meningococcal meningitis by broad-range bacterial PCR with cerebrospinal fluid. J. Clin. Microbiol. 36:2205-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rantakokko-Jalava, K., and J. Jalava. 2001. Development of conventional and real-time PCR assays for detection of Legionella DNA in respiratory specimens. J. Clin. Microbiol. 39:2904-2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rantakokko-Jalava, K., S. Nikkari, J. Jalava, E. Eerola, M. Skurnik, O. Meurman, O. Ruuskanen, A. Alanen, E. Kotilainen, P. Toivanen, and P. Kotilainen. 2000. Direct amplification of rRNA genes in diagnosis of bacterial infections. J. Clin. Microbiol. 38:32-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stoesser, G., W. Baker, A. van den Broek, E. Camon, M. Garcia-Pastor, C. Kanz, T. Kulikova, R. Leinonen, Q. Lin, V. Lombard, R. Lopez, N. Redaschi, P. Stoehr, M. A. Tuli, K. Tzouvara, and R. Vaughan. 2002. The EMBL nucleotide sequence database. Nucleic Acids Res. 30:21-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanner, M. A., B. M. Goebel, M. A. Dojka, and N. R. Pace. 1998. Specific ribosomal DNA sequences from diverse environmental settings correlate with experimental contaminants. Appl. Environ. Microbiol. 64:3110-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Der Zee, A., M. Peeters, C. de Jong, H. Verbakel, J. W. Crielaard, E. C. Claas, and K. E. Templeton. 2002. Qiagen DNA extraction kits for sample preparation for Legionella PCR are not suitable for diagnostic purposes. J. Clin. Microbiol. 40:1126.. [DOI] [PMC free article] [PubMed] [Google Scholar]