Abstract
Tuberculous lymphadenitis (TBLN) is a common form of extrapulmonary tuberculosis with multiple differential diagnoses. Demonstration of the etiologic agent by smear microscopy or culture of fine needle aspirate (FNA) specimens is often unsuccessful. FNA specimens from 40 patients presenting at a rural health center in South Ethiopia and diagnosed as positive for TBLN on the basis of clinical and cytological criteria were analyzed for mycobacterial DNA by PCR. Thirty (75%) had cervical lymphadenitis and 11 (27.5%) were seropositive for human immunodeficiency virus (HIV). Three primer sets were initially used to identify the causative agent at the genus (antigen 85 complex), complex (IS6110 insertion sequence), and species (pncA gene and allelic variation) levels. Among the forty TBLN cases, 35 (87.5%) were positive by PCR at the genus and complex levels. Based on PCR for detection of allelic variation at position 169, 24 (68.6%) of the 35 were positive for Mycobacterium tuberculosis and 6 (17.1%) were positive for M. bovis. These six were positive in additional PCR assays using the JB21-JB22 primer set, which is highly specific for M. bovis. Five (14.1%) showed amplification for both M. tuberculosis and M. bovis with the allele-specific primer set. Cooccurrence of pyrazinamide (PZA)-sensitive and -resistant M. tuberculosis in those five cases was indicated, since all were negative in assays with the JB21-JB22 primer set. This feature was seen in 3 of 11 HIV-positive and 2 of 29 HIV-negative individuals (P < 0.001). Conclusion: among 35 PCR-positive cases of TBLN from southern Ethiopia, 29 (82.9%) were caused by M. tuberculosis and six (17.1%) were caused by M. bovis.
In developing countries with a high incidence of tuberculosis, tuberculous lymphadenitis (TBLN) is one of the most frequent causes of lymphadenopathy (12) and it is the most common form of extrapulmonary tuberculosis (16). TBLN also occurs with increased frequency in human immunodeficiency virus type 1 (HIV-1)-infected individuals (9, 27). Within the Mycobacterium tuberculosis complex, M. tuberculosis and M. bovis are the most common causative agents of TBLN. Which control measures and treatment are to be instituted depends on the most common causative agent in the area. Therefore, species identification is of paramount importance.
Over the past decades, fine needle aspirate (FNA) cytology, an alternative procedure less invasive than excision biopsy, has assumed an important role in the diagnosis of peripheral lymphadenopathy. The cytological criteria for diagnosis of TBLN have been clearly defined (13, 19). However, the amount of material obtained in FNA is usually so small that it is often inadequate for performance of acid-fast smear and culture examinations with reasonable sensitivity.
Introduction of the PCR provided new possibilities for identification of mycobacteria in various types of clinical samples (3, 5); based on the amplification of common sequences, the identification time for detection of mycobacteria in clinical specimens was reduced (4, 8). The M. tuberculosis complex, consisting of M. tuberculosis, M. bovis, M. africanum, and M. microti, has been identified with a number of different targets, including the IS6110 insertion element sequence, using PCR methods (2, 6).
In the present study, PCR was applied for assays of FNAs to identify the causative agent in TBLN in Ethiopia at the genus, complex, and species levels.
MATERIALS AND METHODS
Patients.
The study was initiated after approval by the institutional and national ethical committees. FNAs which were collected between January 2000 and May 2001 at the Butajira health center in southeastern Ethiopia from 72 consecutive patients with a clinical diagnosis of TBLN were studied. Of these, 40 cases of TBLN, which were confirmed on the basis of combined clinical and cytological criteria, were studied further for species identification of the causative agent. FNA was collected from swollen lymph nodes of each subject by using a 21-gauge needle under sterile conditions and was split in two different portions. One part was used for preparation of smears for Ziehl-Neelsen staining for acid-fast bacilli and for hematoxylin and eosin staining for cytological analysis. The rest was stored at −20°C until used for DNA extraction. A clinical diagnosis of TBLN was considered to be appropriate in cases in which the patient had chronic enlarged nontender lymph nodes and lack of response to a 2-week course of broad-spectrum antibiotics. The duration and kind of clinical symptoms and the location and size of affected lymph nodes were recorded on a questionnaire for each patient.
Cytological criteria and demonstration of acid-fast bacteria.
FNAs were considered diagnostic of TBLN in cases in which they contained thick yellowish material showing any combination of the following cytological criteria: existence of a necrotic background associated with the presence of lymphohistiocytic cells and absence of a significant polymorphonuclear cell population; presence of elements of a granulomatous inflammatory reaction consisting of giant cells and/or epithelioid cell clusters and a lymphohistiocytic cell population; and positivity for acid-fast bacilli on Ziehl-Neelsen staining, regardless of the inflammatory cell population.
HIV serology test.
All patients were screened for HIV by using a commercial micro-enzyme-linked immunosorbent assay kit (Vironostika HIV Uni-Form 11 PlusO; Organon Teknika GmbH, Eppelheim, The Netherlands) and following the manufacturer's instructions. Tests were repeated for all cases for confirmation.
DNA extraction.
Mycobacterial genomic DNA was extracted as previously described (32), with minor modifications. Briefly, 200 μl of the FNA material was incubated in a water bath at 80°C for 20 min to inactivate the bacteria and was diluted with 500 μl of Tris-EDTA buffer. The bacteria were thereafter lysed with 50 μl of 10 mg of lysozyme (Sigma, Saint Louis, Mo.)/ml and vortexed before incubation for 1 h at 37°C. The lysozyme-treated samples were incubated at 65°C for 10 min in the presence of 10 mg of proteinase K (GIBCO/BRL-Life Technologies, Gaithersburg, Md.)/ml-10% sodium dodecyl sulfate (Sigma). A 5 M solution of sodium chloride-cetyltrimethylammonium bromide was added to the sample, and phenol-chloroform-isoamyl alcohol (25:24:1) extraction was performed. The DNA precipitate was obtained by adding 0.6 vol of isopropanol to the aqueous phase. After storage for 1 h at −20°C, the DNA was collected by centrifugation at 12,000 rpm (Centrifuge 5415; H. Jurgens & Co., Berman, Germany) for 15 min, washed with 70% ethanol, and resuspended in 30 μl of distilled water. Finally, the sample was treated with a 0.2-mg/ml final concentration of RNase (Sigma) and incubated for 1 h at 37°C before storage at −20°C until the PCR assay was performed.
PCR.
A multiprimer PCR system was used to amplify different targets in the mycobacterial DNA (4). All primers were obtained from TIB Molbiol Syntheselabor, Berlin, Germany, and the reactions were carried out in a Hybaid PCR machine (Omnigene, Teddington, Middlesex, United Kingdom). MT1 (5′-TTCCTGACCAGCGAGCTGCCG-3′) and MT2 (5′-CCCCAGTACTCCCAGCTGTGC-3′) primers (4) were used to amplify equally sized targets of 85a (Rv3804c) and 85b (Rv1886c) genes. No reaction product of the 85c (Rv0129c) gene was obtained with these primers. Amplification of the IS6110 gene was carried out with the IS5 (5′-CGGAGACGGTGCCTAAGTGG-3′) and IS6 (5′-GATGGACCGCCAGGGCTTGC-3′) primers (4).
The PCR mix was prepared in a final volume of 25 μl, using Ready-To-Go beads (Amersham Pharmacia Biotech, Freiburg, Germany). The amplification program for the 85a, 85b, and IS6110 genes included 35 cycles of 94°C for 1 min for denaturation, 71°C for 1.5 min for annealing, and 72°C for 2 min for extension and a final incubation at 72°C for 10 min, with both sets of primers added into the tubes.
An allele-specific PCR system was used for the amplification of the pncA (Rv2043c) gene (8). PncATB-1.2 (5′-ATGCGGGCGTTGATCATCGTC-3′) was used as a forward primer. PncAMT-2 (5′-CGGTGTGCCGGAGAAGCGG-3′) and PncAMB-2 (5′-CGGTGTGCCGGAGAAGCCG-3′) were used as reverse primers. All reactions were performed with the same forward primer and one of the two discriminating reverse primers. The program for the amplification of this gene included 1 cycle of 95°C for 2 min and 30 cycles of 94°C for 1 min, 67°C for 1 min, and 72°C for 1 min.
Using the JB21 (5′-TCGTCCGCTGATGCAAGTGC-3′) and JB22 (5′-CGTCCGCTGACCTCAAGAAG-3′) primers to amplify a 500-bp fragment, an M. bovis-specific PCR assay was carried out to reconfirm the M. bovis-specific PCR results obtained with the PncAMB-2 differential primer. The amplification was done as described previously (24), with cycling conditions of 5 min of denaturation at 94°C and 30 cycles of 94°C for 1 min, 68°C for 1 min, and 72°C for 1 min.
Standardization of the PCR technique and further speciation of mycobacteria.
Using DNA from the reference strain of M. tuberculosis, serial 10-fold dilutions, ranging between 100 ng and 10 fg, were prepared, and PCR was done using multiple primers, including the genus- and complex-specific primers (primer pair MT1 and MT2 and primer pair IS5 and IS6, respectively). The analytical sensitivity of the PCR system was determined by calculating the lowest concentration of DNA that could be amplified and detected. For each sample, original and 1:10-diluted DNA extracts were amplified by PCR to control for false-negative results due to the presence of inhibitors. DNA extracts from cultures of M. tuberculosis (ATCC 35836) and M. bovis (RIVM 12716) isolates from the National Institute of Public Health and Environmental Protection (Bilthoven, the Netherlands) were used as positive controls in each assay. Extract of DNA from an aspirate known to be negative for mycobacterial culture and repeated PCR was used as the internal negative control. Moreover, double-distilled water, instead of template material, was run in parallel in each assay to control for contamination.
Analysis of the amplification products.
Amplified PCR products were analyzed by electrophoresis in 1.5 and 2% agarose gel (Sigma) containing 0.5 ng of ethidium bromide/ml (26). The results were considered positive when a clearly defined DNA band was observed in the agarose gel at the expected position in comparison with those of the molecular weight marker (123 DNA ladder and DNA EcoRI and HindIII digests; Sigma) and a positive control of M. tuberculosis and/or M. bovis DNA.
RESULTS
In this study of 72 consecutive patients with a clinical diagnosis of TBLN, 40 (55.6%) were confirmed TBLN cases, based on combined clinical and cytological criteria. Of the 40 patients, 24 (60%) were male. The mean age was 24.2 years (range, 15 to 60). Eleven (27.5%) of the 40 patients were seropositive for HIV. The most frequently aspirated lymph nodes included cervical (75%), supraclavicular, submandibular, and axillary nodes.
In a separate clinical study of 146 patients suspected of TBLN on the basis of clinically assessed indications, 96 (65%) of the patients were confirmed as TBLN positive by FNA cytology and acid-fast smear examination. FNA cytology revealed that 47 (94%) of the remaining 50 non-TBLN (NTBLN) patients had pyogenic lymphadenitis. For the TBLN and NTBLN patients, the clinical manifestations and percentages of HIV positivity were similar, with 23 of 96 (24%) and 10 of 50 (20%) patients testing as positive, respectively (P > 0.5) (M. A. Yassin and D. Kidane, unpublished observations). Thus, we have no indication that the 32 PCR-negative patients had an atypical presentation or cytology because they were HIV infected.
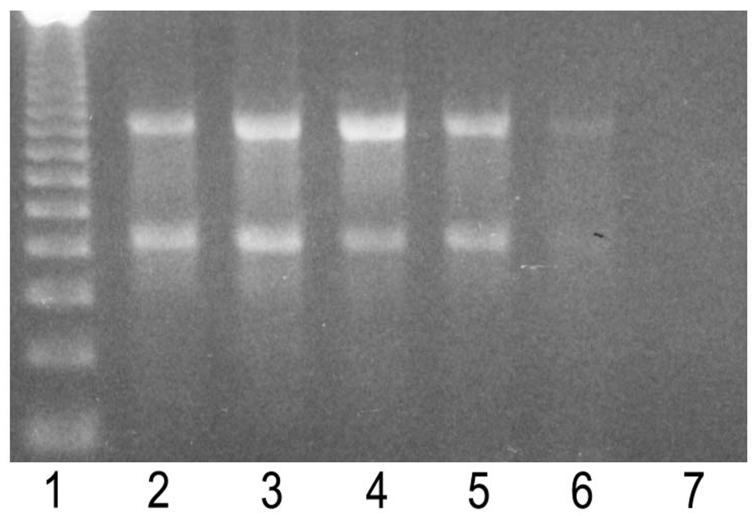
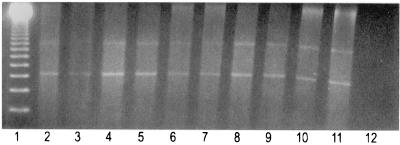
The analytical sensitivity of the multiplex PCR (with primer pair MT1 and MT2 and primer pair IS5 and IS6) was determined using serial dilutions of DNA from reference strains of M. tuberculosis, as shown in Fig. 1. The PCR detected the presence of chromosomal DNA at levels as low as 10 pg. Figure 2 shows the results with clinical samples. Repeat PCR runs on the same samples for control, as described in Material and Methods for species identification, resulted in three more samples being positive on the second run, in addition to confirmation of all previously positive results. Each sample was also tested in duplicate with the original extract and a 1:10 dilution to avoid false-negative results due to inhibitors. Four clinical samples became positive after dilution. On the basis of combined clinical and cytological criteria, five of the confirmed TBLN cases remained negative by PCR. The nature and amount of available FNA material may account for insufficient sensitivity in the PCR assay. Repeat samples were not available for testing.
FIG. 1.
Analytic sensitivity of the multiprimer PCR system for M. tuberculosis DNA. Lane 1, 123 DNA ladder molecular weight markers; lane 2, 100-ng amplification of the 85a, 85b (506 bp), and IS6110 (984 bp) insertion element genes; lane 3, 10 ng; lane 4, 1 ng; lane 5, 100 pg; lane 6, 10 pg; lane 7, 1 pg.
FIG. 2.
Electrophoretic pattern of the amplified 85a, 85b, and IS6110 insertion element genes from clinical samples. Lane 1, 123 DNA ladder molecular weight markers; lanes 2 to 10, clinical samples; lane 11, positive control; lane 12, negative control.
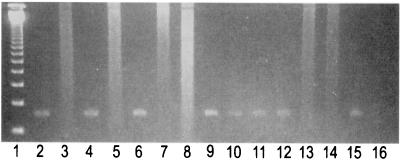
Figure 3 shows that FNA samples containing M. tuberculosis and M. bovis could be distinguished by PCR with the allele-specific primers PncAMT-2 and PncAMB-2, respectively.
FIG. 3.
Electrophoretic pattern of allelic PCR system amplified product of pncA genes (185 bp) from clinical samples in 1.5% agarose gel. Lane 1, 123 DNA ladder molecular weight markers; lanes 2 to 7, clinical samples positive for M. tuberculosis and negative for M. bovis; lanes 8 and 9, clinical sample negative for M. tuberculosis and positive for M. bovis, respectively; lanes 10 and 11, coamplified pncA positive for M. tuberculosis PZA-sensitive and -resistant strains, respectively; lanes 12 and 13, M. tuberculosis reference strain DNA tested for the pncA genes of M. tuberculosis and M. bovis, respectively (positive control); lanes 14 and 15, M. bovis reference strain DNA tested for the pncA genes of M. bovis and M. tuberculosis, respectively (positive control); lane 16, negative control.
Table 1 shows PCR results for aspirates from different sites. Aspirates from 35 of the 40 (87.5%) patients returned positive results with either the M. tuberculosis- or M. bovis-specific PCR system. Of the aspirate samples, 24 (60%) were positive for M. tuberculosis, 6 (15%) were positive for M. bovis, and 5 (12.5%) returned results that were suggestive of coamplification. Table 2 shows that the overall PCR results did not differ for the HIV-negative and HIV-positive patients (86.2 versus 90.9% positive, respectively). However, results suggestive of coamplification of M. tuberculosis and M. bovis were more common for HIV-positive patients (3 of 11 [27.3%]) than for HIV-negative patients (2 of 29 [6.9%]). Although the number of patients with coamplification of the PCR signals was low, it was noted that the difference in results for HIV-positive and HIV-negative patients was statistically significant (P < 0.001).
TABLE 1.
Lymph node involvement and initial speciation of mycobacteria by PCR with primers for the 85a, 85b, and pncA genes in FNAs from 40 patients with a combined clinical and cytological diagnosis of TBLN
| Site (no. of patients) | No. (%) of patients whose isolates were positive by PCR amplification for:
|
||
|---|---|---|---|
| M. tuberculosis | M. bovis | Coamplified M. tuberculosis and M. bovis | |
| Cervical (30) | 20 (66.6) | 4 (13.3) | 3 (10) |
| Others (10)a | 4 (40) | 2 (20) | 2 (20) |
| Total (40) | 24 (60) | 6 (15) | 5 (12.5) |
Supraclavicular, submandibular, or axillary lymph nodes.
TABLE 2.
HIV infection and its association with mycobacterial species as indicated by PCR with primers for the 85a, 85b, and pncA genes
| HIV status (no. of patients) | No. (%) of patients whose isolates were positive by PCR amplification for:
|
|||
|---|---|---|---|---|
| M. tuberculosis | M. bovis | Coamplified M. tuberculosis and M. bovis | Total | |
| Negative (29) | 18 (62.1) | 5 (17) | 2 (6.9) | 25 (86.2) |
| Positive (11) | 6 (54.5) | 1 (9) | 3 (27.3) | 10 (90.9) |
Additional PCR assays with the JB21-JB22 primer set, which was highly specific for M. bovis (24), were done for the five cases that exhibited coamplification and the six cases that were positive for the M. bovis pncA gene. Control samples of M. tuberculosis and M. bovis were negative and positive, respectively, while all five coamplification samples were negative, suggesting that the coamplified products did not have M. bovis DNA. However, the six cases that had been positive for the M. bovis pncA gene again gave positive results with the JB21-JB22 primer set.
DISCUSSION
Extrapulmonary tuberculosis is often difficult to diagnose because of its diverse clinical presentations. In a significant proportion of clinical samples, low numbers and slow growth rates of bacilli limit detection by conventional techniques such as acid-fast staining and bacterial culture (28). For identification of microorganisms at the species level, molecular methods are now extensively used for diagnosis and control of infectious diseases. This approach is fast, sensitive, and of high specificity.
In the present study, we used the PCR technique to identify the causative agent in TBLN. Positive signals at Mycobacterium genus and M. tuberculosis complex levels were obtained in FNAs from 35 (87.5%) of 40 patients with a clinical and cytological diagnosis of TBLN. These results are similar to those presented in other reports, including a report presenting findings in which 83% detection was achieved for lymph node aspirates from 23 patients for whom the cytological diagnosis was consistent with that of TBLN (30), based on amplification of regions in the IS6110 insertion sequence which is present in multiple copies in most strains of M. tuberculosis (31). In a more recent study of TBLN, 55% of the tested cases were PCR positive, based on amplification of the devR gene of M. tuberculosis (28). These primers are from the gene now denoted Rv3133c.
M. tuberculosis, M. bovis, M. africanum, and nontuberculous mycobacteria all induce lymphadenitis, but lymphadenitis due to infection with the M. tuberculosis complex is more chronic in nature, while NTBLN often has a more rapid course (29). However, all the above conditions can be influenced by geographical and ethnic variations. For example, the occurrence of TBLN has been reported to vary considerably, depending on ethnicity, geographical location, age, and gender (7, 17). The group of subjects from the Butajira district included members of a variety of ethnic groups.
Changing trends have been observed for the infectious agent in lymphadenitis, firstly, regarding the relative importance of the M. tuberculosis complex compared with that of other mycobacterial species and, secondly, regarding M. tuberculosis compared with M. bovis.
Changes of the first kind are typical of industrialized countries and are well documented in southeast England (33). From 1980 to 1989, cultures were received from 1,817 patients with mycobacterial lymphadenitis. Of these, 1,677 (92.3%) of the cultures were identified as harboring M. tuberculosis, 25 were M. bovis, 21 were M. africanum, and 94 were “other environmental species.” In comparison with the findings of a survey in the same region in 1973 to 1980, there was a 20% increase in the number of cases due to environmental mycobacteria. In Australia (20) and British Columbia (23), atypical mycobacteria were detected 10 times more frequently than M. tuberculosis mycobacteria in lymphadenitis cases.
In earlier studies from other countries, M. bovis was often considered to be the most common cause of lymph node tuberculosis in humans, mainly acquired through drinking raw milk from tuberculous cattle (10). However, in the United States, by 1910 M. tuberculosis was identified as the cause of 70% of mycobacterial cervical lymphadenitis in children (21), and in 1951, M. tuberculosis was found to be responsible for all except a few cases of tuberculous lymph node disease in adults as well (18). In general, declining rates of M. bovis isolation from human tuberculosis patients have been associated with milk pasteurization and cattle inspection programs in industrialized countries (11, 15).
Changes in the pattern and prevalence of tuberculosis strikingly influence the etiology of mycobacterial lymphadenitis. When mycobacterial infection is highly prevalent, M. tuberculosis complex organisms are the almost exclusive cause of cases of mycobacterial lymphadenitis, the majority being caused by M. tuberculosis (17, 29).
Fine needle aspiration has acquired a prominent place in the diagnostic work on lymphadenitis in developing countries (14). Cytology is more sensitive than culture. PCR is extensively applied. In general, the primers used are designed for demonstration of the presence of the M. tuberculosis complex and very rarely for distinguishing between M. tuberculosis and M. bovis. In the present study, the primers were designed for the latter purpose. M. tuberculosis was found to be the causative agent in most patients, and M. bovis was found to be the causative agent in 6 of the 35 PCR-positive cases.
In the five patients with coamplification of two pncA genes, suggesting a mixed M. tuberculous and M. bovis infection, further investigation to distinguish between coinfection and simultaneous occurrence of pyrazinamide (PZA)-sensitive and -resistant M. tuberculosis was conducted, using the JB21 and JB22 primers to amplify a 500-bp fragment highly specific for M. bovis (24, 25). All of the five samples were negative for the 500-bp fragment of M. bovis, indicating that the coamplification was most likely due to simultaneous occurrence of PZA-sensitive and -resistant M. tuberculosis. It is particularly interesting that this coamplification occurred more frequently in HIV-positive than HIV-negative patients (3 of 11 [27.3%] and 2 of 29 [6.9%], respectively) (P < 0.001). This result is probably related to the occurrence of larger bacillary numbers in HIV-positive than HIV-negative individuals with tuberculous disease. However, the six cases positive for the M. bovis pncA gene alone were positive with the JB21-JB22 primer set, confirming that the infection was due to M. bovis.
Given the rising frequency of HIV infection and its impact on tuberculosis, identification of the etiological agent of TBLN in the HIV-positive host gains particular significance. Coinfection with HIV may influence several clinical and laboratory features among patients with TBLN (22). In this study, TBLN was detected by PCR in 90% of HIV-infected and in 86% of non-HIV-infected subjects. The proportions of TBLN among HIV-infected and non-HIV-infected patients were also found to be similar. This finding might suggest a high rate of mycobacterial transmission in the community. It might also imply that factors other than HIV infection may be more likely to be responsible for the occurrence of TBLN in the population.
This study shows that PCR is a powerful tool for the diagnosis of TBLN at the species level, provided that appropriate steps are followed to recognize and overcome pitfalls such as contamination, low target amplification, and inhibition. We tested all samples at the original dilution and at a 1:10 dilution of the DNA extracts. Four samples which tested as negative in the original extract tested as positive after 1:10 dilution. This shows that inhibition caused by interfering substances in a patient sample may induce false-negative results.
Thus, PCR is a powerful technique for diagnosis and speciation of mycobacterial infection in FNAs from patients with cervical lymphadenopathy. It has been recommended that all patients with suspected TBLN in Africa undergo fine needle aspiration before surgical biopsy or empirical treatment (1).
In low-income, developing countries like Ethiopia, FNA samples can be taken at peripheral health centers and collected for further cytological analysis and PCR in a central laboratory for species identification of the causative agent, thus providing information of importance for treatment and control purposes.
Acknowledgments
This work was supported by the Norwegian Agency for Development Cooperation (NORAD) and the Swedish International Development Cooperation Agency (Sida) through the core budget of the Armauer Hansen Research Institute.
We thank Haimanot Gebrexabher for providing the reference clinical isolates, Mulu Geletu and Azeb Tadesse W/G for technical assistance, Alemayehu Kifle for collecting clinical samples, and Harald G. Wiker for bioinformatic analysis of primers and for preparing the figures for publication.
REFERENCES
- 1.Bem, C., P. S. Patil, A. M. Elliott, K. M. Namaambo, H. Bharucha, and J. D. Porter. 1993. The value of wide-needle aspiration in the diagnosis of tuberculous lymphadenitis in Africa. AIDS 7:1221-1225. [DOI] [PubMed] [Google Scholar]
- 2.Brisson-Noel, A., B. Gicquell, D. Lecossier, V. Levy-Frebault, X. Nassif, and A. J. Hance. 1989. Rapid diagnosis of tuberculosis by amplification of mycobacterial DNA in clinical samples. Lancet ii:1069-1071. [DOI] [PubMed] [Google Scholar]
- 3.Buck, G. E., L. C. O'Hara, and J. T. Summersgill. 1992. Rapid, simple method for treating clinical specimens containing Mycobacterium tuberculosis to remove DNA for polymerase chain reaction. J. Clin. Microbiol. 30:1331-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Del Portillo, P., M. C. Thomas, E. Martinez, C. Maranon, B. Valladares, M. E. Patarroyo, and M. Carlos Lopez. 1996. Multiprimer PCR system for differential identification of mycobacteria in clinical samples. J. Clin. Microbiol. 34:324-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eisenach, K. D., M. D. Cave, J. H. Bates, and J. T. Crawford. 1990. Polymerase chain reaction amplification of a repetitive DNA sequence specific for Mycobacterium tuberculosis. J. Infect. Dis. 161:977-981. [DOI] [PubMed] [Google Scholar]
- 6.Eisenach, K. D., M. D. Sifford, M. D. Cave, J. H. Bates, and J. T. Crawford. 1991. Detection of Mycobacterium tuberculosis in sputum samples using a polymerase chain reaction. Am. Rev. Respir. Dis. 144:1160-1163. [DOI] [PubMed] [Google Scholar]
- 7.Enarson, D., M. J. Ashley, and S. Grzybowski. 1979. Tuberculosis in immigrants to Canada. Am. Rev. Respir. Dis. 119:11-18. [DOI] [PubMed] [Google Scholar]
- 8.Espinosa de los Monteros, L. E., J. C. Galan, M. Gutierrez, S. Samper, J. F. Garcia Marin, C. Martin, L. Dominguez, L. de Rafael, F. Baquero, E. Gomez-Mampaso, and J. Blazquez. 1998. Allele-specific PCR method based on pncA and oxyR sequences for distinguishing Mycobacterium bovis from Mycobacterium tuberculosis: intraspecific M. bovis pncA sequence polymorphism. J. Clin. Microbiol. 36:239-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finfer, M., A. Perchick, and D. E. Burstein. 1991. Fine needle aspiration biopsy diagnosis of tuberculous lymphadenitis in patients with and without the acquired immune deficiency syndrome. Acta Cytol. 35:325-332. [PubMed] [Google Scholar]
- 10.Giffiths, A. S. 1937. Bovine tuberculosis in man. Tubercle 18:529. [Google Scholar]
- 11.Grange, J. M., and C. H. Collins. 1987. Bovine tubercle bacilli and disease in animals and man. Epidemiol. Infect. 92:221-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupta, A. K., M. Nayar, and M. Chandra. 1992. Critical appraisal of fine needle aspiration cytology in tuberculous lymphadenitis. Acta Cytol. 36:391-394. [PubMed] [Google Scholar]
- 13.Gupta, S. K., T. D. Chugh, Z. A. Sheikh, and N. A. al-Rubah. 1993. Cytodiagnosis of tuberculous lymphadenitis: a correlative study with microbiological examination. Acta Cytol. 37:329-332. [PubMed] [Google Scholar]
- 14.Handa, U., A. Palta, H. Mohan, and R. P. Punia. 2002. Fine needle aspiration diagnosis of tuberculous lymphadenitis. Trop. Dr. 32:147-149. [DOI] [PubMed] [Google Scholar]
- 15.Karlson, A. G., and D. T. Carr. 1970. Tuberculosis caused by Mycobacterium bovis. Ann. Intern. Med. 73:979-983. [DOI] [PubMed] [Google Scholar]
- 16.Krishnaswami, H., G. Koshi, K. G. Kulkarni, and C. K. Job. 1972. Tuberculous lymphadenitis in south India, a histological and bacteriological study. Tubercle 53:215-220. [DOI] [PubMed] [Google Scholar]
- 17.Lai, K. K., K. D. Stottmeier, I. H. Sherman, and W. R. McCabe. 1984. Mycobacterial cervical lymphadenopathy. Relation of etiologic agents to age. JAMA 251:1286-1288. [DOI] [PubMed] [Google Scholar]
- 18.Laster, C. W. 1951. Lymph node tuberculosis and treatment of accessible nodes. Am. Rev. Respir. Dis. 64:691-694. [DOI] [PubMed] [Google Scholar]
- 19.Lau, S. K., W. I. Wei, C. Hsu, and U. C. Engzell. 1990. Efficacy of fine needle aspiration cytology in the diagnosis of tuberculosis cervical lymphadenopathy. J. Laryngol. Otol. 104:24-27. [DOI] [PubMed] [Google Scholar]
- 20.Llewelyn, D. M., and D. Dorman. 1971. Mycobacterial lymphadenitis. Aust. Paediatr. J. 7:97-102. [PubMed] [Google Scholar]
- 21.Park, W. H., and C. Krumwiede, Jr. 1911. The relative importance of the bovine and human types of tubercle bacilli in the different forms of human tuberculosis. J. Med. Res. 25:313-333. [PMC free article] [PubMed] [Google Scholar]
- 22.Perenboom, M., C. Richter, A. B. Swai, J. Kitinya, I. Mtoni, H. Chande, and R. R. Kazema. 1995. Clinical features of HIV seropositive and HIV seronegative patients with tuberculous lymphadenitis in Dar es Salaam. Tuberc. Lung Dis. 76:401-406. [DOI] [PubMed] [Google Scholar]
- 23.Roba-Kiewicz, M., and S. Grzybowski. 1974. Epidemologic aspects of nontuberculous mycobacterial diseases and tuberculosis in British Columbia. Am. Rev. Respir. Dis. 109:613-620. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez, J. G., J. C. Fissanoti, P. Del Portillo, M. E. Patarroyo, M. I. Romano, and A. Cataldi. 1999. Amplification of a 500-base-pair fragment from cultured isolates of Mycobacterium bovis. J. Clin. Microbiol. 37:2330-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodriguez, J. G., G. A. Mejia, P. Del Portillo, M. E. Patarroyo, and L. A. Murillo. 1995. Species-specific identification of Mycobacterium bovis by PCR. Microbiology 141:2131-2138. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Shafer, R. W., D. S. Kim, J. P. Weiss, and J. M. Quale. 1991. Extrapulmonary tuberculosis in patients with human immunodeficiency virus infection. Medicine (Baltimore) 70:384-397. [DOI] [PubMed] [Google Scholar]
- 28.Singh, K. K., M. Muralidhar, A. Kumar, T. K. Chattopadhyaya, K. Kapila, M. K. Singh, S. K. Sharma., N. K Jain, and J. S. Tyagi. 2000. Comparison of in house polymerase chain reaction with conventional techniques for the detection of Mycobacterium tuberculosis DNA in granulomatous lymphadenopathy. J. Clin. Pathol. 53:355-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sloane, M. F. 1996. Mycobacterial lymphadenitis, pp. 577-583. In W.N. Rom and S. Garay (ed.), Tuberculosis. Little, Brown and Company, Boston, Mass.
- 30.Thierry, D., M. D. Cave, K. D. Eisenach, J. T. Crawford, J. H. Bates, B. Gicquel, and J. L. Guesdon. 1990. IS6110, an IS-like element of Mycobacterium tuberculosis complex. Nucleic Acids Res. 18:188.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Soolingen, D. 2001. Molecular epidemiology of tuberculosis and other mycobacterial infections: main methodologies and achievements. J. Intern. Med. 249:1-26. [DOI] [PubMed] [Google Scholar]
- 32.Van Sooligen, D., P. E. M. Hermans, P. E. W. de Haas, and J. D. A. van Embden. 1995. RFLP analysis of mycobacteria. Manual for fingerprinting of M. tuberculosis strains. National Institute of Public Health and Environmental Protection, Bilthoven, the Netherlands.
- 33.Yates, M. D., and J. M. Grange. 1992. Bacteriological survey of tuberculous lymphadenitis in southeast England, 1981-1989. J. Epidemiol. Community Health 46:332-335. [DOI] [PMC free article] [PubMed] [Google Scholar]