Abstract
In order to understand genetic polymorphisms among Entamoeba histolytica strains in a limited geographic area and among restricted social populations, we studied nucleotide polymorphism in DNA regions that do not encode proteins (locus 1-2 and locus 5-6) and in genes coding for chitinase and for serine-rich E. histolytica protein. Thirty E. histolytica isolates from domestically infected Japanese amebiasis patients (male homosexuals and residents in institutions for the mentally handicapped) and four reference strains were examined. PCR revealed remarkable polymorphisms in both the number and size of the PCR fragments containing these loci. Polymorphisms in lengths, types, and numbers of internal repeat units were observed in locus 1-2 and the repeat-containing region of serine-rich E. histolytica protein among the Japanese isolates. In contrast, polymorphism at locus 5-6 was observed almost exclusively in the number of repeats of a 16-nucleotide unit. The repeat-containing region of chitinase appeared to be the least polymorphic among the four loci with a single dominant genotype representing 66% (20 out of 30) of all of the isolates. Isolates obtained from male homosexuals showed a more complex genetic polymorphism than those from residents in institutions. Considering all four polymorphic loci together, all 19 Japanese isolates from male homosexuals were distinct. In contrast, all isolates obtained from mass-infection cases at a single institution had an identical genotype, suggesting that these cases were caused by a single E. histolytica strain. No significant correlation was found between genotypes and zymodemes or between genotypes and clinical presentations, e.g., colitis or liver abscess. Certain genotypes were observed with higher frequencies in male homosexuals or residents of institutions. These data indicate that genotyping of the E. histolytica isolates by using these four polymorphic loci could serve as a tool to fingerprint individual isolates. We propose that genotyping of ameba isolates should help to determine geographic origins of isolates and routes of transmission.
The protozoan parasite Entamoeba histolytica causes an estimated 50 million cases of amebiasis and 40,000 to 100,000 deaths annually, placing it second only to malaria as a cause of death resulting from parasitic protozoa (33). Since the first description of amebiasis in 1878 by Lösch (17), we still do not have a proper answer to the question of why disease and symptoms develop in only 5 to 10% of those infected with E. histolytica. It has been speculated that a spectrum of virulence levels among the E. histolytica strains and variability in the host immune response against amebic invasion contribute to the outcome of amebic infection. While variation in human immune responses against amebic infection is not understood, the polymorphic structure of E. histolytica has recently been unveiled (4, 7, 12, 28, 34). These studies have identified and characterized polymorphic DNA loci, including protein-coding sequences, such as those for the serine-rich E. histolytica protein (SREHP) (18) (also described as K2 [16]) and chitinase (12), as well as non-protein-coding regions, including the rRNA genes (4, 28), a strain-specific transcript (6), and loci 1-2 and 5-6 (34). These polymorphic loci have been shown to be potentially useful in investigating the molecular epidemiology of amebiasis. Ghosh et al. (12) and Zaki and Clark (34) showed significant polymorphism among E. histolytica isolates collected from a wide geographic range, e.g., Mexico, Bangladesh, India, Venezuela, and South Africa. However, whether or not genetic polymorphism also exists in an E. histolytica population in a restricted geographic location and, if it does, how extensive the polymorphism is remain unknown.
In contrast to the situation in countries where amebiasis is endemic, where transmission of amebas frequently takes place across wide geographic areas and social populations, amebiasis in Japan is prevalent only in limited social populations, i.e., male homosexuals (21, 22, 32) and institutionalized people, such as residents of institutions for the mentally handicapped (1, 14, 20). The high prevalence of E. histolytica infections in male homosexuals is a unique characteristic of amebiasis in Japan and is in sharp contrast to the dominant Entamoeba dispar infections in male homosexuals in western countries (2, 13, 27). In addition, Japan is one of a few developed countries where numerous E. histolytica isolates are identified in autochthonous amebiasis cases. To advance our understanding of the significance of the polymorphic population structure of E. histolytica, we have analyzed four polymorphic genetic loci of E. histolytica isolates obtained from domestically infected Japanese amebiasis patients. Based on the analysis of these loci, E. histolytica isolates from Japan are very polymorphic, suggesting that E. histolytica has a complex clonal structure even in a limited geographic area and social populations.
MATERIALS AND METHODS
E. histolytica isolates and clinical samples.
A total of 34 E. histolytica samples were analyzed in this study. Twenty-seven Japanese E. histolytica isolates (Table 1) were obtained from clinical specimens collected from amebiasis patients in Japan. Xenic or axenic in vitro cultures were established and maintained in Robinson's medium or Diamond's BI-S-33 medium, respectively, as previously described (10, 25). Nineteen samples (samples 1 to 19) were obtained from male homosexuals, whereas 11 samples (samples 20 to 30) were from mentally handicapped individuals. The samples from the male homosexuals, who visited outpatient clinics, included those from 14 symptomatic cases, three asymptomatic cases, and two cases for which a history was not available. All isolates from mentally handicapped individuals were collected from four institutions geographically separated, i.e., Okayama (institution A), Kanagawa (institution B), and Shizuoka Prefecture (institutions C and D), within a week after mass infection was observed at institutions A to C at different time points (Table 1) (1994 to January 2001). No outbreak was observed at institution D. Most of the xenic and axenic strains were cryopreserved according to Diamond's method (9) immediately after xenic and axenic cultures were established and were revived 1 to 3 months prior to the present study to minimize possible changes, if any, of genotypes. Two E. histolytica-positive fecal samples and one liver aspirate were obtained from human immunodeficiency virus (HIV)-infected patients and kept frozen until DNA extraction. None of the donors had been abroad, and thus they are presumed to have been infected domestically. All cases with intestinal amebiasis or liver abscess were diagnosed by microscopic demonstration of trophozoites or cysts in stool or of trophozoites in liver aspirate, respectively. Past or present history of invasive amebiasis of these patients was verified with serology using the gel diffusion precipitin test (24) and enzyme-linked immunosorbent assay (31), and 25 out of 30 cases were considered invasive amebiasis. E. histolytica reference strains HM-1:IMSS cl6 (11), SAW755CR clB (8), SAW1627, and SAW1453 (5) and E. dispar strain SAW1734R cl AR (19) were used as controls. The E. dispar trophozoites were cultivated as previously described (15). All clinical specimens were collected after obtaining informed consent, and the research described in this paper complied with all relevant institutional guidelines.
TABLE 1.
Background and genotypes of the E. histolytica isolates
| No. | Isolate | Sourcea | Isolation
|
Clinical diagnosis | Serologyc | Zymodeme | DNA origin | Type
|
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locationb | Date | Locus 1-2 | Locus 5-6 | Chitinase | SREHP | |||||||
| 1 | KU 1 | H | Hospital 1 | July 1994 | Colitis | + | II | Xenic | B | A7 | F | Mixd |
| 2 | KU 2 | H | Hospital 2 | December 1988 | Colitis | + | XIX | Axenic | B | A7 | C | Mix |
| 3 | KU 3 | H | Hospital 3 | September 1988 | Colitis | − | II | Axenic | B | A6/Cv | A | M |
| 4 | KU 4 | H | Hospital 4 | July 1994 | NAe | + | XIX | Xenic | B | A8 | C | F |
| 5 | KU 5 | H | Hospital 5 | November 1988 | Asymptomatic | + | II | Axenic | B | A6/Cv | A | M |
| 6 | KU 7 | H | Hospital 6 | March 1995 | Colitis, ALAf | + | II | Xenic | B | A8 | C | Mix |
| 7 | KU 8 | H | Hospital 6 | May 1995 | Colitis, ALA | + | XIV | Xenic | E | A10/A5 | B | Mix |
| 8 | KU 9 | H | Hospital 6 | May 1995 | NA | + | II | Xenic | B | A7 | C | G/M |
| 9 | KU 10 | H | Hospital 4 | October 1991 | Colitis | + | II | Xenic | B | A8/Cv | A/Cg | Mix |
| 10 | KU 11 | H | Hospital 6 | February 1994 | ALA | + | XIV | Xenic | E | A5 | C | B/L |
| 11 | KU 15 | H | Hospital 6 | September 1994 | Colitis | + | XIX | Xenic | E | A5v | B | E |
| 12 | KU 16 | H | Hospital 4 | May 1993 | Colitis | + | XIX | Xenic | D | A5v | C | Mix |
| 13 | KU 23 | H | Hospital 7 | February 2000 | Colitis | + | XIX | Xenic | D | A13/A5 | B | F |
| 14 | KU 30 | H | Hospital 8 | March 2001 | Asymptomatic | + | II | Xenic | G | A5v | B | E |
| 15 | KU 31 | H | Hospital 8 | March 2001 | Asymptomatic | + | II | Xenic | G | A5 | C | E |
| 16 | KU 32 | H | Hospital 9 | June 2001 | Colitis | + | XIV | Xenic | G | A5 | B | E |
| 17 | KH 5 | H | Hospital 6 | 2001 | Colitis, HIV positive | + | NDh | Stool | G | A5v | C | I |
| 18 | KH 9 | H | Hospital 6 | 2001 | Colitis, HIV positive | ND | ND | Stool | B | A8 | ND | ND |
| 19 | KH 15 | H | Hospital 6 | 2001 | ALA, HIV positive | + | ND | ALA | D | A6/Cv | D | ND |
| 20 | KU 13 | M | Institution A | November 1999 | Asymptomatic | + | XIX | Xenic | B | A7 | C | H |
| 21 | KU 14 | M | Institution A | November 1999 | Asymptomatic | + | XIX | Xenic | B | A7 | C | H |
| 22 | KU 18 | M | Institution B | 1994 | NA | + | II | Xenic | F | A5v/Cv | C | K |
| 23 | KU 19 | M | Institution B | 1994 | NA | + | II | Xenic | F | A5v/Cv | C | K |
| 24 | KU 20 | M | Institution B | 1994 | NA | ND | II | Xenic | F | A5v/Cv | C | K |
| 25 | KU 21 | M | Institution B | 1994 | NA | ND | II | Xenic | F | A5v/Cv | C | K |
| 26 | KU 22 | M | Institution B | 1994 | NA | ND | II | Xenic | F | A5v/Cv | C | K |
| 27 | KU 26 | M | Institution C | September 2000 | Asymptomatic | + | II | Xenic | F | A5v/Cv | C | K |
| 28 | KU 27 | M | Institution D | January 2001 | Asymptomatic | + | II | Xenic | C | A7 | C | A |
| 29 | KU 28 | M | Institution D | January 2001 | Asymptomatic | + | II | Xenic | C | A7 | C | A |
| 30 | KU 29 | M | Institution D | January 2001 | Asymptomatic | + | II | Xenic | C | A7 | C | A |
| 31 | HM-1:IMSS cl6i | NA | Mexico | NA | Rectal ulcer, dysentery | NA | II | Axenic | D | A9/A6 | C | O/P |
| 32 | SAW755i | NA | India | NA | Colitis | NA | XIV | Axenic | D | A6 | C | C/N |
| 33 | SAW1627i | NA | India | 1983 | Asymptomatic | NA | IIα- | Axenic | A | A4/Cv | E | J |
| 34 | SAW1453i | NA | NA | NA | NA | NA | XIV | Xenic | A | A7 | C | D |
H, homosexual male; M, mentally handicapped.
Hospitals 1, 2, 4 to 9 are located in Tokyo; hospital 3 is located in Kyoto; institution A is located in Okayama prefecture; institution B is located in Kanagawa prefecture; and institution C and D are located in Shizuoka prefecture.
Serology was done by enzyme-linked immunosorbent assay, gel diffusion precipitin test, and/or indirect fluorescent-antibody test.
Mix, likely a mixture as judged by sequencing.
NA, not available.
ALA, amebic liver abscess.
A mixed culture or heterozyogosity could not be ruled out (see text).
ND, not determined.
Reference strain.
DNA preparation and PCR analysis.
Total genomic DNA from trophozoites and/or cysts was purified from either cultured amebas or clinical specimens by using the QIAamp DNA stool minikit (Qiagen, Tokyo, Japan) according to the manufacturer's directions. We determined DNA concentrations in samples by measuring optical absorbance at 260 and 280 nm spectrophotometrically. To verify that all cultures and samples contained only E. histolytica and not E. dispar, we amplified a 100-bp E. histolytica-specific fragment and a 101-bp E. dispar-specific fragment by PCR with a set of species-specific primers (P11 and P12 for E. histolytica and P13 and P14 for E. dispar) (Table 2) under the conditions described previously (30). We further classified the individual E. histolytica isolates by PCR amplification of four previously described polymorphic loci, i.e., locus 1-2, locus 5-6 (34), chitinase, and SREHP (12), using four sets of oligonucleotides previously described (Table 2). PCR was carried out in a 50-μl reaction mixture containing 0.1 μg of DNA, a 1.5 μM concentration of each primer, 2.5 mM MgCl2, 0.1 μg of bovine serum albumin per μl, a 100 μM concentration of each deoxynucleoside triphosphate, and 1.5 U of HotStar Taq DNA polymerase (Qiagen) with the following cycling parameters: (i) Taq activation at 95°C for 15 min; (ii) 30 cycles of denaturation at 94°C for 30 s, annealing at 50°C (SREHP and locus 5-6) or 45°C (chitinase and locus 1-2) for 30 s, and extension at 72°C for 1 min; and (iii) extension at 72°C for 10 min. PCR products were electrophoresed in 2% NuSieve 3:1 agarose (BioWhittaker Molecular Applications, Rockland, Maine). The results were visualized after staining with ethidium bromide in a UV transilluminator as described previously (26).
TABLE 2.
Oligonucleotide primers
| Primer name | Primer sequence (5′ to 3′) | Nucleotide positiona | Accession no. |
|---|---|---|---|
| P11 (sense) | GGA GGA GTA GGA AAG TTG AC | 214-234 | D10512 |
| P12 (antisense) | TTC TTG CAA TTC CTG CTT CGA | 293-314 | D10512 |
| P13 (sense) | AGG AGG AGT AGG AAA ATT AGG | 213-234 | D00872 |
| P14 (antisense) | TTC TTG AAA CTC CTG TTT CTA C | 292-314 | D00872 |
| R1 (forward) | CTG GTT AGT ATC TTC GCC TGT | 1-21 | AF276055 |
| R2 (reverse) | CTT ACA CCC CCA TTA ACA AT | 383-401 | AF276055 |
| R5A (forward) | CTA AAG CCC CCT TCT TCT ATA ATT | 1-24 | AF276060 |
| R6A (reverse) | CTC AGT CGG TAG AGC ATG GT | 405-424 | AF276060 |
| Chitinase (sense) | GGA ACA CCA GGT AAA TGT ATA | 466-487 | U78319 |
| Chitinase (antisense) | TCT GTA TTG TGC CCA ATT | 799-817 | U78319 |
| SREHP (sense) | GCT AGT CCT GAA AAG CTT GAA GAA GCT G | 258-286 | M80910 |
| SREHP (antisense) | GGA CTT GAT GCA GCA TCA AGG T | 784-806 | M80910 |
Nucleotide positions to which the primer anneals.
Sequence analysis.
PCR products containing locus 1-2, locus 5-6, SREHP, and chitinase were directly sequenced with appropriate primers in both directions. All of the PCR samples that were found to contain single bands on the agarose gels were treated with a Pre-Sequencing kit (USB Corporation, Cleveland, Ohio) before sequencing. Each DNA fragment of the PCR samples that showed double or triple bands by agarose gel electrophoresis was excised and treated using a Geneclean II kit (BIO101, La Jolla, Calif.). Individual PCR products were then sequenced using an ABI PRISM BigDye terminator cycle sequencing ready reaction kit (PE Applied Biosystems, Foster City, Calif.) on an ABI PRISM 310 Genetic Analyzer. The sequences obtained were manually edited and aligned using DNASIS (version 3.7; Hitachi, Yokohama, Japan).
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in the present work have been submitted to the GenBank/EMBL/DDBJ database under accession numbers AB075701 to AB075737.
RESULTS
Polymorphisms in DNA patterns on agarose gels.
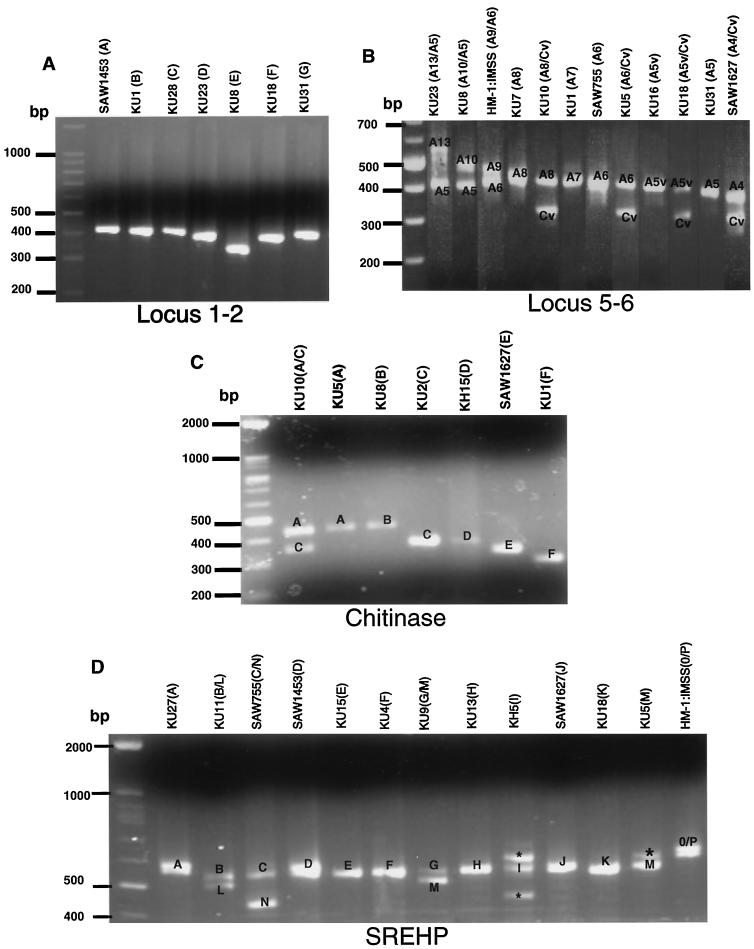
The PCR fragments containing locus 1-2, locus 5-6, chitinase, and SREHP from the 30 Japanese strains and the four reference strains showed remarkable polymorphism in both the number of bands and their sizes (Fig. 1 [only representative isolates that belong to each genotype are shown]), as previously shown (7, 12, 34). The patterns and sizes of amplified fragments corresponding to locus 5-6 or SREHP were highly variable; these PCR fragments consisted of either single or double bands, which is consistent with these loci being either homo- or heterozygous (Fig. 1B and D). In contrast, apparently single PCR fragments corresponding to locus 1-2 and chitinase were observed in all of the isolates tested except for the one described below; only three or four groups among the isolates were distinguishable by the size of the bands (Fig. 1A and C). The presence of two chitinase bands in the KU10 isolate might indicate heterozygosity or a mixed culture (see Discussion). No isolates showed more than two amplified bands of locus 5-6, despite repeated PCR attempts under different conditions, which may indicate that the multiple (>2) bands previously observed (34) may be artifactual. Alternatively, this discrepancy may be due to different clones of HM1:IMSS used.
FIG. 1.
Agarose gel electrophoresis of locus 1-2 (A), locus 5-6 (B), chitinase (C), and SREHP (D) from representative E. histolytica isolates. Only results for representative isolates that belong to each genotype (shown in parentheses) are shown. The individual genotype is designated for each polymorphic DNA fragment. Bands with asterisks are irrelevant PCR fragments (verified by sequencing) observed only in KU1, KU5, KH5, KH9, and KH15, for unknown reasons.
Polymorphism in nucleotide sequences of the noncoding DNA loci among the Japanese isolates.
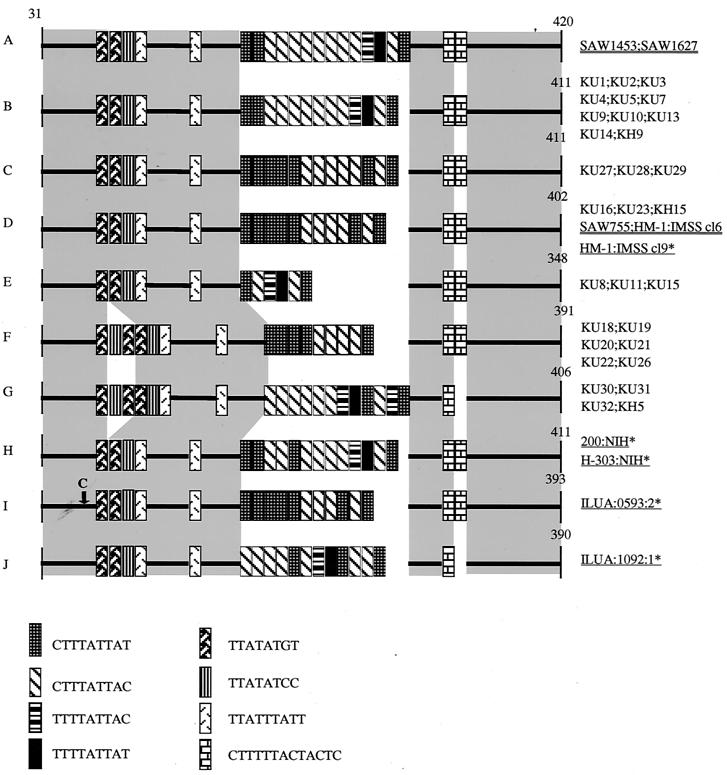
In order to better understand the nature of the polymorphisms among the Japanese strains observed by gel electrophoresis, we sequenced the individual fragments of locus 1-2, locus 5-6, chitinase, and SREHP from all 34 isolates. The nucleotide polymorphisms of these loci were more pronounced than those shown by gel electrophoresis. Although both locus 1-2 and locus 5-6 are present as tandemly linked multiple copies (34), individual DNA fragments in an apparently single band seemed to be homogeneous, suggesting that sequences in these copies are mostly identical. Both locus 1-2 and locus 5-6 contained 6 to 21 copies of 8- to 16-nucleotide repeat units, which is consistent with the previous findings (34). Sequencing of locus 1-2 revealed a complex interisolate polymorphism in length, location, and number of the repeat units (Fig. 2). Based on the nucleotide sequences of locus 1-2, the 30 Japanese isolates were divided into six distinct types (B to G), with a single genotype, B, being the dominant type (37%).
FIG. 2.
Schematic representation of polymorphism in locus 1-2 among the Japanese isolates and reference strains. The numbers shown correspond to nucleotide numbers of locus 1-2 of HM-1:IMSS cl9 (GenBank/EMBL/DDBJ accession number AF276055). Sequences prior to nucleotide 30 are not shown because these nucleotides overlap the PCR primer. Gaps are introduced to optimize alignments. Conserved regions are highlighted with gray rectangles. Names of reference strains previously reported are underlined, and those analyzed in this study are double underlined. Previously unidentified repeats units are also included in this and following figures. A T-to-C nucleotide substitution is indicated by an arrow. An asterisk next to an isolate designation indicates that more information can be found in reference 34.
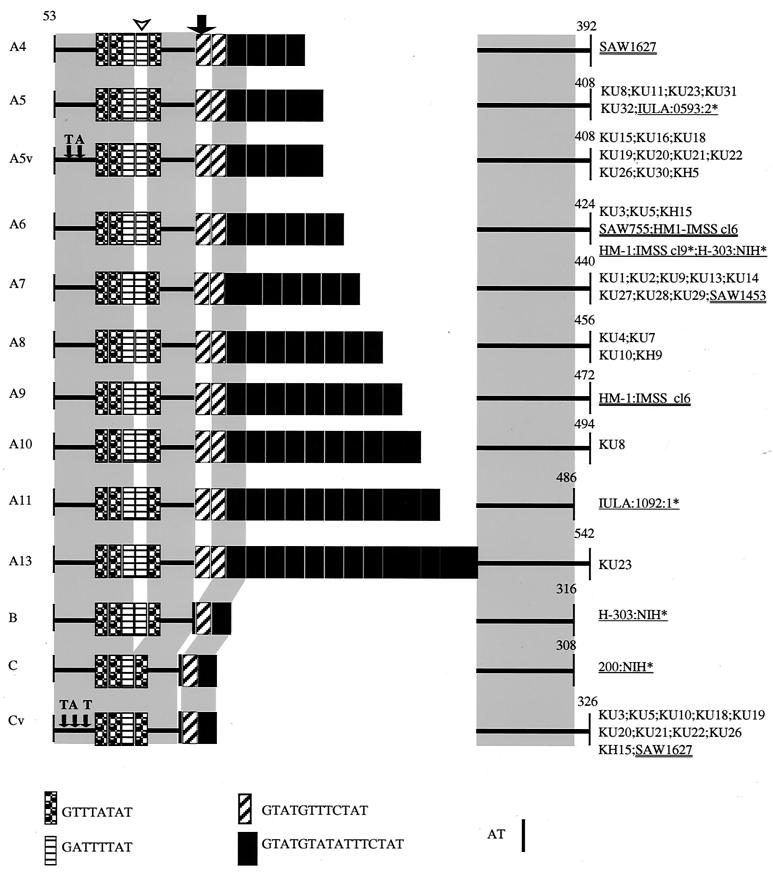
In contrast to the case for locus 1-2, PCR-amplified fragments of locus 5-6 were observed as either single or double bands depending upon the isolate, as mentioned above. Sequencing of the individual bands revealed that polymorphism among these bands was solely due to variations in the number of a 16-nucleotide repeat unit (GTATGTATATTTCTAT; 4 to 13 repeats), with a few exceptions (types A5v, B, C, and Cv). Individual bands of locus 5-6 were designated based primarily on either the presence or absence of the first GTATGTTTCTAT and the second GATTTTAT repeats (Fig. 3) (types A to C), second on the number of the GTATGTATATTTCTAT repeats (A4 to A13), and third on the presence or absence of nucleotide substitutions in the conserved region (A5v and Cv) (Fig. 3). As individual isolates appeared to be either homo- or heterozygous at locus 5-6, we designated the genotype of each isolate, e.g., A7, A13/A5, and A5v/Cv.
FIG. 3.
Schematic representation of polymorphism in locus 5-6 among the Japanese isolates and reference strains. The numbers shown correspond to nucleotide numbers of locus 5-6 of HM-1:IMSS cl9 (GenBank/EMBL/DDBJ accession number AF276060). Sequences prior to nucleotide 52 are not shown for the reason described in the legend to Fig. 2. The first GTATGTTTCTAT and the second GATTTTAT repeats described in text are marked with a thick arrow and an arrowhead, respectively. Variant forms of A5 and C, in which two and three nucleotides were replaced (indicated by thin arrows), are designated A5v and Cv, respectively. An asterisk next to an isolate designation indicates that more information can be found in reference 34.
Polymorphism in the chitinase and SREHP loci among the Japanese isolates.
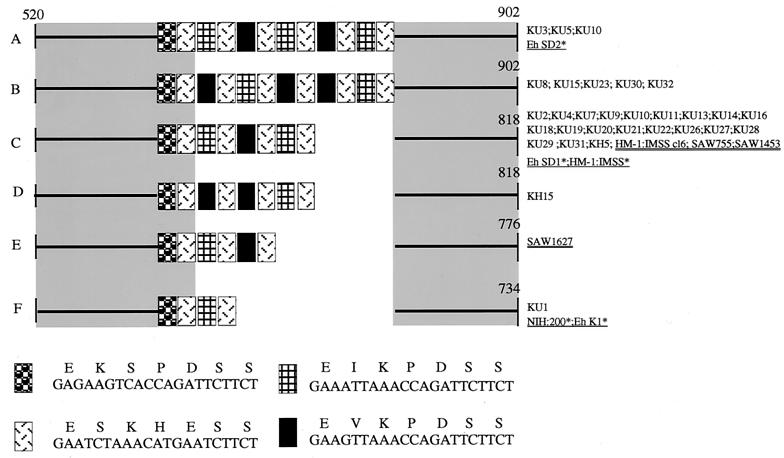
Polymorphisms in the type, location, and number of repeat units were observed in the repeat-containing region of the chitinase gene (Fig. 4). However, the chitinase locus appeared to be the least polymorphic among the four loci, with a single dominant genotype, C (out of five types observed in the Japanese isolates), comprising 66% (20 out of 30) of all of the Japanese isolates. All Japanese and reference isolates, as well as five reported sequences (12), were classified into only six independent types (Fig. 4).
FIG. 4.
Schematic representation of polymorphism in the repeat-containing region of the chitinase gene among the Japanese isolates and reference strains. The numbers shown in this figure correspond to nucleotide numbers of chitinase of HM-1:IMSS (GenBank/EMBL/DDBJ accession number U78319). Nucleotide and deduced amino acid sequences of heptapeptide repeats are also shown. An asterisk next to an isolate designation indicates that more information can be found in reference 12.
The repeat-containing region of SREHP was found to be extensively polymorphic in size, species, number, and order of repeat units among the Japanese isolates (Fig. 5). Consistent with a previous notion that the SREHP locus is either homo- or heterozygous (12), either single or double bands of amplified SREHP fragments were observed, depending upon the isolate (see above). In addition, sequencing of gel-purified PCR bands of the SREHP loci from six isolates that showed an apparently single SREHP band had mixed nucleotide sequences (Table 1), likely due to a mixture in the apparently single PCR bands, which also indicates the heterozygosity of the SREHP locus. The hypothesis that some apparently single SREHP PCR fragments, e.g., HM1:IMSS cl6 (Fig. 1D), consist of two DNA sequences was also confirmed by sequencing individual SREHP fragments from HM-1:IMSS cl6 after cloning into a plasmid (data not shown). Three isolates that showed two SREHP bands that were different in size (KU9, KU11, and SAW755) revealed independent SREHP genotypes, i.e., G/M, B/L, and C/N. In contrast to the previous studies (12), where SREHP fragments corresponding to types P and Q were amplified from HM1:IMSS, two fragments corresponding to types O and P (Fig. 5), which differ only in the presence or absence of a 18-bp repeat, were obtained.
FIG. 5.
Schematic representation of polymorphism in the repeat-containing region of the SREHP gene among the Japanese isolates and reference strains. The numbers shown in this figure correspond to nucleotide numbers of SREHP of HM-1:IMSS (GenBank/EMBL/DDBJ accession number M80910). Nucleotide and deduced amino acid sequences of tetra-, octa-, and nonapeptide repeats are also shown. An asterisk next to an isolate designation indicates that more information can be found in reference 12.
Correlations between genotypes and origins of isolates.
We investigated whether there is any correlation between the genotypes and the origins of the isolates (i.e., from male homosexuals or residents of institutions). The genotypes found in isolates from male homosexuals are highly polymorphic; all of these isolates are independent based on classification using these four loci (Table 1). In contrast, isolates obtained from residents of institutions showed less complex genetic polymorphisms. In addition, the genotypes of isolates obtained from mass infections at a single institution were indistinguishable (e.g., KU13 and -14, KU18 to -22, and KU27 to -29). Although the number of isolates tested was not large enough to enable statistical analysis, locus 1-2/types C and F; locus 5-6/type A5v/Cv; and SREHP/types A, H, and K were found exclusively in the isolates from institutions. In contrast, locus 1-2/types D, E, and G; locus 5-6/types A13/A5, A10/A5, A8, A8/Cv, A6/Cv, A5, and A5v; chitinase/types A, B, D, and F; and SREHP/types B/L, E, F, G/M, I, and M were never found in isolates from institutions but were found exclusively in isolates from male homosexuals. No apparent correlation was found either between genotypes and zymodemes or between genotypes and clinical manifestations (Table 1).
DISCUSSION
In the present study, we identified a large number of novel genotypes of four independent polymorphic loci among the Japanese isolates: 5 for locus 1-2, 6 for locus 5-6, 2 for chitinase, and 10 for SREHP. Combining these four independent polymorphic loci, all of the Japanese isolates were clearly distinct from any of the reference strains and also distinct from one another, except for the cases described below. Although genetic polymorphism among E. histolytica isolates from different geographic areas has been demonstrated (12, 34), the presence of extensive polymorphisms among the Japanese isolates in a limited geographic area and social populations, seen in the present study, reinforces a notion that genotyping of E. histolytica isolates by using these four polymorphic loci could serve as a tool to fingerprint individual isolates.
All 19 isolates obtained from male homosexuals were independent, which strongly indicates that the E. histolytica population in Japanese male homosexuals consists of a complex clonal structure. In contrast, E. histolytica strains from residents of institutions revealed a lower degree of polymorphism than those from male homosexuals. Considering the degree of polymorphism observed among the isolates derived from male homosexuals, it is surprising, but conceivable, that genotypes of isolates obtained from a single mass infection event in an institution for the mentally handicapped (e.g., isolates KU13 and -14, KU18 to -22, and KU27 to -29 [Table 1]) were identical. This fact suggests that a mass infection is likely caused by a single genotypic strain, and this is presumably due to introduction of a single E. histolytica strain into an institution. It is very striking that isolates from six mass infection cases in two independent events (KU18 to -22 and KU26 [Table 1]) that occurred 6 years apart (1994 and 2000) at remote geographic locations (data not shown) revealed an identical genotype.
DNA sequence polymorphisms of a limited number of axenized strains and stool samples have been described previously (12, 34). In the present study, we used a large number of xenic isolates because axenization may select certain genotypes. In addition, most of the strains were cryopreserved immediately after cultures were established and were revived only prior to the present study, as described in Materials and Methods, to minimize possible genotype changes. We also used uncultivated clinical specimens to avoid any selection during in vitro cultivation.
The genotypes of the widely used HM-1:IMSS are virtually indistinguishable in all of the loci tested except for the SREHP locus (7, 12, 34). The fact that the two SREHP sequences obtained from HM1:IMSS varied between Samuelson's and our laboratories, i.e., types O and P in this study and types P and Q previously (12), may indicate that PCR of this locus is prone to artifacts, as previously suggested (12). However, PCR amplification of the SREHP locus from this strain, previously shown by Clark and Diamond (7), apparently revealed no band corresponding to type Q but showed double bands that appeared to be consistent with types O and P in the present study. Although the reasons for this discrepancy are unknown, our demonstration that some of the apparently single PCR bands consist of at least two sequences argues against a proposal (12) that one out of two SREHP fragments was lost during PCR amplification. Thus, we concluded that the genotypes of the widely used reference strain are stable. A recent paper by Zaki et al. (35) also reported that genotypes are stable over time in culture and in the same patient. Homozygosity of the chitinase locus, tentatively deduced from a previous analysis of five isolates (12), was confirmed for all the isolates tested, with one exception (KU10). The presence of two chitinase bands in the KU10 isolate appears to be due to heterozygosity of this locus, rather than indicating the possibility of a mixed culture, for the following reasons. First, the KU10 strain has been xenically maintained for more than 10 years, during which time one strain likely would have outgrown the other if the culture was initiated as a mixture. Second, no mixed genotype of locus 1-2, which is homozygous in all of the isolates tested, was observed in KU10. However, the significance of heterozygosity at the chitinase locus in this isolate is unknown. The finding that the repeat-containing region of chitinase is the least polymorphic among the four loci, together with the fact that chitinase appears to be homozygous in most of our isolates, strongly indicates that there are functional constraints on chitinase polymorphism, which is also consistent with the hypothesis of a bottleneck spread of E. histolytica isolates, proposed by Ghosh et al. (12). Such functional constraints likely include the structural requirements of the enzyme for catalysis and/or multimerization.
The high genetic polymorphism among E. histolytica isolates described in this work, as with polymorphisms described for Leishmania and Trypanosoma cruzi (3, 23, 29), implies similarly diverse biological characteristics such as immunopathological effects, drug sensitivities, and vaccine attributes. The polymorphic nature of SREHP requires further attention because SREHP is being exploited as a protective immunodominant antigen (36, 37). Finally, we propose that molecular typing of ameba isolates by using these polymorphic loci should help in determining geographic origins of isolates and routes of transmission. Analysis of genotypes of E. histolytica isolates from a variety of geographic locations, e.g., Southeast Asia, is in progress.
Acknowledgments
We thank Mehreen Zaki and Graham Clark, London School of Hygiene and Tropical Medicine, for helpful discussions and useful comments on the manuscript; Yumiko Saito-Nakano, National Institute of Infectious Diseases, for technical help; and Shin-ichiro Kawazu and Shigeyuki Kano, International Medical Center of Japan, for technical help in sequencing.
This work was partially supported by a fellowship (no. 200005) from the Japan Science and Technology Corporation to A.H., by a grant for research on emerging and reemerging infectious diseases from the Ministry of Health, Labour, and Welfare of Japan to T.N., and by a grant for research on health sciences focusing on drug innovation (SA14706) from the Japan Health Sciences Foundation to T.N.
REFERENCES
- 1.Abe, N., Y. Nishikawa, A. Yasukawa, and K. Haruki. 1999. Entamoeba histolytica outbreaks in institutions for the mentally retarded. Jpn. J. Infect. Dis. 52:135-136. [PubMed] [Google Scholar]
- 2.Allason-Jones, E., A. Mindel, P. Sargeaunt, and P. Williams. 1986. Entamoeba histolytica as a commensal intestinal parasite in homosexual men. N. Engl. J. Med. 315:353-356. [DOI] [PubMed] [Google Scholar]
- 3.Ayala, F. J. 1993. Trypanosoma and Leishmania have clonal population structures of epidemiological significance. Biol. Res. 26:47-63. [PubMed] [Google Scholar]
- 4.Bhattacharya, S., A. Bhattacharya, and L. S. Diamond. 1992. Entamoeba histolytica extrachromosomal circular ribosomal DNA: analysis of clonal variation in a hypervariable region. Exp. Parasitol. 74:200-204. [DOI] [PubMed] [Google Scholar]
- 5.Blanc, D., and P. G. Sargeaunt. 1991. Entamoeba histolytica zymodemes: exhibition of gamma and delta bands only of glucose phosphate isomerase and phosphoglucomutase may be influenced by starch content in the medium. Exp. Parasitol. 72:87-90. [DOI] [PubMed] [Google Scholar]
- 6.Burch, D. J., E. Li, S. Reed, T. F. Jackson, and S. L. Stanley, Jr. 1991. Isolation of a strain-specific Entamoeba histolytica cDNA clone. J. Clin. Microbiol. 29:696-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark, C. G., and L. S. Diamond. 1993. Entamoeba histolytica: a method for isolate identification. Exp. Parasitol. 77:450-455. [DOI] [PubMed] [Google Scholar]
- 8.Clark, C. G., and L. S. Diamond. 1991. Ribosomal RNA genes of ′pathogenic' and ′nonpathogenic' Entamoeba histolytica are distinct. Mol. Biochem. Parasitol. 49:297-302. [DOI] [PubMed] [Google Scholar]
- 9.Diamond, L. S. 1995. Cryopreservation and storage of parasitic protozoa in liquid nitrogen. J. Eukaryot. Microbiol. 42:585-590. [Google Scholar]
- 10.Diamond, L. S., D. R. Harlow, and C. C. Cunnick. 1978. A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans. R. Soc. Trop. Med. Hyg. 72:431-432. [DOI] [PubMed] [Google Scholar]
- 11.Diamond, L. S., C. F. Mattern, and I. L. Bartgis. 1972. Viruses of Entamoeba histolytica. I. Identification of transmissible virus-like agents. J. Virol. 9:326-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghosh, S., M. Frisardi, L. Ramirez-Avila, S. Descoteaux, K. Sturm-Ramirez, O. A. Newton-Sanchez, J. I. Santos-Preciado, C. Ganguly, A. Lohia, S. Reed, and J. Samuelson. 2000. Molecular epidemiology of Entamoeba spp.: evidence of a bottleneck (demographic sweep) and transcontinental spread of diploid parasites. J. Clin. Microbiol. 38:3815-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldmeier, D., P. G. Sargeaunt, A. B. Price, P. E. Munday, O. Billington, I. Dixon, P. Borriello, J. M. Carder, A. Shaw, J. Hilton, et al. 1986. Is Entamoeba histolytica in homosexual men a pathogen? Lancet i:641-644. [DOI] [PubMed]
- 14.Kaneda, Y., K. Nagakura, H. Tachibana, T. Tanaka, and M. Sasao. 1988. Entamoeba histolytica infection in a rehabilitation center for mentally retarded persons in Japan. Scand. J. Infect. Dis. 20:687.. [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi, S., E. Imai, H. Tachibana, T. Fujiwara, and T. Takeuchi. 1998. Entamoeba dispar: cultivation with sterilized Crithidia fasciculata. J. Eukaryot. Microbiol. 45:3S-8S. [DOI] [PubMed] [Google Scholar]
- 16.Kohler, S., and E. Tannich. 1993. A family of transcripts (K2) of Entamoeba histolytica contains polymorphic repetitive regions with highly conserved elements. Mol. Biochem. Parasitol. 59:49-58. [DOI] [PubMed] [Google Scholar]
- 17.Lesh, F. A. 1975. Massive development of amebas in the large intestine. Fedor Aleksandrovich Lesh (Losch). Am. J. Trop. Med. Hyg. 24:383-392. [DOI] [PubMed] [Google Scholar]
- 18.Li, E., C. Kunz-Jenkins, and S. L. Stanley, Jr. 1992. Isolation and characterization of genomic clones encoding a serine-rich Entamoeba histolytica protein. Mol. Biochem. Parasitol. 50:355-357. [DOI] [PubMed] [Google Scholar]
- 19.Mirelman, D., R. Bracha, A. Wexler, and A. Chayen. 1986. Changes in isoenzyme patterns of a cloned culture of nonpathogenic Entamoeba histolytica during axenization. Infect. Immun. 54:827-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagakura, K., H. Tachibana, T. Tanaka, Y. Kaneda, M. Tokunaga, M. Sasao, and T. Takeuchi. 1989. An outbreak of amebiasis in an institution for the mentally retarded in Japan. Jpn. J. Med. Sci. Biol. 42:63-76. [DOI] [PubMed] [Google Scholar]
- 21.Nozaki, T., S. R. Motta, T. Takeuchi, S. Kobayashi, and P. G. Sargeaunt. 1989. Pathogenic zymodemes of Entamoeba histolytica in Japanese male homosexual population. Trans. R. Soc. Trop. Med. Hyg. 83:525.. [DOI] [PubMed] [Google Scholar]
- 22.Ohnishi, K., and M. Murata. 1997. Present characteristics of symptomatic amebiasis due to Entamoeba histolytica in the east-southeast area of Tokyo. Epidemiol. Infect. 119:363-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oliveira, R. P., A. I. Melo, A. M. Macedo, E. Chiari, and S. D. Pena. 1999. The population structure of Trypanosoma cruzi: expanded analysis of 54 strains using eight polymorphic CA-repeat microsatellites. Mem. Inst. Oswaldo Cruz 94(Suppl. 1):65-70. [DOI] [PubMed] [Google Scholar]
- 24.Ouchterlony, O. 1966. The antigenic pattern of immunoglobulins. G. Mal. Infett. Parassit. 18(Suppl. 1):942-948. [PubMed] [Google Scholar]
- 25.Robinson, G. L. 1968. Laboratory cultivation of some human parasitic amoebae. J. Gen. Microbiol. 53:69-79. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Sargeaunt, P. G., J. K. Oates, I. MacLennan, J. D. Oriel, and D. Goldmeier. 1983. Entamoeba histolytica in male homosexuals. Br. J. Vener. Dis. 59:193-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sehgal, D., A. Bhattacharya, and S. Bhattacharya. 1993. Analysis of a polymorphic locus present upstream of rDNA transcription units in the extrachromosomal circle of Entamoeba histolytica. Mol. Biochem. Parasitol. 62:129-130. [DOI] [PubMed] [Google Scholar]
- 29.Stothard, J., I. Frame, and M. Miles. 1999. Genetic diversity and genetic exchange in Trypanosoma cruzi: dual drug-resistant “progeny” from episomal transformants. Mem. Inst. Oswaldo Cruz. 94(Suppl. 1):189-193. [DOI] [PubMed] [Google Scholar]
- 30.Tachibana, H., S. Kobayashi, M. Takekoshi, and S. Ihara. 1991. Distinguishing pathogenic isolates of Entamoeba histolytica by polymerase chain reaction. J. Infect. Dis. 164:825-826. [DOI] [PubMed] [Google Scholar]
- 31.Takeuchi, T., H. Matsuda, E. Okuzawa, T. Nozaki, S. Kobayashi, and H. Tanaka. 1988. Application of a micro enzyme-linked immunosorbent assay (ELISA) to detection of anti-amoebic antibody in various forms of amoebic infection. Jpn. J. Exp. Med. 58:229-232. [PubMed] [Google Scholar]
- 32.Takeuchi, T., E. Okuzawa, T. Nozaki, S. Kobayashi, M. Mizokami, N. Minoshima, M. Yamamoto, and S. Isomura. 1989. High seropositivity of Japanese homosexual men for amebic infection. J. Infect. Dis. 159:808.. [DOI] [PubMed] [Google Scholar]
- 33.W. H. O./PAHO/UNESCO. 1997. A consultation with experts on amebiasis. Epidemiol. Bull. 18:13-14. [PubMed] [Google Scholar]
- 34.Zaki, M., and C. G. Clark. 2001. Isolation and characterization of polymorphic DNA from Entamoeba histolytica. J. Clin. Microbiol. 39:897-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zaki, M., P. Meelu, W. Sun, and C. G. Clark. 2002. Simultaneous differentiation and typing of Entamoeba histolytica and Entamoeba dispar. J. Clin. Microbiol. 40:1271-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang, T., and S. L. Stanley. 1999. DNA vaccination with the serine rich Entamoeba histolytica protein (SREHP) prevents amebic liver abscess in rodent models of disease. Vaccine 18:868-874. [DOI] [PubMed] [Google Scholar]
- 37.Zhang, T., and S. L. Stanley, Jr. 1997. Expression of the serine rich Entamoeba histolytica protein (SREHP) in the avirulent vaccine strain Salmonella typhi TY2 chi 4297 (delta cya delta crp delta asd): safety and immunogenicity in mice. Vaccine 15:1319-1322. [DOI] [PubMed] [Google Scholar]