Abstract
Pulsed-field gel electrophoresis (PFGE) of XbaI-digested chromosomal DNA was performed on 133 strains of Salmonella enterica serovar Typhi obtained from Papua New Guinea, with the objective of assessing the temporal variation of these strains. Fifty-two strains that were isolated in 1992 and 1994 were of one phage type, D2, and only two predominant PFGE profiles, X1 and X2, were present. Another 81 strains isolated between 1997 and 1999 have shown divergence, with four new phage types, UVS I (n = 63), UVS (n = 5), VNS (n = 4), and D1 (n = 9), and more genetic variability as evidenced by the multiple and new PFGE XbaI profiles (21 profiles; Dice coefficient, F = 0.71 to 0.97). The two profiles X1 and X2 have remained the stable, dominant subtypes since 1992. Cluster analysis based on the unweighted pair group method using arithmetic averages algorithm identifies two main clusters (at 87% similarity), indicating that the divergence of the PFGE subtypes was probably derived from some genomic mutations of the X1 and X2 subtypes. The majority of isolates were from patients with mild and moderate typhoid fever and had various XbaI profiles. A single isolate from a patient with fatal typhoid fever had a unique X11 profile, while four of six isolates from patients with severe typhoid fever had the X1 pattern. In addition, 12 paired serovar Typhi isolates recovered from the blood and fecal swabs of individual patients exhibited similar PFGE patterns, while in another 11 individuals paired isolates exhibited different PFGE patterns. Three pairs of isolates recovered from three individuals had different phage types and PFGE patterns, indicating infection with multiple strains. The study reiterates the usefulness of PFGE in assessing the genetic diversity of S. enterica serovar Typhi for both long-term epidemiology and in vivo stability and instability within an individual patient.
Typhoid fever is a systemic prolonged febrile illness caused by Salmonella enterica serovar Typhi. It continues to be a worldwide health problem, especially in developing countries with their poor sanitation, poor standards of personal hygiene, and contaminated food. Current estimates from the World Health Organization suggest that the worldwide incidence of typhoid fever is approximately 16 million cases annually, with more than 600,000 deaths. Effective epidemiological surveillance is needed to monitor the presence and spread of the disease. For serovar Typhi, the primary tools are serotyping and phage typing (6). However, these phenotypic methods lack discrimination and are often complemented by the more sensitive and discriminative molecular techniques (1). Pulsed-field gel electrophoresis (PFGE) is one of the most common technique used to perform comparative chromosomal DNA analysis of serovar Typhi (6, 15, 21).
Previous studies of serovar Typhi have shown significant genetic diversity, as demonstrated by PFGE differences among 400 isolates obtained from various regions with endemic infection, including Malaysia, Indonesia, Thailand, and Chile (21, 22, 23). In contrast, PFGE analysis of a collection of 52 serovar Typhi isolates obtained in 1992 to 1994 from Papua New Guinea were very homogeneous (25). The present study was carried out to extend the monitoring of genetic diversity to more recent isolations of serovar Typhi strains from Papua New Guinea with respect to the phage types and PFGE profiles.
MATERIALS AND METHODS
Bacterial strains.
Eighty-one serovar Typhi strains obtained from 60 indigenous patients with sporadic cases of typhoid fever in Papua New Guinea between 1997 and 1999 were studied. Among the 81 isolates, 23 paired isolates (i.e., 46 isolates) were from both fecal swab and blood cultures from 23 patients and were analyzed to determine the in vivo stability of the strains. The severity of disease in the patients from whom the isolates were obtained is summarized in Table 1. Random isolates from the 1992 and 1994 collection (25) were also analyzed to confirm the stability of the strains and to compare them with the present strains. The organisms were isolated, maintained, and identified by standard biochemical methods and serotyping. Repeated subculturing of isolates was avoided. Vi phage typing of the isolates was performed by standard procedures by the Salmonella Reference Centre at the Institute of Medical Research, Kuala Lumpur, Malaysia. All the strains were tested for susceptibility to 10 antimicrobial agents by the disk diffusion technique using Mueller-Hinton by the Bauer-Kirby method (3) as recommended by NCCLS (14). These include ampicillin (10 μg), chloramphenicol (30 μg), gentamicin (10 μg), tetracycline (30 μg), co-trimoxazole (25 μg), streptomycin (10 μg), kanamycin (30 μg), ciprofloxacin (5 μg), ceftriazone (30 μg), and nalidixic acid (30 μg). Escherichia coli ATCC 25922 and Staphylococcus aureus ATCC 29213 were used as controls for the potency of antibiotics. Interpretation of the zone of inhibition was made in accordance with the interpretative standards provided by the manufacturer (Diagnostic Pasteur).
TABLE 1.
Association of PFGE patterns of serovar Typhi isolates from Papua New Guinea with disease severity and phage types
| Disease severity | No. of isolates tested | XbaI profiles (no. of isolates) | Phage types (no. of isolates) |
|---|---|---|---|
| Fatal | 1 | X11 (1) | UVS1 (1) |
| Patient dies | |||
| Severe | 6 | X1 (4), X8 (1), X2 (1) | UVS1 (4), VNS (1), D1 (1) |
| Severely ill, with life-threatening complications which include: shock, bowel perforation or hemorrhage diagnosed clinically, neuropsychiatric manifestations, bleeding diathesis or acute renal failure | |||
| Moderate | 27 | X1 (13), X2 (4), X4 (1), X6 (1), X9 (1), X10 (1), X12 (3), X13 (1), X17 (1), X14 (1) | UVS1 (22), UVS (2), D1 (2), VNS (1) |
| Requires hospitalization because of severity, no complications or only mild complications | |||
| Mild | 27X1 (11), X2 (3), X5 (1), X6 (1), X10 (7), X16 (1), X18 (1), X19 (1), X21 (1) | UVS1 (21), UVS (1), VNS (1), D1 (4) | |
| No complications, not severely ill, does not require hospitalization other than supervision of treatment for pediatric cases. | |||
| Unknown statea | 20 | X1 (11), X2 (1), X10 (2), X7 (3), X15 (1), X12 (1), X20 (1) | UVS1 (15), UVS (2), VNS (1), D1 (2) |
Unavailable clinical records.
DNA preparation and PFGE.
Chromosomal DNA for PFGE analysis was prepared in agarose plugs as previously described (21, 24). Slices of DNA-containing agarose plugs were digested overnight with 10 U of XbaI (Promega) at 37°C and then electrophoresed on a CHEF-DR II/III system (Bio-Rad Laboratories) for 26 h at 6 V cm−1, with a ramped pulse time of 1 to 40 s at 12°C. Lambda DNA concatemer PFG marker (New England, Biolabs) was used as a DNA size standard. The gel was stained with ethidium bromide (1 μg ml−1; Sigma) for 15 min, destained in distilled water for 15 min, and photographed under UV illumination. DNA fragment patterns were assessed visually, and distinct profiles were assigned an arbitrary restriction endonuclease analysis pattern. The band differences were interpreted by the method of Tenover et al. (20). By these criteria, isolates that have similar PFGE banding patterns were described as genetically indistinguishable, isolates that gave different banding patterns in fewer than four bands were assumed to be closely related, and isolates with differences in four to six bands might be related. In addition, isolates showing a difference in more than seven bands were considered to be epidemiologically unrelated. Dice coefficients of similarity were calculated to compare the macrorestriction patterns. This coefficient, F, expresses the proportion of shared DNA fragments in two isolates and was calculated by the following formula. F = 2nxy/(nx + ny), where nx is the total number of DNA fragments from isolate X, ny is the total number of DNA fragments from isolate Y, and nxy is the number of DNA fragments that were identical in the two isolates. By this assessment, F = 1.0 indicates complete identity and F = 0 indicates complete dissimilarity. Clustering was based on the unweighted pair group average method (UPGMA) and was performed with GelCompar (Applied Maths, Kontrijk, Belgium).
RESULTS
All strains were susceptible to the antimicrobial agents tested. Among the 81 strains isolated from 1997 to 1999, only four phage types were present: untypeable Vi 1 (UVS1, n = 63), untypeable Vi (UVS, n = 5), Vi negative (VNS, n = 4), and D1 (n = 9). Phage typing was of limited use in subtyping of the strains because it lacked discrimination. Phage type D2, which was found in all 52 strains isolated in 1992 and 1994, was not detected (Table 2).
TABLE 2.
Distribution of PFGE XbaI subtypes of serovar Typhi from Papua New Guinea over 1992 to 1994 and 1997 to 1999
| Yr | Phage types (no. of isolates) | XbaI PFGE patterns (no. of isolates) | F |
|---|---|---|---|
| 1992-1994a | D2 (52) | X1 (27), X2 (23), X3 (2) | 0.94-1.0 |
| 1997 | UVS (2), UVS1 (21), VNS (2), D1 (1) | X1 (18), X2 (3), X4 (1), X5 (1), X6 (1), X7 (1), X21 (1) | 0.85-1.0 |
| 1998 | UVS (2), UVS1 (31), D1 (5), VNS (1) | X1 (14), X2 (3), X6 (1), X7 (1), X8 (1), X9 (1), X10 (8), X11 (1), X12 (4), X13 (1), X14 (1), X15 (1), X16 (1), X20 (1) | 0.71-1.0 |
| 1999 | UVS1 (11), D1 (3), VNS (1), UVS (1) | X1 (7), X2 (3), X7 (1), X10 (2), X17 (1), X18 (1), X19 (1) | 0.78-1.0 |
Data from previous study (25).
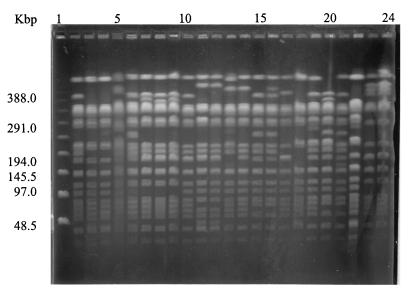
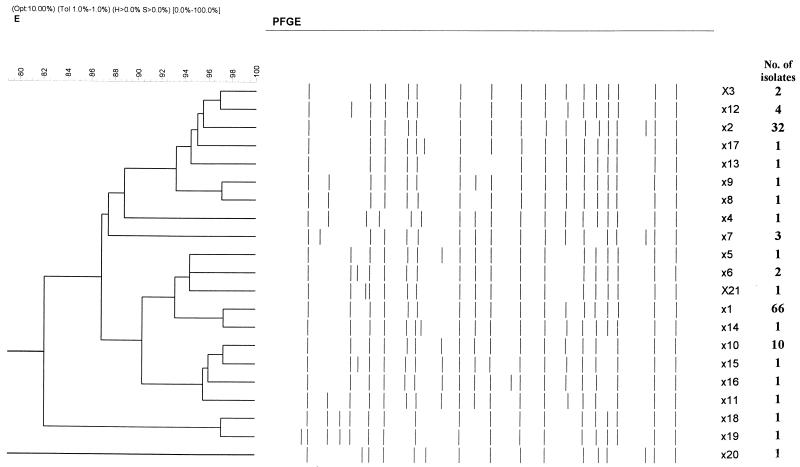
PFGE of XbaI-digested chromosomal DNA from 81 isolates gave stable and reproducible patterns consisting of 15 to 20 fragments. Only XbaI was used in this study since it is more discriminatory and cheaper than AvrII or SpeI and is a more useful restriction endonuclease for serovar Typhi (21). Twenty-one pulsed-field profiles (X1 to X21) were obtained: X1 (n = 39), X2 (n = 9), X6 (n = 2), X7 (n = 3), X10 (n = 10), and X12 (n = 4) were found in multiple isolates, and the other PFPs were represented by one isolate each (F = 0.71 to 1.0). (Table 2; Fig. 1). In comparison with the 52 isolates obtained during 1992 to 1994, which had mostly X1 (52%) and X2 (44%) profiles, the recent isolates (obtained in 1997 to 1999) were genetically more diverse. Nevertheless, X1 and X2 still remained the predominant and stable genotypes (Table 2). The genetic relatedness of the PFGE profiles, as demonstrated by a dendrogram, shows two main clusters (at 87% similarity). Each cluster has either the X1 or the X2 profile, indicating that the divergence of the PFGE subtypes probably occured from these two initial, closely related clones by some genomic mutations (Fig. 2). Each cluster consisted of nine different profiles differing in one to four bands (F = 0.88 to 0.97) and two or three bands (F = 0.92 to 0.94), respectively. Based on the criteria of Tenover et al. (20), these strains can be designated closely related strains.
FIG. 1.
PFGE (XbaI cleavage) patterns of representative isolates from sporadic cases of typhoid fever in Papua New Guinea (1992 to 1999). The lanes contain (from left to right) lambda DNA concatemer marker, UJ19F (X1), UJ42B (X2), UJ164 (X2), UJ5F (X4), UJ6B (X5), UJ6F (X6), UJ49B (X21), UJ495F (6), PNG1 (X1), UJ542F (X7), UJ34F (X7), UJ600F (X8), UJ212F (X9), UJ622B (X10), UJ308B (X11), UJ738B (X12), UJ422B (X13), UJ446B (X14), UJ542F (X15), UJ605F (X16), UJ812F (X17), UJ916 (X18), and UJ934B (X19).
FIG. 2.
Dendrogram showing the cluster analysis of the different PFGE XbaI patterns from 134 strains of serovar Typhi, generated by the GelCompar program using the UPGMA method, based on the matrix of F values.
Fifty-two isolates from patients with mild and moderate typhoid fever had a variety of XbaI patterns. There was only one isolate from a patient with fatal typhoid fever, and this had a unique X11 pattern (differing from X1 in three DNA fragments). The X1 profile was found in 67% of the isolates (four of six) from patients with severe typhoid fever. No distinct correlation between the molecular subtypes with disease severity was found (Table 1).
Twenty-three pairs of isolates obtained from blood and fecal swabs from the 23 individual patients were also compared. Paired blood-fecal isolates from 12 individual patients had indistinguishable patterns. In another 11 patients, the paired blood-fecal isolates differed by one to five bands (F = 0.86 to 0.91). Three of these pairs also had different phage types (VNS-UVS, UVS-UVS1, and D1-UVS1).
DISCUSSION
Strain typing is an integral part of molecular epidemiology and is used to discern the clonality of strains involved in local epidemics or global epidemiology. For serovar Typhi, the primary phenotypic method for subdividing serotypes of clinical or epidemiological importance is phage typing. In Papua New Guinea, the serovar Typhi strains that were isolated in 1992 to 1994 were mostly phage type D2. However, this phage type was replaced by D1, UVS, UVS1, and VNS in the recently isolated strains (1997 and 1999). Most of these new strains belonged to the untypeable Vi 1 type (78%). Changes of phage type could be due to phage type conversion among the Papua New Guinea serovar Typhi strains. Phage conversion is a well-documented phenomenon associated with the loss or acquisition of plasmids, phage conversion by temperate phages, and loss of lipopolysaccharide (2, 4, 17, 18, 26). It remains possible that the replacement of serovar Typhi strains of phage type D2 by strains of phage type D1, UVS, VNS, and UVS1 may represent a posttreatment effect and that strains have been selected following eradication of other competing strains. This is an interesting point to be analyzed in further studies.
Our observations in this study indicate that phage typing remains a useful epidemiological tool, but it must be complemented by molecular methods involving direct chromosomal DNA analysis. These might include plasmid analysis, PFGE, ribotyping, or PCR-based amplification fragment length polymorphism and single-strand conformation polymorphism (13, 19). Among these methods, PFGE is known to be the method of choice for subtyping of S. enterica, since it is very reliable and reproducible and has high discriminatory ability (1, 12). PFGE analysis of serovar Typhi isolates from different parts of Southeast Asia suggests that a wide genetic diversity exists among strains (22).
In our previous study of the serovar Typhi isolates from Papua New Guinea, a limited number of PFGE types were found, with one or two patterns among the 52 isolates studied (25). The limited diversity observed was probably due to the relatively recent introduction of typhoid fever into the country, since the disease was rarely seen before 1985 (16). Typhoid fever was reported to have spread only recently in endemic form in the highland regions and in some of the larger coastal towns. The population is now rather mobile due to improved infrastructure, making it unlikely that isolation or the lack of mobility are the causes of limited diversity among the strains.
Strains that were circulating in Papua New Guinea from 1992 to 1994 were most probably derived from a single introduced clone based on the observation that all the serovar Typhi strains belonging to phage type D2 were sensitive to the same antibiotics and had limited genetic diversity. The present strains (isolated in 1997 to 1999) have now spread and, presumably due to selective pressure, have developed some genetic variability. Since 1997, the number of PFGE subtypes has increased from 2 to 21. The X1 and X2 patterns that were prevalent in 1992 to 1994 could have established themselves in a number of ways. The single clone of X1 could have spread via the movement of people. The assumption is that as time passes and organisms spread, divergence may increase. The PFGE approach is a measurement of diversity within restriction endonuclease recognition sites distributed throughout the entire genome, which may have arisen as a result of mutations which remove or create recognition sites through insertion, deletion, translocation, or inversion or via mobile genetic elements. Some genetic diversity has occurred in serovar Typhi, but the extent to which mutations, deletions, or chromosomal rearrangements were responsible for the divergent XbaI profiles is not clear. Liu and Sanderson (9) suggested that lateral transfer of DNA from an external source is the most probable mechanism for producing genetic changes in serovar Typhi. In addition, it has been demonstrated by Liu and Sanderson (10) that serotype Typhi seems to be more susceptible than other Salmonella serotypes to genetic reorganization and that these genetic changes do not substantially alter the stability and survival of the bacterium. A high natural mutation rate has also been demonstrated in Salmonella (8).
It has been reported that the clinical manifestations differ markedly in different parts of the world where typhoid is endemic. In South America and parts of Southeast Asia (e.g., Malaysia and Thailand), typhoid fever manifests as a relatively mild illness with low fatality rates and minimal complications. In contrast, in Indonesia, severe and often fatal disease is frequently seen, with higher mortality. The reasons for these differences in disease severity are not known but may be related to differences in health care facilities, host immune responses, genetic factors, and perhaps differences in the strains of serovar Typhi circulating in the area of endemicity. Heneine et al. (7) characterized four serovar Typhi isolates from patients with severe and mild typhoid fever by using protein profiles, ribotyping, plasmid analysis, multilocus enzyme electrophoresis, and phage typing. No association was found between the characteristics of the isolates and disease severity. Grossman et al. (5) reported an association between flagellar serotypes and decreased severity of illness and invasiveness.
In our previous study, certain PFGE molecular subtypes of serovar Typhi isolates were associated with the ability to cause fatal or nonfatal disease (25). All 11 isolates obtained from patients with fatal typhoid fever were identical and had the same PFGE pattern combination, whereas the isolates from patients with nonfatal typhoid fever had various PFGE pattern combinations. In the present study, only one isolate from a patient with fatal typhoid fever was available for analysis. This particular isolate had a unique X11 profile, which differed from the X1 pattern in three DNA fragments. At this point, interpretation of the PFGE profiles in terms of disease manifestations is hampered by insufficient clinical information concerning the patients. No association was found between the phage types and PFGE subtypes in that no particular pulsed-field profiles were linked to any phage types. PFGE was definitely more discriminatory than phage typing since it could further differentiate the four phage types into 21 PFGE profiles.
In addition to its use as an epidemiological tool, PFGE is very useful for studying the in vivo stability of a pathogen within an individual. Comparison of pairs of isolates obtained from different body sites from 12 individual patients revealed indistinguishable patterns and phage types. This shows that in these patients, the strains inhabiting different sites are the same. However, the study also indicates that serovar Typhi strains isolated from blood and fecal samples from another 11 patients showed genetic changes that may have occurred in vivo during the course of the infection. These variations in strains from different bodily sites may arise as a result of selective pressure owing to host immune response or antibody therapy. Such variations in the strains indicate that the strains are not independent clones but are so closely related that they are presumed to be the same strain or a clonal variant. An exception was seen in three patients, for whom there is a possibility that they were infected simultaneously by different serovar Typhi strains since these strains not only differed in their PFGE profiles (F = 0.85 to 0.94) but also differed in their phage types. The in vivo instability of the microorganisms was also noted in our previous study (25) and in another study by others (11). The genetic relatedness of strains of Candida obtained from different parts of the body suggested a possibility of “substrain shuffling” within a patient.
In conclusion, we have demonstrated that multiple clones of serovar Typhi are now circulating in the Papua New Guinea population and that the present strains are probably derived from a common ancestral clone. The study reiterates the usefulness of PFGE in assessing the genetic diversity of serovar Typhi for both long-term epidemiology and in vivo stability and instability within an individual patient.
Acknowledgments
This work was supported by IRPA grant 06-02-03-0750, provided by the Ministry of Science, Technology and Environment, Malaysia, and by funding from the Government of Papua New Guinea to the Papua New Guinea Institute of Medical Research.
We thank the research nurses, Paul Kave and Agnes Javati, for collection of specimens and clinical data, and we thank Tilda Wal for preservation and shipping of the isolates from Papua New Guinea to Malaysia and Son Radu, Universiti Putra Malaysia, for useful comments on the manuscript.
REFERENCES
- 1.Arbeit, R. D. 1999. Laboratory procedures for the epidemiologic analysis of microorganisms, p. 116-137. In P. R. Murray, E. J. Barron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, D.C.
- 2.Baggesen, D. L., H. C. Wegener, and M. Madsen. 1997. Correlation of conversion of Salmonella enterica serovar Enteritidis phage type 1, 4, or 6 to phage type 7 with loss of lipopolysaccharide. J. Clin. Microbiol. 35:330-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauer, A. W., M. W. Kirby, J. C. Sherris, and M. Turck. 1966. Antibiotic susceptibility testing by a standardized single disc method. Am. J. Clin. Pathol. 45:493-496. [PubMed] [Google Scholar]
- 4.Frost, J. A., L. R. Ward, and B. Rowe. 1989. Acquisition of a drug resistance plasmid converts Salmonella enteritidis phage type 4 to phage type 24. Epidemiol. Infect. 103:243-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grossman, D. A., N. D. Witham, D. H. Burr, M. Lesmana, F. A. Rubin, G. K. Schoolnik, and J. Parsonnet. 1995. Flagella serotypes of Salmonella typhi in Indonesia: relationships among motility, invasiveness, and clinical illness. J. Infect. Dis. 171:212-216. [DOI] [PubMed] [Google Scholar]
- 6.Hampton, M. D., L. R. Ward, B. Rowe, and E. J. Threlfall. 1998. Molecular fingerprinting of multidrug resistant Salmonella serotype Typhi. Emerg. Infect. Dis. 4:317-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heneine, W., G. Matar, M. Reeves, and B. Swaminathan. 1991. Molecular characterization of Salmonella typhi isolates from patients with severe and mild typhoid fever. Eur. J. Epidemiol. 7:192-193. [DOI] [PubMed] [Google Scholar]
- 8.LeClerc, J. E., B. Li, W. L. Payne, and T. A. Cebula. 1996. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science 274:1208-1211. [DOI] [PubMed] [Google Scholar]
- 9.Liu, S. L., and K. L. Sanderson. 1995. Rearrangements in the genome of the bacterium Salmonella typhi. Proc. Natl. Acad. Sci. USA 92:1018-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu, S. L. and K. L. Sanderson. 1996. Highly plastic chromosomal organisation in Salmonella typhi. Proc. Natl. Acad. Sci. USA 93:10303-10308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lockhart, S. R., B. D. Reed, C. L. Pierson, and D. R. Soll. 1996. Most frequent scenario for recurrent Candida vaginitis is strain maintenance with “substrain shuffling”: demonstration by sequential DNA fingerprinting with probes Ca3, C1, and CARE2. J. Clin. Microbiol. 34:767-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maslow, J. N., A. M. Slutsky, and R. D. Arbeit. 1993. Application of pulsed field gel electrophoresis to molecular epidemiology, p. 563-572. In D. H. Persing, T. F. Smith, F. C. Tenover, and T. J. White (ed.), Diagnostic molecular microbiology: principles and applications. American Society for Microbiology, Washington, D.C.
- 13.Mirza, S., S. Kariuki, M. Z. Mamun, N. J. Beeching, and C. A. Hart. 2000. Analysis of plasmid and chromosomal DNA of multidrug resistant Salmonella enterica serovar Typhi from Asia. J. Clin. Microbiol. 38:1449-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.National Committee for Clinical Laboratory Standards. 1999. Performance standards for antimicrobial susceptibility testing. Ninth informational supplement. M100-S9, vol. 19, no. 1. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 15.Navarra, F., T. Llovet, M. A. Echeita, P. Coll, A. Aladuena, M. A. Usera, and G. Prats. 1996. Molecular typing of Salmonella enterica serovar Typhi. J. Clin. Microbiol. 34:2831-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Passey, M. N. Suve, J. Taime, A. Clegg, R. Lvivi, A. Javati, M. Joannes, K. Karigifa, H. Kave, P. Kave, S. Kimin, S. Lupiwa, P. Namuisi, and M. Alpers. 1995. Highly endemic typhoid fever in Papua New Guinea. Southeast Asian J. Trop. Med. Public Health. 26(suppl. 2):83-84. [Google Scholar]
- 17.Rankin, S., and D. J. Platt. 1995. Phage conversion in Salmonella enterica serotype Enteritidis: implications for epidemiology. Epidemiol. Infect. 114:227-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ridley, A. M., P. Punia, L. R. Ward, B. Rowe, and E. J. Threlfall. 1996. Plasmid characterization and pulsed field gel electrophoretic analysis demonstrate that ampicillin-resistant strains of Salmonella enteritidis phage type 6a are derived from Salmonella enteritidis phage type 4. J. Appl. Bacteriol. 81:613-618. [DOI] [PubMed] [Google Scholar]
- 19.Struelens, M. J., Y. De Gheidre, and A. Deplano. 1998. Comparative and library epidemiological typing systems: outbreak investigations versus surveillance systems. Infect. Control Hosp. Epidemiol. 19:565-569. [DOI] [PubMed] [Google Scholar]
- 20.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thong, K. L., Y. M. Cheong, S. D. Puthucheary, C. L. Koh, and T. Pang. 1994. Epidemiologic analysis of sporadic Salmonella typhi isolates and those from outbreaks by pulsed field gel electrophoresis. J. Clin. Microbiol. 32:1135-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thong, K. L., S. D. Puthucheary, R. M. Yassin, P. Sudarmono, M. Padmidewi, E. Soewandoje, I. Handojo, S. Sarambath, and T. Pang. 1995. Analysis of Salmonella typhi isolates from Southeast Asia by pulsed field gel electrophoresis. J. Clin. Microbiol. 33:1938-1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thong, K. L., A. Cordano, R. M. Yassin, and T. Pang. 1996. Molecular analysis of environmental and human isolates of Salmonella typhi. Appl. Environ. Microbiol. 62:271-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thong, K. L., and T. Pang. 1996. A rapid, simplified method for preparation of chromosomal DNA from pathogenic bacteria for use in pulsed-field gel electrophoresis. Asia Pac. J. Mol. Biol. Biotechnol. 4:59-62. [Google Scholar]
- 25.Thong, K. L., M. Passey, A. Clegg, B. G. Combs, R. M. Yassin, and T. Pang. 1996. Molecular analysis of isolates of Salmonella typhi obtained from patients with fatal and non-fatal typhoid fever. J. Clin. Microbiol. 34:1029-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Threlfall, E. J., H. Chart, L. A. Ward, J. D. H. de Sa, and B. Rowe. 1993. Interrelationships between strains of Salmonella enteritidis belonging to phage types 4, 7, 7a, 8, 13, 13a, 23, 24 and 30. J. Appl. Bacteriol. 75:43-48. [DOI] [PubMed] [Google Scholar]