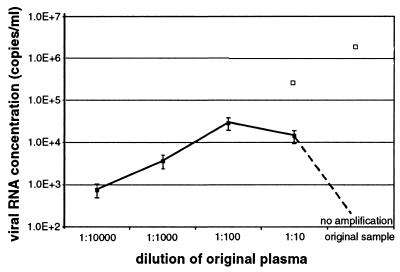
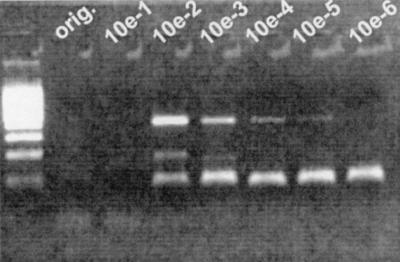
Viral hemorrhagic fevers (VHF) are acute infections with high case fatality rates, associated with the risk of nosocomial transmission (3). A rapid confirmation of the clinical diagnosis is therefore required by methods such as antigen capture enzyme immunoassay and serologic detection of immunoglobulin M. With the development of PCR technology, it has become possible to rapidly test for viruses that cause VHF (4, 5, 10). We have recently confirmed a case of acute yellow fever with fulminating hepatic failure by reverse transcription-PCR (RT-PCR) (2). In spite of the very high viral RNA concentration in the plasma sample, it initially tested negative. A confirmation by PCR would have been missed if we had not tested a duplicate sample, inoculated with in vitro-transcribed yellow fever virus RNA, to detect substances that are inhibitory to RT-PCR (5). When this control reaction failed, we diluted the plasma in log10 intervals, reprepared RNA from the diluted material, and tested it by RT-PCR. The virus was clearly detectable in the patient's plasma diluted 1:100, 1:1,000, and 1:10,000 (Fig. 1). Again, no virus was detectable in the undiluted sample, and detection was also partially inhibited in the 1:10 dilution. Quantitative real-time RT-PCR with the diluted material yielded a projected concentration of >106 copies of viral RNA per ml of the undiluted plasma (Fig. 1). Interestingly, we have observed a similarly strong inhibition phenomenon with plasma from a moribund patient with acute Ebola hemorrhagic fever from Gulu, Uganda (Fig. 2). The viral RNA concentration in this case was 6.9 × 108 copies/ml. No hemolysis, which often causes inhibition of PCR (1), was observed in plasma from the two patients. In both cases, however, the patients were suffering from severe organ manifestation of their disease (aspartate aminotransferase in yellow fever sample, 15,000 IU; alanine aminotransferase, 6,000 IU).
FIG. 1.
Results of quantitative real-time yellow fever virus RT-PCR (5) with original plasma and prediluted plasma. Closed data points depict the viral RNA concentration in plasma, as determined by testing of each dilution step of the patient sample (y axis). In the 1:10,000 to 1:100 dilution steps, the log10 of the determined viral RNA concentration correlated with the dilution factor. Open data points represent the extrapolated RNA concentrations in the undiluted and 1:10 diluted samples, indicating that amplification in these samples was inhibited (note the difference between extrapolated and observed RNA concentration).
FIG. 2.
Result of agarose gel electrophoresis after qualitative Ebola virus RT-PCR (5) with original plasma and prediluted plasma from a patient with acute Ebola fever. The dilution factor is depicted above each gel lane. No amplification was possible when the original plasma (orig.) and the 1:10-diluted plasma (10e-1) were tested. With a dilution of ⩾1:100, amplification was possible due to a decreased concentration of inhibitory substances in the analyzed sample.
Inhibition of RT-PCR in plasma samples has been reported to occur at a frequency of 0.34 to 2.1% of tests (patients infected with human immunodeficiency virus type 1 or hepatitis C virus, respectively [6, 9]). It can usually be circumvented by using standard ETDA sampling tubes (as done in both cases described here) and preparation procedures based on silica column purification (7, 8, 11). Furthermore, at least with plasma, inhibition is usually reversible upon repetition of extraction of nucleic acids from the same sample (6, 9). In these cases, however, the concentration in plasma of substances interfering with PCR appeared to be extraordinarily high: amplification was repeatedly inhibited in a second extraction, even though a reliable silica column purification method (viral RNA kit; Qiagen, Hilden, Germany) was applied. Moreover, partial inhibition occurred even in 1:10-diluted material. Two important consequences can be drawn from these observations.
(i) False-negative RT-PCR results are likely to occur for patients with severe viral hemorrhagic fevers, especially in the acute phase of the disease where a rapid confirmation is required. Their plasma may contain large amounts of RT-PCR inhibitors, probably resulting from the decay of tissue. These inhibitors can be detected by control reactions with spiked samples (low copy numbers of control RNA, 1 log10 above detection limit of the PCR) (5). Control reactions to detect inhibitors of RT-PCR are mandatory for a safe diagnosis for patients with suspected VHF.
(ii) When PCR is used for diagnosis of viral hemorrhagic fevers, dilutions of the test sample should be tested in parallel with the original sample. The high viral RNA concentration in samples from acute VHF cases is likely to facilitate the diagnosis in spite of the dilution factor.
REFERENCES
- 1.Al-Soud, W. A., and P. Radstrom. 2001. Purification and characterization of PCR-inhibitory components in blood cells. J. Clin. Microbiol. 39:485-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anonymous. 2001. Imported case of yellow fever, Belgium. W. H. O. Wkly. Epidemiol. Rep. 76:357-364. [Google Scholar]
- 3.Anonymous. 2001. Outbreak of Ebola haemorrhagic fever, Uganda, August 2000-January 2001. Wkly. Epidemiol. Rec. 76:41-46. [PubMed] [Google Scholar]
- 4.Demby, A. H., J. Chamberlain, D. W. G. Brown, and C. S. Clegg. 1994. Early diagnosis of Lassa fever by reverse transcription-PCR. J. Clin. Microbiol. 32:2898-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drosten, C., S. Göttig, S. Schilling, M. Asper, M. Panning, H. Schmitz, and S. Günther. 2002. Rapid detection and quantification of RNA of Ebola and Marburg viruses, Lassa virus, Crimean-Congo hemorrhagic fever virus, Rift Valley fever virus, Dengue virus, and yellow fever virus by real-time reverse transcription-PCR. J. Clin. Microbiol. 40:2323-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drosten, C., E. Seifried, and W. K. Roth. 2001. TaqMan 5′-nuclease human immunodeficiency virus type 1 PCR assay with phage-packaged competitive internal control for high-throughput blood donor screening. J. Clin. Microbiol. 39:4302-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hale, A. D., J. Green, and D. W. Brown. 1996. Comparison of four RNA extraction methods for the detection of small round structured viruses in faecal specimens. J. Virol. Methods 57:195-201. [DOI] [PubMed] [Google Scholar]
- 8.Lantz, P. G., W. Abu al-Soud, R. Knutsson, B. Hahn-Hagerdal, and P. Radstrom. 2000. Biotechnical use of polymerase chain reaction for microbiological analysis of biological samples. Biotechnol. Annu. Rev. 5:87-130. [DOI] [PubMed] [Google Scholar]
- 9.Nolte, F. S., M. W. Fried, M. L. Shiffman, A. Ferreira-Gonzalez, C. T. Garrett, E. R. Schiff, S. J. Polyak, and D. R. Gretch. 2001. Prospective multicenter clinical evaluation of AMPLICOR and COBAS AMPLICOR hepatitis C virus tests. J. Clin. Microbiol. 39:4005-4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teichmann, D., M. P. Grobusch, H. Wesselmann, B. Temmesfeld-Wollbruck, T. Breuer, M. Dietel, P. Emmerich, H. Schmitz, and N. Suttorp. 1999. A haemorrhagic fever from the Cote d'Ivoire. Lancet 354:1608. [DOI] [PubMed] [Google Scholar]
- 11.Willems, M., H. Moshage, F. Nevens, J. Fevery, and S. H. Yap. 1993. Plasma collected from heparinized blood is not suitable for HCV-RNA detection by conventional RT-PCR assay. J. Virol. Methods 42:127-130. [DOI] [PubMed] [Google Scholar]