Abstract
Based on phage display technology, a peptide-mediated magnetic separation technique was developed to facilitate selective isolation of Mycobacterium avium subsp. paratuberculosis (M. paratuberculosis) from bulk milk of naturally infected dairy herds. Nine recombinant bacteriophages binding to M. paratuberculosis were isolated from a commercial phage-peptide library encoding random 12-mer peptides. Nucleotide sequencing revealed the deduced sequence of the binding peptides. One peptide with the sequence NYVIHDVPRHPA, designated aMP3, was chemically synthesized with an amino-terminal biotin residue attached via an amino-hexacarbonic acid spacer molecule. Paramagnetic beads coated with the phage or with peptide aMP3 enabled the capture of M. paratuberculosis from milk. Combining this peptide-mediated magnetic separation with an ISMav2-based PCR allowed the detection of M. paratuberculosis in artificially spiked milk down to a concentration of 101 ml−1. Experiments using milk from naturally infected cows and bulk milk samples from infected herds demonstrated that the peptide-mediated capture PCR is sufficiently sensitive to detect single strong shedders in pooled milk samples. The method, for the first time, applies phage display technology to microbial diagnostics and has potential value as a completely standardizable tool for the routine M. paratuberculosis screening of bulk milk samples at acceptable costs.
Paratuberculosis or Johne's disease is a chronic and incurable granulomatous bowel disease mainly affecting ruminants. It occurs worldwide with increasing frequency, leading to growing economic losses (16, 20, 31). The disease is caused by Mycobacterium avium subspecies paratuberculosis (M. paratuberculosis), a member of the M. avium complex (25), commonly defined by its slow and mycobactin-dependent growth and the presence of multiple copies of the insertion element IS900 (16). Animals mostly get infected in the first weeks of life via the colostrum or contaminated milk. After an incubation the typical diarrhea may not develop until an age of 3 to 5 years. During the long incubation period M. paratuberculosis is shed intermittently in low numbers; in contrast, clinically infected animals characterized by an incurable diarrhea shed as many as 5 × 1012 M. paratuberculosis bacteria per day (7) while maintaining a regular appetite and normal body temperature. Due to these initially nonalarming clinical findings, milk producers do not commonly isolate these animals immediately. Consequently, a small number of clinically infected animals accounts for the vast majority of M. paratuberculosis introduced into the environment via feces and into the food chain via milk.
Presently, public health concerns about the presence of M. paratuberculosis in milk supplies are increasing due to a reemerging debate potentially linking M. paratuberculosis to human Crohn's disease (6, 17, 27). This is mainly due to the detection of IS900-positive mycobacteria in tissues of Crohn's disease patients (9) and the observation of an extraordinary temperature resistance resulting in M. paratuberculosis not being fully inactivated by pasteurization temperatures (13, 33) and surviving the cheese-manufacturing process (30).
In addition, paratuberculosis eradication programs frequently have not been successful (2) and an M. paratuberculosis reservoir in nonruminant wildlife has been discovered (3, 4). Furthermore, no sufficiently sensitive and rapid diagnostic system for the detection of M. paratuberculosis in bulk milk is commercially available to date. Laboratory methods described are labor intensive and difficult to standardize (14, 15) and, therefore, are not suitable for routine applications to a large number of bulk milk samples.
Phage display technology is a powerful molecular tool. It involves the expression of random peptides or proteins on the surface of a bacteriophage appended to a recombinant viral structural protein (1). The essential feature of the technology is the physical link of the phenotype to the genetic information contained within the phage particle. This allows the purification of single phages with a desired binding specificity from a random phage library encompassing 109 different peptides by a few rounds of biopanning, affinity-based enrichment, and subsequent amplification in Escherichia coli. The technology has been used predominantly to isolate peptides binding specifically to isolated proteins or inorganic material (29, 35). Also, the ability to isolate specific phages by panning from complex target structures has been shown (24). However, despite its enormous potential, the technique has not been used as a tool in routine diagnostic microbiology.
In the paper presented here, we describe the isolation of a peptide from a phage- peptide library, which combined with magnetic separation technology allows the detection of M. paratuberculosis in artificially contaminated milk with a sensitivity comparing favorably to that of immunomagnetic separation with polyclonal antibodies (14). Using this peptide in a peptide-mediated, capture PCR approach with primers specific for the M. paratuberculosis insertion element ISMav2 (32), we were able to reproducibly detect M. paratuberculosis in bulk milk samples from M. paratuberculosis-infected dairy herds. As the test is based solely on defined reagents, it might be suitable for routine testing of pooled herd milk samples. This would allow the detection and removal of strong shedders from the herd at acceptable costs and, therefore, reduce environmental contamination, lower the infection pressure in the herd, and decrease public health concerns.
MATERIALS AND METHODS
Bacterial strains, bacteriophages, plasmids, primers, and growth conditions.
The bacterial strains, bacteriophages, plasmids, and primers used in this study are listed in Table 1. Mycobacteria were grown on Middlebrook 7H10 agar (Difco Laboratories, Detroit, Mich.) supplemented with 100 ml of oleic acid-albumin-dextrose-catalase enrichment (sodium chloride [145 mM], bovine serum albumin [fraction V; 0.5 g], dextrose [1.1 M], catalase [3 mg], oleic acid [60 μl], glycerol [0.2%]) and 2 μg of mycobactin ml−1 (Symbiotics, Lyon, France). For further use mycobacteria were harvested by careful removal from the agar and resuspended in phosphate-buffered saline (PBS) buffer. Homogenization of the suspension was done by vortexing for 5 min with glass beads (30 beads of 2-mm diameter per 5 ml of bacterial suspension in a polypropylene tube).
TABLE 1.
Strains, phages, and primers used in this study
| Strain, primer, or phage | Characteristics | Source |
|---|---|---|
| Strains | ||
| M. avium subsp. paratuberculosis | Strain 6783 (DSM 44135) | Laboratory reference strain (clinical isolate) |
| M. avium subsp. avium | DSM 44157 | Reference strain obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany |
| E. coli Top 10 F' | F' mcrA Δ(mrr-hsdRMS-mcrBC) Φ80/acZΔM15 ΔlacX74 recA1 deoR araD139 Δ(araleu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| Primers | ||
| −96 M13gIII | 5′-CCC TCA TAG TTA GCG TAA CG-3′ | New England Biolabs |
| +130 M13gIII | 5′-TCA CCT CGA AAG CAA GCT GA-3′ | New England Biolabs |
| ISMav1 | 5′-GTA TCA GGC CGT GAT GGC GG-3′ | This work |
| ISMav2 | 5′-CGC GAC CAG CGC TCG ATA CA-3′ | This work |
| Phage library | ||
| Ph.D.-12 | New England Biolabs |
E. coli strains were grown on Luria-Bertani medium; for the plating of phages isopropyl β-d-thisqogalactopyranoside (IPTG; 1 mM final concentration) and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 60 mM final concentration) were added to the top agarose.
Selection of bacteriophages—biopanning.
Biopanning was performed based on the Ph.D.-12 phage display library (New England Biolabs, Beverly, Mass.). The library contains random 12-mer peptides with a complexity of 1.9 × 109and has a concentration of 3.8 × 1012 PFU per ml (PFU ml−1). In the first round of panning, 10 μl of the library (3.8 × 109 PFU ml−1) was incubated with 1 ml of M. paratuberculosis strain 6783 (optical density at 660 nm = 0.5) for 1 h at room temperature. Mycobacteria were washed eight times with Tris-buffered saline-Tween buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, and 0.05% Tween 20) by centrifugation (5 min, room temperature, 16,000 × g). Phage particles were eluted with 1 ml of a glycine-bovine serum albumin buffer (0.2 M glycine-HCl [pH 2.2] and 1 mg of bovine serum albumin ml−1). The eluate was neutralized immediately with 150 μl of Tris-HCl buffer (1 M [pH 9.1]). The PFU titer was determined by serial 10-fold dilution and plating on E. coli strain Top 10 F' (Invitrogen, Carlsbad, Calif.), and bacteriophages were amplified for the second round of biopanning on the same strain. Amplified phage particles were precipitated using polyethylene glycol (20%) and sodium chloride (2.5 M) and were resuspended in PBS (150 mM NaCl, 1.5 mM KH2PO4, 9 mM Na2HPO4, and 2.5 mM KCl [pH 7.2]). In the following rounds of biopanning an average of 3.75 × 1011 PFU of the amplified phages was used. Biopanning was repeated five times; in the final round of panning Tris-buffered saline-Tween buffers containing either guanidine hydrochloride (4 M) or urea (6 M) were used. After the final round of biopanning 30 single plaques were picked; specific DNA was amplified by PCR using primers −96 M13gIII and +130 M13gIII (Table 1) and was subjected to DNA-sequencing analysis.
Labeling of phage particles.
Selected phages were amplified, precipitated, and resuspended in PBS to a final concentration of 1012 PFU ml−1. For labeling with fluorescein isothiocyanate (FITC; Pierce Inc., Rockford, Ill.), 500 μl of carbonate buffer (50 mM [pH 9.8]), 1 ml of PBS, and 400 μl of FITC (1 mg ml of PBS−1) was added to 500 μl of phage suspension and incubated overnight at 4°C. Unbound FITC was removed by precipitating phage particles as described above. For labeling with biotin, N-hydroxysuccinimide coupled to biotin via a long spacer molecule (NHS-LC biotin; Molecular Biosciences Inc., Boulder, Colo.) was dissolved in dimethyl sulfoxide at 5 mg ml−1; 10 μl of NHS-LC biotin solution was added to 100 μl of phage suspension and incubated on ice for 1 h. Unbound NHS-LC biotin was removed by precipitating phage particles as described above. For labeling with alkaline phosphatase, the biotinylated phage (1011 PFU) was incubated with 50 μl of streptavidin-alkaline phosphatase (93 μg ml−1) for 1 h at room temperature. Unbound enzyme was removed by 15 min of dialysis on floating membranes with a pore size of 0.025 μm (Millipore, Eschborn, Germany). For the coupling of phages to paramagnetic beads, 100 μg of streptavidin MagneSphereparamagnetic particles (Promega Inc., Madison, Wis.) was incubated with 0.5 × 1012 PFU in PBS for 6 h at 4°C. To remove unbound phage, the particles were washed three times with PBS.
Plate binding assay.
A mycobacterial suspension was prepared in carbonate buffer (35 mM NaHCO3, 15 mM Na2CO3 [pH 9.8]) and adjusted to an optical density of 1.0 at 660 nm, corresponding to approximately 108 bacteria per ml. Maxisorp microtiter plates (Nunc, Roskilde, Denmark) were coated using 100 μl of bacterial suspension per well and incubated overnight at 4°C. Blocking was done overnight at 4°C with the gelatin (0.5%)-supplemented supernatant of an E. coli strain Top 10 F' culture infected with the whole phage library. To investigate the binding of alkaline phosphatase-labeled phages to a variety of M. paratuberculosis isolates and to an M. avium reference strain, 100 μl of serial twofold dilutions (starting concentration, 1012 PFU ml−1) of the phage clones was incubated on the plates for 1 h at room temperature, washed with 0.1× PBS containing 0.05% Tween 20, and developed with p-nitrophenyl phosphate.
Synthesis and coupling of peptides.
Based on the deduced amino acid sequence obtained from the isolated phage particles, peptides were synthesized with amino terminal biotinylation using amino-hexacarbonic acid as spacer (Affina Immuntech, Berlin, Germany). The peptide was dissolved in 50 μl of dimethyl sulfoxide and 950 μl of distilled water in a stock concentration of 10 mg ml−1. For coupling to paramagnetic particles 100 μg of streptavidin MagneSphereparamagnetic particles (Promega) was incubated with 20 μg of biotinylated peptide in PBS for 6 h at 4°C. To remove unbound peptide, the particles were washed three times with PBS and resuspended in 100 μl of PBS.
Enrichment of M. paratuberculosis from milk.
One-milliliter samples (i) of pasteurized whole milk spiked with 100 to 105 M. paratuberculosis PFU, (ii) of milk from fecal culture-positive cows, and (iii) of bulk milk from herds in eastern Germany that had tested positive in a milk serum enzyme-linked immunosorbent assay (ELISA) (Svanovir, Göteborg, Sweden [36]) were incubated with 10 μl of the phage- or peptide-coated paramagnetic particles at 4°C with slight agitation overnight. The particles were isolated as recommended by the manufacturer, washed three times with 1 ml of 0.1× PBS containing 0.05% Tween 20, and resuspended in 50 μl of Tris-EDTA (TE) buffer. To visualize the binding of M. paratuberculosis to phage- or peptide-coated paramagnetic particles, 5 μl of the suspension was transferred to a microscopic slide, air dried, and stained with the acridine orange stain for mycobacteria (28). Slides were examined using a fluorescent microscope; acid-fast bacilli showed a bright orange fluorescence against a dark background, whereas the paramagnetic particles did not fluoresce.
PCR on peptide-captured mycobacteria.
After peptide capture the mycobacterium-containing paramagnetic particles were resuspended in 50 μl of 0.1× TE buffer (1× TE buffer is 10 mM Tris-HCl [pH 8.0] and 1 mM EDTA) and boiled in a microwave oven for 8 min at 180 W. DNA was purified using the Gene Clean Kit (Q-biogene Inc., Carlsbad, Calif.) according to the manufacturer's instructions. PCR was performed using primers ISMav1 and ISMav2 (Table 1), based on the insertion sequence-like element ISMav2 (32). Briefly, the purified DNA was divided into four aliquots of 5 μl each, and one aliquot of the purified DNA was spiked with M. paratuberculosis DNA to serve as positive control. Twenty microliters of master mix (1.5 mM MgCl2, 1× PCR buffer [Invitrogen], a 0.2 mM concentration of each deoxnynucleoside triphosphate, a 0.5 μM concentration of each primer, and Taq DNA polymerase [0.02 U μl−1] [Invitrogen]) was added to each tube; after an initial denaturation step (94°C, 3 min) 32 PCR cycles (94°C, 30 s; 64.5°C, 30 s; and 72°C, 30 s) were performed, followed by a final extension step (72°C, 10 min). The PCR products were analyzed on a 1.5% agarose gel containing ethidium bromide (0.2 μg ml−1). To assess the minimum detection limit of the capture PCR, a serial 10-fold dilution of an M. paratuberculosis suspension with an initial concentration of 108 ml−1 (determined by counting in a Thoma chamber [Roth, Karlsruhe, Germany]) was prepared in pasteurized milk. Milk samples (1 ml) were inoculated with M. paratuberculosis to achieve final concentrations of calculated 100, 101, 102, 103, 104, and 105 bacteria ml of milk−1. Each sample was incubated with peptide-coated paramagnetic beads (10 μl) and processed as described above.
RESULTS
Isolation and characterization of recombinant bacteriophages binding to M. paratuberculosis.
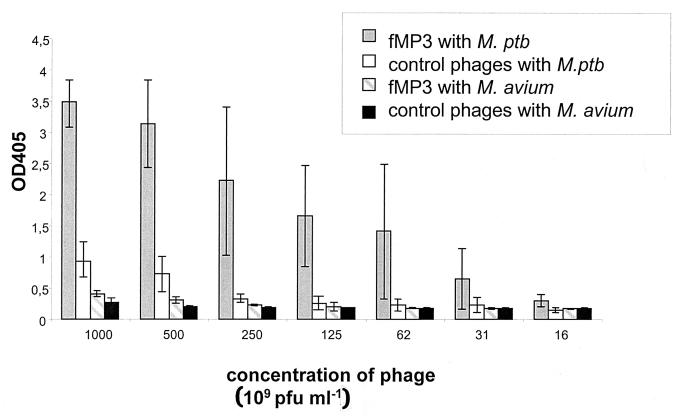
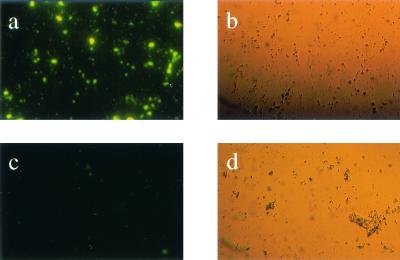
After five rounds of biopanning, with chaotropic buffers used in the last round, 30 single plaques were picked and subjected to nucleotide sequencing. Nine different sequences were obtained (Table 2). The quality of M. paratuberculosis binding was confirmed in a plate binding assay with a twofold titration of alkaline phosphatase-labeled phages and M. paratuberculosis or M. avium serving as solid-phase antigen. All isolated phages showed a binding to M. paratuberculosis at least twice as high as that for the phage library used as control (Fig. 1). To further assess the quality of binding, phages were labeled with FITC and incubated with M. paratuberculosis. It was shown that the specific phages bound to M. paratuberculosis, whereas the original phage library did not (Fig. 2). Phage isolate fMP3 consistently reacted strongly with M. paratuberculosis in both tests and was used for further analyses. To prove the specificity of fMP3 for M. paratuberculosis, the plate binding assay was repeated using M. avium as solid-phase antigen. In this test phage fMP3 bound only poorly to M. avium (Fig. 1); a reaction twice as strong as that of the original phage library serving as control could not be shown.
TABLE 2.
Isolated phages and deduced peptide sequences
| Phage or peptide | Sequence |
|---|---|
| Phages | |
| fMP1 | 5′-TCT ACT GTG CCG CGT ATG CCG CTT TCT CCG CCT AAT-3′ |
| fMP2 | 5′-TTT ATG AAT CCT CTT CAT CCG AGG GTG TTG CGG CCT-3′ |
| fMP3 | 5′-AAT TAT GTG AAT CAT GAT GTG CCG CGT CAT CCT GCG-3′ |
| fMP4 | 5′-GTG AAT ATG GGG AAT TAT AGG CAG CAG CAG CCG CTG-3′ |
| fMP5 | 5′-AGT ATG CTG GAT CTT TTT CCG CGT GCT GCT TCT TAT-3′ |
| fMP6 | 5′-TCT GTG TAT AAT GTT CGG CCT TCT TCG CTG TCT GCT-3′ |
| fMP7 | 5′-ACT GTG ATG GCT TCT CCG ACG ATG AAG AGT AAT TCT-3′ |
| fMP8 | 5′-GTG AAT ATG GGG AAT TAT AGG CAG CAG CAG CCC CAG-3′ |
| fMP9 | 5′-AAG TTT CCG GGT CCT AAT TGT TGT CAT GCT CTG CCG-3′ |
| Peptides | |
| aMP1 | S T V P R M P L S P P N |
| aMP2 | F M N P L H P R V L R P |
| aMP3 | N Y V I H D V P R H P A |
| aMP4 | V N M G N Y R Q Q Q P L |
| aMP5 | S M L D L F P R A A S Y |
| aMP6 | S V Y N V R P S S L S A |
| aMP7 | T V M A S P T M K S N S |
| aMP8 | V N M G N Y R Q Q Q P Q |
| aMP9 | K F P G P N C C H A L P |
| aMPr (control peptide) | H S Q P K Q V K K A S R |
FIG. 1.
Plate binding assay using M. paratuberculosis (M. ptb) and M. avium as solid-phase antigen and serial twofold dilutions of alkaline phosphatase-labeled phage fMP3 and control phages (initial phage library). The bars are the arithmetic means of the normalized optical densities at 405 nm (OD405) obtained in three independent experiments, each performed in duplicate with the lines indicating the standard deviation.
FIG. 2.
Binding of phage fMP3 to M. paratuberculosis. Phage fMP3 (a and b) and the original phage library as control phages (c and d) were labeled with FITC and incubated with M. paratuberculosis. Specific binding of phage fMP3 to M. paratuberculosis was shown by comparing fluorescence (a and c) and light microscopy (b and d).
Binding of M. paratuberculosis to paramagnetic particles coated with recombinant phage fMP3 or fMP3-derived peptide.
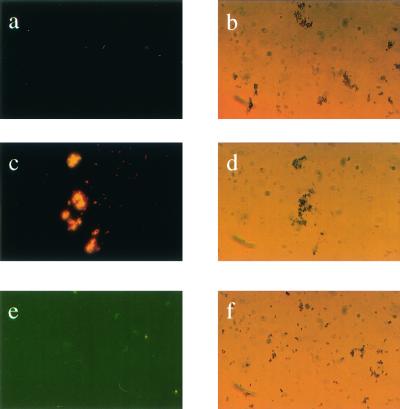
Phage fMP3 and peptide aMP3 synthesized based on the deduced amino acid of fMP3 were coupled to paramagnetic beads; the initial phage library and a random peptide (fMPr) were used as a control. An acridine orange stain and subsequent fluorescent microscopy revealed that M. paratuberculosis bound specifically to the paramagnetic beads coated with phage fMP3 or peptide aMP3 (Fig. 3). Paramagnetic particles coated with the phage library or with the control peptide aMPr did not facilitate attachment of M. paratuberculosis (Fig. 3).
FIG. 3.
Capture of M. paratuberculosis using peptide-coated paramagnetic beads. Acridine orange stain of paramagnetic beads alone (a and b) and M. paratuberculosis incubated with coated paramagnetic beads carrying peptide aMP3 (c and d) or peptide aMPr (negative control; e and f). Specific binding of peptide aMP3 to M. paratuberculosis was shown by comparing fluorescence microscopy (a, c, and e) and light microscopy (b, d, and e).
Phage- and peptide-mediated capture PCR for the detection of M. paratuberculosis in milk.
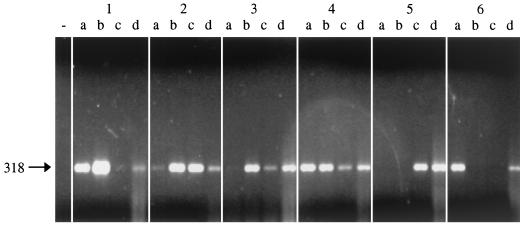
Pasteurized milk was spiked with M. paratuberculosis at concentrations of 100 to 105 PFU ml−1. When a capture assay with paramagnetic beads coated with phage fMP3 or peptide aMP3 followed by boiling and DNA purification was performed, M. paratuberculosis was detected in a PCR with ISMav2 primers. By using phage-mediated capture, M. paratuberculosis could be detected at a concentration of 102 PFU ml−1. The peptide-mediated capture resulted in the detection of 101 M. paratuberculosis PFU ml−1. The results were highly reproducible with usually at least two of the three parallel reactions being positive (Fig. 4).
FIG. 4.
Peptide-mediated capture PCR of M. paratuberculosis from milk spiked with M. paratuberculosis using ISMav2-derived primers. Blocks 1 to 6 correspond to milk samples containing 105 to 100 ml of M. paratuberculosis−1. Lanes a to c contain the products of three parallel PCRs of the respective sample; lanes d contain the internal positive control (sample spiked with M. paratuberculosis DNA), and − indicates the position of the negative control (coated beads and milk only). The arrow to the left indicates the expected position of the PCR product of 318 bp in length.
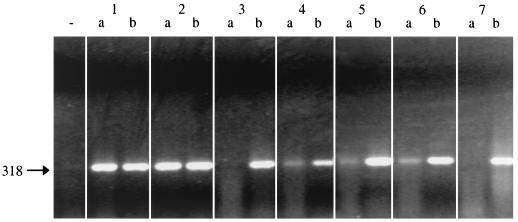
Peptide-mediated capture PCR for the detection of M. paratuberculosis in milk samples from seropositive cows and bulk milk samples from seropositive herds.
In order to test the peptide-mediated capture PCR on naturally infected milk, samples from nine cows that reacted serologically positive for M. paratuberculosis in milk or blood ELISA were tested with the method described above. Seven of the nine milk samples revealed positive results by PCR (data not shown). As these findings implied a high sensitivity of the capture PCR developed, milk samples from serological paratuberculosis-positive herds were tested. The method was applied to seven bulk milk samples obtained from seropositive herds. It was found that five of seven samples reacted positively in the peptide-mediated capture PCR (Fig. 5).
FIG. 5.
Peptide-mediated capture PCR of M. paratuberculosis from bulk milk of ELISA-positive herds using ISMav2-derived primers. Blocks 1 to 7 correspond to milk samples from seven different ELISA-positive herds. Lanes a contain the products of the PCRs of the respective sample; lanes b contain the internal positive control (sample spiked with M. paratuberculosis DNA), and − indicates the position of the negative control (coated beads and milk only). The arrow to the left indicates the expected position of the PCR product of 318 bp in length.
DISCUSSION
This study describes a peptide-mediated capture PCR assay for the specific detection of M. paratuberculosis in milk and is the first utilization of phage display technology for developing a diagnostic test. The procedure is simple and rapid and can be standardized and automated and is, therefore, suitable for routine application in diagnostic laboratories. Such a diagnostic test is urgently required to implement effective control measures, since the prevalence of M. paratuberculosis in dairy cattle is increasing and since there are rising concerns over putative wildlife reservoirs (3, 4) and the zoonotic potential of the organism (17). As voluntary eradication and certification programs instituted in several European countries (22, 26) and in many states within the United States have not improved the general situation (34), mandatory control programs need to be devised (19). For dairy cattle, the most economic and efficient control regime would involve regular monitoring of herd milk samples for the presence of M. paratuberculosis.
Present techniques for the milk-based diagnosis of paratuberculosis mainly rely on the detection of antibodies in milk (21). However, as a positive antibody response in a herd does not necessarily indicate shedding of the organism, mandatory control measures should be based on the direct detection of the organism. Recent studies have shown that this can be done effectively using immunomagnetic separation (IMS) and subsequent PCR analyses (14, 15). Since, however, the method was performed with a polyclonal antibody, it would be difficult to standardize and certify it for routine applications.
In order to solve the problem of standardization, we propose the use of high-affinity peptides as an alternative to antibodies in routine diagnostic microbiology. In this study we used a commercially available phage-peptide library to identify a peptide that would specifically bind M. paratuberculosis. However it was necessary to introduce a final high-stringency wash with a chaotropic buffer during biopanning in order to identify specific phages with sufficient affinity to be used for mycobacterial capture in milk. This might have been due to the fact that milk contains a large variety of different proteins and peptides that likely coat the mycobacterial surface. This hypothesis is supported by the fact that IMS of M. paratuberculosis in whole milk has been shown to require a longer time than in PBS (14).
A special processing of milk samples is necessary to gain optimal results for PCR-based assays. Milk is a substrate that is known to be inhibitory to the PCR (5, 23); therefore, an elimination of milk constituents facilitates the prevention of false negative results. When the Gene Clean Kit is used, those constituents are mostly removed, and false-negative results are eliminated by incorporating an internal control as is recommended for diagnostic PCR (18). The failure of one of three PCRs in spiked milk samples containing 105 bacteria per ml as well as the differences in strength of the PCR products are likely to be caused by residual inhibitory substances commonly observed in milk (23). The failure of two of three PCRs in spiked milk containing 101 and 100 bacteria per ml are likely due to the lack of bacterial DNA in these vials.
The sensitivity that we obtained with a peptide-mediated capture PCR in artificially contaminated milk was comparable to the IMS results obtained with a polyclonal serum and subsequent culture of the organism (15). Thus, for the IMS PCR using IS900 primers, a detection limit of 103 mycobacteria per 50 ml was estimated, and we could detect 10 mycobacteria per ml. We used primers directed against a different insertion element of M. paratuberculosis, ISMav2, as unspecific reactions have been reported for IS900 (8, 10). Furthermore, ISMav2 is present in only three copies on the genome, and therefore, positive results obtained with this element potentially allow a detection of the organism also when using single-copy genes.
Binding of a peptide or an antibody to cultured bacteria does not necessarily ensure that it will detect bacteria shed by the infected animal, as the synthesis of surface structures is dependent on the environment (12). We showed that the peptide-mediated capture PCR also could be used successfully on milk samples of naturally infected animals as well as on bulk milk samples. Therefore, the peptide apparently binds to a structure expressed on the mycobacterial surface upon growth in vitro and in vivo. Due to the lack of toxic reagents, the preparation protocol is likely to facilitate not only PCR detection but also culturing of the affinity-purified pathogen.
In summary, we have, for the first time, applied phage display technology to microbial diagnostics, developing a peptide-mediated capture PCR for the detection of M. paratuberculosis in milk. Due to the defined sequence of the synthetic peptide, this assay can be standardized completely, allowing certification for routine applications. In addition, all steps of the method can be automated such that it could be applied to high-throughput screening when combined with a molecular beacon approach as described recently (11).
Acknowledgments
This work was supported by EU grant QLK 2-CT-2001-01420.
REFERENCES
- 1.Adda, C. G., R. F. Anders, L. Tilley, and M. Foley. 2002. Random sequence libraries displayed on phage: identification of biologically important molecules. Comb. Chem. High Throughput Screen 5:1-14. [DOI] [PubMed] [Google Scholar]
- 2.Bakker, D., P. T. Willemsen, and F. G. van Zijderveld. 2000. Paratuberculosis recognized as a problem at last: a review. Vet. Q. 22:200-204. [DOI] [PubMed] [Google Scholar]
- 3.Beard, P. M., M. J. Daniels, D. Henderson, A. Pirie, K. Rudge, D. Buxton, S. Rhind, A. Greig, M. R. Hutchings, I. McKendrick, K. Stevenson, and J. M. Sharp. 2001. Paratuberculosis infection of nonruminant wildlife in Scotland. J. Clin. Microbiol. 39:1517-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beard, P. M., K. Stevenson, A. Pirie, K. Rudge, D. Buxton, S. M. Rhind, M. C. Sinclair, L. A. Wildblood, D. G. Jones, and J. M. Sharp. 2001. Experimental paratuberculosis in calves following inoculation with a rabbit isolate of Mycobacterium avium subsp. paratuberculosis. J. Clin. Microbiol. 39:3080-3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bickley, J., J. K. Short, D. G. McDowell, and H. C. Parkes. 1996. Polymerase chain reaction (PCR) detection of Listeria monocytogenes in diluted milk and reversal of PCR inhibition caused by calcium ions. Lett. Appl. Microbiol. 22:153-158. [DOI] [PubMed] [Google Scholar]
- 6.Chamberlin, W., D. Y. Graham, K. Hulten, H. M. El Zimaity, M. R. Schwartz, S. Naser, I. Shafran, and F. A. El Zaatari. 2001. Review article: Mycobacterium avium subsp. paratuberculosis as one cause of Crohn's disease. Aliment. Pharmacol. Ther. 15:337-346. [DOI] [PubMed] [Google Scholar]
- 7.Clarke, C. J. 1997. The pathology and pathogenesis of paratuberculosis in ruminants and other species. J. Comp. Pathol. 116:217-261. [DOI] [PubMed] [Google Scholar]
- 8.Cousins, D. V., R. Whittington, I. Marsh, A. Masters, R. J. Evans, and P. Kluver. 1999. Mycobacteria distinct from Mycobacterium avium subsp. paratuberculosis isolated from the faeces of ruminants possess IS900-like sequences detectable by IS900 polymerase chain reaction: implications for diagnosis. Mol. Cell Probes 13:431-442. [DOI] [PubMed] [Google Scholar]
- 9.El-Zaatari, F. A., M. S. Osato, and D. Y. Graham. 2001. Etiology of Crohn's disease: the role of Mycobacterium avium paratuberculosis. Trends Mol. Med. 7:247-252. [DOI] [PubMed] [Google Scholar]
- 10.Englund, S., G. Bolske, and K. E. Johansson. 2002. An IS900-like sequence found in a Mycobacterium sp. other than Mycobacterium avium subsp. paratuberculosis. FEMS Microbiol. Lett. 209:267-271. [DOI] [PubMed] [Google Scholar]
- 11.Fang, Y., W.-H. Wu, J. L. Pepper, J. L. Larsen, S. A. E. Marras, E. A. Nelson, W. B. Epperson, and J. Christopher-Hennings. 2002. Comparison of real-time, quantitative PCR with molecular beacons to nested PCR and culture methods for detection of Mycobacterium avium subsp. paratuberculosis in bovine fecal samples. J. Clin. Microbiol. 40:287-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finlay, B. B., and S. Falkow. 1997. Common themes in microbial pathogenicity revisited. Microbiol. Mol. Biol. Rev. 61:136-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grant, I. R., H. J. Ball, and M. T. Rowe. 1998. Effect of high-temperature, short-time (HTST) pasteurization on milk containing low numbers of Mycobacterium paratuberculosis. Lett. Appl. Microbiol. 26:166-170. [DOI] [PubMed] [Google Scholar]
- 14.Grant, I. R., H. J. Ball, and M. T. Rowe. 1998. Isolation of Mycobacterium paratuberculosis from milk by immunomagnetic separation. Appl. Environ. Microbiol. 64:3153-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grant, I. R., C. M. Pope, L. M. O'Riordan, H. J. Ball, and M. T. Rowe. 2000. Improved detection of Mycobacterium avium subsp. paratuberculosis in milk by immunomagnetic PCR. Vet. Microbiol. 77:369-378. [DOI] [PubMed] [Google Scholar]
- 16.Harris, N. B., and R. G. Barletta. 2001. Mycobacterium avium subsp. paratuberculosis in veterinary medicine. Clin. Microbiol. Rev. 14:489-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hermon-Taylor, J., and T. Bull. 2002. Crohn's disease caused by Mycobacterium avium subspecies paratuberculosis: a public health tragedy whose resolution is long overdue. J. Med. Microbiol. 51:3-6. [DOI] [PubMed] [Google Scholar]
- 18.Ieven, M., and H. Goossens. 1997. Relevance of nucleic acid amplification techniques for diagnosis of respiratory tract infections in the clinical laboratory. Clin. Microbiol. Rev. 10:242-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kennedy, D. J., and G. Benedictus. 2001. Control of Mycobacterium avium subsp. paratuberculosis infection in agricultural species. Rev. Sci. Tech. 20:151-179. [DOI] [PubMed] [Google Scholar]
- 20.Kreeger, J. M. 1991. Ruminant paratuberculosis—a century of progress and frustration. J. Vet. Diagn. Investig. 3:373-382. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen, S. S., C. Gronbaek, J. F. Agger, and H. Houe. 2002. Maximum-likelihood estimation of sensitivity and specificity of ELISAs and faecal culture for diagnosis of paratuberculosis. Prev. Vet. Med. 53:191-204. [DOI] [PubMed] [Google Scholar]
- 22.Paisley, L. G. 2001. Economic aspects of disease monitoring with special reference to bovine paratuberculosis. Acta Vet. Scand. Suppl. 94:17-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riffon, R., K. Sayasith, H. Khalil, P. Dubreuil, M. Drolet, and J. Lagace. 2001. Development of a rapid and sensitive test for identification of major pathogens in bovine mastitis by PCR. J. Clin. Microbiol. 39:2584-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodi, D. J., R. W. Janes, H. J. Sanganee, R. A. Holton, B. A. Wallace, and L. Makowski. 1999. Screening of a library of phage-displayed peptides identifies human bcl-2 as a taxol-binding protein. J. Mol. Biol. 285:197-203. [DOI] [PubMed] [Google Scholar]
- 25.Saito, H., H. Tomioka, K. Sato, H. Tasaka, M. Tsukamura, F. Kuze, and K. Asano. 1989. Identification and partial characterization of Mycobacterium avium and Mycobacterium intracellulare by using DNA probes. J. Clin. Microbiol. 27:994-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scientific Committee on Animal Health and Animal Welfare, European Commission. 2000. Possible links between Crohn's disease and paratuberculosis. [Online.] http://europa.eu.int/comm/food/sc/scah/out38_en.pdf.
- 27.Sechi, L. A., M. Manuela, T. Francesco, L. Amelia, S. Antonello, F. Giovanni, and Z. Stefania. 2001. Identification of Mycobacterium avium subsp. paratuberculosis in biopsy specimens from patients with Crohn's disease identified by in situ hybridization. J. Clin. Microbiol. 39:4514-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smithwick, R. W., M. R. Bigbie, Jr., R. B. Ferguson, M. A. Karlix, and C. K. Wallis. 1995. Phenolic acridine orange fluorescent stain for mycobacteria. J. Clin. Microbiol. 33:2763-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smothers, J. F., and S. Henikoff. 2001. Predicting in vivo protein peptide interactions with random phage display. Comb. Chem. High Throughput Screen. 4:585-591. [DOI] [PubMed] [Google Scholar]
- 30.Spahr, U., and K. Schafroth. 2001. Fate of Mycobacterium avium subsp. paratuberculosis in Swiss hard and semihard cheese manufactured from raw milk. Appl. Environ. Microbiol. 67:4199-4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stabel, J. R. 1998. Johne's disease: a hidden threat. J. Dairy Sci. 81:283-288. [DOI] [PubMed] [Google Scholar]
- 32.Strommenger, B., K. Stevenson, and G. F. Gerlach. 2001. Isolation and diagnostic potential of ISMav2, a novel insertion sequence-like element from Mycobacterium avium subspecies paratuberculosis. FEMS Microbiol. Lett. 196:31-37. [DOI] [PubMed] [Google Scholar]
- 33.Sung, N., and M. T. Collins. 1998. Thermal tolerance of Mycobacterium paratuberculosis. Appl. Environ. Microbiol. 64:999-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wells, S. J., and B. A. Wagner. 2000. Herd-level risk factors for infection with Mycobacterium paratuberculosis in US dairies and association between familiarity of the herd manager with the disease or prior diagnosis of the disease in that herd and use of preventive measures. J. Am. Vet. Med. Assoc. 216:1450-1457. [DOI] [PubMed] [Google Scholar]
- 35.Whaley, S. R., D. S. English, E. L. Hu, P. F. Barbara, and A. M. Belcher. 2000. Selection of peptides with semiconductor binding specificity for directed nanocrystal assembly. Nature 405:665-668. [DOI] [PubMed] [Google Scholar]
- 36.Winterhoff, C., M. Beyerbach, M. Homuth, K. Strutzberg, and G.-F. Gerlach. 2002. Etablierung und Evaluation eines ELISA zum Nachweis von Antikörpern gegen Mycobacterium avium subspecies paratuberculosis in Milch. Dtsch. Tieraerztl. Wochenschr. 109:217-252. [PubMed] [Google Scholar]