Abstract
Streptococcus pneumoniae serotype 5 is the third most common capsular type causing invasive diseases in children younger than 5 years in Latin America. Preliminary data on Colombian serotype 5 isolates indicated a common clonal origin associated with resistance to tetracycline (TET) and chloramphenicol (CHL). We studied 172 S. pneumoniae serotype 5 invasive isolates from Argentina, Brazil, Colombia, Guatemala, Mexico, and Uruguay and confirmed the presence of the Colombia5-19 clone throughout Latin America. Fifteen subtypes of a pulsed-field gel electrophoresis pattern and 4 electrophoretic types (ET) were obtained. Most of the isolates from different geographical regions belonged to pattern A (34.3%), subtype A5 (41.9%), and ET1 (91.1%). The A pattern (n = 59) was resistant to TET and had variable resistance to CHL; it was present in Brazil (10.2%), Colombia (78%), Guatemala (8.5%), and Mexico (3.4%). Subtype A5 with variable susceptibility to TET and sensitive to CHL was found in Argentina (29.2%), Mexico (8.3%), and Uruguay (62.5%). Subtypes A1-A4, A7-A8, and A9-A11 (closely related to A) also shared ET1, while subtype A6 was assigned to ET1, ET2, and ET3. Eleven subtypes (n = 21) were found to be specific for one country each. In summary, the S. pneumoniae serotype 5 isolates from Latin American are genetically closely related but show different patterns of antibiotic resistance, probably as a result of horizontal transfer.
Streptococcus pneumoniae is present in the bacterial flora of the human upper respiratory tract. S. pneumoniae is the most important cause of community-acquired pneumonia, meningitis, otitis media, and bacteremia, particularly in the extremes of life (1, 23). It is estimated that more than a million children younger than 5 years die each year of pneumococcal pneumonia (11, 22). The principal S. pneumoniae virulence factor is the capsular polysaccharide. Only 10 to 12 of the 90 capsular serotypes described are responsible for most invasive illness, but their distribution follows different geographic patterns (1, 15, 30). Additionally, resistance to different antimicrobials has been widely documented and distributed through serotypes and countries (1, 9, 25, 35).
An S. pneumoniae surveillance program was started in 1994 in six Latin American countries (SIREVA-Vigía project) coordinated by the Pan-American Health Organization and cosponsored by the Canadian International Development Agency (6). This regional initiative was designed to obtain information about the S. pneumoniae serotype distribution in order to determine the ideal composition for a conjugate vaccine that could be useful for the region. Additionally, the project was aimed at monitoring the rates of S. pneumoniae serotype distribution and antimicrobial resistance (3, 4, 6, 8, 16, 17, 28). After a 5-year surveillance, a high prevalence of serotype 5 (9.6%) was documented in Latin America. In Argentina and Uruguay serotype 5 ranked second (14.1 and 14.8% respectively), in Chile it ranked third (11.7%), in Colombia it ranked fourth (7.9%), and in Brazil it ranked fifth (6.7%) (7).
In Colombia the serotype 5 isolates showed a unique pulsed-field gel electrophoresis (PFGE) pattern with resistance to tetracycline and chloramphenicol, suggesting the circulation of a specific clone (33). The pneumococcal molecular epidemiology network (21) recently recognized the clone as Colombia5-19 (K. Klugman, Minutes of the Fifth Meeting of PMEN, p. 2, 2001).
The aim of this study was to determine the genetic relatedness among S. pneumoniae serotype 5 invasive isolates recovered from children younger than 5 years from Argentina, Brazil, Colombia, Guatemala, Mexico, and Uruguay and to compare these results with the antibiotic resistance patterns of the isolates.
MATERIALS AND METHODS
Bacterial isolates.
S. pneumoniae capsular type 5 invasive isolates recovered from children younger than 5 years from Argentina, Brazil, Mexico, Guatemala, and Uruguay were submitted to the Microbiology Group at the Instituto Nacional de Salud in Colombia. The identities of all isolates were confirmed by standard methods (10). The serotyping was performed using the Quellung reaction with antisera produced by the Statens Seruminstitut (Copenhagen, Denmark) (32). Of 53 Colombian isolates analyzed, 43 had been previously studied by PFGE (33). Isolate INS-Sp Col 106 was the strain submitted to identify the clone Colombia5-19 (Klugman, 5th Meet. PMEN, 2001). Laboratory strain R6, kindly provided by Alexander Tomasz from The Rockefeller University, was included as a molecular weight marker.
Antimicrobial susceptibility.
Inhibition zones were determined by the Kirby-Bauer method, and MICs were determined by broth microdilution; the results were interpreted on the basis of the National Committee for Clinical Laboratory Standards (NCCLS) tables for the following antimicrobials: penicillin, ceftriaxone, chloramphenicol, tetracycline, trimethoprim-sulfamethoxazole, erythromycin, and vancomycin (24).
MLEE.
Serotype 5 isolates were typed by multilocus enzyme electrophoresis (MLEE) in Montevideo, Uruguay, as described elsewhere (29). Some modifications were made to the technique. Briefly, each isolate was grown in 5% sheep blood agar plates. The whole growth from the plates was pelleted and resuspended in 0.5 ml of lysis buffer (5 mM EDTA, 50 mM Tris [pH 7.5]) and frozen at −20°C for a minimum of 48 h. The cell debris was removed by centrifugation at 15,000 × g for 15 min at 4°C. Thawed lysates were absorbed into paper wicks and inserted into vertical slits cut in a 12% starch gel (Connaught Laboratories, Swiftwater, Pa.). After electrophoresis for 6 h, gel slices were stained for specific enzyme activities. The following enzymes were examined after electrophoresis in buffer system A: 6-phosphogluconate dehydrogenase, glutamate dehydrogenase, nucleoside phosphorylase, esterase, and phosphoglucose isomerase. The following enzymes were examined after electrophoresis in buffer system B: adenylate kinase, leucine aminopeptidase, l-lactate dehydrogenase, and glucose 6-phosphate dehydrogenase. The following enzymes were examined after electrophoresis in buffer system G: leucyl-alanine peptidase, leucyl-glycyl-glycine peptidase, and phenyl-leucine peptidase.
Each unique combination of migration patterns (equated with alleles at the corresponding gene loci) for the 12 enzymes studied was called an electrophoretic type (ET). All ETs were compared with each other to determine genetic relatedness (29).
PFGE.
S. pneumoniae isolates were grown in supplemented Todd-Hewitt broth (Difco, Becton Dickinson, Sparks, Md.) (37). S. pneumoniae chromosomal DNA embedded in agarose disks was prepared by a previously described method (31, 37). The disks were digested with 20 U of SmaI (Promega, Madison, Wis.). The molecular weight marker used was the lambda ladder (New England Biolabs, Beverly, Mass.) and the R6 strain. The SmaI macrorestriction fragments were separated by electrophoresis (CHEF DRII apparatus; Bio-Rad Laboratories, Richmond, Calif.) at 6 V/cm with switch times ramped from 1 to 30 s over a 23-h period at 11.3°C. PFGE patterns were classified according to Tenover's criterion. Isolates that had the same number of bands with the same molecular sizes were designated genetically indistinguishable and assigned a single pattern; an isolate was considered to be closely related to the pattern when its PFGE profile differed in two or three bands, indicating a change by a single event; and an isolate was considered to be possibly related when the PFGE profile differed in four to six bands, meaning a change by two independent genetic events (34).
To analyze the PFGE results, Diversity Database (Bio-Rad Laboratories) was used to automatically identify band positions and compare two patterns by calculating the Dice coefficient of similarity, SD. Dendrograms were generated using the unweighted pair group method of average linkage (UPGMA).
RESULTS
Bacterial isolates.
A total of 172 isolates were studied. Of these, 165 (96%) were recovered between 1993 and 1999, 23 from Argentina, 12 from Brazil, 53 from Colombia, 10 from Guatemala, 10 from Mexico, and 57 from Uruguay. These isolates corresponded to 100% of the serotype 5 isolates recovered in Colombia, Mexico, Uruguay, and Guatemala and to 50 and 40% from Argentina and Brazil, respectively. Additionally, four isolates recovered in 2000 from Colombia and three isolates recovered in 1988 and 1989 from Uruguay were studied.
In Table 1 the isolates are listed by general code, country code, and year of isolation. Isolates were recovered from children suffering from meningitis (n = 59), pneumonia (n = 101), arthritis (n = 2), sepsis (n = 6), or abscess (n = 1) or from sources without data (n = 3).
TABLE 1.
Serotype 5 S. pneumoniae isolates from Argentina, Brazil, Colombia, Guatemala, Mexico, and Uruguay
| Code | Country codea | Yr | MIC (μg/ml) ofb:
|
ET | PFGE type | ||
|---|---|---|---|---|---|---|---|
| TET | CHL | STX | |||||
| 1032 | ARG708 | 1995 | 0.12 | 4 | 1/19 | 1 | A5 |
| 1033 | ARG741 | 1995 | 4 | 4 | 1/19 | 1 | A5 |
| 1034 | ARG746 | 1995 | 0.25 | 4 | 1/19 | 1 | A5 |
| 1035 | ARG793 | 1995 | 0.12 | 4 | 1/19 | 1 | A5 |
| 1036 | ARG1033 | 1996 | 0.12 | 4 | 1/19 | 1 | A5 |
| 1037 | ARG1043 | 1996 | 0.25 | 4 | 2/38 | 1 | A5 |
| 1038 | ARG1061 | 1996 | 0.12 | 4 | 1/19 | 1 | A5 |
| 1039 | ARG1096 | 1996 | 0.5 | 4 | 1/19 | 1 | A5 |
| 1040 | ARG1105 | 1996 | 0.25 | 4 | 1/19 | 1 | A5 |
| 1041 | ARG1107 | 1996 | 0.25 | 4 | 2/38 | 1 | A5 |
| 1042 | ARG1108 | 1996 | 16 | 2 | 1/19 | 2 | A6 |
| 1043 | ARG1119 | 1996 | 0.25 | 4 | 2/38 | 1 | A5 |
| 1044 | ARG1127 | 1996 | 0.25 | 4 | 2/38 | 1 | A5 |
| 1045 | ARG1128 | 1996 | 0.25 | 4 | 2/38 | 1 | A5 |
| 1046 | ARG1152 | 1996 | 0.25 | 4 | 2/38 | 1 | A5 |
| 1047 | ARG1157 | 1996 | 0.25 | 4 | 2/38 | 1 | A5 |
| 1048 | ARG1164 | 1996 | 0.25 | 4 | 1/19 | 1 | A5 |
| 1049 | ARG1179 | 1997 | 16 | 2 | 0.25/4.7 | 1 | A5 |
| 1050 | ARG1180 | 1997 | 8 | 2 | 4/76 | 2 | A6 |
| 1051 | ARG1182 | 1997 | 16 | 2 | 0.25/4.7 | 1 | A5 |
| 1153 | ARG1206 | 1997 | 32 | 4 | 1/19 | 1 | A5 |
| 1054 | ARG1224 | 1997 | 0.12 | 4 | 2/38 | 1 | A5 |
| 1055 | ARG1240 | 1997 | 0.12 | 4 | 1/19 | 1 | A5 |
| 1010 | BRA56 | 1996 | 2 | 4 | 0.25/4.7 | 2 | A6 |
| 1011 | BRA57 | 1996 | 8 | 4 | 8/152 | 1 | A |
| 1012 | BRA71 | 1996 | 0.5 | 2 | 0.25/4.7 | 2 | A6 |
| 1013 | BRA102 | 1996 | 16 | 4 | 8/152 | 1 | A |
| 1015 | BRA169 | 1997 | 16 | 4 | 8/152 | 1 | A |
| 1016 | BRA171 | 1997 | 32 | 4 | 8/152 | 1 | A2 |
| 1017 | BRA210 | 1997 | 0.25 | 4 | 1/19 | 1 | A7 |
| 1009 | BRA39 | 1997 | 0.12 | 4 | 0.25/4.7 | 1 | A7 |
| 1020 | BRA410 | 1997 | 16 | 4 | 8/152 | 1 | A |
| 1018 | BRA213 | 1998 | 16 | 4 | 16/302 | 1 | A |
| 1019 | BRA291 | 1998 | 16 | 4 | 8/152 | 1 | A |
| 1007 | BRA5 | 1998 | 0.25 | 4 | 1/19 | 1 | A7 |
| 7 | COL7 | 1994 | 32 | 32 | 2/38 | 1 | A |
| 12 | COL12 | 1994 | 32 | 16 | 4/76 | 1 | A |
| 24 | COL24 | 1994 | 32 | 32 | 2/38 | 1 | A |
| 30A | COL30A | 1994 | 16 | 16 | 4/76 | 1 | A |
| 30B | COL30B | 1994 | 32 | 32 | 4/76 | 1 | A |
| 47 | COL47 | 1994 | 32 | 32 | 0.5/9.5 | 1 | A |
| 49A | COL49A | 1994 | 32 | 16 | 1/19 | 1 | A |
| 49C | COL49C | 1994 | 32 | 16 | 1/19 | 1 | A |
| 72 | COL72 | 1994 | 0.12 | 2 | 0.12/2.3 | 3 | A6 |
| 86 | COL86 | 1994 | 32 | 32 | 8/152 | 1 | A |
| 106 | COL106 | 1994 | 32 | 32 | 2/38 | 1 | A |
| 123 | COL123 | 1994 | 32 | 16 | 0.25/4.7 | 1 | A |
| 133 | COL133 | 1994 | 32 | 16 | 0.06/1.1 | 1 | A |
| 135 | COL135 | 1994 | 0.25 | 4 | 2/38 | 1 | A4 |
| 136 | COL136 | 1994 | 32 | 16 | 2/38 | 1 | A1 |
| 161 | COL161 | 1995 | 16 | 32 | 2/38 | 1 | A |
| 175 | COL175 | 1995 | 32 | 32 | 8/152 | 1 | A |
| 177 | COL178 | 1995 | 32 | 32 | 8/152 | 1 | A |
| 189 | COL190 | 1995 | 16 | 32 | 1/19 | 1 | A |
| 190 | COL191 | 1995 | 32 | 32 | 1/19 | 1 | A |
| 201 | COL202 | 1995 | 32 | 16 | 2/38 | 1 | A |
| 212 | COL213 | 1995 | 8 | 16 | 8/152 | 1 | A |
| 214 | COL215 | 1995 | 32 | 16 | 2/38 | 1 | A1 |
| 221 | COL222 | 1995 | 32 | 16 | 2/38 | 1 | A |
| 222 | COL223 | 1995 | 32 | 4 | 0.5/9.5 | 1 | A |
| 224 | COL225 | 1995 | 0.12 | 4 | 0.12/2.3 | 2 | A6 |
| 226 | COL227 | 1995 | 32 | 2 | 0.25/4.7 | 1 | A |
| 227 | COL228 | 1995 | 32 | 16 | 2/38 | 1 | A |
| 252 | COL253 | 1995 | 16 | 16 | 2/38 | 1 | A |
| 255 | COL256 | 1995 | 16 | 32 | 1/19 | 1 | A |
| 261 | COL262 | 1995 | 16 | 16 | 2/38 | 1 | A/PICK> |
| 282 | COL283 | 1995 | 16 | 16 | 0.06/1.1 | 1 | A |
| 283 | COL284 | 1995 | 16 | 16 | 1/19 | 1 | A |
| 299 | COL300 | 1995 | 16 | 16 | 0.06/1.1 | 1 | A |
| 303 | COL304 | 1995 | 16 | 16 | 0.25/4.7 | NDc | A |
| 316 | COL317 | 1996 | 32 | 32 | 8/152 | 1 | A |
| E1H | COLE1H | 1996 | 32 | 8 | 2/38 | 1 | A |
| E1LP | COLE1LP | 1996 | 32 | 32 | 2/38 | 1 | A |
| E16 | COLE16 | 1996 | 32 | 32 | 1/19 | 1 | A |
| E18 | COLE18 | 1996 | 32 | 32 | 4/76 | 1 | A |
| E45 | COLE45 | 1996 | 32 | 32 | 2/38 | 1 | A |
| E68 | COLE68 | 1996 | 32 | 32 | 2/38 | 1 | A1 |
| E72 | COLE72 | 1996 | 32 | 4 | 1/19 | 1 | A3 |
| E73 | COLE73 | 1996 | 32 | 32 | 2/38 | 1 | A |
| E86 | COLE86 | 1997 | 0.25 | 4 | 2/38 | 1 | A4 |
| E105 | COLE105 | 1997 | 32 | 32 | 2/38 | 1 | A |
| E149 | COLE145 | 1997 | 32 | 32 | 2/38 | 1 | A |
| E161 | COLE157 | 1997 | 0.12 | 4 | 2/38 | 1 | A8 |
| E178 | COLE174 | 1997 | 32 | 32 | 2/38 | 1 | A |
| E196 | COLE192 | 1998 | 0.25 | 4 | 2/38 | 1 | A8 |
| E225 | COLE220 | 1998 | 32 | 16 | 2/38 | 1 | A |
| E236 | COLE231 | 1998 | 32 | 16 | 8/152 | 1 | A |
| E324 | COLE318 | 1999 | 0.12 | 4 | 0.5/9.5 | 1 | A8 |
| E386 | COLE386 | 2000 | 32 | 16 | 32/608 | ND | A |
| E387 | COLE387 | 2000 | 32 | 16 | 16/304 | ND | A |
| E390 | COLE390 | 2000 | 16 | 16 | 2/38 | ND | A |
| E391B | COLE391B | 2000 | 16 | 16 | 1/19 | ND | A |
| 1878 | GUA168 | 1997 | 4 | 4 | 1/19 | ND | A |
| 1879 | GUA187 | 1997 | 8 | 4 | 4/76 | ND | A |
| 1880 | GUA753 | 1998 | 8 | 4 | 1/19 | ND | A14 |
| 1881 | GUA773 | 1998 | 8 | 4 | 1/19 | ND | A14 |
| 1882 | GUA783 | 1998 | 16 | 4 | 1/19 | ND | A15 |
| 1883 | GUA808 | 1998 | 8 | 4 | 1/19 | ND | A |
| 1884 | GUA811 | 1998 | 8 | 4 | 1/19 | ND | A |
| 1885 | GUA824 | 1998 | 4 | 4 | 1/19 | ND | A14 |
| 1886 | GUA1348 | 1998 | 8 | 4 | 1/19 | ND | A14 |
| 1887 | GUA1424 | 1998 | 16 | 4 | 1/19 | ND | A |
| 1025 | MEX49 | 1994 | 4 | 16 | 2/38 | 1 | A |
| 1026 | MEX110 | 1995 | 0.25 | 4 | 2/38 | 1 | A5 |
| 1027 | MEX111 | 1995 | 0.25 | 4 | 2/38 | 1 | A5 |
| 1028 | MEX112 | 1995 | 0.25 | 4 | 2/38 | 1 | A5 |
| 1029 | MEX115 | 1995 | 16 | 16 | 2/38 | 1 | A3 |
| 1023 | MEX10 | 1996 | 0.25 | 4 | 2/38 | 1 | A5 |
| 1024 | MEX16 | 1996 | 16 | 16 | 2/38 | 1 | A |
| 1030 | MEX146 | 1966 | 8 | 2 | 2/38 | 1 | A3 |
| 1021 | MEX2NM | 1998 | 0.25 | 4 | 0.5/9.5 | 1 | A5 |
| 1022 | MEX2PS | 1998 | 0.25 | 4 | 1/19 | 1 | A5 |
| 1215 | URU112 | 1988 | 8 | 2 | 0.25/4.7 | 2 | A12 |
| 1217 | URU185 | 1988 | 2 | 2 | 0.25/4.7 | 2 | A7 |
| 1216 | URU332 | 1989 | 8 | 4 | 0.25/4.7 | 2 | A12 |
| 1118 | URU96 | 1993 | 0.25 | 4 | 1/19 | 1 | A5 |
| 1119 | URU N48 | 1993 | 0.12 | 4 | 0.25/4.7 | 1 | A10 |
| 1120 | URU N649 | 1993 | 8 | 2 | 0.25/4.7 | 1 | A6 |
| 1121 | URU114 | 1994 | 0.25 | 4 | 1/19 | 1 | A5 |
| 1122 | URU139 | 1994 | 0.25 | 4 | 8/152 | 1 | A5 |
| 730 | URU168 | 1994 | 0.12 | 4 | 8/152 | 1 | A5 |
| 731 | URU173 | 1994 | 0.25 | 4 | 1/19 | 1 | A5 |
| 1123 | URU176 | 1994 | 0.25 | 4 | 8/152 | 1 | A5 |
| 1124 | URU181 | 1994 | 0.12 | 4 | 1/19 | 1 | A5 |
| 1125 | URU183 | 1994 | 0.25 | 4 | 2/38 | 1 | A5 |
| 732 | URU204 | 1994 | 0.25 | 4 | 2/38 | 1 | A5 |
| 733 | URU212 | 1994 | 4 | 2 | 0.12/2.3 | 1 | A5 |
| 1126 | URU216 | 1994 | 8 | 4 | 0.25/4.7 | 2 | A6 |
| 734 | URU227 | 1994 | 8 | 2 | 0.12/2.3 | 1 | A6 |
| 1127 | URU237 | 1994 | 8 | 2 | 0.25/4.7 | 2 | A6 |
| 735 | URU243 | 1994 | 0.25 | 4 | 1/19 | 1 | A9 |
| 1128 | URU252 | 1994 | 0.25 | 4 | 2/38 | 1 | A5 |
| 1129 | URU284 | 1995 | 0.12 | 4 | 0.5/9.5 | 1 | A5 |
| 1130 | URU297 | 1995 | 0.12 | 4 | 1/19 | 1 | A5 |
| 1131 | URU304 | 1995 | 0.25 | 4 | 2/38 | 1 | A5 |
| 736 | URU305 | 1995 | 8 | 4 | 0.12/2.3 | 1 | A5 |
| 1132 | URU316 | 1995 | 0.25 | 4 | 2/38 | 1 | A5 |
| 737 | URU325 | 1995 | 0.25 | 4 | 2/38 | 1 | A5 |
| 738 | URU350 | 1995 | 0.25 | 4 | 2/38 | 1 | A5 |
| 1133 | URU391 | 1995 | 8 | 4 | 0.25/4.7 | 2 | A6 |
| 1134 | URU392 | 1995 | 0.25 | 4 | 1/19 | 1 | A5 |
| 1135 | URU393 | 1995 | 0.12 | 4 | 2/38 | 1 | A5 |
| 739 | URU420 | 1996 | 8 | 4 | 0.25/4.7 | 1 | A6 |
| 740 | URU427 | 1996 | 0.12 | 4 | 4/76 | 1 | A5 |
| 742 | URU480 | 1996 | 0.12 | 4 | 1/19 | 1 | A5 |
| 743 | URU499 | 1996 | 0.12 | 4 | 1/19 | 1 | A5 |
| 1136 | URU504 | 1996 | 0.25 | 4 | 0.5/9.5 | 1 | A5 |
| 1137 | URU519 | 1996 | 0.25 | 4 | 1/19 | 1 | A5 |
| 1138 | URU524 | 1996 | 0.25 | 4 | 2/38 | 1 | A9 |
| 1139 | URU708 | 1997 | 0.5 | 4 | 2/38 | 1 | A13 |
| 744 | URU812 | 1997 | 0.25 | 4 | 1/19 | 1 | A5 |
| 745 | URU887 | 1997 | 0.25 | 4 | 1/19 | 1 | A5 |
| 746 | URU888 | 1997 | 0.25 | 4 | 2/38 | 1 | A5 |
| 1140 | URU910 | 1997 | 0.25 | 4 | 2/38 | 1 | A5 |
| 747 | URU954 | 1997 | 0.25 | 4 | 2/38 | 1 | A5 |
| 1141 | URU973 | 1998 | 0.25 | 4 | 1/19 | 1 | A5 |
| 1142 | URU988 | 1998 | 0.25 | 4 | 1/19 | 1 | A5 |
| 1143 | URU1016 | 1998 | 0.25 | 2 | 0.5/9.5 | 4 | A11 |
| 1144 | URU1033 | 1998 | 0.25 | 4 | 2/38 | 1 | A5 |
| 1145 | URU1040 | 1998 | 0.25 | 4 | 1/19 | 1 | A5 |
| 1146 | URU1042 | 1998 | 0.25 | 4 | 1/19 | 1 | A5 |
| 1147 | URU1060 | 1998 | 0.25 | 4 | 1/19 | 1 | A5 |
| 1148 | URU1093 | 1999 | 0.25 | 4 | 2/38 | 1 | A5 |
| 1149 | URU1113 | 1999 | 0.25 | 4 | 1/19 | 1 | A5 |
| 1150 | URU1123 | 1999 | 0.25 | 4 | 1/19 | 2 | A6 |
| 1151 | URU1127 | 1999 | 0.25 | 4 | 1/19 | 1 | A5 |
| 1152 | URU1161 | 1999 | 0.25 | 4 | 1/19 | 1 | A5 |
| 1153 | URU1193 | 1999 | 0.25 | 4 | 0.5/9.5 | 1 | A5 |
| 1154 | URU1205 | 1999 | 0.25 | 4 | 1/19 | 1 | A5 |
| 1155 | URU1208 | 1999 | 0.25 | 4 | 1/19 | 1 | A5 |
| 1156 | URU1213 | 1999 | 0.25 | 4 | 1/19 | 1 | A5 |
| 1157 | URU1215 | 1999 | 0.25 | 4 | 1/19 | 1 | A5 |
ARG: Argentina; BRA: Brazil; COL: Colombia; GUA: Guatemala; MEX: Mexico; URU: Uruguay.
All 172 isolates were susceptible to penicillin, ceftriaxone, erythromycin, and vancomycin. TET, tetracycline resistant (≥8 μg/ml); CHL, chloramphenicol resistant (≥4 μg/ml); STX, trimethoprim-sulfamethoxazole resistant (≥4 μg/ml) and intermediate (1 and 2 μg/ml) (25).
ND, not determined.
Phenotypic marker and antimicrobial susceptibility.
All 172 isolates were susceptible to penicillin, ceftriaxone, erythromycin, and vancomycin, except for one isolate (ARG 1206) from Argentina which was resistant to erythromycin (MIC, 32 μg/μl). Antimicrobial resistance to chloramphenicol was present in 50 (29%) isolates, resistance to tetracycline was present in 82 (47.7%), high resistance to trimethoprim-sulfamethoxazole was present in 25 (14.5%), and intermediate resistance was expressed in 114 of 172 isolates (66.3%) (Table 1).
MLEE.
Four different, closely related ETs were found among 157 isolates analyzed (Table 1). Variability in electrophoretic mobility was limited to three enzymes (esterase, l-lactate dehydrogenase and leucyl-glycyl-glycine peptidase), while the remaining nine were monomorphic. ET1 was the most common, represented by 143 isolates (91.1%). ET2 was shared by 11 isolates (7.0%), and ET3 and ET4 were represented by one isolate each. In Argentina, 21 isolates (91.3%) were ET1 and 2 (8.7%) were ET2. In Brazil, 10 (83.3%) were ET1 and 2 (16.7%) were ET2. In Colombia, 50 (96.2%) were ET1, 1 (1.9%) was ET2, and 1 (1.9%) was ET3. In Mexico, all 10 (100%) isolates were ET1. In Uruguay, 53 (88.3%) were ET1, 6 (10%) were ET2, and 1 (1.6%) was ET4.
Genotypic marker and PFGE.
The 172 invasive isolates analyzed belonged to pattern A. Based on the number of bands and their molecular weights, 15 PFGE subtypes (A1 to A15) were classified (Table 1). Pattern A, with 59 isolates, had 13 bands ranging from 340 to 25 kb. Subtypes with two or three band differences (closely related) with respect to 109 isolates were assigned to A1 to A8, A10, A12, A14, and A15; and subtypes with four to six band differences (possibly related) with respect to 4 isolates were assigned to A9, A11 and A13. PFGE patterns A, A1, A3, A5, A6, and A12 are shown in Fig. 1.
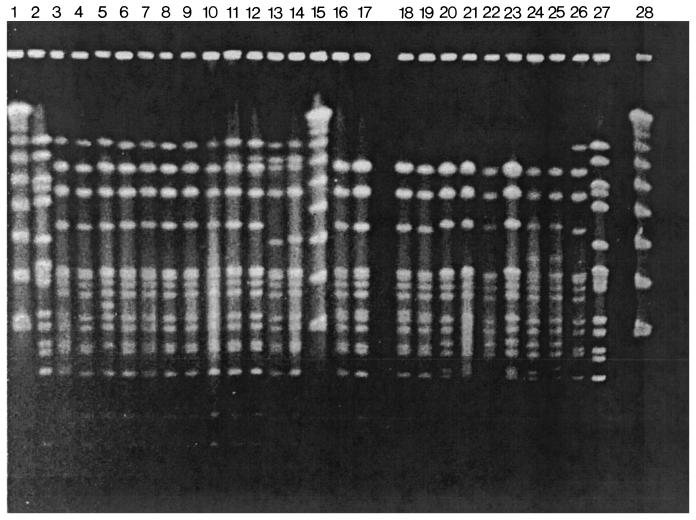
FIG. 1.
PFGE subtype patterns of S. pneumoniae serotype 5 invasive isolates. Chromosomal DNA fragments were digested with SmaI. Lanes 1, 15, and 28 (counting from the left) correspond to Lambda ladder; lanes 2 and 27 contain strain R6. Lanes 3 to 10 and 26 show PFGE pattern A: Col 7, Col E330, Col E390, Bra 102, Bra 291, Gua 168, Gua811, Mex 49, and Col 133. Lanes 11 and 12 show PFGE pattern A1: Col E68 and Col 136. Lanes 13 and 14 show PFGE pattern A3: Col E72 and Mex 146. lanes 16 to 19 show PFGE pattern A5: Arg 1061, Arg 1105, Uru 1215, and Mex 110. Lanes 20 to 23 show PFGE pattern A6: Uru 216, Arg 1108, Col 72, and Bra 71. Lanes 24 and 25 show PFGE pattern A12: Uru 332 and Uru 112.
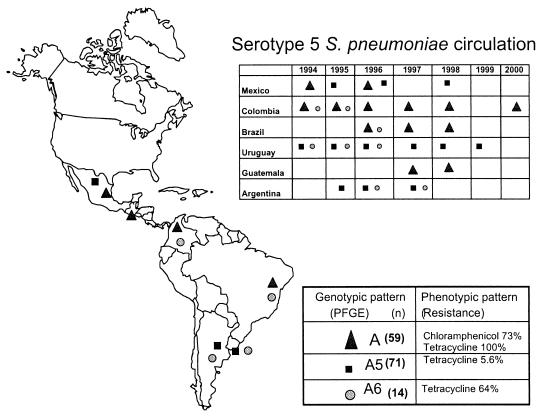
Most of the isolates (83.7%) were concentrated into three patterns: A (n = 59), A5 (n = 72) and A6 (n = 13). The prevalence of PFGE subtypes was determined per country and year. Pattern A circulated in Mexico, Guatemala, Colombia, and Brazil. Subtype A5 circulated in Argentina, Uruguay, and Mexico, while subtype A6 circulated in Colombia, Brazil, Uruguay, and Argentina (Table 1; Fig. 2). Two other subtypes, A3 (n = 3) and A7 (n = 4), were present in two countries. The remaining 11 subtypes (n = 21), with four isolates or fewer each, were identified in only one country.
FIG. 2.
Geographic and temporal distribution of the S. pneumoniae serotype 5 clone.
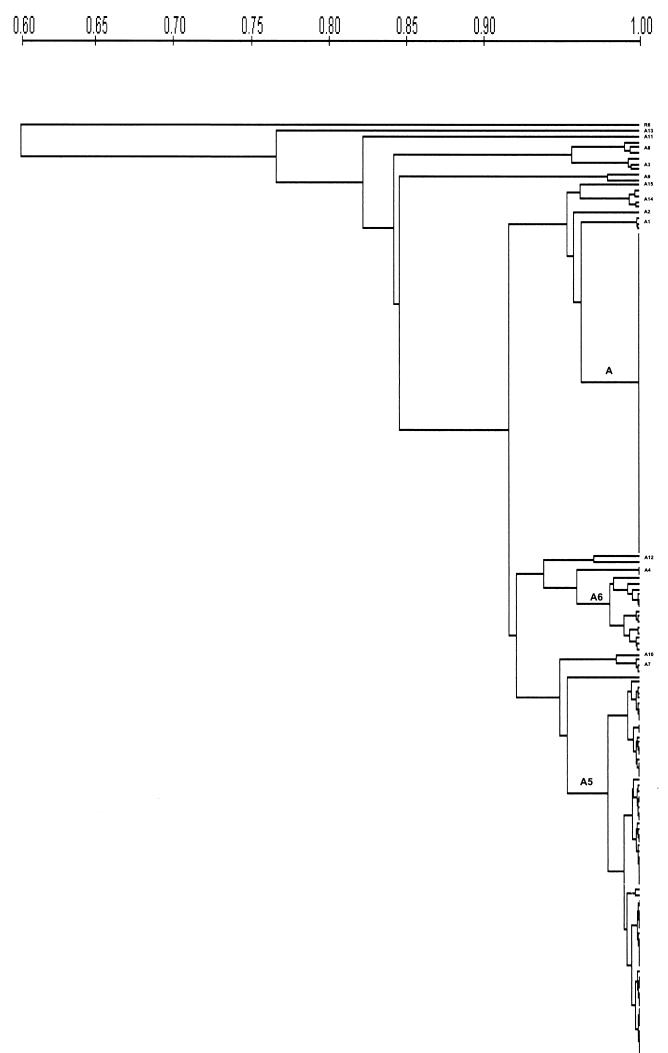
The similarity between all subtypes was more than 80% by Dice coefficient, while subtype A13 had 77% similarity to pattern A. The R6 reference strain had 60% similarity (Fig. 3).
FIG. 3.
Phylogenetic tree of Latin American serotype 5 S. pneumoniae isolates on the basis of PFGE patterns (UPGMA).
Correlation between the phenotypic and genotypic markers.
Patterns A, A1, A2, A3, and A12 grouped 68 isolates from Brazil, Colombia, Guatemala, Mexico, and Uruguay that were resistant to tetracycline and had a 340-kb characteristic PFGE band. Resistance to tetracycline was observed with lower incidences in subtypes A5 (5.5%) and A6 (61.5%) isolated from Argentina and Uruguay.
Resistance to chloramphenicol was observed exclusively in patterns A (46 of 59), A1 (3 of 3), and A3 (1 of 3). All these isolates except one were also resistant to tetracycline. In pattern A1, all three Colombian isolates were resistant, and in pattern A3, 50% of Mexican isolates were resistant. On the other hand, the isolates resistant only to tetracycline were pattern A (12 of 59) and subtypes A2 (1 of 1), A3 (2 of 3), A5 (4 of 72), A6 (8 of 13), A12 (2 of 2), A14 (3 of 4), and A 15 (1 of 1). Isolates of subtypes A7 to A11 and A13 were all susceptible to chloramphenicol and tetracycline.
DISCUSSION
As previously reported, S. pneumoniae serotype 5 is one of the most frequent causes of invasive pneumococcal disease in Latin American children younger than 5 years (3, 4, 7, 8, 16, 28). In a recent review by Hausdorff et al. (14), in more than 70 data sets originating worldwide, between five and eight serogroups comprised at least 75% of pneumococcal isolates from young children. Moreover, they found that throughout Asia serotype 5 ranked fourth, ranking second in China (12.8%) and third in Israel (13.4%). In Africa, serotype 5 ranked sixth, comprising 14.6% of isolates in Rwanda, 9.5% in Gambia, and 14.3% in Kenya. In Europe, serotype 5 ranked 10th, with the most representative country being Spain (5.7%) (14). However, in the United States and Canada, serotype 5 does not appear among the 12 most important circulating serotypes. In a subsequent analysis, Hausdorff et al. reported that serotype 5 was isolated in third place from middle ear fluid and blood and second from cerebrospinal fluid in Asia (13).
In Colombia, where serotypes 5, 14, and 23F have the highest incidence in invasive disease among children (5), the circulation of the major clones Spain23F-1 and Spain9v-3, as well as a unique clone 23F associated with resistance to penicillin, has been described (37). In 1999, we reported the presence of a Colombian serotype 5 that was later confirmed as a specific clone (Colombia5-19) (33; Klugman, 5th Meet. PMEN, 2001). A study done in Israel showed the presence of serotype 5 isolates, which have the same PFGE pattern as Latin American isolates (R. Dagan, personal communication).
The isolates analyzed in the present study had the same PFGE A pattern and 91% showed the same ET, suggesting a common origin, previously described as the Colombian5-19 clone. Moreover, 86% of isolates belonged to only 3 of the 15 subtypes identified (A, A5, and A6), each showing a different geographic distribution. Pattern A was isolated in Colombia, Guatemala, Mexico, and Brazil, while subtype A5 circulated in Argentina, Uruguay and Mexico. These subtypes have been disseminating in the region at least since 1994. Although, in Colombia the pattern A frequency had its highest peak in 1995, it is still present until today. Interestingly, none of the three Uruguayan isolates from 1988 and 1989 belonged to the predominant subtypes or had the predominant ET1. The similarity between pattern A and the two other most common subtypes, A5 and A6, which are closely related, was higher than 92%. Subtypes A11 and A13, possibly related to pattern A, showed less than 85% similarity (Fig. 3).
Correlation between PFGE and MLEE results was 100% with respect to the two most common PFGE subtypes (A and A5), but a lower correlation was observed among other PFGE types and ETs. One possible explanation is that the two methods detect different genetic events. PFGE identifies changes in restriction sites, which, according to Hall and Duke (12), would be due to DNA insertion or deletion arrangements of mobile elements, instead of point mutations. On the other hand, MLEE analyzes the allelic variation of enzymatic loci, which have a low mutation rate (18). Spratt et al. analyzed three serotype 5 Colombian isolates by multilocus sequence typing (MLST) and reported identical profiles for two isolates while the third isolate showed a single locus variant of the clone (personal communication). Additionally, one Uruguayan serotype 5 isolate had a single locus variant with the clone's profile but at a different locus from the Colombian isolate (http://www.MLST.net). MLST provides a highly discriminating typing method to analyze closely related genetic population and could be useful to identify differences between the Latin-American serotype 5 PFGE subtypes and ETs.
The close genetic relatedness between S. pneumoniae serotype 5 isolates suggests two possible explanations. The first is that this serotype is infrequently isolated from healthy carriers (36) and thus lacks the opportunities for genetic exchange with its own or other related species (20). In Latin America and Israel, serotypes 5 and 1 are some of the most common causes of invasive disease, but they are rarely isolated from the nasopharynges of healthy children (26, 27). A second possibility is that the clone may have been recently established and there has not been enough time for differentiation or dissemination.
The presence of a 340-kb DNA band from Colombian serotype 5 isolates has been associated with high resistance to tetracycline and chloramphenicol (19, 33). Our findings showed that all the isolates with this SmaI DNA fragment were resistant to tetracycline and 71.4% were resistant to chloramphenicol, showing an indirect correlation between the presence of the 340-kb band and resistance to chloramphenicol and tetracycline among Latin American serotype 5 isolates. These results suggest that there could be a different insertion site of the mobile elements along the chromosomal DNA. Most tetracycline- and chloramphenicol-resistant isolates shared the A pattern, suggesting clonal dissemination, while the presence of tetM and cat genes among other PFGE subtypes suggests horizontal transfer or differentiation events that occurred after the common lineage became established in the region. It is also important to take into account the observation that antibiotic use differs among Latin American countries (16).
It is important to point out that serotype 5 has not been included in the heptavalent conjugate vaccine. The formulation was made based on studies in North America, where serotype 5 is not as frequent as in Latin America or Asia (2, 13, 14, 30). Thus, it would be beneficial if, when formulating a new vaccine, the geographic distribution and the serotypes prevalent worldwide are considered.
The results of this study provide strong support for a unique genetic origin of the Latin American serotype 5 invasive S. pneumoniae isolates. It is important to perform molecular studies with serotype 5 isolates from other countries to analyze relatedness. Additionally, continuing surveillance to explore predominant clones would be helpful to monitor the influence of selective pressure when conjugate vaccines are introduced into the Latin American population (2).
Acknowledgments
Support for this study was provided by COLCIENCIAS (2104-04-302-98).
We acknowledge Maria Claudia Vela, Instituto Nacional de Salud, for her valuable help in developing this project. We also thank Maria Mercedes Zambrano for her help in the revision of the manuscript.
REFERENCES
- 1.Appelbaum, P. C. 1992. Antimicrobial resistance in Streptococcus pneumoniae: an overview. Clin. Infect. Dis. 15:77-83. [DOI] [PubMed] [Google Scholar]
- 2.Black, S., H. Shinefield, B. Fireman, E. Lewis, P. Ray, J. R. Hansen, L. Elvin, K. M. Ensor, J. Hackell, G. Siber, F. Malinoski, D. Madore, I. Chang, R. Kohberger, W. Watson, R. Austrian K. Edwards, and the Northern California Kaiser Permanente Vaccine Study Center Group. 2000. Efficacy, safety and immunogenicity of heptavalent pneumococcal conjugate vaccine in children. Pediatr. Infect. Dis. J. 19:187-195. [DOI] [PubMed] [Google Scholar]
- 3.Brandileone, M. C. C., V. S. Dias Vieira, S. T. Casagrande, R. C. Zanella, M. L. Guerra, S. Bokermann, J. L. Di Fabio, and the Pneumococcal Study Group in Brazil for the SIREVA Project. 1997. Prevalence of serotypes and antimicrobial resistance of Streptococcus pneumoniae strains isolated from Brazilian children with invasive infections. Microb. Drug Resist. 3:141-146. [DOI] [PubMed] [Google Scholar]
- 4.Castañeda, E., A. L. Leal, O. Castillo, F. De la Hoz, M. C. Vela, M. Arango, H. Trujillo, A. Levy, M. E. Gama, M. Calle, M. L. Valencia, W. Parra, N. Agudelo, G. I. Mejia, S. Jaramillo, F. Montoya, H. Porras, A. Sánchez, D. Saa, J. L. Di Fabio, A. Homma, and the Pneumococcal Study Group in Colombia. 1997. Distribution of capsular types and antimicrobial susceptibility of invasive isolates of Streptococcus pneumoniae in Colombian children. Microb. Drug Resist. 3:147-152. [DOI] [PubMed] [Google Scholar]
- 5.Castañeda, E., I. Peñuela, M. C. Vela, the Colombian Pneumococcal Study Group, and A. Tomasz. 1998. Penicillin-resistant Streptococcus pneumoniae in Colombia: presence of international epidemic clones. Microb. Drug Resist. 4:233-236. [DOI] [PubMed] [Google Scholar]
- 6.DiFabio J. L., A. Homma, and C. De Quadros. 1997. Pan American Health Organization epidemiological surveillance network for Streptococcus pneumoniae. Microb. Drug Resist. 3:131-133. [DOI] [PubMed] [Google Scholar]
- 7.DiFabio, J. L., E. Castañeda, C. I. Agudelo, F. De La Hoz, M. Hortal, T. Camou, G. Echániz-Avilés, M. N. Carnalla Barajas, I. Heitmann, J. C. Hormazabal, M. C. Brandileone, V. S. Dias Vieira, M. Regueira, R. Ruvinski, A. Corso, M. Lovgren, J. A. Talbot, C. De Quadros, and the PAHO Sireva-Vigia study group. 2001. Evolution of Streptococcus pneumoniae serotypes and penicillin susceptibility in Latin America, Sireva-Vigia group, 1994 to 1999. Pediatr. Infect. Dis. J. 20:959-967. [DOI] [PubMed] [Google Scholar]
- 8.Echániz-Aviles, G., M. E. Veláquez-Meza, M. N. Carnalla-Barajas, A. Soto-Noguerón, F. Solórzano-Santos, A. Pérez Miravete, R. Gatica-Marquina, and J. L. Di Fabio. 1997. Antimicrobial susceptibilities and capsular types of invasive Streptococcus pneumoniae isolated in children in Mexico city. Microb. Drug Resist. 3:153-157. [DOI] [PubMed] [Google Scholar]
- 9.Fenoll, A., I. Jado, D. Vicioso, A. Perez, and J. Casal. 1998. Evolution of Streptococcus pneumoniae serotypes and antibiotic resistance in Spain: Update (1990 to 1996). J. Clin. Microbiol. 36:3447-3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Facklam, R. R., and J. A. Washington. 1991. Streptococcus and related catalase-negative gram-positive cocci, p. 238-257. In A. Balows, W. J. Hausler, Jr., K. L. Hermann, H. D. Isenberg, and H. J. Shadomy (ed.), Manual of clinical microbiology, 5th ed. American Society for Microbiology, Washington, D.C.
- 11.Garenne, M. M., and C. Ronsmaus. 1992. The magnitude of mortality from acute respiratory infectious in children under 5 years in developing countries. World Health Stat. Q. 45:180-191. [PubMed] [Google Scholar]
- 12.Hall, L. M., and B. Duke. 1998. Conservation of restriction sites in isolates of Streptococcus pneumoniae with diverse restriction fragment pattern. J. Clin. Microbiol. 36:1805-1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hausdorff W. P., J. Bryant, C. Kloek, P. R. Paradiso, and G. R. Siber. 2000. The contribution of specific pneumococcal serogroups to different disease manifestation: implications for conjugate vaccine formulation and use. Part II. Clin. Infect. Dis. 30:122-140. [DOI] [PubMed] [Google Scholar]
- 14.Hausdorff, W. P., J. Bryant, P. R. Paradiso, and G. R. Siber. 2000. Which pneumococcal serogroups cause the most invasive disease: implications for conjugate vaccine formulation and use. Part I. Clin. Infect. Dis. 30:100-121. [DOI] [PubMed] [Google Scholar]
- 15.Henrichsen, J. 1995. Six newly recognized types of Streptococcus pneumoniae. J. Clin. Microbiol. 33:2759-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hortal, M., and the Pneumococcus Study Group (G. Algorta, I. Bianchi, G. Borthagaray, I. Cestau, T. Camou, M. Castro, M. De Los Santos, R. Diez, L. DellÁcqua, A. Galiana, A. Giordano, P. Giordano, G. Lopez-Ghemi, N. Milanese, C. Mogdasy, R. Palacio, W. Pedreira, A. Pisano, and L. Pivel). 1997. Capsular type distribution and susceptibility to antibiotics of Streptococcus pneumoniae clinical strains isolated from Uruguayan children with systemic infections. Microb. Drug Resist. 3:159-163. [DOI] [PubMed] [Google Scholar]
- 17.Kertesz D. A., J. L. Di Fabio, M. C. Brandileone, E. Castañeda, G. Echániz-Aviles, I. Heitmann, A. Homma, M. Hortal, M. Lovgren, R. O. Ruvinsky, J. A. Talbot, J. Weekes, J. S. Spika, and the PAHO Pneumococcal Surveillance Study Group. 1998. Invasive Streptococcus pneumoniae infection in Latin American children: results of the Pan-American Health Organization surveillance study. Clin. Infect. Dis. 26:1355-1361. [DOI] [PubMed] [Google Scholar]
- 18.Lefevre, J. C., G. Faucon, A. M. Sicard, and A. M. Gasc. 1993. DNA fingerprinting of Streptococcus pneumoniae strains by pulse-field gel electrophoresis. J. Clin. Microbiol. 31:2724-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marchese, A., M. Ramirez, G. C. Schito, and A. Tomasz. 1998. Molecular epidemiology of penicillin-resistant Streptococcus pneumoniae isolates recovered in Italy from 1993 to 1996. J. Clin. Microbiol. 36:2944-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGee, L., K. P. Klugman, and A. Tomasz. 2000. Serotypes and clones of antibiotic-resistant pneumococci. p. 375-379. In A. Tomasz (ed.), Streptococcus pneumoniae: molecular biology and mechanisms of disease. Mary Ann Liebert, Inc., New York, N.Y.
- 21.McGee, L., L. McDougal, J. Zhou, B. G. Spratt, F. C. Tenover, R. Geoge, R. Hakenbeck, W. Hryniewicz, J. L. Lefévre, A. Tomasz, and K. P. Klugman. 2001. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the pneumococcal molecular epidemiology network. J. Clin. Microbiol. 39:2565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moreno, F., C. Crisp, J. H. Jorgensen, and J. E. Patterson. 1995. The clinical and molecular epidemiology of bacteremia at a university hospital caused by pneumococci not susceptible to penicillin. J. Infect. Dis. 172:427-432. [DOI] [PubMed] [Google Scholar]
- 23.Musher D. M. 1995. Streptococcus pneumoniae, In p. 1811-1826. G. Mandell, J. E., Bennett, and R. Dolin (ed.), Principles and practice of infectious disease, 4th ed. Churchill Livingstone, Inc., New York, N.Y.
- 24.National Committee for Clinical Laboratory Standards. 2000. Supplemental tables. Disk diffusion, MIC. NCCLS publications M100-S10(M2) and M100-S10(M7). National Committee for Clinical Laboratory Standards, Villanova, Pa.
- 25.Paris, M. M., O. Ramilo, and G. H. McCracken, Jr. 1995. Management of meningitis caused by penicillin-resistant Streptococcus pneumoniae. Antimicrob. Agents Chemother. 39:2171-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peñuela, I. J., A. L. Leal, and E. Castañeda. 1999. Colonizatión nasofaríngea y resistencia antimicrobiana de Streptococcus pneumoniae en niños de una guarderia en Santa Fe de Bogotá. Biomédica 19:214-222. [Google Scholar]
- 27.Porat N., R. Trefler, and R. Dagan. 2001. Persistence of two invasive Streptococcus pneumoniae clones of serotypes 1 and 5 in comparison to that multiple clones of serotypes 6B and 23F among children in Southern Israel. J. Clin. Microbiol. 39:1827-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rossi, A., R. Ruvinsky, M. Regueira, A. Corso, J. Pace, A. Gentile, J. L. Di Fabio, and the Streptococcus pneumoniae Working Group. 1997. Distribution of capsular types and penicillin-resistance of strains of Streptococcus pneumoniae causing systemic infections in Argentinean children under 5 years of age. Microb. Drug Resist. 3:135-140. [DOI] [PubMed] [Google Scholar]
- 29.Selander, R. K., D. A. Caugant, H. Ochman, J. M. Musser, M. N. Gilmour, and T. S. Whittam. 1986. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl. Environ. Microbiol. 51:873-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sniadack, D. H., B. Schwartz, and H. Kipman. 1995. Potential interventions for the prevention of childhood pneumonia: geographic and temporal differences in serotype and serogroup distribution of sterile site pneumococcal isolates from children- implications for vaccine strategies. Pediatr. Infect. Dis. J. 14:503-511. [PubMed] [Google Scholar]
- 31.Soares, S., K. G. Kristinsson, J. M. Musser, and A. Tomaz. 1993. Evidence for the introduction of a multiresistant clone of serotype 6B Streptococcus pneumoniae from Spain to Iceland in the late 1980s. J. Infect. Dis. 168:158-163. [DOI] [PubMed] [Google Scholar]
- 32.Sorensen, U. B. S. 1993. Typing of pneumococci by using 12 pooled antisera. J. Clin. Microbiol. 31:2097-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamayo M, R. Sa Leao, I. Santos ´Sanchez, E. Castañeda, and H. De Lancastre. 1999. Dissemination of a chloramphenicol-and tetracycline-resistant but penicillin-susceptible invasive clone of serotype 5 Streptococcus pneumoniae in Colombia. J. Clin. Microbiol. 37:2337-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tenover, F., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulse-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tomasz A., A. Corzo, and members of the PAHO/Rockefeller University Workshop (E. P. Severina, G. Echániz-Aviles, M. C. Brandileone, T. Camou, E. Castañeda, O. Figueroa, A. Rossi, and J. L. Di Fabio). 1998. Molecular epidemiologic characterization of penicillin-resistant Streptococcus pneumoniae invasive pediatric isolates recovered in six Latin American countries: an overview. Microb. Drug Resist. 4:195-207. [DOI] [PubMed] [Google Scholar]
- 36.Tuomanen, E. I., and H. R. Masure. 2000. Molecular and cellular biology of pneumococcal infection, p. 295-308. In A. Tomasz (ed.), Streptococcus pneumoniae: molecular biology and mechanisms of disease. Mary Ann Liebert, Inc., New York, N.Y.
- 37.Vela, M. C., N. Fonseca, J. L. Di Fabio, and E. Castañeda. 2001. Presence of international multiresistant clones of Streptococcus pneumoniae in Colombia. Microb. Drug Resist 7:153-164. [DOI] [PubMed] [Google Scholar]