The ubiquitous feline intraerythrocytic pathogen Bartonella henselae causes a range of important human infections, and seroprevalence in blood donors in Australia is around 5% (6). Two antigenically distinct subtypes have been described, named for the type strain (Houston-1) and the more recently recognized Marseille strain (5), which have characteristic DNA sequences in the 16S rRNA gene (rDNA) and the gltA (citrate synthase) gene (2, 3, 5). Culture of this fastidious, gram-negative bacterium is difficult, and serology is a primary diagnostic approach. A recent report that Houston-like strains (including Houston-1, 90-615, and SA-2) may be distinguished from serologically distinct Marseille-like strains (including URBHLLY-8, URBHLIE-9, Fizz, and CAL-1) by the faster migration of a characteristic band (ca. 29 kDa) on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis and immunoblotting of whole-cell lysates is therefore particularly noteworthy (7).
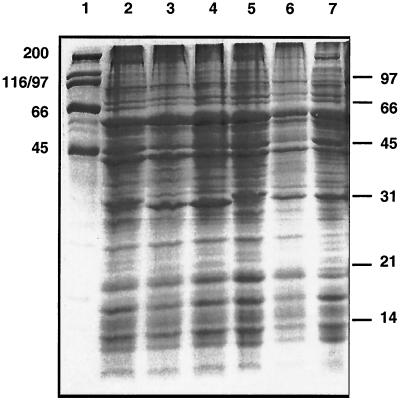
However, the SDS-PAGE data appear to contrast with previous publications from the same laboratory (8, 9) and others (11). We therefore compared local human isolates with the Houston-1 and Marseille strains and also found two closely related genotypes with corresponding patterns on SDS-PAGE of whole-cell lysates (Fig. 1). All human strains appear to have either the 30-kDa band (Houston) or the 29-kDa band (Marseille), in agreement with earlier studies (10, 11). The agar-pitting colony phenotype and number of in vitro passages have no apparent influence (Fig. 1 and Table 1). However, a local feline strain of the type II (Marseille-like) 16S rDNA genotype has the faster-migrating (29-kDa) band and a gltA gene sequence typical of Houston strains, consistent with the considerable diversity known to exist in feline strains (1, 10) and illustrating a dissociation of the accepted genotype-phenotype relationships. The strains in the recent study (7) were all human isolates and were assigned to genotypes by individual sequencing of several genes, including groEL (12) and the 16S rDNA (3). While the 29- to 30-kDa antigen(s) may yet prove useful for serotyping of human isolates, it thus seems unlikely that reported inconsistencies in the data can be easily explained by phase variation or genotype. In addition, we have no reason to assume that the more diverse feline strains are all nonpathogenic in humans. For both of thesereasons, we therefore urge caution in adopting this promising target as a basis for serotype distinction until the picture is clearer.
FIG. 1.
Marseille and Houston types. Coomassie-stained SDS-PAGE of whole-cell lysates harvested from chocolate agar plates (grown at 35°C in 5% CO2). High-molecular-weight (lane 1 and left border) and low-molecular-weight (right border) markers are shown, with values in kilodaltons. Lanes: 2, BH5; 3, BH4; 4, URBHLLY-8 Marseille; 5, ATCC 49882 Houston-1; 6, HC35 (S4); 7, HC35 (S20).
TABLE 1.
B. henselae isolates used in this study
| Isolate | 16S (Bergmans) typea | gltA varianta | Colonies (piliation) | Subculture (passage) no. | Source |
|---|---|---|---|---|---|
| H-1 | I | H | SNP (+) | >3 | ATCC 49882 (Houston-1 type strain) |
| 49793 | I | H | SNP (−) | >6 | ATCC 49793 |
| BH5 | I | H | DAP (+) | 2 | Cat scratch disease patient (this study) |
| M-1 | II | M | SNP (+) | >3 | URBHLLY-8 (Marseille); D. Raoult (5) |
| BH4 | II | M | SNP (+) | 1 | Cat scratch disease patient (this study) |
| HC35 | II | H | DAP (+) | 4 | T. Gottlieb (4); feline isolate |
| HC35 | II | H | DAP (+) | 20 | This study |
16S and gltA types were determined by direct sequencing of PCR products as previously described (2, 3). Two variants (designated H and M) of the citrate synthase gene (gltA) were compared with previously published sequences (3); the BH5 gltA sequence was identical to that of HC35, ATCC 49793, and Houston-1; the BH4 gltA sequence was identical to that of URBHLLY-8 and was submitted to the EMBL database and assigned accession no. AJ439406. Colonies grew as dry-agar-pitting (DAP) or smooth, nonpitting (SNP) forms on chocolate agar (37°C; 5% CO2); piliation was determined by transmission electron microscopic examination of late-logarithmic-phase cultures grown on chocolate agar (+ = detected; − = not detected). Some strains imported or acquired from other labs had been subcultured in vitro prior to arrival. BH4 and BH5 were grown directly from fresh human lymph nodes in our laboratory.
Acknowledgments
P.K. and D.B. are recipients of Australian Postgraduate Awards.
REFERENCES
- 1.Arvand, A., A. J. Klose, D. Schwartz-Porsche, H. Hahn, and C. Wendt. 2001. Genetic variability and prevalence of Bartonella henselae in cats in Berlin, Germany, and analysis of its genetic relatedness to a strain from Berlin that is pathogenic for humans. J. Clin. Microbiol. 39:743-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergmans, A. M. C., J. F. P. Schellekens, J. D. A. van Embden, and L. M. Schouls. 1996. Predominance of two Bartonella henselae variants among cat-scratch disease patients in The Netherlands. J. Clin. Microbiol. 34:254-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birtles, R. J., and D. Raoult. 1996. Comparison of partial citrate synthase gene (gltA) sequences for phylogenetic analysis of Bartonella species. Int. J. Syst. Bacteriol. 46:891-897. [DOI] [PubMed] [Google Scholar]
- 4.Branley, J., C. Wolfson, P. Waters, T. Gottlieb, and R. Bradbury. 1996. Prevalence of Bartonella henselae bacteremia, the causative agent of cat-scratch disease, in an Australian cat population. Pathology 28:262-265. [DOI] [PubMed] [Google Scholar]
- 5.Drancourt, M., R. Birtles, G. Chaumentin, F. Vandenesch, J. Etienne, and D. Raoult. 1996. New serotype of Bartonella henselae in endocarditis and cat-scratch disease. Lancet 347:441-443. [DOI] [PubMed] [Google Scholar]
- 6.Flexman, J. P., S. C. Chen, D. J. Dickeson, J. W. Pearman, and G. L. Gilbert. 1997. Detection of antibodies to Bartonella henselae in clinically diagnosed cat-scratch disease. Med. J. Aust. 166:532-535. [DOI] [PubMed] [Google Scholar]
- 7.La Scola, B., Z. Liang, Z. Zeaiter, P. Houpikian, P. A. D. Grimont, and D. Raoult. 2002. Genotypic characteristics of two serotypes of Bartonella henselae. J. Clin. Microbiol. 40:2002-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang, Z., and D. Raoult. 2000. Differentiation of Bartonella species by a microimmunofluorescence assay, sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and Western immunoblotting. Clin. Diagn. Lab. Immunol. 7:617-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang, Z., B. La Scola, H. Lepidi, and D. Raoult. 2001. Production of Bartonella genus-specific monoclonal antibodies. Clin. Diagn. Lab. Immunol. 8:847-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sander, A., M. Ruess, S. Bereswill, M. Schuppler, and B. Steinbrueckner. 1998. Comparison of different DNA fingerprinting techniques for molecular typing of Bartonella henselae isolates. J. Clin. Microbiol. 36:2973-2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamamoto, K., B. B. Chomel, R. W. Kasten, C. M. Hew, D. K. Weber, W. I. Lee, S. Droz, and J. K. Koehler. 2002. Experimental infection of domestic cats with Bartonella koehlerae and comparison of protein and DNA profiles with those of other Bartonella species infecting felines. J. Clin. Microbiol. 40:466-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeaiter, Z., P. E. Fournier, H. Ogata, and D. Raoult. 2002. Phylogenetic classification of Bartonella species by comparing groEL sequences. Int. J. Syst. E vol. Microbiol. 52:165-171. [DOI] [PubMed] [Google Scholar]