Abstract
The detection of the human RNA viruses, calicivirus and astrovirus, requires high sensitivity and broad reactivity. A novel single-tube nested reverse transcription-PCR (RT-PCR) method is described here, in which all of the required reagents are included in the one tube; however, those required for the nested amplification are separated in a “hanging drop” in the cap to be introduced by centrifugation after the RT and first-round cDNA amplification steps. Broad reactivity was obtained by using primer cocktails covering the published sequence variation in the primer targets. The method was evaluated with clinical fecal samples from outbreak and sporadic cases. Norwalk-like virus types 1 and 2 and rotavirus were the causal agents in 10 of 12 outbreaks. A viral agent was detected in 44% of 197 samples from sporadic infections in patients presenting to community health centers and a children's hospital. Interestingly, whereas rotavirus was more common than astrovirus in patients presenting to the hospital (33 and 7.6%, respectively), the reverse was true for patients presenting to community health centers (4.2 and 34%, respectively).
The cause of many acute gastroenteritis cases remains unknown, even in developed countries where comprehensive diagnostic facilities are common (10), but there is growing evidence that viruses such as rotavirus, human calicivirus, and astrovirus are the predominant cause of gastroenteritis. Rotaviruses are recognized as the most common cause of pediatric diarrheal disease in the world (9, 11, 14). Astroviruses are second only to rotaviruses as a cause of hospitalization for childhood viral gastroenteritis (9). Two calicivirus species, Norwalk-like viruses (NLVs) and Sapporo-like viruses (SLVs), have also been recognized as significant. NLVs, also known as small round structured viruses, are a major cause of epidemic nonbacterial gastroenteritis (3, 8, 10, 16, 35) and are a significant cause of foodborne outbreaks, which are often caused by contaminated shellfish (22, 29). Infection with SLV, also known as classic calicivirus, appears to be associated with infantile gastroenteritis but may be a less important cause of infection in older children and adults and less frequently associated with foodborne outbreaks (4, 23, 30).
The low infectious dose of such viruses (2) requires that investigative methods be able to detect very low numbers of virions in patient samples, contaminated ground and estuarine water, and foodstuffs such as shellfish. This high level of sensitivity remains a challenge even for nucleic acid amplification methods, especially for the detection of RNA viruses, which require an additional reverse transcription (RT) step. Nested sequential amplification has improved sensitivity, but cross-contamination of the nested amplification step by primary amplification products is a significant cause of false-positive results, diminishing the reliability of the results. The combination of both amplification reactions of the nested-PCR into a “single-tube” nested-PCR to reduce the risk of contamination has been reported. The use of differential annealing temperatures to sequentially select amplification primed by first outer (high melting temperature [Tm]) primer pairs, followed by inner (low Tm) primer pairs, is one such method (17, 19). Alternative approaches have physically separated the primers and additional reagents required for the nested reaction from those of the first round within the one tube in a manner that allows them to be introduced into the amplification mix when required without opening the tube. For example, nested reaction reagents have been suspended in modified pipette tips within the reaction tube (26) or dried in the lid, stabilized in a trehalose matrix (34). These procedures have their disadvantages. Annealing temperature-differentiated nested reactions can produce nonspecific amplification products from the activity of the outer primers during the nested amplification cycles. Physical devices used to separate the nested reaction reagents can be awkward and complicated to use, and although overcoming contamination of the second nested reaction, increase the risk of contamination from other sources while preparing and dispensing the reactions.
Additional complexity results from the increased genome variation within RNA viruses caused by the lack of genomic error correction during replication. Even focusing on the more conserved, functionally constrained regions rarely eliminates sequence diversity among strains at the primer target sites. The broad specificity for all strains required in investigative methods usually requires either separate amplification reactions specific for each strain type or the inclusion of a cocktail of primers that encompass the known primer target variation in one reaction.
We describe here a very simple method for performing single-tube nested-PCR amplification wherein the nested reaction reagents are located in the cap as a “hanging drop” to be introduced by centrifugation after the first-round RT-PCR amplification step. Walsh et al. have recently reported a similar method to detect respiratory syncytial virus, but only the two amplification reactions are included, with the preliminary RT step being performed separately (31). In the method reported here all three steps are included in the one tube. Further, the present study presents “universal” inner and outer primer cocktails designed from the GenBank-registered sequences of NLV (types 1 and 2), SLV, and astrovirus strains, and the component titrations used to optimize the method for the four viruses. Genome regions similar to those already published were targeted so that sequence comparison of future positives could be made with published strain genotypes (1, 25, 35). Universality of the primer sets was obtained by use of a combination of multiple primers and primers with mixed bases. To evaluate the method, RNA extracts of known positive material, 197 randomly chosen fecal samples from symptomatic children and adults, and 122 samples from 12 suspected outbreaks were examined for evidence of viral presence.
MATERIALS AND METHODS
Virus strains.
Known positive materials were kindly donated as follows: NLV-2 from P. J. Wright, Department of Microbiology, Monash University, Melbourne, Australia; NLV-1 and SLV from J. A. Marshal of the Victorian Infectious Diseases Reference Laboratory, North Melbourne, Australia; and cultures of the prototype astrovirus strains from the Department of Gastroenterology and Clinical Nutrition, Royal Children's Hospital, Melbourne, Australia, with the permission of J. P. Kurtz, John Radcliffe Hospital, Oxford, United Kingdom (the original source of the strains). Additional control material used for the comparison of single- and two-tube nested RT-PCR was obtained from fecal samples positive for one of the four viruses from specimens submitted to the Infectious Diseases Laboratories, Institute of Medical and Veterinary Science.
Clinical samples.
Fecal samples were obtained from outbreaks notified to health authorities within South Australia or were randomly selected from specimens routinely submitted for microbiological testing to the Infectious Diseases Laboratories, Institute of Medical and Veterinary Science (IMVS), and Microbiology and Infectious Diseases, Women's and Children's Hospital (WCH), both in Adelaide, Australia. The former (IMVS) represents samples collected from diarrheal cases, both pediatric and adult, presenting to community medical practices, whereas the latter (WCH) represents samples collected from diarrheal cases presenting to a pediatric hospital.
RNA extraction.
A 100-μl aliquot of a 10% fecal suspension in Dulbecco phosphate-buffered saline (pH 7.4) (7) was extracted by using the Qiagen RNeasy Plant Mini kit (Qiagen GmbH, Hilden, Germany) in accordance with the manufacturer's instructions, with RLT buffer, to obtain 50 μl of RNA sample.
Primer design.
From alignments of complete and large partial sequences of genomes obtained from GenBank for NLV-1, NLV-2, SLV, and astrovirus constructed by using GeneBase 1.0 (Applied Maths, Sint-Martens-Latem, Belgium), primers were chosen to target four nonoverlapping sites suitable for nested amplification. The polymerase-encoding and the capsid-encoding regions were chosen for the caliciviruses (NLV-1, NLV-2, and SLV) and the astrovirus, respectively, based on a previously published sequence to enable comparative analysis (1, 30, 35). Using both primers with mixed bases and additional variant primers, primer cocktails accommodating all known variation at the target site were selected and manufactured (Table 1). More primers were required to cover the published target sequence diversity among strains for NLV-1 and SLV than was necessary for NLV-2 and astrovirus. The complete cocktail of primers for each outer and inner primer pair were then used together for each of the four amplifications targeting the four viruses.
TABLE 1.
Primary and nested primer sets used for the detection of calicivirus and astrovirus in the nested PCR systema
| Virus | Primer | Sequence | Primer target (nucleotide range)b |
|---|---|---|---|
| NLV-2 | NLV-2-OF | TAGCWGCAGCMCTWGARATCATGG | 4335-4358 |
| NLV-2-OR | TTSAGYTTTGCWGTYARTTTYTCTGGG | 4633-4659 | |
| NLV-2-IF | AGTGTGRTKGATGWGGGTGAC | 4418-4438 | |
| NLV-2-IR | TTATRTCWGTACTSACWATYTCATCATC | 4598-4625 | |
| NLV-1 | NLV-1-OFa | ATGAYRGAGTCWTTYTCAATCATGTGYCG | 4592-4620 |
| NLV-1-OFb | ATGATGGAGTCCTTTAACATCATGTG | 4592-4617 | |
| NLV-1-ORa | CTAAGGACCTGGGTGAGTCGAG | 4896-4917 | |
| NLV-1-ORb | CCTAAGAACTTGTGTCAGTTTTGCTGG | 4892-4918 | |
| NLV-1-ORc | TTTGAGCACTTGTGTCAATTTAATTGGG | 4891-4918 | |
| NLV-1-IFa | AGCCCCTTCCGAAATGGATGTGG | 4669-4691 | |
| NLV-1-IFb | CACCCTCAGAGATGGATGTTGG | 4671-4692 | |
| NLV-1-IFc | ACTTTCTCCTTCTGAAATGGATGTTGG | 4666-4692 | |
| NLV-1-IRa | TCTGTGGACACGATCTCATCGTC | 4856-4877 | |
| NLV-1-IRb | CTATGTCATTTGACACTATTTCATCATC | 4856-4883 | |
| NLV-1-IRc | CAACATCTGTGGAGACTATCTCATCATC | 4856-4883 | |
| SLV | SLV-OFa | ACTSCAAATGGGATTCCACWCAAMACC | 4358-4384 |
| SLV-OFb | ACWCTAAATGGGAYTCCACACAGAATCC | 4358-4385 | |
| SLV-ORa | GTGRAAGATKGAWGCRGTGGCAGG | 4693-4716 | |
| SLV-ORb | TCAAAGATGGAAGCGGTTGCCG | 4694-4715 | |
| SLV-IF | TGGHCTMCCWTCWGGSATGCC | 4518-4537 | |
| SLV-IRa | CACACRCTGTASATGCAGTCATCACC | 4666-4691 | |
| SLV-IRb | CACAAGGAGTATATGCAATCATCACC | 4666-4691 | |
| Astrovirus | ASTRO-OF | TCACAGAAGAGCAACTCCATCGC | 4274-4296 |
| ASTRO-OR | CWGGTTTWGGTCCTGTGACACC | 4541-4562 | |
| ASTRO-IF | GACCAAAGAAGTGYGATGGCTAGC | 4310-4333 | |
| ASTRO-IR | GTTGBTTAKTGACAATKTTACGGACACG | 4502-4529 |
Primer nomenclature was as follows. Primary amplification primers: OF, outer forward; OR, outer reverse. Nested amplification primers: IF, inner forward; IR, inner reverse. Suffixes a, b, and c indicate the multiple primers included in the reaction to account for known nucleotide base variation in the target sequences. Mixed base codes conform to standard IUB/IUPAC nomenclature: R(AG), Y(CT), M(AC), K(GT), S(GC), W(AT), H(ACT), and B(GCT).
The primer target location is given for a representative strain (with the genome completely sequenced) for each group as follows: NLV-2 (Camberwell AF145896), NLV-1 (Norwalk M87661), SLV (Manchester X86560), and astrovirus (Oxford NC_002597). However, many primers contain variations found only in related strains.
One-step nested RT-PCR method.
In outline, the primary amplification mix, containing the necessary components to perform the RT and cDNA amplification steps, both of which are primed with the outer primers, was located in the bottom of the tube and overlaid with paraffin oil. The secondary amplification mix, containing the inner primers, additional DNA polymerase, and deoxynucleoside triphosphates (dNTPs), was suspended in the cap as a hanging drop contained solely by surface tension. The RT and PCR steps were performed without a heated lid. The tubes were then centrifuged to mix in the secondary amplification reagents, and the nested amplification cycle performed. Separate RT and PCR steps were not considered as the contamination risk during routine diagnostic testing was considered too high.
After a number of RT and PCR enzyme combinations (see Results) were tested, the final method details were determined to be as follows. A separate reaction was performed for each of the four virus types, each containing the complete set of outer and inner primers for each virus, as listed in Table 1. The primary RT-PCR step was based on the Titan One Tube RT-PCR System (Roche Molecular Biochemicals, Basel, Switzerland), modified to facilitate the second amplification reaction. The primary RT-PCR mix contained (i) 5 μl of Titan 5× PCR buffer that contains MgCl2 (1.5 mM final concentration), (ii) 200 μM concentrations of each dNTP (Amersham Pharmacia Biotech, Inc., San Francisco, Calif.), (iii) 5 mM dithiothreitol (Titan), (iv) 10 U of RNase inhibitor (Ambion, Inc., Austin, Tex.), (v) 2 pmol of each forward and reverse outer primer (Table 1), (vi) 1 μl of enzyme mix (Titan), and (vii) 2.5 μl of RNA extract in a final aqueous volume of 25 μl and was overlaid with paraffin oil. The secondary PCR mix (25 μl) containing (i) 2.5 μl of GeneAmp 10× PCR Buffer II (Applied Biosystems, Inc. [ABI], Foster City, Calif.) and 5.5 mM MgCl2 (ABI) to yield 3.5 mM MgCl2 during secondary amplification, (ii) 200 μM concentrations of each dNTP, (iii) 10 pmol of each forward and reverse inner primer (Table 1), and (iv) 1.25 U of AmpliTaq Gold (ABI) was located in the cap. In practice, to reduce contamination risk, enzyme-reagent mixes were made and dispensed in a “no DNA/RNA” clean room, and the PCR tubes were closed before transfer to a second clean room for the addition of the RNA sample into the primary RT-PCR mix under the oil. Only each respective sample and PCR tube were open at any one time. Using a PTC-200 thermocycler (MJ Research, Watertown, Mass.), primary RT-PCR amplification consisted of first-strand cDNA synthesis at 50°C for 30 min, immediately followed by denaturation (94°C for 2 min) and 40 cycles of amplification (94°C for 30 s, 55°C for 30 s, and 68°C for 45 s) without a heated lid. A final incubation (68°C for 5 min) was included to ensure that all amplification products were full length, and the tubes were then held at 12°C. After centrifugation at ≥8,000 × g for 15 s to combine the primary and nested amplification mixes (final volume, 50 μl) under the oil, the tubes were returned to the thermocycler for the nested amplification. The tubes were never left overnight between the two amplification cycles. Amplification conditions identical to that for the primary amplification were used except that the preamplification denaturation at 94°C was extended to 10 min to activate the AmpliTaq Gold.
After completion of the amplification, 10 μl of the amplification product was electrophoresed in 2% agarose (analytical grade; Promega Corp, Madison, Wis.) in 0.5× Tris-borate-EDTA buffer (pH 8.0). Bands were visualized by staining with 0.25 mg of ethidium bromide/liter and then inspected under UV light. The amplicon product size was estimated by mobility comparison with 500 ng of HpaII-digested pUC19 DNA marker (GeneWorks, Adelaide, South Australia, Australia). Products of the predicted size were verified by sequencing in both forward and reverse directions by using the PRISM Ready Reaction DyeDeoxy Terminator Cycle Sequencing Big Dye II Kit (ABI), after the amplicons had been purified by using the QIAquick PCR Purification Kit (Qiagen GmbH, Hilden, Germany). Sequencing reactions were purified by using isopropanol precipitation according to the manufacturer's recommendations, and DNA sequencing was performed on a 3700 DNA sequence analyzer (Applied Biosystems). Sequence alignments, error correction, and similarity analysis (unweighted pair group method using arithmetic averages [UPGMA]) were performed by using GeneBase 1.0.
Controls.
For each of the four viruses, positive RNA extracts were diluted to 10 times the titrated amplification endpoint and then stored as frozen aliquots to be used in each assay as a positive control. Two “no RNA” water controls were included in every assay batch: one inoculated when the reagents were dispensed and one inoculated after the RNA extracts had been added. In addition, all extracts were tested for inhibition in a separate amplification reaction spiked with 2.5 μl of an NLV-2-positive sample extract, diluted to 10 times the titrated amplification endpoint. If inhibition was detected, the sample was repeat tested at a 1:10 dilution or greater until inhibition could not be detected. The sample extract was then retested for the presence of calicivirus and astrovirus at that dilution.
Stability at −70°C.
An RNA extract from a known NLV-2-positive sample was divided into aliquots and frozen at −70°C. The titrated nested RT-PCR endpoint of the fresh extract was 10−4. Twelve sets of PCR tubes were dispensed with outer and inner amplification reagents as described above, and the tubes were stored at −70°C. Each month for 12 months an NLV-2 RNA aliquot and a set of PCR tubes were thawed, and the RNA aliquot was diluted serially to 10−5. Each 10-fold dilution, plus negative controls, was then inoculated into the thawed PCR tubes and amplified as normal to determine the nested RT-PCR endpoint.
Enzyme immunoassays.
The presence of rotavirus and adenovirus was detected by using an in-house enzyme immunoassay on 50 μl of a 10% fecal suspension in phosphate-buffered saline that was clarified by centrifugation (1,000 × g for 5 min) (15).
RESULTS
Optimization of method. (i) RT-PCR step.
Using serial 10-fold dilutions of RNA extracted from faces known to contain NLV-2, the performance of three manufacturer's reagents for single-tube RT-PCR (not nested) were compared: MuLV RT and AmpliTaq Gold (ABI); Omniscript and HotStarTaq, with or without Q-Solution (Qiagen); and the Titan One Tube RT-PCR System (Roche). The Titan System consistently produced amplification product at dilutions 100 to 1,000 times more dilute than the other two (data not shown) and was chosen as the most sensitive method for the one-tube RT-PCR step.
Nested amplification step.
The efficiency of the nested amplification step after the primary RT-PCR was similarly determined. Expand High Fidelity PCR System enzyme mix (Roche), AmpliTaq (ABI), or AmpliTaq Gold were tested as the enzyme in the nested reagent mix in the single-tube RT-PCR with serially diluted NLV-2 RNA. Nested product was obtained at lower template dilutions when AmpliTaq Gold was used compared with the other two (data not shown).
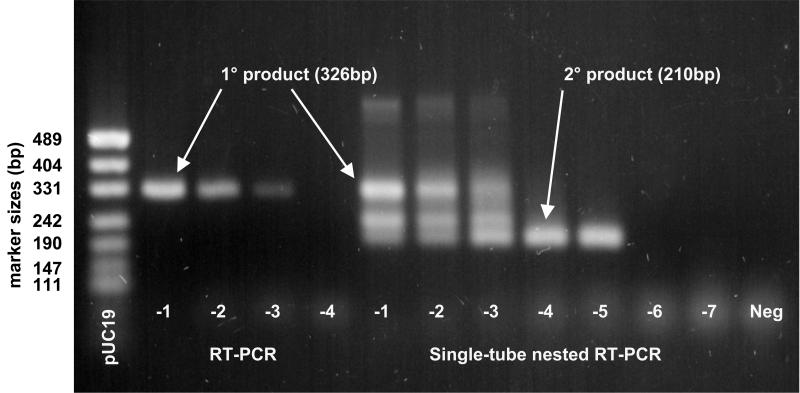
A typical amplification of serially diluted NLV-2 RNA from both single and nested RT-PCR is presented in Fig. 1. Typically, a 100-fold increase, but occasionally a 1,000-fold increase, of sensitivity was observed with the addition of the nested step. The nested RT-PCR yielded products from both outer and inner primers when the number of target templates was high. It was also common to obtain a third intermediate product as well, the result of amplification with an outer and inner primer pair. At a high dilution, near the endpoint dilution, where target templates were few, only the second amplification product was visible.
FIG. 1.
Typical amplification patterns for 10-fold serially diluted fecal RNA extracts containing NLV-2 by using RT-PCR (lanes 2 to 5) and one-tube nested RT-PCR (lanes 6 to 13), showing a 100-fold increase in sensitivity with the latter method. See Table 1 for primers; RT-PCR was performed with NLV-2-OF and NLV-2-OR to produce a 326-bp product. Nested RT-PCR was performed with NLV-2-OF and NLV-2-OR priming the RT and primary PCR steps and with NLV-2-IF and NLV-2-IR priming the secondary nested amplification to produce a 210-bp nested product. The lanes are marked with the RNA extract dilution; “−1” means “10−1,” etc. Note also in the latter method the presence of an intermediate-length product produced by amplification from inner and outer primer pairs.
Primer optimization.
Primer concentrations in the primary RT-PCR were titrated for maximum amplification performance for the least amount of each primer. For each virus, performance was measured by using serial 10-fold dilutions of RNA extracts of feces known to contain the virus. Primer concentrations for the nested reaction were then similarly titrated for maximum nested product amplification by using the complete single-tube nested RT-PCR method. Although several strains for each virus were used, the unavailability of some variant strains precluded the titration of primers against every possible variant strain. Consistently, 2 pmol (in 25 μl) and 10 pmol (in 50 μl) of each primer in each set for the RT-PCR and nested amplification steps, respectively, were the lowest concentrations that provided the maximum sensitivity measured with serially diluted RNA fecal extracts for each virus. Very rarely, a particular primer inhibited maximum amplification when used within a cocktail of primers, presumably because it produced dimers with another primer. Without exception, redesigning the offending primer with a reduced GC clamp removed the inhibitory effect.
Comparison with two-tube nested RT-PCR.
Although the high contamination risk of two-tube nested RT-PCR was thought to preclude the technique from routine diagnostic use, a comparison was performed to determine the relative sensitivity of the “hanging-drop” single-tube nested method with traditional two-tube nested amplification. Using fresh 10-fold serial dilutions of an RNA extract from a sample containing astrovirus, the sensitivity was compared in 12 replicates, with no more than four replicates being tested on any one day. Both methods gave an identical sensitivity with 10 of the replicates, and each outperformed the other by one 10-fold dilution for one replicate each. In addition, extracts from 12 samples known to contain NLV-2 were similarly tested. Identical sensitivity was obtained with six of the samples, and each method outperformed the other by one dilution in three extracts. Therefore, the mean sensitivity of the two nested amplification methods is equivalent.
Stability at −70°C.
The amplification endpoint dilution with freshly extracted RNA and nonfrozen reagents was 10−4. The identical amplification endpoint was obtained after both RNA and dispensed reagents were frozen for periods of 1 to 12 months, indicating that no loss of reagent potency occurred during this time.
Clinical evaluation.
A clinical evaluation method was used to examine fecal samples from both outbreak and sporadic diarrheal cases. Twelve outbreaks were reported between April 1999 and February 2001, resulting in 122 samples being collected for testing. A causative agent was detected in 10 of the outbreaks; NLV-2 was detected in 8 outbreaks, and NLV-1 and rotavirus were detected in 1 outbreak each. (Table 2). In one other outbreak, only 1 of 28 specimens was rotavirus positive, but this was not considered to be the cause of the outbreak. No etiological agent could be determined for the remaining outbreak. In total, 70 of 122 (57%) of outbreak samples were positive, but 60 of 81 (74%) of specimens from the NLV-2 outbreaks were positive.
TABLE 2.
Analysis of 12 gastroenteritis outbreaks and the implicated viral agent
| Outbreak | Type | No. of samples | No. positive | % Positive | Agent |
|---|---|---|---|---|---|
| 1 | Retirement home | 11 | 10 | 92 | NLV-2 |
| 2 | Fast food outlet | 5 | 4 | 80 | NLV-2 |
| 3 | Hospital | 4 | 2 | 50 | NLV-2 |
| 4 | Private function | 4 | 4 | 100 | NLV-1 |
| 5 | Restaurant | 4 | 0 | 0 | NDa |
| 6 | Retirement home | 26 | 24 | 92 | NLV-2 |
| 7 | Child care centre | 5 | 5 | 100 | Rotavirus |
| 8 | Retirement home | 18 | 8 | 44 | NLV-2 |
| 9 | Retirement home | 5 | 4 | 80 | NLV-2 |
| 10 | Retirement home | 8 | 5 | 62 | NLV-2 |
| 11 | Hospital | 4 | 3 | 75 | NLV-2 |
| 12 | Hospital | 28 | 1 | 4 | NDb |
ND, not detected.
Rotavirus was detected in one sample but was not considered to be the cause of the outbreak.
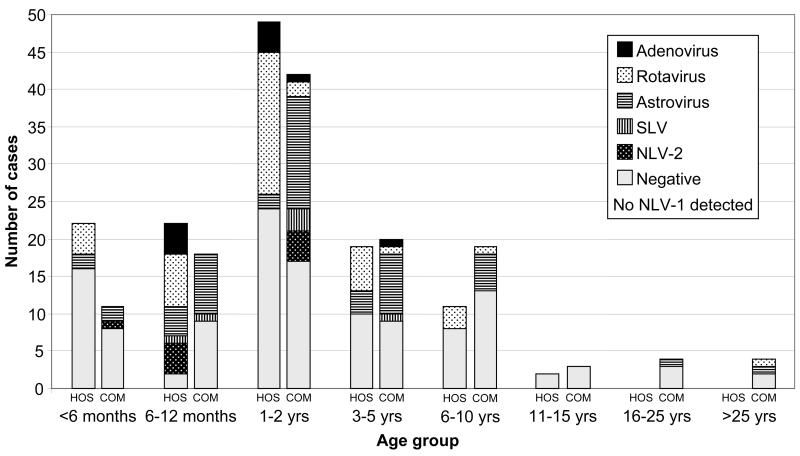
In addition, 197 randomly selected fecal samples submitted from sporadic cases of diarrhea were examined in two separate series. In the first series, from November 1999 to March 2000 (summer), 79 samples referred to the IMVS from cases presenting to a children's hospital with diarrhea and 70 samples referred from cases (both children and adults) of diarrhea presenting to community medical practices were tested for caliciviruses (NLV-1, NLV-2, and SLV) and astrovirus by nested RT-PCR and for the presence of rotavirus and adenovirus antigens by enzyme immunoassay. Viruses were detected in 66 of 149 (44%) of the samples from both sources, and the prevalence rates for each virus are presented in Table 3. Ten cases were mixed infections as follows: three astrovirus-SLV, two astrovirus-rotavirus, two NLV-2-rotavirus, and one each NLV-2-adenovirus, astrovirus-adenovirus, SLV-rotavirus, and astrovirus-rotavirus-adenovirus. The high prevalence of astrovirus in the community cases was thought to be unusual, and a second series of 48 community specimens referred to the IMVS during September and October (spring) of 2000 was examined. Viruses were detected in 19 of 48 (40%) of the cases, two with mixed infections with astrovirus and either rotavirus or adenovirus. The virus prevalence rates for this series are similarly presented in Table 3. The high rate of astrovirus was repeated with a combined community astrovirus rate of 40 of 118 samples (33.8%) compared with 5 of 118 samples (4.2%) for rotavirus, a virtual reversal of the rates for the two viruses from the hospital samples. Figure 2 graphically summarizes the prevalence rates for the hospital and combined community samples for each age group. The different rates for astrovirus and rotavirus in the two populations is statistically significant (chi square, P < 0.001). The mean ages were 2.4 and 2.2 years for astrovirus- and rotavirus-infected patients under 10 years of age, respectively. In all, a viral agent was detected in 85 of 197 (43%) of the samples.
TABLE 3.
Prevalence rates for each virus in 197 samples of sporadic infection referred from a children's hospital and community health centersa
| Source (no.) | No. of positive samples (%) as determined by:
|
|||||
|---|---|---|---|---|---|---|
| NLV-1 PCR | NLV-2 PCR | HAstV PCR | SLV PCR | ROTA EIA | ADENO EIA | |
| Community (70)b | 0 | 4 | 27* | 3 | 2† | 0 |
| Hospital (79)b | 0 | 3 | 6* | 1 | 26† | 5 |
| Total (149)b | 0 (0) | 7 (5) | 33 (22) | 4 (2) | 28 (19) | 5 (3) |
| Community (48)c | 0 (0) | 1 (2) | 13* (27) | 2 (4) | 3† (6) | 2 (4) |
| Combined totals | 0 (0) | 8 (4) | 46 (23) | 6 (3) | 31 (16) | 7 (4) |
Note the different detection rates of astrovirus * and rotavirus † in samples from community and hospital sources. EIA, enzyme immunoassay; HAstV, astrovirus; ROTA, rotavirus; ADENO, adenovirus.
Collected from November 1999 to March 2000 (summer).
Collected in September and October 2000 (spring).
FIG. 2.
Detection rates of each virus for each patient age group referred from a children's hospital (HOS) or from community health centers (COM).
Only the inner amplification product was visually detected in 32 of 60 (53%) of all positive findings from sporadic sources, suggesting a >2-fold increase in positive results through the use of a nested reaction. Six samples produced exclusively outer amplification product (the three NLV-1 outbreak samples and one NLV-2 and two astrovirus sporadic samples), indicating that the viral RNA in these samples was too variant from published sequences for the inner primers to bind. Inhibition was detected in 14 (7.1%) of the samples but could be removed at a dilution of 1:10, with the exception of one sample, which required a dilution of 1:30. Virus (NLV-2) was detected in only one (13%) of these samples, suggesting that the rate of virus detection in these inhibitory samples may have been reduced either by the dilution effect or by persisting inhibition. No evidence of contamination in the negative controls was observed in any of the assay runs.
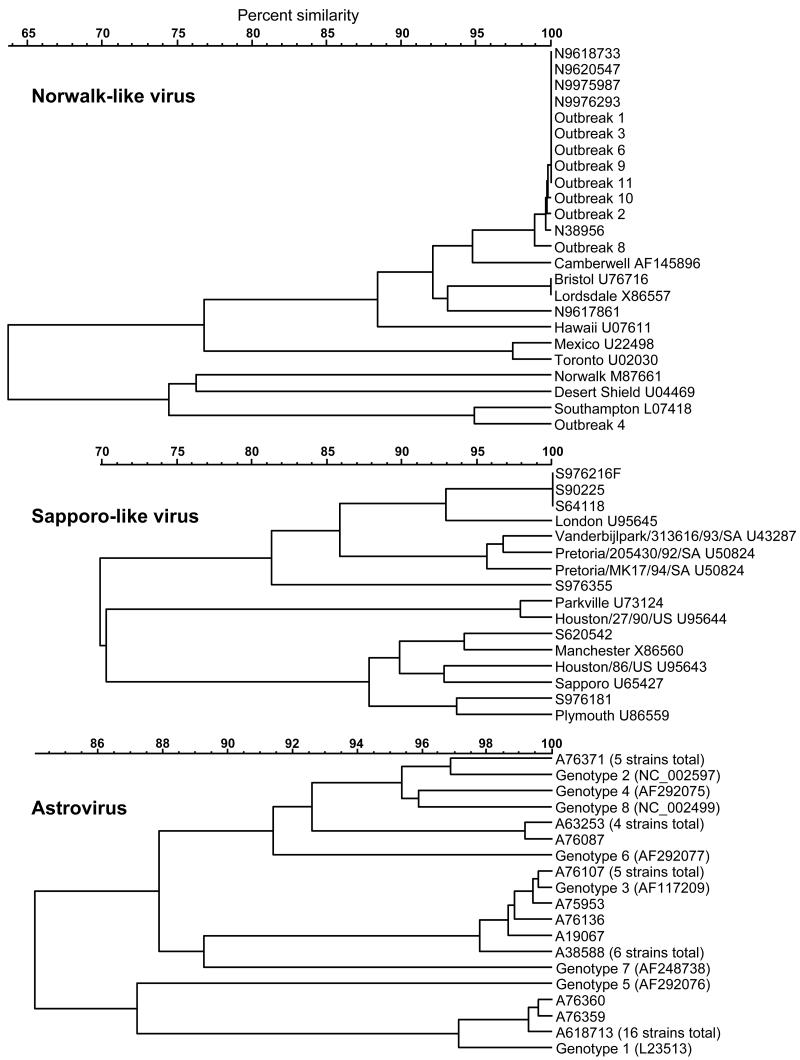
Insufficient product was produced in two NLV-2- and four astrovirus-positive samples (all from sporadic cases) to enable sequencing to be performed. The phylogenetic analysis of the remainder is presented in Fig. 3. The sequences obtained from all positive samples within each of the nine NLV-associated outbreaks were identical, supporting a causal association for each outbreak. The NLV-2 strains detected in both sporadic and outbreak samples were all closely related to the Camberwell strain, with one exception (sporadic) which was more similar to the Bristol and Lordsdale strains. The sole outbreak-associated NLV-1 strain was related to the Southampton strain. The astrovirus strains were predominantly genotype 1 (16 with identical sequence) and less frequently genotypes 2 and 3. A cluster of four strains (represented by A63253 and A76087) were unrelated to currently well-characterized genotypes. The SLV strains, although fewer in number, were much more diverse and related to the Manchester, London, and Plymouth strains. One strain (S976355) appeared to be novel.
FIG. 3.
UPGMA dendrograms of sequence similarities found among sporadic and outbreak-associated strains detected in this study compared to some published reference strains for NLV, SLV, and astrovirus. The vertical bar joining two strains or clusters indicates level of similarity. GenBank accession numbers are given for the reference strains.
DISCUSSION
Nested PCR has rightfully earned a reputation of being at risk of contamination with primary amplification product. For routine diagnostic use, the risk of such false-positive reactions has precluded its wide use, except where purpose-designed facilities are available. The novel but very simple one-tube nested RT-PCR described here eliminates contamination by primary product, reducing the risk of contamination to that equivalent for single-round amplification. It does not require the use of additional reagents or purpose-designed consumables. In addition, because the only manipulation required between the primary and secondary amplification cycles is a simple centrifugation to combine the reagents, the method is very labor efficient. Both the 0.2- and 0.6-ml tubes with flat or domed lids perform equally well. No evidence of premature mixing of the secondary amplification reagents from the lid into the primary amplification mix was ever observed, except when the tubes were subject to jolting, such as by being dropped onto the floor. The droplet of secondary amplification reagents remained in the lid during repeated opening and closing of the cap during setup, throughout amplification, and even during freezing to −70°C. Once frozen, the droplet was quite stable, but it did dislodge if jolted during thawing. This was not a problem if the tubes were left to thaw completely for 10 min. The ability to freeze preprepared tubes for up to 12 months with no loss of potency greatly simplifies routine use if sample numbers are few and the screening for several viruses is required.
The optimized higher concentration of second-round “nested” primers compared to that for first round primers, in order to preferentially favor the formation of nested product during nested amplification, has been reported elsewhere (26).
Primer homology.
An additional advantage was recognized when fecal extracts from some of the sporadic cases were tested. It was occasionally observed that only product from outer primers or only intermediate-length product from an inner and outer primer pair was generated after nested amplification, implying that some primers had failed to bind and prime a true nested amplification. Such samples may have been erroneously reported as negative by a nonnested RT-PCR method. Exhaustive attempts had been made to design universal primer sets for each virus. However, because of the frequency with which mutational substitutions can be introduced into the genome during replication, the primer target sequences in strains will continue to change. The sequencing of the primer target regions within these variant new strains and the incorporation of additional target sequence variation into the primer sets will ensure the spectrum of detectable strain variability remains broad.
Clinical samples.
That 9 of the 12 outbreaks examined were caused by NLV is a clear indication of the importance of monitoring for this virus in outbreak samples and is consistent with published reports that NLV is the most common cause of gastroenteritis outbreaks (6, 10, 12, 16, 21).
In the samples from sporadic cases, the high prevalence of rotavirus observed in cases presenting to the hospital compared to those presenting to the community health centers, with the opposite true for astrovirus, was unexpected. The latter high prevalence of astrovirus and low prevalence of rotavirus in cases presenting to community health centers was not seasonal or age related, based on the small sample size reported here. Given that both groups ultimately represent community-acquired infection, a possible explanation for differing prevalence rates is that although astrovirus infection is common in the community, the disease severity is such that patients are managed by their local health practitioners (32). In comparison, rotavirus infection may be more severe, and patients are more likely to require hospitalization. The high prevalence of rotavirus as a cause of severe gastroenteritis is well documented (9, 11, 33), and the increasing recognition of the role of calicivirus and astrovirus has been reported (13, 16, 27, 28), with astrovirus commonly being associated with a less severe illness. High prevalence rates for astrovirus have also been previously reported, but usually in regions where unsanitary living conditions are common (5, 18). However, published surveys are typically hospital based, which may bias the prevalence rates toward agents that cause more severe symptoms. In a study of outbreaks of gastroenteritis in U.S. daycare centers, astrovirus was implicated in 6 of 81 (7%) of outbreaks, and 52% of infected children were asymptomatic. Both prevalence the rate and the symptom severity were higher in children under 12 months of age (20). Such a finding suggests that disease severity in symptomatic astrovirus infections may be less severe.
In this survey, a viral etiological agent was detected in 43% of samples. Based on the presence of a first-round product compared to only the nested product as a crude comparison of single-round amplification and nested amplification, the use of nested amplification in this survey more than doubled the detection rates for calicivirus and astrovirus. This finding suggests that published prevalence rates, which are commonly based on normal nonnested amplification methods, significantly underestimate the actual rate of viral carriage. Finally, the present study suggests the frequent presence of a range of viruses in patients with gastroenteritis but whether they are causally related is unclear. A prospective case control study of hospital- and community-acquired diarrhea by these methods is currently being performed in order to investigate the association with disease.
The benefit of sequence-based phylogenetic analysis was evident in this study. Sequence identity among strains detected in the outbreaks confirmed a causal association. The high level of sequence similarity detected in NLV-2 strains from both outbreak and sporadic episodes suggests that a single dominant epidemic strain is currently circulating within the community examined here and most likely being disseminated by direct or indirect human-to-human spread. Strains of the same clonal origin, based on high sequence similarity, have been reported to have attained dominance in both the United States and Europe during 1995 and 1996 (24). Interestingly, the domination was reported to be short-lived, with a greater variety of NLV strains again being detected during the following season. Whether a similar loss of domination, as evidenced by increasing sequence diversity among detected strains causing infection, occurs in this community remains to be seen. In contrast, the SLV strains appear to be much more genetically diverse, although the number of strains detected is small. This suggests that the transmission routes and origin of such strains is more diverse. A level of strain clonality is also evident among the astrovirus strains, with identical genotype 1 sequence in 38% of the astrovirus strains detected.
In summary, a novel, but very simple and robust nested RT-PCR method has been successfully evaluated for four clinically relevant viruses causing gastroenteritis. The sensitivity and broad reactivity of this approach is suggested by the high rates of viral detection in fecal samples from 10 outbreaks and from an uncontrolled survey of sporadic infection. The method will be useful for detecting these viruses in both sporadic and outbreak gastroenteritis but may also be applicable to the testing of food and environmental samples either implicated in outbreaks or thought to be contaminated and pose an infectious risk. The additional sensitivity obtained from the nested amplification would be especially useful in such specimens, where virion numbers can be very low. A higher than expected prevalence rate for astrovirus was detected in fecal samples from nonhospital sources, which requires further examination with a comprehensive prospective controlled study of community-acquired gastroenteritis to properly assess comparative prevalence rates and disease severity.
Acknowledgments
We acknowledge the cooperation of the Department of Microbiology and Infectious Diseases, Women's and Children's Hospital, and the Communicable Diseases Control Unit of the South Australian Department of Human Resources for assistance in obtaining some of the patient samples used in this study, as well as the generous financial support of Medvet Science Pty., Ltd., Thebarton, South Australia, Australia, and the Clive and Vera Ramaciotti Foundation.
REFERENCES
- 1.Ando, T., J. S. Noel, and R. L. Fankhauser. 2000. Genetic classification of “Norwalk-like viruses.” J. Infect. Dis. 181:S336-S348. [DOI] [PubMed] [Google Scholar]
- 2.Berg, D. E., M. A. Kohn, T. A. Farley, and L. M. McFarland. 2000. Multi-state outbreaks of acute gastroenteritis traced to fecal-contaminated oysters harvested in Louisiana. J. Infect. Dis. 181:S381-S386. [DOI] [PubMed] [Google Scholar]
- 3.Caul, E. O. 1996. Viral gastroenteritis: small round structured viruses, caliciviruses and astroviruses. I. The clinical and diagnostic perspective. J. Clin. Pathol. 49:874-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiba, S., S. Nakata, K. Numata-Kinoshita, and S. Honma. 2000. Sapporo virus: history and recent findings. J. Infect. Dis. 181:S303-S308. [DOI] [PubMed] [Google Scholar]
- 5.Cruz, J. R., A. V. Bartlett, J. E. Herrmann, P. Caceres, N. R. Blacklow, and F. Cano. 1992. Astrovirus-associated diarrhea among Guatemalan ambulatory rural children. J. Clin. Microbiol. 30:1140-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deneen, V. C., J. M. Hunt, C. R. Paule, R. I. James, R. G. Johnson, M. J. Raymond, and C. W. Hedberg. 2000. The impact of foodborne calicivirus disease: the Minnesota experience. J. Infect. Dis. 181:S281-S283. [DOI] [PubMed] [Google Scholar]
- 7.Dulbecco, R., and M. Vogt. 1954. Plaque formation and isolation of pure lines with poliomyelitis viruses. J. Exp. Med. 98:167-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fankhauser, R. L., J. S. Noel, S. S. Monroe, T. Ando, and R. I. Glass. 1998. Molecular epidemiology of “Norwalk-like viruses” in outbreaks of gastroenteritis in the United States. J. Infect. Dis. 178:1571-1578. [DOI] [PubMed] [Google Scholar]
- 9.Glass, R. I., P. E. Kilgore, R. C. Holman, S. Jin, J. C. Smith, P. A. Woods, M. J. Clarke, M. S. Ho, and J. R. Gentsch. 1996. The epidemiology of rotavirus diarrhea in the United States: surveillance and estimates of disease burden. J. Infect. Dis. 174:S550-S511. [DOI] [PubMed] [Google Scholar]
- 10.Glass, R. I., J. Noel, T. Ando, R. Fankhauser, G. Belliot, A. Mounts, U. D. Parashar, J. S. Bresee, and S. S. Monroe. 2000. The epidemiology of enteric caliciviruses from humans: a reassessment using new diagnostics. J. Infect. Dis. 181:S254-S261. [DOI] [PubMed] [Google Scholar]
- 11.Hafferjee, J. E. 1995. The epidemiology of rotavirus infections: a global perspective. J. Paediatr. Gastroenterol. Nutr. 20:275-276. [DOI] [PubMed] [Google Scholar]
- 12.Hedlund, K. O., E. Rubilar-Abreu, and L. Svensson. 2000. Epidemiology of calicivirus infections in Sweden, 1994-1998. J. Infect. Dis. 181:S275-S280. [DOI] [PubMed] [Google Scholar]
- 13.Inouye, S., K. Yamashita, S. Yamadera, M. Yoshikawa, N. Kato, and N. Okabe. 2000. Surveillance of viral gastroenteritis in Japan: pediatric cases and outbreak incidents. J. Infect. Dis. 181:S270-S274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kapikian, A. Z., and R. M. Chanock. 1996. Rotaviruses, p. 1657-1708. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa.
- 15.Kok, T.-W., A. Sharley, L. Payne, R. B. Johnson, and J. Pacina. 1998. A composite enzyme immunoassay for detection of rotavirus and adenovirus in fecal samples. Aust. J. Med. Sci. 19:56-59. [Google Scholar]
- 16.Koopmans, M., J. Vinje, M. de Wit, I. Leenen, W. van der Poel, and Y. van Duynhoven. 2000. Molecular epidemiology of human enteric caliciviruses in The Netherlands. J. Infect. Dis. 181:S262-S269. [DOI] [PubMed] [Google Scholar]
- 17.Llop, P., A. Bonaterra, J. Penalver, and M. M. Lopez. 2000. Development of a highly sensitive nested-PCR procedure using a single closed tube for detection of Erwinia amylovora in asymptomatic plant material. Appl. Environ. Microbiol. 66:2071-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maldonado, Y., M. Cantwell, M. Old, D. Hill, M. L. Sanchez, L. Logan, F. Millan-Velasco, J. L. Valdespino, J. Sepulveda, and S. Matsui. 1998. Population-based prevalence of symptomatic and asymptomatic astrovirus infection in rural Mayan infants. J. Infect. Dis. 178:334-339. [DOI] [PubMed] [Google Scholar]
- 19.Mathis, A., R. Weber, H. Kuster, and R. Speich. 1997. Simplified sample processing combined with a sensitive one-tube nested PCR assay for detection of Pneumocystis carinii in respiratory specimens. J. Clin. Microbiol. 35:1691-1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitchell, D. K., R. Van, A. L. Morrow, S. S. Monroe, R. I. Glass, and L. K. Pickering. 1993. Outbreaks of astrovirus gastroenteritis in day care centers. J. Pediatr. 123:725-732. [DOI] [PubMed] [Google Scholar]
- 21.Monroe, S. S., T. Ando, and R. I. Glass. 2000. Human enteric caliciviruses: an emerging pathogen whose time has come. J. Infect. Dis. 181:S249-S251. [DOI] [PubMed] [Google Scholar]
- 22.Murphy, A. M., G. S. Grohmann, P. J. Christopher, W. A. Lopez, G. R. Davey, et al. 1979. An Australia-wide outbreak of gastroenteritis from oysters caused by Norwalk virus. Med. J. Aust. 2:329-333. [DOI] [PubMed] [Google Scholar]
- 23.Nakata, S., S. Honma, K. K. Numata, K. Kogawa, S. Ukae, Y. Morita, N. Adachi, and S. Chiba. 2000. Members of the family caliciviridae (Norwalk virus and Sapporo virus) are the most prevalent cause of gastroenteritis outbreaks among infants in Japan. J. Infect. Dis. 181:2029-2032. [DOI] [PubMed] [Google Scholar]
- 24.Noel, J. S., R. L. Fankhauser, T. Ando, S. S. Monroe, and R. I. Glass. 1999. Identification of a distinct common strain of “Norwalk-like viruses” having a global distribution. J. Infect. Dis. 179:1334-1344. [DOI] [PubMed] [Google Scholar]
- 25.Noel, J. S., T. W. Lee, J. B. Kurtz, R. I. Glass, and S. S. Monroe. 1995. Typing of human astroviruses from clinical isolates by enzyme immunoassay and nucleotide sequencing. J. Clin. Microbiol. 33:797-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olmos, A., M. Cambra, O. Esteban, M. T. Gorris, and E. Terrada. 1999. New device and method for capture, reverse transcription, and nested PCR in a single closed-tube. Nucleic Acids Res. 27:1564-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palombo, E. A., and R. F. Bishop. 1996. Annual incidence, serotype distribution, and genetic diversity of human astrovirus isolates from hospitalized children in Melbourne, Australia. J. Clin. Microbiol. 34:1750-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pang, X. L., S. Honma, S. Nakata, and T. Vesikari. 2000. Human caliciviruses in acute gastroenteritis of young children in the community. J. Infect. Dis. 181:S288-S294. [DOI] [PubMed] [Google Scholar]
- 29.Stafford, R., D. Strain, M. Heymer, C. Smith, M. Trent, and J. Beard. 1997. An outbreak of Norwalk virus gastroenteritis following consumption of oysters. Commun. Dis. Intell. 21:317-320. [DOI] [PubMed] [Google Scholar]
- 30.Vinje, J., H. Deijl, R. van der Heide, D. Lewis, K. O. Hedlund, L. Svensson, and M. P. Koopmans. 2000. Molecular detection and epidemiology of Sapporo-like viruses. J. Clin. Microbiol. 38:530-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walsh, E. E., A. R. Falsey, I. A. Swinburne, and M. A. Formica. 2001. Reverse transcription polymerase chain reaction (RT-PCR) for diagnosis of respiratory syncytial virus infection in adults: use of a single-tube “hanging droplet” nested PCR. J. Med. Virol. 63:259-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walter, J. E., and D. K. Mitchell. 2000. Role of astroviruses in childhood diarrhea. Curr. Opin. Pediatr. 12:275-279. [DOI] [PubMed] [Google Scholar]
- 33.Waters, V., E. L. Ford-Jones, M. Petric, M. Fearon, P. Corey, and R. Moineddein. 2000. Etiology of community-acquired pediatric viral diarrhea: a prospective longitudinal study in hospitals, emergency departments, pediatric practices and child care centers during the winter rotavirus outbreak, 1997 to 1998. Pediatr. Infect. Dis. J. 19:843-848. [DOI] [PubMed] [Google Scholar]
- 34.Wolff, C., D. Hornschemeyer, D. Wolff, and K. Kleesiek. 1995. Single-tube nested PCR with room-temperature-stable reagents. PCR Methods Appl. 4:376-379. [DOI] [PubMed] [Google Scholar]
- 35.Wright, P. J., I. C. Gunesekere, J. C. Doultree, and J. A. Marshall. 1998. Small round-structured (Norwalk-like) viruses and classical human caliciviruses in southeastern Australia, 1980-1996. J. Med. Virol. 55:312-320. [PubMed] [Google Scholar]