Abstract
Quantitation of cytomegalovirus (CMV) DNA in plasma and serum by PCR is increasingly used to identify patients at risk for developing CMV disease and to monitor the efficacy of antiviral therapy. Although CMV DNA levels are generally interpreted as viral loads, the exact nature of the viral DNA in these specimens is unknown. We studied the state of CMV DNA in plasma and serum specimens obtained from three renal transplant recipients at peak viral DNA levels during primary CMV infection. For this purpose, DNA isolated from these specimens was fractionated by size, and CMV DNA levels in the resulting DNA fractions were measured by quantitative PCR targeted at large (578-bp) and small (134-bp) amplicons. These experiments showed that the molecular sizes of DNA fragments from which CMV DNA is amplified were small (<2,000 bp), indicating that CMV DNA in plasma and serum is highly fragmented. Furthermore, CMV DNA levels were consistently higher when targeted at the smaller amplicon, providing additional evidence for the fragmentation of viral DNA. In conclusion, the first results with three patients have shown that CMV DNA in plasma and serum is highly fragmented and does not necessarily reflect the amount of infectious virus. These observations have potential consequences for understanding CMV pathogenesis and interpreting CMV DNA levels in individual patient management.
Infection with cytomegalovirus (CMV) (human herpesvirus 5) is an important cause of morbidity and mortality in immunocompromised individuals, such as transplant recipients and AIDS patients. In the management of CMV infection, preemptive treatment strategies, aimed at preventing CMV disease in high-risk patients, are receiving increasing attention. For the benefit of such strategies, quantitative detection of CMV DNA in the blood compartment by PCR is increasingly used to identify patients at risk for CMV disease. In addition, measurements of viral DNA load may be important for monitoring the efficacy of antiviral treatment and predicting the development of drug resistance (3, 22, 27).
The genome of human CMV consists of a large (about 230,000-bp) double-stranded linear DNA molecule which is encapsidated within a double protein shell and a lipid envelope (24). Most of the CMV DNA in the blood compartment is present in abortively infected polymorphonuclear leukocytes (4, 12, 16, 26); less DNA is found in peripheral blood mononuclear cells, part of which, upon differentiation into macrophages, support viral replication (12, 26, 28). In addition, circulating, productively infected endothelial cells may be a source of CMV DNA (16, 18). CMV DNA can also be detected in serum and plasma, which are convenient specimens for CMV DNA load measurement (10, 15, 19, 29). In recent years, many studies on the qualitative and quantitative detection of CMV DNA in these specimens have been reported, which generally show that levels of CMV DNA found in plasma or serum are significantly lower than those found in white blood cells (4, 5, 13, 15, 33).
At present, it is unclear whether CMV DNA present in serum and plasma actually represents circulating infectious virus carrying an intact DNA genome. Nevertheless, CMV DNA loads measured in plasma and serum are generally interpreted as the burden of infectious virus (5, 8, 10, 11, 19, 20, 31), although some authors have raised questions regarding the state of CMV DNA in these specimens (21, 26). It may well be that CMV DNA is present in these specimens in an unprotected form, vulnerable to DNA-degrading enzymes (DNases), rather than in a protected form within the enveloped viral capsid. Since serum is rich in DNases (17, 23), such unprotected DNA may be degraded further during and following the clotting process. On the other hand, unprotected CMV DNA present in plasma that is anticoagulated with EDTA is expected to be stabilized, since DNases need bivalent cations for activity.
We studied the state of CMV DNA in serum and EDTA-anticoagulated plasma specimens from renal transplant recipients suffering from a primary CMV infection. For this purpose, we fractionated CMV DNA isolated from clinical specimens by agarose gel electrophoresis in the presence of a DNA molecular size marker. After electrophoresis, slices were cut from the gel and DNA was isolated from the slices together with a known amount of internal control (IC) DNA that served as a PCR quantitation reference (8). Then, for each slice the amount of PCR target was determined by competitive PCR using each of two primer pairs, one pair resulting in a small, 138-bp amplicon (either IC or CMV specific) and the other pair resulting in a more-than-four-times-larger, 578-bp amplicon. If CMV DNA was intact, the same quantitation results and size distributions of target DNA would be expected for either primer pair. If, on the other hand, CMV DNA was highly fragmented (e.g., containing many single-stranded or double-stranded breaks), CMV loads based on the large amplicon could well be lower than those calculated from the small amplicon, and different size distributions of target CMV DNA could be found. By these experiments we show that the majority of CMV DNA found in plasma and serum obtained at peak CMV DNA levels is highly fragmented.
MATERIALS AND METHODS
Patient specimens.
Clotted and EDTA-anticoagulated blood specimens were obtained from three renal transplant recipients (recipients A to C) suffering from a symptomatic primary CMV infection, as diagnosed by seroconversion of anti-CMV antibodies. Written informed consent was obtained from all patients prior to blood sampling. The clinical syndrome consisted of fever and malaise in recipients A and B and esophagitis and duodenitis in patient C. The blood specimens from recipients A to C that were analyzed were obtained around peak CMV DNA loads as measured in whole blood by quantitative PCR (QPCR) (8). Recipient A had received 4 days of antiviral therapy with ganciclovir at this time; recipients B and C did not receive treatment. For culture of CMV, patient leukocytes were cocultivated with human diploid cells and were tested on the next day for CMV immediate-early (IE) antigen expression by immunofluorescent staining with a monoclonal antibody (anti-human CMV-IEA; Argene Biosoft).
Human CMV DNA.
To study the behavior of intact viral DNA targets, CMV DNA templates from three different origins were used: (i) sucrose density-gradient purified human CMV strain AD 169 (Advanced Biotechnologies Inc., Columbia, Md.); (ii) Rat-9G cells, which carry approximately 50 to 60 copies of the CMV IE regions 1 and 2 stably integrated into chromosomal DNA (6, 32); and (iii) plasmid pES, which contains CMV IE regions 1 and 2 as a 7-kb EcoRI-SalI fragment (6) (in the experiments described here, the 7-kb CMV DNA insert was released from the plasmid by EcoRI and SalI cleavage). Extraction of CMV DNA and Rat-9G cellular DNA was done by a silica-guanidinium thiocyanate (Si-GuSCN) procedure (7).
IC DNA.
The construction of IC DNA has been described previously (7). A solution containing 14 molecules of IC DNA (as a linearized plasmid) and 20 ng calf thymus DNA (Sigma Chemical Company) per μl of Tris-EDTA (TE) buffer was used in this study as a quantitation reference.
DNA purification and DNA fractionation.
DNA was purified from 200 μl of serum or plasma or from 50 μl of EDTA-anticoagulated whole blood by a Si-GuSCN procedure using 20 μl of size-fractionated silica particles (7); DNA was eluted in 100 μl of TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0). DNA (25 μl) was electrophoresed through horizontal 1% agarose (Agarose MP; Roche Diagnostics Corp., Mannheim, Germany) slab gels (5 to 10 V/cm) in a buffer system, described previously (1), containing 1 μg of ethidium bromide per ml. The equivalent of 12.5 μl of whole blood or 50 μl of plasma was loaded onto the gel in the presence of 1 μg of phage lambda DNA (HindIII digested; Gibco BRL) and 4 μg of a 100-bp ladder (Gibco BRL). Following electrophoresis, approximately thirty slices (2 by 5 by 5 mm) of decreasing molecular size were cut from the gel. For DNA purification, each agarose slice was dissolved in 900 μl of lysis buffer L6 (7) in the presence of 280 molecules (in the case of a final elution volume of 100 μl of TE buffer) or 420 molecules (in the case of a final elution volume of 150 μl) of IC DNA for 30 min at 37°C in an Eppendorf Thermomixer (type 5436) rotating at 800 rpm. Next, 20 μl of silica particles was added, and DNA was purified after a 10-min binding step as described previously (7). In short, silica-DNA complexes were washed twice with wash buffer L2, twice with 70% ethanol, and once with acetone. After drying, DNA was eluted in 100 or 150 μl of TE buffer, and 25 μl was used for PCR. In addition, 25 μl of DNA was electrophoresed again and visualized under UV illumination to correlate slices with DNA fragment size.
Quantitation procedure.
Quantitation of CMV DNA in clinical specimens was done as described previously (8). Quantitation of CMV DNA purified from agarose slices was done in a similar way. In short, DNA was isolated from the slices in the presence of a known amount of IC DNA. Twenty-five microliters of the eluates was subjected to PCR and hybridized with CMV-specific and IC-specific probes, and the amount of hybrids was measured by electrochemiluminescence (ECL). The ratio of virus-specific signal to IC-specific signal was calculated after correction for background. The amount of CMV DNA targets present in the slice was then calculated by multiplying this ratio by a factor of 420 or 280 (for final elution volumes of 150 and 100 μl, respectively). For a detailed description of the algorithm, see reference 8.
PCR.
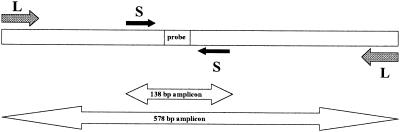
Amperase (uracil-N-glycosylase) and primers were from Perkin-Elmer (Nieuwerkerk aan de IJssel, The Netherlands). Primer pair L (for large) consisted of CMV-531 (5′ ACA AGG TGC TCA CGC ACA TTG ATC 3′; nucleotide positions 2034 to 2057) and Bio-CMV-1107 (5′CAC TGG CTC AGA CTT GAC AGA CAC 3′; 5′ biotinylated; nucleotide positions 2588 to 2611). Primer pair S (for small) consisted of CMV-S5 (5′ CCA AGC GGC CTC TGA TAA CCA A 3′; nucleotide positions 2223 to 2244) and Bio-CMV-S6 (5′ GGT CAT CCA CAC TAG GAG AGC AGA C 3′; 5′ biotinylated; nucleotide positions 2336 to 2360). Nucleotide numbering was as described by Akrigg et al. (2). Primer pair L generates a 578-bp amplicon from CMV DNA and IC DNA, whereas primer pair S generates a 138-bp amplicon from these templates (Fig. 1). The primer pairs were chosen in perfectly conserved areas of exon 4 of the major IE gene. PCR conditions were identical for both primer pairs and have been described previously (8), except that alpha casein was present during PCR at 400 ng/μl of reaction mixture (9). Excess primer was removed from PCR products as described previously (8), and DNA was eluted in 100 μl of 1× PCRII buffer (Perkin-Elmer) (10 mM Tris-HCl [pH 8.3], 50 mM KCl). The purified PCR products were subsequently used for hybridization.
FIG. 1.
Schematic representation (not to scale) of primer pair L and primer pair S, used for amplification of 578- and 138-bp amplicons, respectively. The 25-bp region in which the CMV-specific probe and the IC-specific probe are located is indicated. Except for the probe region, CMV and IC sequences are similar. For details, see Materials and Methods.
Hybridization and ECL measurement.
For QPCR, the purified PCR product was diluted 5 times (for primer pair L) or 40 times (for primer pair S) in 1× PCR II buffer, and 30 μl was used in hybridization reactions with CMV- and IC-specific Tris(2,2′-bipyridine) ruthenium(II) chelate (TBR)-labeled probes. TBR-labeled probes were TBR-CMV-1 (CMV-specific probe, 5′-TGA AGG TCT TTG CCC AGT ACA TTC T 3′; nucleotide positions 2292 to 2316) and TBR-CMV-2 (IC-specific probe, 5′ CCC TTT ACA TCT TTC TGA AGT AGG G 3′). Both probes contained a single TBR label at the 5′ end and were obtained from Perkin-Elmer; they are now available from IGEN International, Inc. (Gaithersburg, Md.). Following hybridization, 3 μl of streptavidin-coated magnetic beads (Dynabeads M280 Streptavidin, 10 mg/ml; Dynal Biotech, Oslo, Norway) and 7 μl of 1× PCR II buffer were added, and the mixture was left for 15 min at ambient temperature. Fifty microliters of the bead-hybrid suspension was added to 100 μl of water, and the ECL signal, expressed in relative luminosity units, was measured in an M8 ECL detection system according to the instructions of the manufacturer (IGEN International, Inc). In this device, excess TBR-labeled probe is automatically removed by washing, and the amount of labeled hybrids is determined after excitation by applying an electric field.
RESULTS
In Fig. 1, the primer pairs used in this study and the resulting amplicons are shown. Primer pair L generates a 578-bp amplicon from CMV and IC DNAs. Primer pair S generates another, 138-bp amplicon (more than fourfold smaller) from these templates. Following PCR, the amplicons were hybridized with CMV-specific and IC-specific TBR-labeled probes, and the amount of hybrids was determined by ECL. A known amount of IC DNA was coextracted from the specimen (agarose gel slices or clinical specimens) and was coamplified with CMV target DNA. The amount of target CMV DNA was then calculated from the ratio of the CMV-specific and IC-specific hybridization signals.
Validity of quantitation for both primer pairs.
For our analysis, it was essential that quantitations based on large (578-bp) and small (138-bp) amplicons were accurate. For intact CMV DNA targets, both primer pairs should yield the same quantitation results.
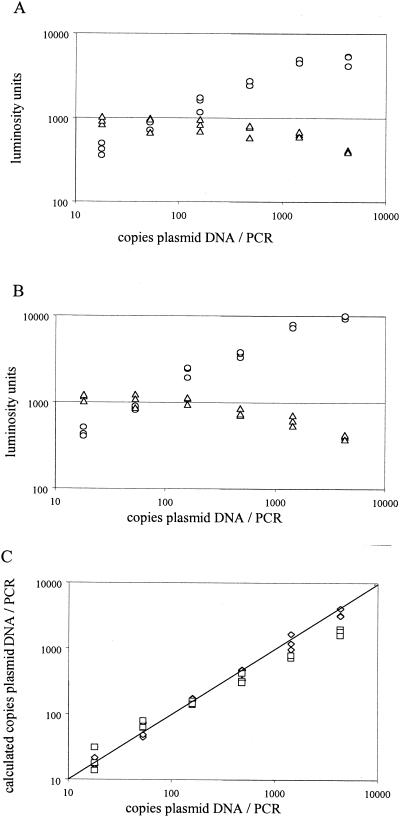
When serially diluted plasmid pES DNA (containing the CMV IE region) was used as input (in triplicate) for QPCR, the same hybridization kinetics were observed for either primer pair (Fig. 2A and B). This resulted in a linear dynamic range for quantitation of about 20 to 500 DNA molecules input per PCR irrespective of the primer pair that was used (Fig. 2C). At above 500 molecules per PCR, CMV DNA loads calculated for the small amplicon were underestimated. Loads calculated for the large amplimer were linear in the range of about 20 to 4,300 molecules per PCR. All experiments described below were within the linear dynamic range of the assays.
FIG. 2.
Validation of QPCR for 138- and 578-bp amplicons. (A and B) A serial threefold dilution of plasmid pES was subjected in triplicate to QPCR with the S (A) and L (B) primer pairs. Raw data (in luminosity units) obtained after hybridization with the IC-specific probe (triangles) and the CMV-specific probe (circles) are given. (C) From the raw data, the number of copies target DNA was calculated from the ratio of CMV-specific and IC-specific signals. Diamonds, 578-bp amplicon; squares, 138-bp amplicon.
Size distribution of intact CMV DNA.
In the experiments described below, DNA was purified from clinical specimens and from agarose gel slices by a procedure in which DNA was bound to silica particles. This inevitably resulted in some shearing (introduction of double-stranded and single-stranded breaks) of CMV DNA, since a single molecule of CMV DNA will be bound by several silica particles. During the wash steps (which involve vigorous vortexing) of the purification procedure, double-stranded breaks will be introduced by centrifugal forces, resulting in a rather broad size distribution of DNA fragments (7). Since CMV DNA is a very large molecule (approximately 230,000 bp), it was expected to have suffered from at least some shearing during DNA isolation. To study the behavior of intact CMV DNA, DNA was extracted from two different sources which were considered to be representative for high-molecular-size CMV DNA.
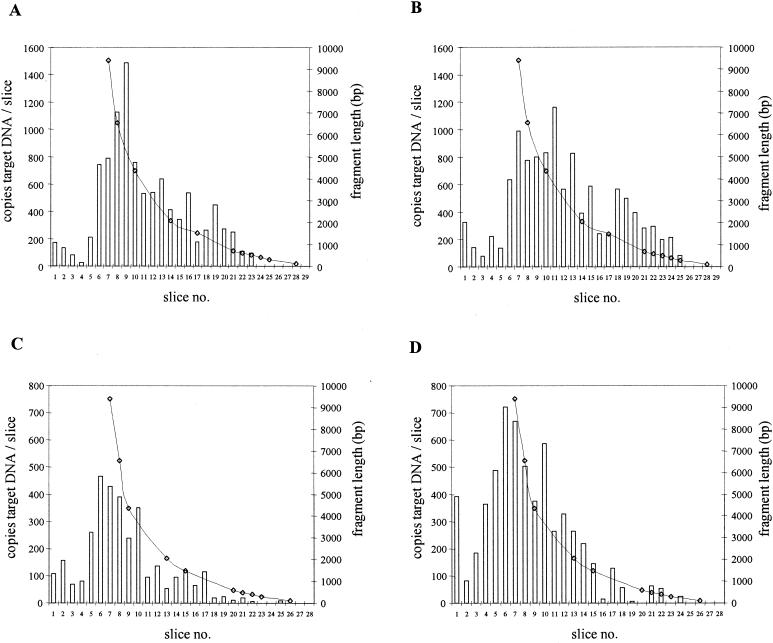
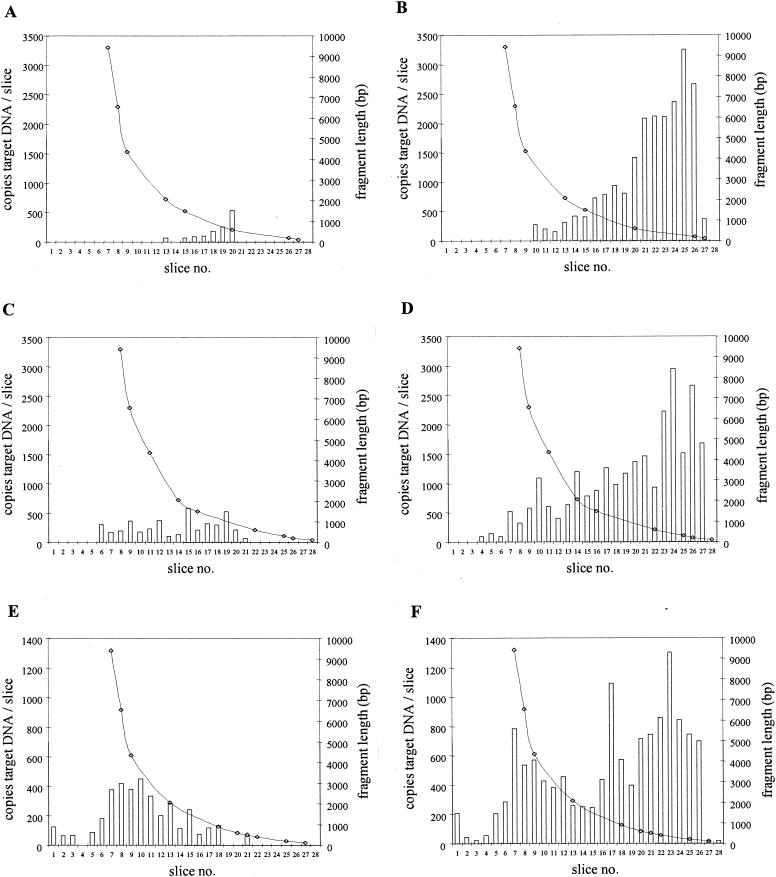
DNA was extracted from purified CMV and was size fractionated by gel electrophoresis. Next, the gel was sliced, the amount of CMV target DNA for both primer pairs was determined for each slice, and this amount was correlated to the size of the DNA fragment from which the target was amplified. Figure 3A and B show that the majority of DNA fragments from which CMV DNA was amplified were larger than 2,000 bp for each primer pair. The calculated amount of CMV DNA applied to the gel (13,750 copies, based on QPCR with L primers) was similar to the total amounts of CMV DNA in the slices for the 578-bp amplimer (10,138 copies) and for the 138-bp amplimer (11,468 copies).
FIG. 3.
Size distributions of PCR targets for CMV DNA isolated from purified CMV virions (A and B) and from Rat-9G cells (C and D) for the 578-bp (A and C) and 138-bp (B and D) amplicons. Slice 1 represents the slot to which DNA was applied in the agarose gel. Bars indicate the number of copies of CMV DNA molecules per agarose slice. Lines represent the lengths of the DNA fragments from which CMV DNA was amplified.
As an additional control for the behavior of intact CMV DNA following isolation and fractionation, the same experiment as described above was performed with DNA purified from a known number of Rat-9G cells (6, 32). Figure 3C and D show that for this CMV DNA template also, similar distributions were found for either primer pair, with the majority of DNA fragments being larger than 2,000 bp. The total amount of CMV DNA applied to the gel as determined by QPCR with the L primers (6,206 copies) correlated well with the total amount expected from the Rat-9G cell counts (6,250 copies). Furthermore, the total amounts of target DNA in the slices for the 578-bp amplimer (3,323 copies) and the 138-bp amplimer (5,936 copies) were in good accordance with the expected values given the variation coefficient of 25% for the assay (8).
From the experiments described above, it was concluded that for intact CMV DNA, quantitation results were the same for either primer pair. The size distributions of the two CMV DNA PCR targets were similar, and the majority of the DNA targets were larger than 2,000 bp in length.
Size distribution of CMV DNA targets in clinical specimens.
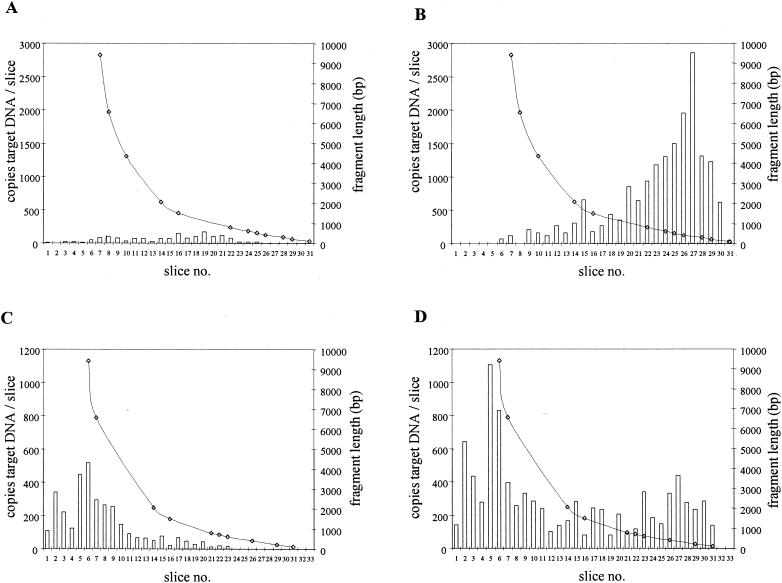
DNA was isolated from EDTA-anticoagulated whole blood within 30 min, and from the corresponding plasma within 50 min, after venipuncture of recipient A. DNA was then size fractionated, and the number of CMV DNA targets for each primer pair in each fraction was determined. Figure 4A and B show that there were two major differences between the size distributions observed for plasma DNA and those observed for intact CMV DNA. First, the majority of the CMV DNA targets were small (<2,000 bp). Second, the total amount of CMV targets found in the slices for the 138-bp amplicon was 12 times higher than the total amount of targets found for the 578-bp amplicon (17,679 and 1,496 copies, respectively). This difference can be explained by the physical inability of the L primer pair to amplify targets smaller than about 578 bp. Such small targets can be amplified only by the S primer pair. These observations indicate that plasma CMV DNA was highly fragmented.
FIG. 4.
Size distributions of CMV DNA targets in plasma (A and B) and in whole blood (C and D) from a renal transplant recipient (recipient A) for the 578-bp (A and C) and 138-bp (B and D) amplicons. The amounts of DNA applied to the gel corresponded to 12.5 μl of whole blood and 50 μl of plasma. Slice 1 represents the slot in the agarose gel. Bars indicate the number of copies of CMV DNA molecules per agarose slice. Lines represent the lengths of the DNA fragments from which CMV DNA was amplified.
The corresponding whole-blood DNA from this patient analyzed in the same way showed a size distribution that was similar to that expected for intact CMV DNA when tested for the 578-bp amplicon (Fig. 4C). The total amount of CMV DNA targets found in the slices for the 138-bp amplicon was about three times higher than the total amount of targets found for the 578-bp amplicon (9,023 and 3,308 copies, respectively). This was at least partly due to the small DNA fragments present in the plasma fraction which could not be detected by the L primer pair (Fig. 4D). Note that in all figures, the data presented for plasma and serum correspond to 50 μl of plasma and serum, whereas those presented for blood correspond to 12.5 μl of whole blood.
DNA was isolated from whole blood, EDTA-plasma, and serum obtained from another transplant recipient (recipient B) and was analyzed as described above. At the time of venipuncture, cocultivation of white blood cells with human diploid cells was strongly positive (150 positive nuclei per 106 peripheral blood mononuclear cells) for CMV IE antigen expression, indicating the presence of infectious virus.
For serum DNA, there was a sharp cutoff for positivity for DNA fragments smaller than about 600 bp when the 578-bp amplicon was measured (Fig. 5A), and the total quantity of targets in the slices was about 16 times lower (1,304 copies) than that of targets found for the 138-bp amplicon (21,395 copies) (Fig. 5B). This difference was mainly due to the presence of large amounts of small DNA targets that could not be amplified by the L primer pair.
FIG. 5.
Size distributions of CMV DNA targets in serum (A and B), EDTA-plasma (C and D), and whole blood (E and F) from a renal transplant recipient (recipient B) for the 578-bp (A, C, and E) and 138-bp (B, D, and F) amplicons. The amounts of DNA applied to the gel corresponded to 12.5 μl of whole blood and 50 μl of plasma or serum. Slice 1 represents the slot in the agarose gel. Bars indicate the number of copies of CMV DNA molecules per agarose slice. Lines represent the lengths of the DNA fragments from which CMV DNA was amplified.
Similar distributions were found for the corresponding plasma specimen (Fig. 5C and D); that is, the total amounts of DNA targets in the slices obtained for the 138-bp amplicon were about sixfold higher than those obtained for the 578 bp amplicon (25,486 and 4,366 copies, respectively). DNA fragments from which CMV targets were amplified appeared to be larger in plasma than in serum (Fig. 5A and C).
Whole-blood DNA showed a distribution that was similar to that expected for intact virus DNA when tested for the 578-bp amplicon (Fig. 5E). For the 138-bp amplicon (Fig. 5F), the total amount of targets in the slices was about fourfold larger than that observed for the 578-bp amplicon (13,159 and 3,697 copies, respectively), which was mainly due to the presence of small (<2,000-bp) CMV DNA targets. The amount of these small targets could not be explained from the amount of small targets present in the corresponding plasma fraction.
Stability of CMV DNA in EDTA-anticoagulated blood.
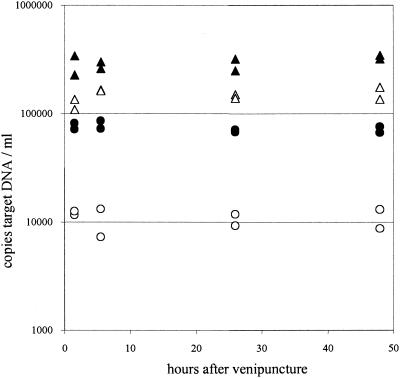
To exclude the possibility that the differences in CMV DNA loads between large and small amplicons were secondary to degradation of DNA during storage of blood, DNA was purified from whole blood and the corresponding plasma of recipient C at different time points (1.5 to 48 h at 4°C) after venipuncture. Next, the amount of CMV DNA was determined with each primer pair. The results showed that CMV DNA loads measured with either primer pair were constant upon storage of blood for at least 48 h at 4°C. This suggests that steady-state levels of CMV DNA as present in vivo were fixed at the time of venipuncture for EDTA-anticoagulated whole blood and the corresponding plasma (Fig. 6). For whole blood, CMV DNA loads calculated for the 138-bp amplicon were twice as high as those calculated for the 578-bp amplicon (295,630 and 146,531 copies/ml of blood, respectively). Plasma loads calculated for the 138-bp amplicon were about sevenfold higher than those calculated for the 578-bp amplicon (74,222 and 11,088 copies/ml of plasma, respectively).
FIG. 6.
Stability of CMV DNA targets in whole blood and plasma prepared at different time points after venipuncture of recipient C. Results are from duplicate extractions. Triangles, whole blood; circles, EDTA-plasma. Open symbols, loads obtained for the 578-bp amplicon; closed symbols, loads obtained for the 138-bp amplicon.
DISCUSSION
We have shown with three renal transplant recipients that during primary CMV infection, the majority of CMV DNA detected in plasma and serum obtained at peak viral DNA loads is highly fragmented. This conclusion was reached from two lines of evidence. First, CMV DNA levels measured in plasma and serum were consistently and significantly higher (up to 16 times) when a small (138-bp) CMV DNA amplicon was amplified than when a four-times-larger (578-bp) amplicon was tested. Second, by experiments in which DNA purified from these specimens was fractionated by size and the amounts of large and small targets were determined in relation to the molecular size of the DNA fragments from which the CMV DNA was amplified, it was shown that the majority of DNA targets in plasma and serum was significantly smaller than 2,000 bp.
In our experiments, DNA was purified from clinical specimens and agarose gel slices by an Si-GuSCN procedure (7), which is widely used for extraction of CMV DNA from clinical specimens (8, 9, 15, 30). It is known that shearing of large DNA molecules occurs during this procedure (7). For this reason, it was important to show that the DNA purification procedure itself did not result in the extent of fragmentation of CMV DNA that we observed in plasma and serum. We therefore analyzed two DNA templates that were considered to be representative of intact high-molecular-size CMV DNA: DNA purified from sucrose density-gradient purified CMV and DNA purified from Rat-9G cells, which carry the CMV IE gene stably integrated into chromosomal DNA at a known copy number. For both DNA templates, no significant differences in CMV DNA loads were observed between large and small amplicons. Moreover, gel profiles were similar for either template irrespective of the amplicon length, with the majority of DNA targets being larger than 2,000 bp. In addition, the amounts of targets applied to the gel were in good accordance with the total amounts recovered from the gel slices, indicating that no PCR targets were destroyed upon DNA extraction from the gel slices. From these experiments, it was concluded that the DNA purification procedure did not significantly contribute to the fragmentation of CMV DNA observed in plasma and serum.
Serum and plasma are not true body fluids, since neither exists in vivo. Serum is formed after a complex clotting process following venipuncture and is expected to contain DNases, which may degrade unprotected CMV DNA (17, 23). Therefore, DNA loads measured in serum may not represent the in vivo situation. On the other hand, in blood anticoagulated with EDTA (which efficiently complexes bivalent cations, thereby preventing DNase activity), DNA loads may be fixed at the time of venipuncture. For renal transplant patients, Schäfer et al. recently showed that CMV DNA loads in leukocytes isolated from EDTA-anticoagulated blood were constant for up to 72 h of storage (25). Together with our finding that, irrespective of the target size, CMV DNA loads in EDTA-anticoagulated whole blood and plasma did not change during storage, these data strongly suggest that the state of CMV DNA in blood was indeed fixed at the time of venipuncture. This indicates that the extent of fragmentation of CMV DNA that we observed in plasma and whole blood already existed in vivo.
Our observation that serum and plasma specimens of our three patients contain mainly highly fragmented CMV DNA, which is preferentially detected when aiming at small PCR targets, implies that this DNA does not represent infectious virus. This is in accordance with the notion that CMV usually cannot be cultured from CMV DNA-positive plasma specimens (13, 15, 29). Therefore, a rise in CMV DNA load in plasma and serum does not necessarily represent a rise in virus production but may also reflect an active immune response leading to degradation of CMV DNA. Alternatively, the presence of CMV DNA fragments may represent disordered production of viral DNA. More studies are needed to address the question of in which form the CMV DNA fragments are present in plasma, e.g., as naked (or partially protected, nucleosomal) DNA, in apoptotic bodies, or within virus particles rendered susceptible to DNases.
There is no doubt that infectious CMV is present in blood at some stages of infection, as evidenced by the recovery of infectious virus by cocultivation of white blood cells with cells permissive for CMV replication. Therefore, at least some of the CMV DNA detected in whole blood may have been derived from infectious virus. Indeed, the size distribution of whole-blood CMV DNA as detected by large amplicons was similar to that found for intact virions or Rat-9G cells. However, when testing whole blood for small PCR targets, a clear biphasic size distribution of DNA targets was found: one population consisted of large DNA targets (greater than approximately 2,000 bp), while another population consisted of small targets (less than approximately 2,000 bp). In addition, whole-blood CMV DNA loads found for the small amplicon were two to four times higher than those found for the large amplicon. For one patient (recipient A), this difference could partially be explained by the amount of small targets present in the plasma fraction of blood. However, for another patient (recipient B), the amount of small targets present in blood was about three times higher than expected from the amount present in the plasma fraction. In other experiments, we have found up to 17-fold-higher levels of CMV DNA in whole blood for the small targets relative to large targets (data not shown). These observations suggest that a significant proportion of fragmented CMV DNA may also be present in blood cells, possibly in polymorphonuclear leukocytes since these have been found to harbor the majority of CMV DNA detected in blood (4, 5, 13). Further studies are needed to assess whether this indeed is the case.
It is beyond doubt that CMV DNA load measurements are important for prediction and diagnosis of CMV disease. However, proposed nominal CMV DNA loads as guidance for treatment initiation or prediction of disease development vary substantially between different studies, even in similar patient groups (3). Some of this variability may be explained by our observation that the levels of CMV DNA measured by PCR in plasma, serum, and, to a lesser extent, whole blood are highly dependent on the length of the amplicon. In addition, the choice of specimen influences the levels of viral DNA detected. We have recently shown that CMV DNA loads in serum specimens of renal transplant recipients were significantly lower (up to 27-fold) than those observed in paired EDTA-anticoagulated plasma specimens, most likely due to continued degradation of CMV DNA by DNases in serum, which are inactivated in EDTA-plasma (8). In contrast, Patel et al. reported similar levels of CMV DNA in serum and EDTA-plasma specimens from liver transplant recipients (21). This discrepancy may be explained by the fact that the PCR target used in the study by Patel et al. (152 bp) was much smaller than our PCR target (578 bp), thus enabling amplification of highly fragmented CMV DNA fragments in serum which could not be amplified by our primers. Target size-dependent differences in CMV DNA levels are less likely when high-molecular-size CMV DNA is prevalent, as seems to be the case in whole blood.
Beside the influence of the PCR target size on the level of CMV DNA, it should also be realized that a different DNA population will be detected when targeting is at large amplicons (e.g., 578 bp) than when the PCR is targeted at a small amplicon (e.g., 138 bp), since the larger amplicon excludes detection of highly fragmented CMV DNA. This may be of special importance when the CMV DNA load is measured in whole blood. In this respect, caution is warranted when calculating half-lives and doubling rates of the virus from the kinetics of CMV DNA loads in the presence and absence of antiviral treatment, respectively, especially when small PCR targets are studied (14).
In conclusion, our studies with three renal transplant recipients showed that CMV DNA as detected in plasma and serum is highly fragmented and therefore does not represent circulating infectious virus. As a consequence, levels of CMV DNA as measured by PCR are highly dependent on the amplicon length. While further studies with larger numbers of patients are needed to confirm our conclusions, as well as to assess whether the same extent of CMV DNA fragmentation in the various blood compartments is observed in patient groups other than renal transplant recipients, these findings may have important consequences for understanding CMV pathogenesis and for the interpretation of CMV DNA load measurements in individual patient management.
Acknowledgments
We thank the members of the Clinical Virology section, Zelleke Ayde, Ingrid Gomes, Paul Appelman, Wim van Est, Tonnie Walraven, Gerrit Klingers, and Ed Plasman for their enthusiastic and skillful contributions to this study. We acknowledge S. Surachno and L. E. Gamadia for their help in collecting patient blood specimens and providing clinical data.
REFERENCES
- 1.Aaij, C., and P. Borst. 1972. The gel electrophoresis of DNA. Biochim. Biophys. Acta 269:192-200. [DOI] [PubMed] [Google Scholar]
- 2.Akrigg, A., G. W.G. Wilkinson, and J. D. Oram. 1985. The structure of the major immediate early gene of human cytomegalovirus strain AD169. Virus Res. 2:107-121. [DOI] [PubMed] [Google Scholar]
- 3.Boeckh, M., and G. Boivin. 1998. Quantitation of cytomegalovirus: methodologic aspects and clinical applications. Clin. Microbiol. Rev. 11:533-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boivin, G., J. Handfield, E. Toma, G. Murray, R. Lalonde, and G. Bergeron. 1998. Comparative evaluation of the cytomegalovirus DNA load in polymorphonuclear leukocytes and plasma of human immunodeficiency virus-infected subjects. J. Infect. Dis. 177:355-360. [DOI] [PubMed] [Google Scholar]
- 5.Boivin, G., J. Handfield, E. Toma, R. Lalonde, and M. G. Bergeron. 1999. Expression of the late cytomegalovirus (CMV) pp150 transcript in leukocytes of AIDS patients is associated with a high viral load in leukocyte and presence of CMV DNA in plasma. J. Infect. Dis. 179:1101-1107. [DOI] [PubMed] [Google Scholar]
- 6.Boom, R., J. L. Geelen, C. J. Sol, A. K. Raap, R. P. Minnaar, B. P. Klaver, and J. van der Noordaa. 1986. Establishment of a rat cell line inducible for the expression of human cytomegalovirus immediate-early gene products by protein synthesis inhibition. J. Virol. 58:851-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boom, R., C. J. A. Sol, M. M. Salimans, C. L. Jansen, P. M. E. Wertheim-van Dillen, and J. van der Noordaa. 1991. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boom, R., C. Sol, J. Weel, Y. Gerrits, M. De Boer, and P. Wertheim-van Dillen. 1999. A highly sensitive assay for the detection and quantitation of human cytomegalovirus DNA in serum and plasma by PCR and electrochemiluminescence. J. Clin. Microbiol. 37:1489-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boom, R., C. Sol, J. Weel, K. Lettinga, Y. Gerrits, A. Van Breda, and P. Wertheim-van Dillen. 2000. Detection and quantitation of human cytomegalovirus DNA in faeces. J. Virol. Methods 84:1-14. [DOI] [PubMed] [Google Scholar]
- 10.Brytting, M., W. Xu, B. Wahren, and V. Sundqvist. 1992. Cytomegalovirus DNA detection in sera from patients with active cytomegalovirus infections. J. Clin. Microbiol. 30:1937-1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caliendo, A. M., R. Schuurman, B. Yen-Lieberman, S. A. Spector, J. Andersen, R. Manjiry, C. Crumpacker, N. S. Lurain, and A. Erice. 2001. Comparison of quantitative and qualitative PCR assays for cytomegalovirus in plasma. J. Clin. Microbiol. 39:1334-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dankner, W., J. A. McCutchan, D. D. Richman, K. Hirata, and S. Spector. 1990. Localization of human cytomegalovirus in peripheral blood leukocytes by in situ hybridization. J. Infect. Dis. 161:31-36. [DOI] [PubMed] [Google Scholar]
- 13.Drouet, E., S. Michelson, G. Denoyel, and R. Colimon. 1993. Polymerase chain reaction detection of human cytomegalovirus in over 2000 blood specimens correlated with virus isolation and related to urinary virus excretion. J. Virol. Methods 45:259-276. [DOI] [PubMed] [Google Scholar]
- 14.Emery, V. C., A. V. Cope, E. F. Bowen, D. Gor, and P. Griffiths. 1999. The dynamics of human cytomegalovirus replication in vivo. J. Exp. Med. 190:177-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerna, G., M. Furione, F. Baldanti, and A. Sarasini. 1994. Comparative quantitation of human cytomegalovirus DNA in blood leukocytes and plasma of transplant and AIDS patients. J. Clin. Microbiol. 32:2709-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerna, G., E. Percivalle, F. Baldanti, S. Sozzani, P. Lanzarini, E. Genini, D. Lilleri, and M. G. Revello. 2000. Human cytomegalovirus replicates abortively in polymorphonuclear leukocytes after transfer from infected endothelial cells via transient microfusion events. J. Virol. 74:5629-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibson, U. E. M., A. B. Chen, D. L. Baker, and D. V. Sinicropi. 1992. An antibody capture bioassay (ACB) for DNAse in human serum samples. J. Immunol. Methods 155:249-256. [DOI] [PubMed] [Google Scholar]
- 18.Grefte, A., N. Blom, M. Van der Giessen, W. Van Son, and T. H. The. 1995. Circulating cytomegalovirus-infected endothelial cells after renal transplantation: possible clue to pathophysiology? Transplant. Proc. 27:939-942. [PubMed] [Google Scholar]
- 19.Ishigaki, S., M. Takeda, T. Kura, et al. 1991. Cytomegalovirus DNA in the sera of patients with cytomegalovirus pneumonia. Br. J. Haematol. 79:198-204. [DOI] [PubMed] [Google Scholar]
- 20.Limaye, A. P., M. L. Huang, W. Leisenring, L. Stensland, L. Corey, and M. Boeckh. 2001. Cytomegalovirus (CMV) DNA load in plasma for the diagnosis of CMV disease before engraftment in hematopoietic stem-cell transplant recipients. J. Infect. Dis. 183:377-382. [DOI] [PubMed] [Google Scholar]
- 21.Patel, R., T. F. Smith, M. Espy, R. H. Wiesner, R. A. Krom, D. Portela, and C. V. Paya. 1994. Detection of cytomegalovirus DNA in sera of liver transplant recipients. J. Clin. Microbiol. 32:1431-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pellegrin, I., I. Garrigue, E. Ekouevi, et al. 2000. New molecular assays to predict occurrence of CMV disease in renal transplant recipients. J. Infect. Dis. 182:36-42. [DOI] [PubMed] [Google Scholar]
- 23.Prince, W. S., D. L. Baker, A. H. Dodge, A. E. Ahmed, R. W. Chestnut, and D. V. Sinicropt. 1998. Pharmacodynamics of recombinant human DNAse I in serum. Clin. Exp. Immunol. 113:289-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roizman, B. 1990. Herpesviridae: a brief introduction, p. 1787-1793. In B. N. Fields, D. M. Knipe, R. M. Chanock, et al. (ed.), Fields virology, vol. 2. Raven Press, New York, N.Y.
- 25.Schäfer, P., W. Tenschert, K. Gutensohn, and R. Laufs. 1998. Minimal effects of delayed sample processing on results of quantitative PCR for cytomegalovirus DNA in leukocytes compared to results of an antigenemia assay. J. Clin. Microbiol. 35:741-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schäfer, P., W. Tenschert, L. Cremaschi, K. Gutensohn, and R. Laufs. 1998. Utility of major leukocyte populations for monitoring secondary cytomegalovirus infections in renal allograft recipients by PCR. J. Clin. Microbiol. 36:1008-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sia, I. G., and R. Patel. 2000. New strategies for prevention and therapy of cytomegalovirus infection and disease in solid-organ transplant patients. Clin. Microbiol. Rev. 13:83-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Söderberg-Nauclér, C., K. N. Fish, and J. A. Nelson. 1997. Reactivation of latent human cytomegalovirus by allogeneic stimulation of blood cells from healthy donors. Cell 91:119-126. [DOI] [PubMed] [Google Scholar]
- 29.Spector, S. A., R. Merrill, D. Wolf, and W. M. Dankner. 1992. Detection of human cytomegalovirus in plasma of AIDS patients during acute visceral disease by DNA amplification. J. Clin. Microbiol. 30:2359-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spector, S. A., K. Hsia, M. Crager, M. Pilcher, S. Cabral, and M. J. Stempien. 1999. Cytomegalovirus (CMV) DNA load is an independent predictor of CMV disease and survival in advanced AIDS. J. Virol. 73:7027-7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tong, C. Y. W., L. E. Cuevas, H. Williams, and A. Bakran. 2000. Prediction and diagnosis of cytomegalovirus disease in renal transplant recipients using qualitative and quantitative polymerase chain reaction. Transplantation 69:985-991. [DOI] [PubMed] [Google Scholar]
- 32.Van de Corput, M. P., R. W. Dirks, R. P. Van Gijlswijk, F. M. Van de Rijke, and A. K. Raap. 1998. Fluorescence in situ hybridization using horseradish peroxidase-labeled oligodeoxynucleotides and tyramide signal amplification for sensitive DNA and mRNA detection. Histochem. Cell. Biol. 110:431-437. [DOI] [PubMed] [Google Scholar]
- 33.Zipeto, D., S. Morris, C. Hong, A. Dowling, R. Wolitz, T. C. Merigan, and L. Rasmussen. 1995. Human cytomegalovirus (CMV) DNA in plasma reflects quantity of CMV DNA present in leukocytes. J. Clin. Microbiol. 33:2607-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]