Abstract
We show here that the radiosensitive Chinese hamster cell mutant (V-C8) of group XRCC11 is defective in the breast cancer susceptibility gene Brca2. The very complex phenotype of V-C8 cells is complemented by a single human chromosome 13 providing the BRCA2 gene, as well as by the murine Brca2 gene. The Brca2 deficiency in V-C8 cells causes hypersensitivity to various DNA-damaging agents with an extreme sensitivity toward interstrand DNA cross-linking agents. Furthermore, V-C8 cells show radioresistant DNA synthesis after ionizing radiation, suggesting that Brca2 deficiency affects cell cycle checkpoint regulation. In addition, V-C8 cells display tremendous chromosomal instability and a high frequency of abnormal centrosomes. The mutation spectrum at the hprt locus showed that the majority of spontaneous mutations in V-C8 cells are deletions, in contrast to wild-type V79 cells. A mechanistic explanation for the genome instability phenotype of Brca2-deficient cells is provided by the observation that the nuclear localization of the central DNA repair protein in homologous recombination, Rad51, is reduced in V-C8 cells.
V-C8 is a Chinese hamster cell mutant that represents the XRCC11 complementation group, among X-ray-sensitive rodent cell mutants, as well as a distinguished group among mitomycin C (MMC)-sensitive rodent cell mutants (for a review, see reference 46). This mutant is extremely sensitive to cross-linking agents, but it also shows an increased sensitivity toward many other DNA-damaging agents, such as methyl methanesulfonate (MMS) and UV light (19, 48). This suggests that the XRCC11 gene is involved in a wide-ranging cellular response induced by various types of DNA damage. V-C8 cells display radioresistant DNA synthesis (RDS) after ionizing irradiation (36), which is indicative of a defect in DNA damage recognition or cell cycle checkpoint regulation. However, the high level of spontaneous and cross-link-induced chromosomal aberrations manifested by V-C8 cells (19) indicates a possible defect in DNA repair. Indeed, V-C8 cells have an impaired capacity for repair of DNA double-strand breaks (DSBs) after irradiation (36).
In mammalian cells, DSBs are repaired via either nonhomologous end joining (NHEJ) or homologous recombination (HR) (reviewed in reference 14). Genetic complementation studies have determined that V-C8 cells are not defective in DNA-PKcs, Ku80, or Xrcc4, key components of NHEJ, nor in Xrcc2 or Xrcc3, proteins involved in HR, and that this mutant represents a separate complementation group, XRCC11 (36, 46). When compared to cell lines defective in the above-mentioned proteins, the overall phenotype of V-C8 cells more closely resembles those of the Xrcc2- and Xrcc3-defective hamster cell lines irs1 and irs1SF, respectively (8, 12, 16). In common with V-C8, irs1 and irs1SF exhibit an extreme sensitivity to cross-linking agents that is not observed in cell lines defective in NHEJ proteins. This favors the hypothesis that the XRCC11 gene defective in V-C8 cells might be involved in HR. A key player in DSB repair through HR is the Rad51 protein (reviewed in reference 33). Rad51 is a homolog of the Escherichia coli RecA protein. Rad51 facilitates strand invasion of the broken DNA into a homologous double-stranded DNA by forming nucleoprotein filaments. In response to treatment with a number of DNA-damaging agents, Rad51 exhibits a dynamic redistribution within cells, localizing into nuclear foci at sites of DNA damage (28). In mammals, Rad51 belongs to a family of homologous proteins—Rad51, Rad51B, Rad51C, Rad51D, Xrcc2, Xrcc3, and Dmc1—of which the last mainly functions in meiotic cells (reviewed in reference 31). Impaired formation of Rad51 foci, in response to DNA damage, has been demonstrated in hamster or chicken cells defective in the Rad51 paralogs Xrcc2, Xrcc3, Rad51B, Rad51C, and Rad51D, as well as in mammalian Brca1- or Brca2-defective cells (1, 3, 25, 26, 44, 45).
Here we demonstrate that V-C8 is defective in Rad51 foci formation in response to DNA damage, suggesting that the XRCC11 gene is required for the assembly or stabilization of the Rad51 protein complex. We also show that the phenotype of V-C8 cells is due to a deficiency in Brca2 and that this Brca2 deficiency leads to RDS after gamma-irradiation. Furthermore, we found that V-C8 cells exhibit a spontaneous mutation spectrum at the hprt locus predominated by deletions, unlike wild-type V79 cells. Finally, we demonstrate that the Brca2 protein is required for efficient nuclear localization of the Rad51 protein.
MATERIALS AND METHODS
Cells and culture conditions.
The V-C8 mutant cell line derived from Chinese hamster V79 cells has been described previously (19, 36, 48), as has the ataxia telangiectasia (AT)-like mutant cell line V-E5 (47). Cells were routinely cultured in 10-cm dishes (Greiner) in Ham’s F-10 (Life Technologies) supplemented with 10% fetal bovine serum (FBS; Life Technologies) and PS (100 U of penicillin and 1 mg of streptomycin/ml), without hypoxanthine and thymidine. Monochromosomal MCH204.3 human-mouse hybrid cells, containing human chromosome 13, tagged with a single neomycin resistance gene, were cultured in Dulbecco’s modified Eagle medium-F-10 medium, supplemented with 10% FBS and PS, and 800 μg of G418 (Life Technologies)/ml. The simian virus 40-transformed wild-type human fibroblast cell line MRC5V1 (11) was cultured in Dulbecco’s modified Eagle medium-F-12 medium supplemented with 10% FBS and PS. Cells were maintained at 37°C in a 5% CO2 atmosphere humidified to 95 to 100%.
Microcell-mediated chromosome transfer and transfection of exogenous DNA.
Introduction of the human chromosome 13 into V-C8 cells by microcell-mediated chromosome transfer was performed basically as described previously (15). As a donor for human chromosome 13, monochromosomal MCH204.3 hybrid cells were used. Microcells were obtained by a 48-h incubation of the MCH204.3 cells with Colcemid (0.05 μg/ml; Life Technologies), followed by 60 min of centrifugation at 34°C (fixed-angle Sorvall GSA rotor, 7,500 rpm, relative centrifugal force value of 5761) in the presence of 20 μg of cytochalasin B (Sigma)/ml. The microcells were filtered in series through 8-, 5-, and 5-μm-pore-size polycarbonate membrane filters (Nuclepore); subsequently added to a monolayer of recipient cells; and finally fused by treatment with 47% polyethylene glycol (Sigma). After 24 h, the cells were cultured under selective pressure of 400 μg of Geneticin (G418 sulfate; Life Technologies)/ml, and microcell hybrid clones were isolated after 10 to 14 days.
Transfections (of the bacterial artificial chromosome [BAC] containing murine Brca2, pBAC421-Neo, or of human RAD51-green fluorescent protein [GFP] cDNA) were performed by using the GenePORTER transfection reagent according to the manufacturer’s protocol (BIOzym). pBAC421-Neo contains a 220-kb genomic sequence inserted as an EcoRI-EcoRI fragment in pBACe3.6 (i.e., pBAC108L into which a neomycin resistance gene has been inserted). This insert includes the full-length murine Brca2 gene. In addition to containing the 60 kb of the gene it contains 70 kb of upstream sequences and 90 kb of downstream sequences. This BAC has been used to completely rescue the embryonic lethality associated with the Brca2 mutation in mice (S. Swaminathan and S. K. Sharan, unpublished data).
Clonogenic survival assays.
Cell cultures in exponential growth were trypsinized, and 300 to 700 cells (3,000 to 28,000 cells for the highest doses) were seeded into 10-cm dishes (two dishes per dose; three dishes for the untreated control) and left to attach for 4 h. The cells were irradiated with X rays (dose rate, ∼2.8 Gy/min; 200 kV; 4 mA; 0.78 mm Al), treated with MMC (continuous), or treated with MMS (1 h). After MMS treatment, the cells were rinsed twice with phosphate-buffered saline (PBS) and fresh medium was added. After 8 to 12 days, the cells were rinsed with 0.9% NaCl and stained with 0.25% methylene blue, and visible colonies were counted. Each survival curve represents the mean of at least three independent experiments. Error bars represent the standard errors of the mean.
Immunofluorescence labeling and microscopy.
To examine centrosomes or Rad51 focus formation, cells were seeded 1 day prior to analysis onto glass slides, giving subconfluent cells at time of fixation. For centrosome analysis, the cells were fixed with methanol-acetone (7:3). For Rad51 focus analysis, cells were either mock treated or else irradiated with 12 Gy of X rays (dose rate, ∼2.8 Gy/min; 200 kV; 4 mA; 0.78 mm Al) or incubated with 2.4 μg of MMC/ml for 1 h, after which the cells were washed with PBS (10 mM Na2HPO4, 0.14 mM NaCl; pH 7.4), and fresh medium was given. Eight hours after (mock) treatment, the cells on the slides were fixed with 2% paraformaldehyde.
After fixation, cells were permeabilized with 0.1% Triton X-100 in PBS and blocked with PBS+ (20 mM glycine [J. T. Baker]-0.5% [wt/vol] BSA [Sigma] in PBS). The cells were incubated with rabbit antiserum against γ-tubulin (Sigma) for centrosome analysis or against human Rad51 (FBE-2) and subsequently with Alexa 488-conjugated goat anti-rabbit immunoglobulin G (IgG) (Molecular Probes). Cell nuclei were stained with DAPI (4′,6′-diamidino-2-phenylindole; Sigma). Centrosomes and Rad51 foci were examined under a Leitz Axioplan fluorescence microscope.
Measurement of RDS after gamma-irradiation.
Measurement of RDS was performed essentially as described but without the trichloroacetic acid precipitation step (47). In short, cells were seeded in duplicate 30-mm dishes (four for the unirradiated control) and were prelabeled overnight with [14C]thymidine (Amersham) in HEPES-buffered thymidine-free Ham’s F-10 medium and then exposed to graded doses of 137Cs gamma-irradiation (1.2 Gy/min) and subsequently labeled with [3H]thymidine (Amersham) for 4 h. Free thymidine pools were chased by a further 30- to 45-min incubation in unlabeled medium. Scintillation-counted 3H/14C radioactivity ratios of alkali-lysed cells were taken as a measure of DNA synthesis rates and plotted as percentages of unirradiated cells.
Analysis of chromosomal aberrations and SCEs.
Frequencies of spontaneous or MMC-induced chromosomal aberrations or sister chromatid exchanges (SCEs) were determined in exponentially growing cell cultures. The cells were either mock treated or treated for 24 h (chromosomal aberrations) or 2 h (SCEs) with 0.25, 0.5, and 1 ng of MMC/ml (V-C8) or with 1, 15, 30, 40, or 80 ng of MMC/ml (V79 and V-C8 with human chromosome 13 or the BAC containing murine Brca2). For SCE analysis, 5-bromo-2′-deoxyuridine (Sigma) was then added to the medium (10 μM final concentration) to enable sister chromatid differentiation. The cells were harvested by trypsinization 24 to 28 h after (mock) treatment, after 2 h of incubation with 1 μg of Colcemid/ml. The cells were fixed, after treatment with hypotonic solution (0.6% sodium citrate), in ethanol-glacial acetic acid (3:1). Air-dried preparations were made and stained with Giemsa or with fluorochrome plus Giemsa. For chromosomal aberrations, 100 mitotic cells were analyzed at each dose, and SCEs were scored in 25 cells. All experiments were repeated at least once.
Immunoblot analysis.
For immunoblot analysis, whole-cell extracts were made by resuspending cell pellets in lysis buffer (10 mM Tris [pH 7.4], 150 mM NaCl, 1% Igepal CA-630, 0.5% deoxycholic acid, 1 mM EDTA [pH 7.4], 1 mM dithiothreitol, 0.5 mg of Pefabloc/ml, 1 μg of aprotinin/ml, 1 μg of leupeptin/ml), followed by sequential snap-freezing on dry ice and thawing at 30°C three times. Cell debris was removed by centrifugation at 14,000 rpm at 4°C for 15 min. For determination of the subcellular localization of Rad51-GFP, cellular fractionation was performed as described previously (4). The quality of the fractions was checked by using a monoclonal antibody against the nuclear protein p62 (6). Protein concentrations were determined by using the Bradford protein assay with bovine serum albumin as the standard. Protein (100 μg) was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride membrane (Bio-Rad), and probed with polyclonal antiserum against human Rad51 (FBE-2) or mouse Brca2 (pep-4) or with monoclonal antiserum against human BRCA2 (Ab-1; Oncogene) or GFP (Roche). The membranes were then probed with horseradish peroxidase-conjugated goat anti-rabbit IgG or goat anti-mouse IgG, and antibody binding was detected by enhanced chemiluminescence (Amersham). The equality of loading was confirmed by probing with monoclonal antiserum against actin (Santa Cruz).
Determination of the spontaneous mutation spectrum at the hprt locus.
For the isolation of spontaneous hprt mutants, V-C8 cells were divided among 25 independent cultures of 103 cells per dish. Each population was propagated separately to ensure that all hprt mutants obtained were independent. From each population of ca. 3 × 107 cells, 2 × 106 cells were plated in 6-thioguanine-containing medium (5 μg/ml) at a density of 105 cells per dish. After 10 days, only one 6-thioguanine-resistant colony was isolated per culture. In all, 21 independent spontaneous hprt mutants were isolated. PCR analysis of hprt cDNA and genomic DNA, as well as sequence analysis of hprt cDNA, was performed as described earlier (30).
RESULTS
Impaired Rad51 focus formation in V-C8 cells.
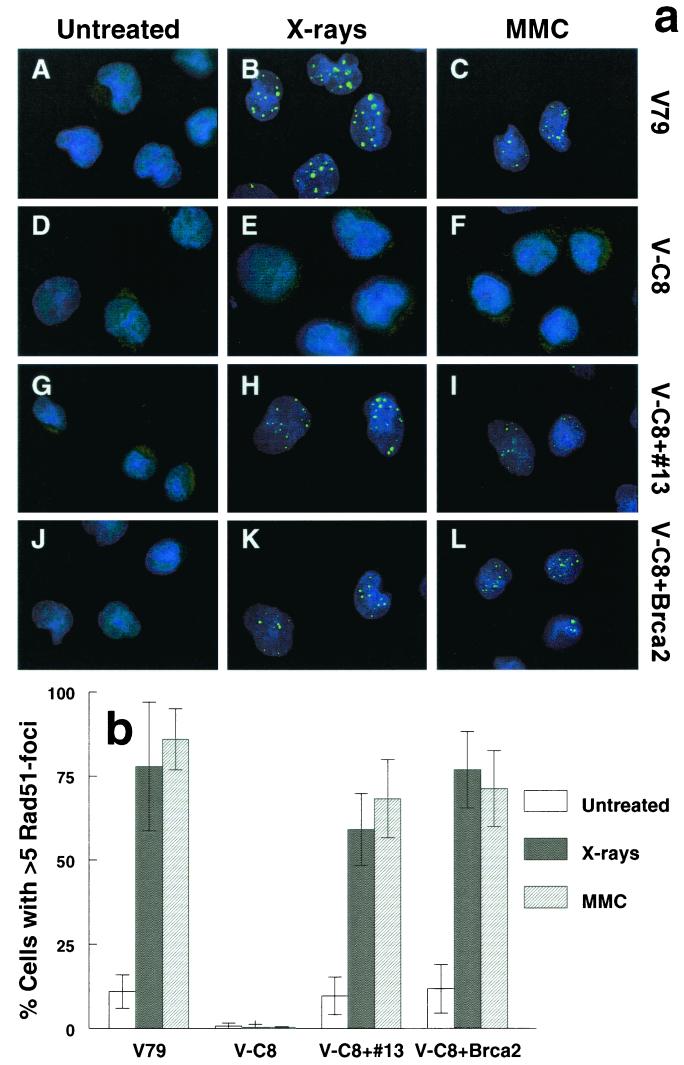
To investigate whether V-C8 cells are defective in HR, we examined the ability of these cells to form Rad51 foci after DNA damage induction. V-C8 cells were treated with X rays or MMC. After 8 h, the cells were fixed, and Rad51 foci were visualized microscopically by labeling the cells with rabbit polyclonal antiserum against Rad51, followed by labeling with a rabbit specific fluorophore-conjugated antibody. We found that V-C8 cells were clearly defective in Rad51 focus formation after X-ray or MMC treatment (Fig. 1). While treatment with X rays or MMC induced Rad51 foci in ∼80% of wild-type V79 cells after 8 h, in V-C8 cells the percentage of cells positive for Rad51 foci remained at a background level. In untreated cells it has been found that Rad51 foci form typically in S phase (27). The lack of detection of Rad51 foci in V-C8 cells cannot be due to differences in cell cycle distribution, especially in S phase, because this was found to be similar in V79 and V-C8 cells after treatment with MMC or X-ray irradiation (20; unpublished results). Immunoblot analysis of whole-cell extracts of V-C8 cells showed normal levels of Rad51 protein (data not shown). Taken together, these results suggest that the XRCC11 gene is involved in the assembly and/or stabilization of the Rad51 protein complex.
FIG. 1.
(a) Immunofluorescent visualization of Rad51 nuclear foci: wild-type V79 (A to C), V-C8 (D to F), V-C8 cells with human chromosome 13 providing the BRCA2 gene (V-C8+#13) (G to I), and V-C8 containing a BAC with the murine Brca2 gene (V-C8+Brca2) (J to L). Cells were analyzed for 8 h after treatment with either 12 Gy of X rays (B, E, H, and K) or 2.4 μg of MMC/ml for 1 h (C, F, I, and L). (b) Quantification of Rad51 focus-positive cells. A cell containing more than five distinct foci was considered positive. Each bar represents the result of scoring at least 100 cells. The error bars represent the standard error of the mean (SEM).
Complementation of cross-sensitivity and Rad51 focus formation by human chromosome 13 and by a BAC containing the murine BRCA2 gene.
To identify the defective gene in V-C8 cells, we examined candidate genes involved in Rad51 complex assembly for their ability to complement the defects of these cells. Transfection of hRad51 cDNA, as well as transfer of a single human chromosome 15 (providing hRAD51), did not complement the MMC sensitivity of V-C8 cells. Furthermore, sequencing of V-C8 Rad51 cDNA revealed no mutations (data not shown). No complemention was found after transfection of V-C8 cells with cDNA of the Rad51 paralogs hRAD51B, hRAD51C, and hRAD51D (data not shown). As mentioned in the introduction, it had already been determined by previous genetic complementation studies that V-C8 cells are not defective in Xrcc2 or Xrcc3 (46). Therefore, the Brca1 and Brca2 genes were considered as candidate defective genes.
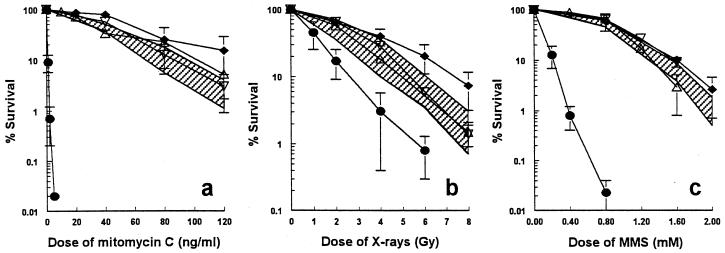
To examine the effect of the BRCA2 gene on V-C8 cells, a single human chromosome 13 providing this gene (39) was transferred into these cells by microcell-mediated chromosome transfer. Several V-C8+#13 microcell hybrid clones were obtained and examined for complementation of the defect in V-C8 cells. The sensitivities of V-C8 to MMC, X rays, MMS (Fig. 2), and UV light (data not shown) were largely complemented by human chromosome 13. Transfer of human chromosome 17 providing the BRCA1 gene, as well as transfer of several other single human chromosomes (8, 9, 11, 12, 18, 19, 21), did not result in any complementation of V-C8 cells (data not shown). Taken together, these results indicate that only human chromosome 13 complements the defect in V-C8 cells, suggesting that the BRCA2 gene might be responsible. To confirm this, we transfected V-C8 cells with a BAC containing the murine Brca2 gene and found complementation of clonogenic survival, similar to that observed with V-C8+#13 microcell hybrids (Fig. 2). Both the human chromosome 13 and the murine Brca2 gene restored the ability of Rad51 focus formation in response to X-ray irradiation or treatment with MMC (Fig. 1).
FIG. 2.
Cell survival after exposure to MMC (a), X rays (b), or MMS (c) of wild-type V79 (⧫), V-C8 (•), V-C8 cells with human chromosome 13 providing the BRCA2 gene (V-C8+#13) (shaded area, four independent clones), and V-C8 containing a BAC with the murine Brca2 gene (V-C8+Brca2, two independent clones; ▵ and ▿). The error bars represent the SEM.
Complementation of RDS.
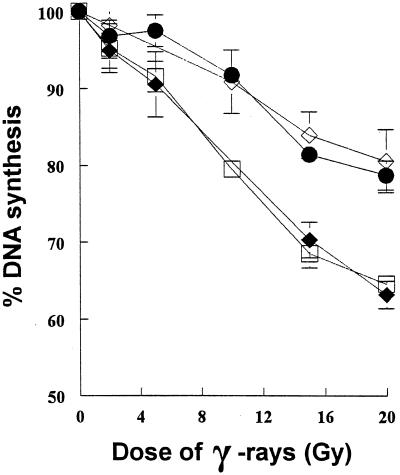
V-C8 cells display RDS (36), a phenomenon characteristic of cells derived from patients with AT and the AT-like hamster mutant cell lines of group XRCC8 (13, 32, 47). To investigate whether the RDS phenotype seen in V-C8 cells is due to a defective Brca2 protein, we examined whether the BRCA2 gene restored DNA synthesis inhibition in this cell line after ionizing irradiation. Data presented in Fig. 3 show that the dose response of DNA synthesis following gamma-irradiation was restored by the addition of a single human chromosome 13, providing the BRCA2 gene, suggesting that the RDS phenotype of V-C8 cells is due to a defect in Brca2.
FIG. 3.
Dose-response curve of the rate of DNA synthesis after gamma-irradiation in V-C8 cells with human chromosome 13 providing the BRCA2 gene. The curves shown are for wild-type V79 cells (⧫), V-C8 cells (•), V-C8+#13 (□, two independent clones), and the AT-like hamster cell line V-E5 (⧫). The error bars represent the SEM.
Complementation of spontaneous and MMC-induced chromosomal aberrations.
V-C8 cells display tremendous genomic instability (Tables 1 and 2) (19). V-C8 cells show ca. 25-fold more spontaneous aberrations than wild-type V79 cells. The majority of spontaneous chromosomal exchanges in V-C8 were of the chromatid type (Table 1), which are usually lethal. In V79 these were rarely found. After treatment with MMC, the relative chromosomal sensitivity of V-C8 was increased ca. 600-fold compared to V79 cells (Table 2). Cells in which the human or murine Brca2 gene was expressed had clearly reduced levels of spontaneous and MMC-induced chromosomal aberrations, indicating that defective Brca2 in V-C8 cells is responsible.
TABLE 1.
Spontaneous and MMC-induced chromosomal aberrations per 100 cells
| Cell line | Treatmenta (ng/ml) | Chromatid
|
Chromosome
|
Total no. of breaksc ± SEM | ||
|---|---|---|---|---|---|---|
| No. of breaks | No. of exchangesb | No. of breaks | No. of exchanges | |||
| V79 | Spontaneous | 6 | 0 | 1 | 0 | 7 ± 2 |
| MMC (80) | 26 | 4 | 7 | 1 | 43 ± 7 | |
| V-C8 | Spontaneous | 134 | 12 | 19 | 3 | 183 ± 20 |
| MMC (0.5) | 241 | 29 | 5 | 4 | 312 ± 39 | |
| V-C8+#13 | Spontaneous | 29 | 0 | 5 | 0 | 34 ± 4 |
| MMC (80) | 75 | 14 | 10 | 1 | 115 ± 4 | |
| V-C8+Brca2 | Spontaneous | 32 | 1 | 5 | 1 | 41 ± 4 |
| MMC (80) | 277 | 58 | 9 | 1 | 404 ± 135 | |
The depicted frequencies of MMC-induced chromosomal aberrations are after treatment with equitoxic doses, which give 30 to 40% survival.
Chromatid exchanges are interchanges, intrachanges, triradials, or subchromatid exchanges.
For calculations of total breaks, the exchanges were counted as two breaks.
TABLE 2.
Relative sensitivities for chromosomal aberrationsa
| Cell line | Relative sensitivity
|
|||
|---|---|---|---|---|
| V79 | V-C8 | V-C8+#13 | V-C8+Brca2 | |
| Spontaneous | 1b | 26 | 5 | 6 |
| MMC induced | 1c | 635 | 3 | 11 |
Relative sensitivities were calculated from the slopes of the dose-response curves for MMC-induced chromosomal aberrations.
The frequency of spontaneous aberrations per 100 cells in wild type V79 was 7 ± 2, and this was set at 1.
The slope of the induction curve of chromosomal aberrations after MMC treatment in wild-type V79 cells was 0.4 aberration per 100 cells per ng of MMC/ml, and this was set at 1.
Expression of Brca2 in V-C8 cells.
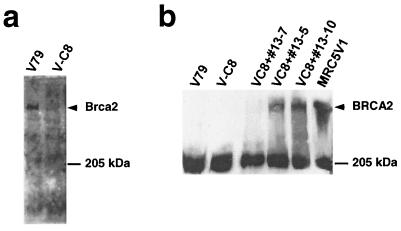
The data presented above show that the Brca2 gene complements the cross-sensitivity to DNA-damaging agents, Rad51 focus formation, RDS, and chromosomal aberrations. We conclude therefore that V-C8 is defective in Brca2. To verify this conclusion, the expression of Brca2 was examined in V-C8 cells by immunoblot analysis. Significantly, Brca2 protein was not detected in V-C8 cells, whereas V79 cells clearly expressed Brca2 (Fig. 4a). The antibody used, pep-4, has been raised against the 13 most carboxy-terminal amino acids of the mouse Brca2 protein. These results suggest that in V-C8 cells Brca2 is not expressed, is truncated, or is unstable. The data presented in Fig. 4b show that in V-C8 cells complemented by a single human chromosome 13 (clones #13-5 and #13-10), the BRCA2 protein could be detected, as in wild-type human MRC5V1 cells, whereas clone #13-7, which is not complemented for focus formation and clonogenic survival, does not express the BRCA2 protein. The antibody used for this immunoblot, Ab-1, has been raised against amino acids 1651 to 1821 of human BRCA2 and does not recognize the hamster protein (Fig. 4b, lanes 1 and 2). Equality of loading was confirmed by probing with monoclonal antiserum against actin (not shown). Taken together, these results strengthen our conclusion that the phenotype of V-C8 cells is indeed due to defective Brca2.
FIG. 4.
Immunoblot analysis of the hamster Brca2 protein in V-C8 and V79 cells with antibody pep-4 raised against the 13 most carboxy-terminal amino acids of the mouse Brca2 protein (a) and the human BRCA2 protein in V-C8+#13 microcell hybrids with antibody Ab-1, raised against amino acids 1651 to 1821 of human BRCA2 (b). The wild-type simian virus 40-transformed human fibroblast cell line MRC5V1 was used as a positive control.
Spontaneous and MMC-induced SCEs.
Chicken cells defective in one of the paralogs of Rad51 all show reduced frequencies of both spontaneous and MMC-induced SCEs (23, 25, 26). To determine whether Brca2 deficiency also leads to reduced levels of SCEs, the frequency of SCEs was measured in V-C8 cells. The number of spontaneous SCEs per cell in V-C8 was found to be similar to that in V79 cells (Table 3). In contrast, after treatment with different doses of MMC, a clear induction of SCE levels with dose was seen in V79 cells (15.0 ± 0.7 at 15 ng/ml and 21.3 ± 1.3 at 30 ng/ml) but not in V-C8 of cells. Due to the MMC toxicity for V-C8 cells, measurable metaphase spreads could be obtained only with doses of up to 0.5 ng of MMC/ml. With these low doses, however, SCE levels were found to be similar to the spontaneous level (Table 3). Interestingly, in V-C8 cells with an additional human or mouse Brca2 gene, SCEs were induced after MMC treatment, although at a lower frequency than in V79 cells. These results suggest that V-C8 cells are defective in MMC-induced SCE formation and that the human or mouse Brca2 gene partially complements this defect. Therefore, Brca2 might play a role in the formation of SCEs in the normal cellular response to cross-links.
TABLE 3.
Frequencies of spontaneous and MMC-induced SCEs
| Dose of MMC (ng/ml) | No. of SCEs per cell ± SD
|
|||
|---|---|---|---|---|
| V79 | V-C8 | V-C8+#13 | V-C8+Brca2 | |
| 0 | 6.6 ± 0.4 | 5.4 ± 0.4 | 6.3 ± 0.4 | 5.5 ± 0.3 |
| 0.25 | NDa | 4.8 ± 0.4 | ND | ND |
| 0.5 | ND | 6.9 ± 0.5 | ND | ND |
| 1 | 5.9 ± 0.4 | – | ND | ND |
| 5 | ND | – | 6.9 ± 0.4 | 6.5 ± 0.3 |
| 15 | 15.0 ± 0.7 | – | 8.4 ± 0.5 | 8.5 ± 0.5 |
| 30 | 21.3 ± 1.3 | – | – | – |
ND, not determined; -, not measurable.
Abnormal centrosomes in V-C8 cells.
Besides defective HR, chromosomal instability can also be caused by incorrect segregation of chromosomes into daughter nuclei during cell division. Chromosome segregation is mediated by the spindle poles, which nucleate from the centrosome. The centrosome normally duplicates once per cell cycle, at the G1-S boundary, and the two daughter centrosomes migrate to opposite poles prior to chromosome segregation (50). Brca2-defective mouse embryonic fibroblasts (MEFs) show abnormal centrosome duplication, resulting in multiple centrosomes per cell (34). We investigated whether V-C8 cells displayed aberrant centrosome duplication as well (Fig. 5). Centrosomes were visualized microscopically by labeling the cells with rabbit antiserum against γ-tubulin, followed by labeling with a rabbit specific fluorophore-conjugated antibody. The data presented in Fig. 5 show that in 89% of V79 cells the centrosomes were visible as clearly defined, dot-shaped structures, while in the majority of V-C8 cells (64%) the γ-tubulin staining was scattered and diffuse. This was partially complemented by human chromosome 13 and the murine Brca2 gene. These results suggest that Brca2 deficiency leads to the formation of aberrant centrosomes. In order to find out whether this scattered γ-tubulin staining could represent functional multiple centrosomes, we labeled V-C8 cells with β-tubulin to visualize the spindles in mitotic cells. We found that only 1 of 18 mitotic V-C8 cells examined with scattered γ-tubulin staining showed a multipolar spindle (data not shown). This suggests that, despite the aberrant centrosome structures, bipolar cell division still takes place in most V-C8 cells.
FIG. 5.
(a) Centrosome analysis in interphase cells of wild-type V79, Brca2-deficient V-C8, V-C8 with human chromosome 13 providing the BRCA2 gene (V-C8+#13, three independent clones), and V-C8 containing a BAC with the murine Brca2 gene (V-C8+Brca2, two independent clones). (b) Immunofluorescent visualization of normal centrosomes in V79 cells (left panel) and abnormally structured centrosomes in V-C8 cells (right panel).
Molecular analysis of hprt mutations in V-C8.
To investigate the molecular nature of hprt mutations in V-C8, 21 independent spontaneously arisen hprt mutants were isolated. From every mutant clone, cytoplasmic RNA was isolated, allowing hprt cDNA synthesis, amplification of the hprt coding region, and direct sequencing of the PCR-amplified cDNA. In 10 of the cases, no detectable amounts of hprt cDNA could be obtained after PCR. Therefore, genomic DNA was isolated and used for PCR with exon-specific primers. The nature of the molecular changes of the spontaneous hprt mutants of V-C8 is shown in Table 44. The mutation spectrum is predominated by deletions (11 mutants; 52%), which are mainly large deletions of one or more exons, while base substitutions were found in only two mutants (10%). This mutation spectrum significantly differs from that of V79 cells (49). In V79 cells, mainly base substitutions have been found (43% transversions, 21% transitions), while deletions were found in only 13% of the hprt mutants. This suggests that the Brca2 protein is involved in a specific process that prevents the formation of deletions rather than protecting against mutations in general.
TABLE 4.
Types and locations of spontaneous mutations in the hprt gene of Brca2-deficient V-C8 cells
| Mutation type | Mutant | Positiona | Exon | Base change | Sequence change | Amino acid |
|---|---|---|---|---|---|---|
| Transversion | SP-17 | 617 | 9 | GC→TA | ATTT(G)TGTC | Val→Phe |
| Double mutation | SP-7 | 530 | 7 | AT→GC | CCAG(A)CTTT | Asp→Gly |
| 531 | 7 | CG→AT | CAGA(C)TTTG | Asp→Glu | ||
| Insertion | SP-23 | 438–441 | 6 | Insertion of 21 bp | ||
| Frameshift | SP-5 | 156 | 3 | Del. of 1 bp | GAGA(T)GTCA | |
| Deletion | SP-21 | 137 | 3 | Del. of 8 bp | ||
| SP-11 | Del. ex. 2 | Del. ex. 2 | ||||
| SP-12 | Del. ex. 4 | Del. ex. 4 | ||||
| SP-14 | Del. ex. 4 | Del. ex. 4 | ||||
| SP-20 | Del. ex. 2, 3, and 4 | Del. ex. 2, 3, and 4 | ||||
| SP-2 | No cDNA | Del. of entire gene | ||||
| SP-8 | No cDNA | Del. of entire gene | ||||
| SP-13 | No cDNA | Del. of entire gene | ||||
| SP-15 | No cDNA | Del. of entire gene | ||||
| SP-16 | No cDNA | Del. of entire gene | ||||
| SP-19 | No cDNA | Del. of entire gene | ||||
| Other | SP-3 | No cDNA | All ex. present | |||
| SP-10 | No cDNA | All ex. present | ||||
| SP-22 | No cDNA | All ex. present | ||||
| SP-24 | No cDNA | All ex. present | ||||
| SP-25 | Del. ex. 7 | Splice | ||||
| SP-18 | No mutation found |
Del., deletion; ex., exon(s).
Subcellular localization of Rad51.
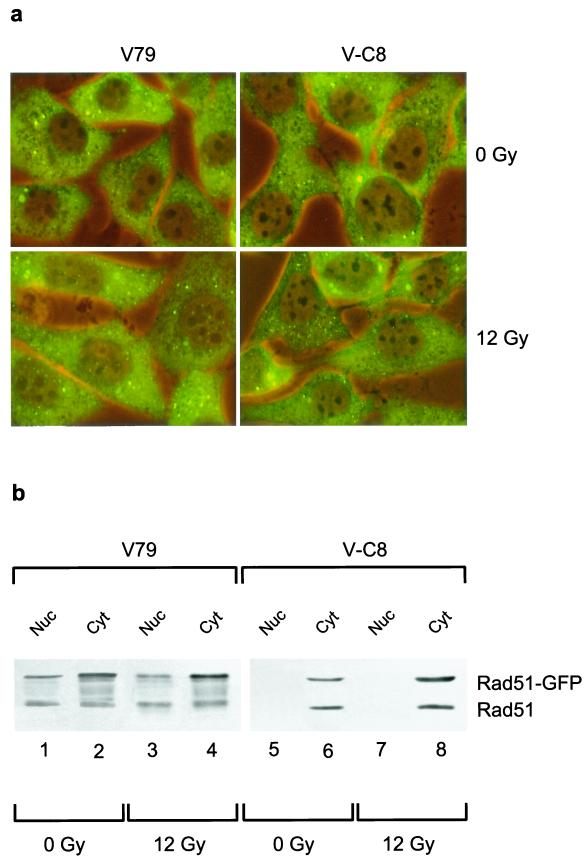
To gain insight into the underlying reason for the failure to detect efficient DNA damage-induced Rad51 focus formation in V-C8 cells, we investigated Rad51 focus formation in living cells. To this end, stable transfectants of parental wild-type hamster V79 and the Brca2-deficient V-C8 derivative, expressing a fusion between Rad51 and GFP, were generated. Upon treatment of the cells with ionizing radiation, Rad51-GFP accumulated into nuclear foci in V79 cells but not in V-C8 cells (Fig. 6a), just as was observed for the endogenous Rad51 protein (Fig. 1). Interestingly, although the immunofluorescence protocol for the detection of endogenous Rad51 did not detect the presence of Rad51 in the cytoplasm (Fig. 1) (10), the majority of Rad51-GFP was located there. Immunoblot analysis of nuclear and cytoplasmic fractions of the Rad51-GFP-expressing cells confirmed that the majority of the Rad51-GFP was located in the cytoplasm, in both V79 and V-C8 cells (Fig. 6b). In addition, the level of Rad51-GFP present in the nucleus was reduced in V-C8 cells compared to V79 cells (Fig. 6b). Immunoblotting with an anti-Rad51 antibody showed that the endogenous and GFP-tagged variant of Rad51 behaved similarly. Our results are in agreement with a recent study that demonstrates the presence of Rad51 in the cytoplasm and shows a reduction in the nuclear protein level of RAD51 in BRCA2-deficient human CAPAN-1 cells (4). The failure to detect efficient DNA damage-induced Rad51 focus formation in Brca2-deficient cells might be related to a reduction in the level of Rad51 that is present in the nucleus.
FIG. 6.
Subcellular localization of Rad51-GFP in V-C8 cells. Stable transfectants of wild-type V79 or V-C8 cells expressing Rad51-GFP were analyzed by immunofluorescence (a) or immunoblotting with anti-Rad51 antibodies (b) at 6 h after mock treatment or irradiation with 12 Gy of gamma-irradiation.
DISCUSSION
We have found that the Chinese hamster cell mutant, V-C8, of group XRCC11 is defective in Rad51 focus formation after DNA damage. This, as well as cross-sensitivity to DNA-damaging agents, RDS, and chromosomal aberrations, could be rescued by a functional Brca2 gene, suggesting that Brca2 is defective in these cells. This was supported further by immunoblot analysis which showed that the expression of Brca2 was severely reduced or absent in V-C8 cells, indicating that the phenotype of V-C8 cells is indeed due to a deficiency in Brca2.
The human BRCA2 gene has been identified as a tumor suppressor gene (29), of which germ line mutations are responsible for about one-third of the familial breast cancer cases (7). The BRCA2 protein has been found to play an important role in cellular responses to DNA damage and in maintaining genomic integrity (35). How this protein performs these cellular functions remains largely unknown. The human BRCA2 gene has 27 exons, encoding a protein of 3,418 amino acids. The large central exon 11 contains eight copies of 30- to 80-amino-acid BRC repeats, which are conserved across mammalian Brca2 proteins (2). Through these repeats Brca2 interacts with Rad51 (38). An additional Rad51-binding domain has been identified in the C-terminal region of mouse Brca2, which is largely conserved in the human sequence (22). Defective Rad51 focus formation has been demonstrated in the BRCA2-deficient human pancreatic adenocarcinoma cell line CAPAN-1, as well as in Brca2-deficient MEFs. Both of these cell lines encode Brca2 proteins truncated within exon 11, deleting the C-terminal Rad51 binding domain and part of the BRC repeats (44, 45).
To date, the majority of identified human germ line mutations in BRCA2 are small deletions, which can be found along the whole protein (see the Human Gene Mutation Database [http:/www.uwcm.ac.uk/uwcm/mg/hgmd0.html]). Nearly all mutations have been predicted to result in a truncated BRCA2 protein, causing loss of the nuclear localization signals, possibly resulting in a defect in the subcellular localization of the protein (24). Besides aberrant subcellular localization of Brca2 itself, defects in Brca2 could affect the localization of Rad51, as we have found in V-C8 cells (Fig. 6). The amount of nuclear Rad51 is reduced in V-C8 compared to V79 cells. In accordance with this, it has been demonstrated in CAPAN-1 cells that Rad51 is localized mainly in the cytoplasm (4).
The interaction of BRCA2 with Rad51 (4, 38) and its effect on Rad51 localization (Fig. 6) (4) suggest that BRCA2 is specifically involved in DNA repair through HR. Indeed, by using the I-SceI endonuclease to induce a DSB into a recombination repair substrate, which had been integrated in Brca2-deficient human CAPAN-1 or mouse embryonic stem (ES) cells, it has been shown that Brca2 is essential for DSB repair via HR (18). Brca2 deficiency also leads to impaired spontaneous HR (18, 40), for which the interaction of BRCA2 with RAD51 appears to be necessary (40).
The large size of the BRCA2 protein suggests the existence of several additional functional domains able to interact with other proteins than Rad51, which act in various cellular processes (37). Consistent with this notion is the sensitivity of V-C8 cells toward a variety of DNA-damaging agents including, e.g., UV light and alkylating agents, which induce DNA lesions that are mostly not repaired by HR. Thus, BRCA2 is likely also involved in a general cellular response to DNA damage. Importantly, we found that V-C8 cells display RDS (Fig. 3) (36). RDS is a hallmark of cells derived from patients with AT, AT-like disorder, and Nijmegen breakage syndrome, which are defective in ATM, MRE11, and NBS1, respectively (21). Since the dose response of DNA synthesis inhibition in hamster cells differs from that in human cells, we used the AT-like hamster cell line V-E5 (47) to compare RDS levels. We found that the RDS level of V-C8 cells is similar to that seen in V-E5 cells and that the BRCA2 gene corrects this phenotype (Fig. 3). Interestingly, it has been shown that the human BRCA1-deficient HCC1937 cells also display RDS (41). These results indicate that, in addition to ATM, MRE11, and NBS1, both BRCA1 and BRCA2 function in the S-phase checkpoint, giving a new view on this checkpoint, but exactly how the BRCA proteins function in this respect still needs to be elucidated.
V-C8 cells display enormous chromosomal instability (Tables 1 and 2) (19). Our data are in agreement with recent results showing that BRCA2 deficiency leads to chromosome breakage and gross chromosomal rearrangements (44), as well as to centrosome abnormalities (34). Abnormal centrosomes, potentially giving rise to incorrect chromosome segregation, have also been found in Brca1-deficient MEFs (42) in the hamster cell mutants irs1 and irs1SF, which are defective in Xrcc2 and Xrcc3, respectively (9), and in Mre11-deficient chicken cells (43), suggesting a link between DSB repair mechanisms and centrosome functioning, but the exact correlation is far from clear. Nevertheless, our results indicate that the centrosome abnormalities found in V-C8 cells are most probably not the main cause of the observed chromosomal instability, since only 1 of 18 cells with aberrant centrosomes had a multipolar spindle. The relatively poor complementation by either the human or the mouse Brca2 gene could indicate differences between the Brca2 proteins of these species. However, this can be better addressed once the hamster cDNA is available.
We found a change in the spontaneous mutation spectrum of V-C8 cells compared to that of normal V79 cells. The majority of spontaneous mutations in V-C8 cells at the hprt locus are deletions (Table 4), while in wild-type V79 cells mainly base substitutions are found (49). This suggests that Brca2 is involved in a process preventing formation of deletions during replication. This phenotype is reminiscent of HR deficiency in Rad54 knockout mouse ES cells, in which DSB repair through deletion formation is increased (5). The observation of chromosomal instability and the much higher frequency of spontaneous deletions provide a mechanistic basis for cancer development initiated by Brca2 deficiency.
The tremendous sensitivity of V-C8 cells toward cross-links indicates that Brca2 is important in protecting cells against these agents. Sensitivity to cross-links has also been found in Brca2-deficient mouse lymphocytes (44) and is displayed by hamster or chicken cell mutants defective in one of the Rad51 paralogs (16, 26) and by Brca1-deficient mouse ES cells (1, 17). In parallel, as in Brca2-deficient cells, defects in the above genes lead to chromosomal instability and impaired damage-induced Rad51 focus formation, suggesting a role for all of these proteins in maintaining genomic integrity through Rad51-dependent HR.
Our results indicate that hamster V-C8 cells offer a useful tool for studying the functions of the Brca2 gene. Unlike the Brca2-deficient MEFs and CAPAN-1 cells, V-C8 cells grow well in culture, and large amounts of cells can be relatively quickly obtained. In addition, they have a high cloning efficiency (25%) and can easily be transfected with exogenous DNA. The very high MMC sensitivity of V-C8 cells provides an easy readout in, for example, functional complementation studies. Currently, we cannot exclude the possibility that the extreme sensitivity of V-C8 cells to cross-links results from a specific mutation in Brca2 in these cells. However, this finding might have important consequences for the therapy of some BRCA2-deficient breast cancer patients because they could be more responsive to treatment with cross-linking agents.
Acknowledgments
We thank H. Vrieling for providing the primers for the PCR of hprt cDNA and genomic hprt DNA, L. van Veelen for valuable experimental help, E. J. Stanbridge for providing the chromosome 13 donor cell line MCH204.3, and F. E. Benson for providing the antibody against Rad51, FBE-2.
This work was supported by grant 9.0.14 from the J. A. Cohen Institute, Interuniversity Research Institute for Radiopathology and Radiation Protection, Leiden, The Netherlands; by NWO grants 901-01-190 and 901-01-097; by the European Union grant FIGH-CT1999-00010; by the Association for International Cancer Research; and by the National Cancer Institute, U.S. Department of Health and Human Services.
REFERENCES
- 1.Bhattacharyya, A., U. S. Ear, B. H. Koller, R. R. Weichselbaum, and D. K. Bishop. 2000. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J. Biol. Chem. 275:23899–23903. [DOI] [PubMed] [Google Scholar]
- 2.Bignell, G., G. Micklem, M. R. Stratton, A. Ashworth, and R. Wooster. 1997. The BRC repeats are conserved in mammalian BRCA2 proteins. Hum. Mol. Genet. 6:53–58. [DOI] [PubMed] [Google Scholar]
- 3.Bishop, D. K., U. Ear, A. Bhattacharyya, C. Calderone, M. Beckett, R. R. Weichselbaum, and A. Shinohara. 1998. Xrcc3 is required for assembly of Rad51 complexes in vivo. J. Biol. Chem. 273:21482–21488. [DOI] [PubMed] [Google Scholar]
- 4.Davies, A. A., J. Y. Masson, M. J. McIlwraith, A. Z. Stasiak, A. Stasiak, A. R. Venkitaraman, and S. C. West. 2001. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol. Cell 7:273–282. [DOI] [PubMed] [Google Scholar]
- 5.Dronkert, M. L., H. B. Beverloo, R. D. Johnson, J. H. Hoeijmakers, M. Jasin, and R. Kanaar. 2000. Mouse RAD54 affects DNA double-strand break repair and sister chromatid exchange. Mol. Cell. Biol. 20:3147–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fischer, L., M. Gerard, C. Chalut, Y. Lutz, S. Humbert, M. Kanno, P. Chambon, and J. M. Egly. 1992. Cloning of the 62-kilodalton component of basic transcription factor BTF2. Science 257:1392–1395. [DOI] [PubMed] [Google Scholar]
- 7.Ford, D., D. F. Easton, M. Stratton, S. Narod, D. Goldgar, P. Devilee, D. T. Bishop, B. Weber, G. Lenoir, J. Chang-Claude, H. Sobol, M. D. Teare, J. Struewing, A. Arason, S. Scherneck, J. Peto, T. R. Rebbeck, P. Tonin, S. Neuhausen, R. Barkardottir, J. Eyfjord, H. Lynch, B. A. Ponder, S. A. Gayther, and M. Zelada-Hedman. 1998. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The Breast Cancer Linkage Consortium. Am. J. Hum. Genet. 62:676–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuller, L. F., and R. B. Painter. 1988. A Chinese hamster ovary cell line hypersensitive to ionizing radiation and deficient in repair replication. Mutat. Res. 193:109–121. [DOI] [PubMed] [Google Scholar]
- 9.Griffin, C. S., P. J. Simpson, C. R. Wilson, and J. Thacker. 2000. Mammalian recombination-repair genes XRCC2 and XRCC3 promote correct chromosome segregation. Nat. Cell Biol. 2:757–761. [DOI] [PubMed] [Google Scholar]
- 10.Haaf, T., E. I. Golub, G. Reddy, C. M. Radding, and D. C. Ward. 1995. Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc. Natl. Acad. Sci. USA 92:2298–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huschtscha, L. I., and R. Holliday. 1983. Limited and unlimited growth of SV40-transformed cells from human diploid MRC-5 fibroblasts. J. Cell Sci. 63:77–99. [DOI] [PubMed] [Google Scholar]
- 12.Jones, N. J., R. Cox, and J. Thacker. 1987. Isolation and cross-sensitivity of X-ray-sensitive mutants of V79-4 hamster cells. Mutat. Res. 183:279–286. [DOI] [PubMed] [Google Scholar]
- 13.Jones, N. J., S. A. Stewart, and L. H. Thompson. 1990. Biochemical and genetic analysis of the Chinese hamster mutants irs1 and irs2 and their comparison to cultured ataxia telangiectasia cells. Mutagenesis 5:15–23. [DOI] [PubMed] [Google Scholar]
- 14.Karran, P. 2000. DNA double strand break repair in mammalian cells. Curr. Opin. Genet. Dev. 10:144–150. [DOI] [PubMed] [Google Scholar]
- 15.Koi, M., M. Shimizu, H. Morita, H. Yamada, and M. Oshimura. 1989. Construction of mouse A9 clones containing a single human chromosome tagged with neomycin-resistance gene via microcell fusion. Jpn. J. Cancer Res. 80:413–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu, N., J. E. Lamerdin, R. S. Tebbs, D. Schild, J. D. Tucker, M. R. Shen, K. W. Brookman, M. J. Siciliano, C. A. Walter, W. Fan, L. S. Narayana, Z. Q. Zhou, A. W. Adamson, K. J. Sorensen, D. J. Chen, N. J. Jones, and L. H. Thompson. 1998. XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol. Cell 1:783–793. [DOI] [PubMed] [Google Scholar]
- 17.Moynahan, M. E., T. Y. Cui, and M. Jasin. 2001. Homology-directed DNA repair, mitomycin C resistance, and chromosome stability is restored with correction of a Brca1 mutation. Cancer Res. 61:4842–4850. [PubMed] [Google Scholar]
- 18.Moynahan, M. E., A. J. Pierce, and M. Jasin. 2001. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell 7:263–272. [DOI] [PubMed] [Google Scholar]
- 19.Overkamp, W. J., M. A. Rooimans, I. Neuteboom, P. Telleman, F. Arwert, and M. Z. Zdzienicka. 1993. Genetic diversity of mitomycin C-hypersensitive Chinese hamster cell mutants: a new complementation group with chromosomal instability. Somat. Cell Mol. Genet. 19:431–437. [DOI] [PubMed] [Google Scholar]
- 20.Papouli, E., C. Lafon, A. Valette, M. Z. Zdzienicka, M. Defais, and F. Larminat. 2000. Involvement of apoptosis in mitomycin C hypersensitivity of Chinese hamster cell mutants. Biochem. Pharmacol. 59:1101–1107. [DOI] [PubMed] [Google Scholar]
- 21.Petrini, J. H. 2000. The Mre11 complex and ATM: collaborating to navigate S phase. Curr. Opin. Cell Biol. 12:293–296. [DOI] [PubMed] [Google Scholar]
- 22.Sharan, S. K., M. Morimatsu, U. Albrecht, D. S. Lim, E. Regel, C. Dinh, A. Sands, G. Eichele, P. Hasty, and A. Bradley. 1997. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature 386:804–810. [DOI] [PubMed] [Google Scholar]
- 23.Sonoda, E., M. S. Sasaki, C. Morrison, Y. Yamaguchi-Iwai, M. Takata, and S. Takeda. 1999. Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol. Cell. Biol. 19:5166–5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spain, B. H., C. J. Larson, L. S. Shihabuddin, F. H. Gage, and I. M. Verma. 1999. Truncated BRCA2 is cytoplasmic: implications for cancer-linked mutations. Proc. Natl. Acad. Sci. USA 96:13920–13925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takata, M., M. S. Sasaki, E. Sonoda, T. Fukushima, C. Morrison, J. S. Albala, S. M. Swagemakers, R. Kanaar, L. H. Thompson, and S. Takeda. 2000. The Rad51 paralog Rad51B promotes homologous recombinational repair. Mol. Cell. Biol. 20:6476–6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takata, M., M. S. Sasaki, S. Tachiiri, T. Fukushima, E. Sonoda, D. Schild, L. H. Thompson, and S. Takeda. 2001. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol. Cell. Biol. 21:2858–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tashiro, S., N. Kotomura, A. Shinohara, K. Tanaka, K. Ueda, and N. Kamada. 1996. S phase specific formation of the human Rad51 protein nuclear foci in lymphocytes. Oncogene 12:2165–2170. [PubMed] [Google Scholar]
- 28.Tashiro, S., J. Walter, A. Shinohara, N. Kamada, and T. Cremer. 2000. Rad51 accumulation at sites of DNA damage and in postreplicative chromatin. J. Cell Biol. 150:283–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tavtigian, S. V., J. Simard, J. Rommens, F. Couch, D. Shattuck-Eidens, S. Neuhausen, S. Merajver, S. Thorlacius, K. Offit, D. Stoppa-Lyonnet, C. Belanger, R. Bell, S. Berry, R. Bogden, Q. Chen, T. Davis, M. Dumont, C. Frye, T. Hattier, S. Jammulapati, T. Janecki, P. Jiang, R. Kehrer, J. F. Leblanc, and D. E. Goldgar. 1996. The complete BRCA2 gene and mutations in chromosome 13q-linked kindreds. Nat. Genet. 12:333–337. [DOI] [PubMed] [Google Scholar]
- 30.Telleman, P., W. J. Overkamp, and M. Z. Zdzienicka. 1996. Spectrum of spontaneously occurring mutations in the HPRT gene of the Chinese hamster V79 cell mutant V-H4, which is homologous to Fanconi anemia group A. Mutagenesis 11:155–159. [DOI] [PubMed] [Google Scholar]
- 31.Thacker, J. 1999. A surfeit of RAD51-like genes? Trends Genet. 15:166–168. [DOI] [PubMed] [Google Scholar]
- 32.Thacker, J., and A. N. Ganesh. 1990. DNA-break repair, radioresistance of DNA synthesis, and camptothecin sensitivity in the radiation-sensitive irs mutants: comparisons to ataxia-telangiectasia cells. Mutat. Res. 235:49–58. [DOI] [PubMed] [Google Scholar]
- 33.Thompson, L. H., and D. Schild. 2001. Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat. Res. 477:131–153. [DOI] [PubMed] [Google Scholar]
- 34.Tutt, A., A. Gabriel, D. Bertwistle, F. Connor, H. Paterson, J. Peacock, G. Ross, and A. Ashworth. 1999. Absence of Brca2 causes genome instability by chromosome breakage and loss associated with centrosome amplification. Curr. Biol. 9:1107–1110. [DOI] [PubMed] [Google Scholar]
- 35.Venkitaraman, A. R. 1999. Breast cancer genes and DNA repair. Science 286:1100–1102. [DOI] [PubMed] [Google Scholar]
- 36.Verhaegh, G. W., W. Jongmans, B. Morolli, N. G. Jaspers, G. P. van der Schans, P. H. Lohman, and M. Z. Zdzienicka. 1995. A novel type of X-ray-sensitive Chinese hamster cell mutant with radioresistant DNA synthesis and hampered DNA double-strand break repair. Mutat. Res. 337:119–129. [DOI] [PubMed] [Google Scholar]
- 37.Welcsh, P. L., K. N. Owens, and M. C. King. 2000. Insights into the functions of BRCA1 and BRCA2. Trends Genet. 16:69–74. [DOI] [PubMed] [Google Scholar]
- 38.Wong, A. K. C., R. Pero, P. A. Ormonde, S. V. Tavtigian, and P. L. Bartel. 1997. RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene brca2. J. Biol. Chem. 272:31941–31944. [DOI] [PubMed] [Google Scholar]
- 39.Wooster, R., S. L. Neuhausen, J. Mangion, Y. Quirk, D. Ford, N. Collins, K. Nguyen, S. Seal, T. Tran, and D. Averill. 1994. Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12-13. Science 265:2088–2090. [DOI] [PubMed] [Google Scholar]
- 40.Xia, F., D. G. Taghian, J. S. DeFrank, Z. C. Zeng, H. Willers, G. Iliakis, and S. N. Powell. 2001. Deficiency of human BRCA2 leads to impaired homologous recombination but maintains normal nonhomologous end joining. Proc. Natl. Acad. Sci. USA 98:8644–8649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu, B., S. Kim, and M. B. Kastan. 2001. Involvement of Brca1 in S-phase and G2-phase checkpoints after ionizing irradiation. Mol. Cell. Biol. 21:3445–3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu, X., Z. Weaver, S. P. Linke, C. Li, J. Gotay, X. W. Wang, C. C. Harris, T. Ried, and C. X. Deng. 1999. Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol. Cell 3:389–395. [DOI] [PubMed] [Google Scholar]
- 43.Yamaguchi-Iwai, Y., E. Sonoda, M. S. Sasaki, C. Morrison, T. Haraguchi, Y. Hiraoka, Y. M. Yamashita, T. Yagi, M. Takata, C. Price, N. Kakazu, and S. Takeda. 1999. Mre11 is essential for the maintenance of chromosomal DNA in vertebrate cells. EMBO J. 18:6619–6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu, V. P., M. Koehler, C. Steinlein, M. Schmid, L. A. Hanakahi, A. J. van Gool, S. C. West, and A. R. Venkitaraman. 2000. Gross chromosomal rearrangements and genetic exchange between nonhomologous chromosomes following BRCA2 inactivation. Genes Dev. 14:1400–1406. [PMC free article] [PubMed] [Google Scholar]
- 45.Yuan, S. S., S. Y. Lee, G. Chen, M. Song, G. E. Tomlinson, and E. Y. Lee. 1999. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 59:3547–3551. [PubMed] [Google Scholar]
- 46.Zdzienicka, M. Z. 1996. Mammalian X ray sensitive mutants: a tool for the elucidation of the cellular response to ionizing radiation. Cancer Surv. 28:281–293. [PubMed] [Google Scholar]
- 47.Zdzienicka, M. Z., N. G. Jaspers, G. P. van der Schans, A. T. Natarajan, and J. W. Simons. 1989. Ataxia-telangiectasia-like Chinese hamster V79 cell mutants with radioresistant DNA synthesis, chromosomal instability, and normal DNA strand break repair. Cancer Res. 49:1481–1485. [PubMed] [Google Scholar]
- 48.Zdzienicka, M. Z., and J. W. Simons. 1987. Mutagen-sensitive cell lines are obtained with a high frequency in V79 Chinese hamster cells. Mutat. Res. 178:235–244. [DOI] [PubMed] [Google Scholar]
- 49.Zhang, L. H., H. Vrieling, A. A. van Zeeland, and D. Jenssen. 1992. Spectrum of spontaneously occurring mutations in the hprt gene of V79 Chinese hamster cells. J. Mol. Biol. 223:627–635. [DOI] [PubMed] [Google Scholar]
- 50.Zimmerman, W., C. A. Sparks, and S. J. Doxsey. 1999. Amorphous no longer: the centrosome comes into focus. Curr. Opin. Cell Biol. 11:122–128. [DOI] [PubMed] [Google Scholar]