Abstract
Multiple genetic pathways have been shown to regulate life span and aging in the yeast Saccharomyces cerevisiae. Here we show that loss of a component of the RNA polymerase II complex, Hpr1p, results in a decreased life span. Although hpr1Δ mutants have an increased rate of recombination within the ribosomal DNA (rDNA) array, this is not accompanied by an increase in extrachromosomal rDNA circles (ERCs). Analyses of mutants that affect replication of the rDNA array and suppressors that reverse the phenotypes of the hpr1Δ mutant show that the reduced life span is associated with increased genomic instability but not with increased ERC formation. The hpr1Δ mutant acts in a pathway distinct from previously described mutants that reduce life span.
Cellular or organismal life span is determined by genetic and extrinsic factors. Toward the end of the life span, cells often deteriorate, giving rise to the phenotypes associated with aging. Recently, experimental systems have been developed that now permit the identification of genes that influence life span. Often, although not always, mutations that reduce life span result in the phenotype of an early appearance of aging features (32). The ability to tolerate stress and the effects of caloric restriction are correlated with increased life span in mice, Caenorhabditis elegans, Drosophila melanogaster, and Saccharomyces cerevisiae (for reviews, see references 18, 23, 33, 51, and 57). Recognition of the contribution that these processes have on life span has led to an identification of some of the genetic components underlying life span and senescence in various experimental organisms (12, 19).
In addition to the above genetic factors, in the yeast S. cerevisiae life span is influenced by gene silencing. This is thought to act by silencing within the ribosomal DNA (rDNA) array, which is composed of 100 to 200 tandem repeats. The Sir2 protein, encoding an NAD-dependent histone deacetylase (22), is localized to the silenced genomic regions that are the telomeres (16), the HML and HMR mating-type genes (40), and the rDNA array to form a silenced state of chromatin (4, 25, 50, 52). The silenced rDNA array is repressed for recombination (15). The products of rDNA recombination, excised circles with rDNA repeats, which are termed ERCs (for extrachromosomal rDNA circles), have been suggested to be correlated with aging, but this metric of aging has been challenged in recent studies (21, 27, 28). Nonetheless, instability within the rDNA has been correlated with reduced life span in a number of mutants, including the sgs1 and sir2 mutants (9, 24, 31, 42, 48). Sgs1p is the yeast homolog of the Escherichia coli RecQ helicase and the human BLM and WRN proteins. Since the topoisomerase genes TOP1, TOP2, and TOP3 function in maintaining wild-type levels of recombination within the rDNA and the Top2 and Top3 proteins interact with the Sgs1 protein (13, 56), other mutants that affect rDNA stability may have an interaction with the topoisomerase genes. To further explore the relationship between rDNA instability and life span, we examined the hpr1Δ mutant of S. cerevisiae.
The yeast hyperrecombination mutant hpr1Δ increases intrachromosomal recombination between non-rDNA direct repeats by almost 1,000-fold (2). In those studies we noted a slight enhancement of recombination within the rDNA array in the hpr1Δ mutant strain (2). Other phenotypes of the hpr1Δ mutant also suggested that Hpr1p had a function in the stabilization of the rDNA array. First, a full complement of topoisomerase activities is necessary for viability in an hpr1Δ background (2). Second, hpr1Δ cells have aberrant ring-shaped nucleoli and general defects in rRNA processing when shifted to a nonpermissive growth temperature (37°C) (46). Although the exact biochemical function of Hpr1p remains unclear, it has a functional role in RNA polymerase II transcription, and most genetic suppressors of hpr1Δ-stimulated hyperrecombination are mutations in components of the RNA polymerase II transcription machinery (10, 11, 37, 38, 45, 54). Furthermore, the hyperrecombination phenotype of hpr1Δ is dependent on RNA polymerase II transcription (10), specifically transcriptional elongation (6). Hpr1p has recently been identified as a component of at least two unique protein complexes, each with both biochemical and genetic associations with RNA polymerase II transcription. One biochemically defined complex containing Hpr1p, Paf1p, Ccr4p, and Cdc73p appears to be a subunit of an alternative RNA polymerase II holoenzyme with distinct regulatory functions (5), while another biochemically defined complex is composed of Hpr1p, Tho2p, Mft1p, and Thp2 and functions in transcriptional elongation (7).
In this study we have reinvestigated the effect of the hpr1Δ mutation on rDNA recombination. We demonstrate that hpr1Δ does indeed affect the stability of the rDNA repeat array and life span in yeast in an ERC-independent process. The hpr1Δ effect on life span is independent of other mutants known to affect yeast life span and suggests that transcription effects have an important influence on cellular life span.
MATERIALS AND METHODS
Strains and growth media.
The S. cerevisiae strains used in this study are listed in Table 1. All strains are isogenic and have the W303 RAD5+ background. The W303R strain contains a single copy of ADE2 inserted into the rDNA repeat array and is described elsewhere (24). The strains with the ADE2 marker inserted into the rDNA array were used for obtaining rDNA recombination rates. The non-ADE2 marker strains were used for life span analysis. The fob1::URA3 disruption was generated by a targeted PCR method described earlier (3, 41). URA3 was amplified from the pRS316 vector. Primers were designed such that 40 nucleotides of homology to FOB1 were incorporated at the ends of the URA3 amplification. The resulting disruption deleted the entire FOB1 gene except for the first and last 40 nucleotides. Descriptions of the hpr1 and soh1 null allele strains can be found elsewhere (2, 10). The sgs1Δ::URA3 disruption and the W303R strain were kindly provided by R. Rothstein and L. Guarente, respectively. Strains were cultured in standard yeast extract-peptone-dextrose (YPD) medium containing 2% glucose at 30°C unless otherwise noted (20).
TABLE 1.
Yeast strainsa
| Strain | Genotype | Source |
|---|---|---|
| 580-10D | MATaRAD5 in W303 background | H. Klein |
| HFY824-1A | MATα hpr1Δ::HIS3 RAD5 W303 | H. Fan |
| RMY212-12D | MATacdc73Δ::TRP RAD5 W303 | This study |
| RMY059-2B | MATasgs1Δ::URA3 RAD5 W303 | This study |
| RMY058-5A | MATahpr1Δ::HIS3 sgs1Δ::URA3 RAD5 W303 | This study |
| HFY1002-22A | MATasoh1Δ::TRP1 RAD5 W303 | H. Fan |
| RMY125-7A | MATahpr1Δ::HIS3 soh1Δ::TRP1 RAD5 W303 | This study |
| RMY178-1A | MATafob1Δ::URA3 RAD5 W303 | This study |
| RMY151-11B | MATα hpr1Δ::HIS3 fob1Δ::URA3 RAD5 W303 | This study |
| RMY206-5B | MATα sir2Δ::HIS3 RAD5 W303 | This study |
| RMY207-3A | MATα hpr1Δ::HIS3 sir2Δ::HIS3 RAD5 W303 | This study |
| W303R | MATaRDN1::ADE2 RAD5 W303 | L. Guarente |
| RMY184-8C | MATahpr1Δ::HIS3 RDN1::ADE2 RAD5 W303 | This study |
| RMY194-12D | MATacdc73Δ::TRP RDN1::ADE2 RAD5 W303 | This study |
| RMY186-9A | MATasgs1Δ::URA3 RDN1::ADE2 RAD5 W303 | This study |
| RMY189-7B | MATahpr1Δ::HIS3 sgs1Δ::URA3 RDN1::ADE2 RAD5 W303 | This study |
| RMY175-11C | MATafob1Δ::URA3 RDN1::ADE2 RAD5 W303 | This study |
| RMY176-4A | MATahpr1Δ::HIS3 fob1Δ::URA3 RDN1::ADE2 RAD5 W303 | This study |
| RMY170-2C | MATasoh1Δ::TRP1 RDN1::ADE2 RAD5 W303 | This study |
| RMY172-5C | MATahpr1Δ::HIS3 soh1Δ::TRP1 RDN1::ADE2 RAD5 W303 | This study |
| RMY192-4A | MATα sir2Δ::HIS3 RDN1::ADE2 RAD5 W303 | This study |
| RMY193-12C | MATα hpr1Δ::HIS3 sir2Δ::HIS3 RDN1::ADE2 RAD5 W303 | This study |
All strains possess the W303 genetic background (ura3-1 leu2-3,112 his3-11,15 trp1-1 ade2-1 can1-100).
Life span analysis.
The methods for life span analysis are similar to those described by Kennedy et al. (26). Life spans were determined by counting the number of daughter cells generated by a single mother cell. A micromanipulator was used to tease away and remove each daughter cell that emerged as a bud from its mother cell. Mother cells are larger than daughter cells and are easily differentiated. Virgin cells to be used as mother cells for life span analysis were generated by randomly picking cells from a log-phase liquid culture that had been streaked onto a solid YPD plate. The first virgin granddaughter from each of the randomly chosen cells was isolated as a bud from a mother cell and subsequently used as the mother cell for further life span analysis.
At least 50 mother cells were used from each strain to generate the life span curves and to determine average longevity. The Mann-Whitney test was used to determine statistical significance between the different average life spans. Statistical differences in mortality rates at specified generations were analyzed by using the log-rank test (35, 36).
Determination of rDNA recombination rates.
Mitotic rDNA recombination rates were determined as described previously (8). The loss of an ADE2 marker integrated into the rDNA array was used to measure recombination. Strains were grown to mid-log phase in liquid YPD medium, diluted, and plated onto solid YPD. Colonies were grown at 30°C for 2 days and then transferred to 4°C for 4 days prior to analysis. Half-sectored red and white colonies indicate that the ADE2 marker was lost in the first cell division after plating. Partially red colonies indicate that the marker was lost after the first cell division after plating. Fully red colonies can only be the result of marker loss prior to plating and are not used in the analysis. The recombination rate is determined by considering only the first cell division after plating and is calculated by dividing the total number of half-sectored colonies by the total number of colonies (white plus half-sectors plus partial sectors). The Student’s t test was used to analyze significance between rDNA recombination rates.
Segregation of old cells.
The segregation of old cells from the majority of young cells in a mid-log phase culture was accomplished by using the techniques described in detail elsewhere (47, 49). Briefly, cells were cultured at 30°C in liquid YPD medium to mid-log phase (optical density at 600 nm [OD600] = 0.7). Cells were collected by centrifugation, washed twice in phosphate-buffered saline (PBS) buffer, and resuspended in 1 ml of PBS. Approximately 10 mg of sulfo-NHS-LC-biotin (Pierce) was added to the cells, and the mixture as gently shaken at room temperature for 15 min. Cells were washed eight times in 1 ml of PBS and then resuspended in 1 ml of YPD liquid medium. Then, 108 cells from this biotin-labeled culture were resuspended into 1 liter of YPD containing 3% glucose, followed by growth at 30°C overnight for ca. 12 to 13 h (cultures were not allowed to exceed an OD600 of 1.0). Cells were collected by centrifugation and resuspended in 40 ml of cold PBS. Next, 300 μl of streptavidin-coated magnetic beads (PerSeptive Biosystems; 5 μg/μl, washed and equilibrated with PBS) was added to the cells, and the cells were kept on ice for 4 h with occasional mixing. Cells were distributed to 10-ml test tubes and placed in a magnetic sorter for 15 min at 4°C. Free cells were carefully removed with a pipette. The bead-bound cells were then resuspended in 10 ml of ice-cold YPD and placed back into the magnetic sorter at 4°C. This washing procedure was repeated eight times before the cells were pooled and stored on ice prior to DNA isolation. An aliquot of cells was used for staining with Calcofluor and examination with a fluorescence microscope to count the number of bud scars per cell. Cell from the old cell fraction contained 8 to 10 staining bud scars per cell, indicating that the age of this cell fraction was 8 to 10 generations.
Old and young cell DNA isolation and ERC quantitation.
DNA preparations from old cells were performed immediately after sorting. Old and young cells were washed in 1 ml of double-distilled water, centrifuged, and resuspended in 0.5 ml of sorbitol solution (1 M sorbitol, 0.1 EDTA [pH 8.0]) containing 30 μg of zymolyase 100T (U.S. Biological)/ml. Then, 0.05 ml of 0.28 M β-mercaptoethanol was added, and the cells were incubated at 37°C for 25 min. The resulting spheroplasts were gently centrifuged and resuspended in 0.5 ml of 50 mM Tris (pH 7.8)-20 mM EDTA. The spheroplasts were lysed by adding 0.05 ml of 10% sodium dodecyl sulfate and incubating them at 65°C for 20 min. Then, 0.2 ml of 5 M potassium acetate was added, followed by gentle mixing and incubation on ice for 30 min. Tubes were centrifuged at top speed for 5 min, the supernatant was collected, and the DNA was precipitated with an equal volume of isopropanol. The DNA pellet was resuspended in 0.05 ml of TE. ERCs were resolved by loading equal volumes of DNA on 0.7% agarose gels without ethidium bromide at 1 V/cm for 20 to 24 h. rDNA species were detected by Southern blotting and hybridization to the radiolabeled rDNA fragment, RDN25. Autoradiographs were visualized by using a Bio-Rad phosphorimager, and quantitation was performed by using NIH Image 1.6. ERC levels in old cells were normalized against the chromosomal rDNA. ERC levels in young cells were quantitated by normalizing against a single-copy chromosomal sequence, ACT1. Equal volumes of DNA were digested with EcoRI and resolved on a 1% agarose gel. ACT1 was detected by Southern blotting and hybridization to a radiolabeled probe generated from the 0.6-kb internal ClaI fragment of ACT1.
Terminal morphologies.
Terminal morphologies were determined as described earlier (34). Small budded cells were defined as budding cells where the bud was less than one-fourth the diameter of the adjoining mother cell. Large budded cells were defined as budding cells where the bud was greater than or equal to one-fourth the diameter of the adjoining mother cell.
RESULTS
hpr1Δ cells have an increased rate of rDNA mitotic intrachromosomal recombination.
Loss-of-function mutations in the HPR1 gene result in a dramatic increase in mitotic intrachromosomal recombination between direct repeats in non-rDNA chromosomal locations (2). At the time, we had a suggestive effect of a hpr1 mutation on recombination within the rDNA repeat array, but the recombination reporter was not optimal. To more accurately determine whether a hpr1Δ mutant had an increased recombination rate in the rDNA, we measured rDNA recombination by monitoring the loss of an ADE2 marker inserted into the repeat array, by using a half-sector protocol that measures recombination only in the first cell division after plating (see Materials and Methods). We observed that the hpr1Δ strain has a significant increase in rDNA recombination compared to the HPR1 control strain (Table 2). hpr1Δ loses the ADE2 marker at a rate that is threefold higher than the rate with HPR1 (P < 0.005).
TABLE 2.
rDNA recombination ratesa
| Genotype | Recombination rate (103) (SD) | Fold difference vs wild type |
|---|---|---|
| Wild type | 1.3 (0.5) | |
| hpr1 | 4.0 (0.9) | 3.1 |
| cdc73 | 1.5 (0.03) | 1.1 |
| fob1 | 0.5 (0.2) | 0.4 |
| hpr1 fob1 | 5.6 (1.6) | 4.3 |
| sgs1 | 7.4 (2.6) | 5.8 |
| hpr1 sgs1 | 11.9 (2.6) | 9.2 |
| soh1 | 1.6 (0.4) | 1.2 |
| hpr1 soh1 | 1.7 (0.5) | 1.3 |
| sir2 | 6.3 (0.3) | 4.8 |
| hpr1 sir2 | 8.3 (0.9) | 6.4 |
The rDNA recombination rates was measured by determining the loss of an ADE2 marker (see Materials and Methods). Average rates were calculated from at least four independent experiments totaling at least 20,000 colonies.
The average longevity of hpr1Δ cells is reduced.
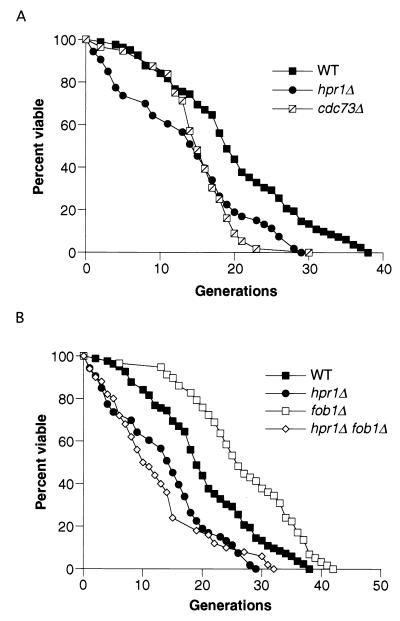
Since the hpr1Δ mutant has an increased level of rDNA recombination, we next determined whether hpr1Δ had an effect on life span. Yeast life span is measured by counting the number of daughter cells generated from an individual mother cell (see Materials and Methods). The average life span for the W303-RAD5 strain background is 20.1 generations, while the average life span for the hpr1Δ mutant in this background is 13.7 generations (Fig. 1A), which is a significant reduction in life span (P < 0.001) (Table 3). It is interesting that at least some of the difference in the average life span derives from an increased mortality rate in young hpr1Δ cells. Since Hpr1p has been identified as part of a complex containing Cdc73p, Paf1p, and Ccr4p interacting with RNA polymerase II, we examined additional mutants to see whether there was a consistent effect on life span. We chose a cdc73Δ strain since this null allele mutant has a greater hyperrecombination phenotype than mutants of other components in this complex and we wanted to determine whether there was a correlation between the hyperrecombination phenotype and life span (5). Similar to hpr1Δ, cdc73Δ has a decreased average life span compared to wild type (P < 0.001). However, the life span curve differs from that of the hpr1Δ strain, particularly in the susceptibility of young cells to an early death.
FIG. 1.
hpr1Δ cells have a reduced average life span that is FOB1 independent. (A) The life spans for wild-type (WT; 580-10D), hpr1Δ (HFY824-1A), and cdc73Δ (RMY212-12D) cells were determined. The mean life spans for the wild-type (n = 82), hpr1Δ (n = 53), and cdc73Δ (n = 56) strains were 20.1, 13.7, and 15.2 generations, respectively. (B) The life spans for the fob1Δ (RMY178-1A) and hpr1Δ fob1Δ (RMY151-11B) strains were determined and are shown with the wild-type and hpr1Δ data from panel A. The mean life spans for the fob1Δ (n = 56) and hpr1Δ fob1Δ (n = 50) strains were 26.9 and 12.4 generations, respectively. The life spans of individual haploid mother cells were determined as described in Materials and Methods.
TABLE 3.
Average life spans
| Genotype | Avg life span(no. of generations)a | % Wild-type life span |
|---|---|---|
| Wild type | 20.1 | |
| hpr1 | 13.7 | 68 |
| cdc73 | 15.2 | 76 |
| fob1 | 26.9 | 134 |
| hpr1 fob1 | 12.4 | 62 |
| sgs1 | 8.3 | 41 |
| hpr1 sgs1 | 5.0 | 25 |
| soh1 | 15.4 | 77 |
| hpr1 soh1 | 15.2 | 76 |
| sir2 | 12.8 | 64 |
| hpr1 sir2 | 8.2 | 41 |
Average life spans for all the strains in the present study.
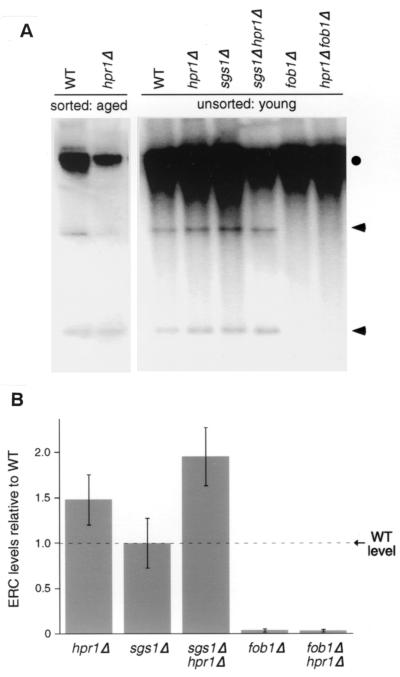
Since hpr1Δ cells display an increase in rDNA recombination and a decrease in life span, we investigated whether these phenotypes could be attributed to an overabundance of ERCs. To accomplish this, we tested both unsorted logarithmically growing cells (young cells) and aged mother cells that had been sorted away from younger cells. We examined ERCs in young cells because, thus far, any factor suggested to have a positive or negative effect on rDNA circle formation in aged cells also shows the same effect in young unsorted cells (9, 21, 24). ERC accumulation in young cells is a simple measure to determine the relative abundance of ERCs, regardless of whether or not they accumulate prematurely in old cells. Figure 2A shows the levels of ERCs in unsorted young cells. There is no obvious difference in the abundance of the two visible ERC species observed in young wild-type and young hpr1Δ cells. Quantitating these results by normalizing ERC levels against a single-copy chromosomal sequence, ACT1, confirmed this observation (Fig. 2B). We found no significant differences between ERC levels present in wild-type and hpr1Δ cells when the levels of the two visible ERC species were combined and averaged from three independent experiments.
FIG. 2.
ERC accumulation in young and old cells. (A) DNA was isolated from young cells (logarithmically growing cultures) and old cells (ca. 8 to 10 generations old) to determine the levels of ERCs in each strain: wild type (580-10D), hpr1Δ (HFY824-1A), sgs1Δ (RMY059-2B), hpr1Δ sgs1Δ (RMY058-5A), fob1Δ (RMY178-1A), and hpr1Δ fob1Δ (RMY151-11B). Old cells were sorted, and DNA was electrophoresed as described in Materials and Methods. Transfers were probed with rDNA sequence RDN25. The circle indicates chromosomal rDNA; the arrows indicate two identifiable ERC species. (B) ERC levels of each mutant relative to the wild type in young cells. The two visible ERC species were averaged and quantitated against a single-copy chromosomal sequence (ACT1). The levels indicated are the averages from three additional independent experiments. The error bars represent the standard deviations from each set of experiments.
We also examined aged wild-type and hpr1Δ cells and observed slight differences in the relative distribution between the two observable ERC species (Fig. 2A). The lowest-molecular-weight ERC species is enriched twofold in hpr1Δ cells at the expense of the higher-molecular-weight ERCs. However, there is no indication of major ERC accumulation in hpr1Δ. Therefore, although hpr1Δ is hyperrecombinogenic for rDNA recombination, this recombination does not produce detectable ERCs, and the shortened life span does not seem to be a direct result of an overabundance of ERCs.
hpr1Δ-mediated rDNA hyperrecombination and life span is FOB1-independent.
FOB1 is required for the replication fork block that is observed within the rDNA array during replication (29, 30). A fob1 mutation results in a decreased rDNA recombination rate, a decreased amount of ERCs, and an increased life span (9). This indicates that the replication block mediated by Fob1p is necessary for the majority of the rDNA recombination events and consequently for ERC formation in wild-type cells. We used the hpr1Δ fob1Δ double mutant to determine whether the rDNA recombination events observed in hpr1Δ cells are dependent on the Fob1p-mediated replication fork block. The rDNA recombination rate in the hpr1Δ fob1Δ double mutant is statistically the same as that obtained for the hpr1Δ single mutant (Table 2). This suggests that the majority of rDNA recombination events in hpr1Δ are independent of FOB1.
Since the hpr1Δ fob1Δ double mutant has levels of rDNA recombination similar to those of the hpr1Δ strain, we examined whether the double mutant could generate ERCs in a FOB1-independent manner. The hpr1Δ fob1Δ double mutant has almost no detectable ERCs in unsorted young cells and is indistinguishable from the fob1Δ single mutant (Fig. 2A). Quantitation of the ERC levels in these two strains confirms this observation (Fig. 2B). Therefore, the formation of ERCs in hpr1Δ cells is dependent on FOB1, in spite of rDNA recombination being FOB1 independent. This reinforces the observation that hpr1Δ-mediated rDNA recombination is not conservative (44) and does not give reciprocal products expected from a crossing over. Nonconservative recombination would result in the absence of ERCs. Other types of nonconservative recombination include expansions and contractions of repeat sequence tracts during replication slippage events and changes in the number of rDNA array units through gene conversion events (14).
As fob1Δ suppresses the formation of ERCs in hpr1Δ cells, we examined whether a fob1Δ mutation would suppress the decreased life span of the hpr1Δ mutant. Figure 1B and Table 3 show that the life span of the hpr1Δ fob1Δ double mutant is not significantly different from that of the hpr1Δ single mutant. Taken together, these data indicate that the increased rDNA recombination rate and the decreased life span observed in hpr1Δ are independent of FOB1 and are not correlated with ERC formation.
hpr1Δ has additive effects with both sgs1Δ and sir2Δ for rDNA recombination and life span.
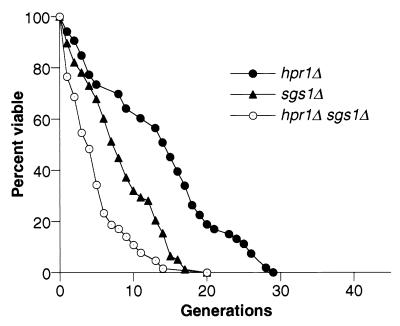
The decreased life span of the hpr1Δ mutant is most likely due to the combined effects of genomic instability both within the rDNA array and in non-rDNA chromosomal regions. Another mutant, sgs1Δ, also displays genetic instability within and outside of the rDNA repeat array (13, 55), as well as a decrease in life span (48). We examined the hpr1Δ sgs1Δ double mutant to determine if these two genes acted together or separately in determining life span. The effect of both mutations on the rDNA recombination rate of the hpr1Δ sgs1Δ double mutant was additive (P < 0.05) (Table 2). The hpr1Δ sgs1Δ double mutant also has a significantly decreased life span compared to the life spans of the sgs1Δ and hpr1Δ single mutants (P < 0.001) (Fig. 3). The reduction in average life span of the hpr1Δ sgs1Δ mutant is the product of the reductions in life span of the hpr1Δ and sgs1Δ mutants, showing that these genes act independently on life span (Table 3).
FIG. 3.
Additive effects of the sgs1Δ and hpr1Δ mutations on life span. The life spans for the sgs1Δ (RMY059-2B) and hpr1Δ sgs1Δ (RMY058-5A) strains were determined and are shown with the hpr1Δ data from Fig. 1. The mean life spans of the sgs1Δ (n = 78), sgs1Δ hpr1Δ (n = 64), and hpr1Δ (n = 53) strains were 8.3, 5.0, and 13.7 generations, respectively.
ERCs in young unsorted cells were also examined in the hpr1Δ sgs1Δ double mutant. No significant difference was observed in the levels of the two detectable ERC species when hpr1Δ sgs1Δ was compared to wild type (Fig. 2). We also examined ERC levels in old (8 to 10 generations) sgs1Δ and hpr1Δ sgs1Δ strains. The levels of ERCs in these strains were not in excess compared to age-matched wild-type and hpr1Δ strains (data not shown). Therefore, similar to hpr1Δ, we can find no evidence that suggests sgs1Δ leads to the accumulation of an overabundance of ERCs at any stage in its life span. These data are consistent with recently published results that also failed to reveal an increase in the levels of ERCs in old sgs1Δ strains compared to age-matched wild-type strains (21).
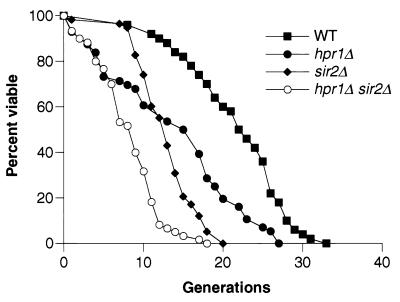
The sir2Δ mutant has increased genetic instability within the rDNA as well as a decreased life span. In addition, the sir2Δ mutant displays a general decrease in transcriptional silencing of RNA polymerase II-transcribed genes when inserted into the rDNA repeat array. Since HPR1 has a role in transcription and in the maintenance of rDNA stability and wild-type life spans, the relationship between HPR1 and SIR2 in controlling rDNA stability, life span, and transcriptional silencing was examined. The results of hpr1Δ sir2Δ mutant analyses show that HPR1 functions independently of SIR2 in maintaining rDNA stability and normal life span. The hpr1Δ sir2Δ double mutant displays a small but significant (P < 0.05) increase in rDNA recombination over the sir2Δ single mutant (Table 2), indicating separate contributions of hpr1Δ and sir2Δ to rDNA instability. These data suggested that the hpr1Δ sir2Δ double mutant would have a shorter life span than either single mutant. Figure 4 shows that this is indeed the case. The hpr1Δ sir2Δ life span is significantly shorter than both the sir2Δ life span (P < 0.001) and the hpr1Δ life span (P < 0.001), indicating that HPR1 and SIR 2 promote wild-type life spans through different pathways. Table 3 shows that the reduction in the average life span of the hpr1Δ sir2Δ double mutant is the product of the reduction in average life spans of the hpr1Δ and sir2Δ mutants, indicating independent influences on life span.
FIG. 4.
Additive effects of the sir2Δ and hpr1Δ mutations on life span. The life spans for wild-type (WT; 580-10D), hpr1Δ (HFY824-1A), sir2Δ (RMY206-5B), and hpr1Δ sir2Δ (RMY207-3A) strains were determined. The mean life spans for the wild-type (n = 50), hpr1Δ (n = 56), sir2Δ (n = 58), and hpr1Δ sir2Δ (n = 60) strains were 21.6, 13.8, 12.8, and 8.2 generations, respectively.
We have not observed any effect of a hpr1Δ mutation on silencing of the HML or HMR loci or subtelomeric reporter genes. Nevertheless, since hpr1Δ displays significant effects on rDNA recombination, we tested whether hpr1Δ had any effect on transcriptional silencing within the rDNA. Using a CAN1 reporter (42), we have observed that hpr1Δ strains show no discernible difference in transcriptional silencing compared to wild-type strains. In addition, the hpr1Δ sir2Δ double mutant displays the same rDNA silencing characteristics as that of the sir2Δ single mutant (data not shown).
Old hpr1Δ cells display terminal phenotypes characteristic of old wild-type cells.
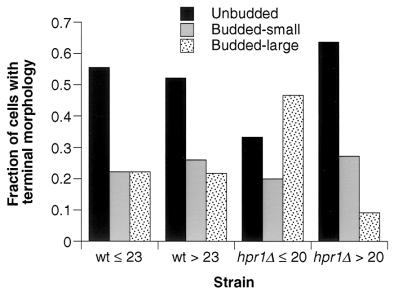
The terminal morphology of the dividing yeast mother cell can give information on whether cell death is stochastic and age independent or whether it is related to the aging processes (34). Mother cells from wild-type yeast strains usually cease division as unbudded cells in G1 at the end of their life span. This unbudded terminal morphology comprises the majority of cell deaths for both short-lived (young, ≤75% of the maximal life span) and long-lived (old, >75% of the maximal life span) yeast cells (34) (Fig. 5). Analysis of the terminal morphologies of young and old hpr1Δ cells showed that the majority of terminal phenotypes of young hpr1Δ cells were as budded cells. Since this terminal G2 arrest is not as common in young wild-type cells, young hpr1Δ cells may be dying due to stochastic and age-independent mechanisms. However, the majority of old hpr1Δ cells display an unbudded terminal phenotype similar to that of old wild-type cells. Therefore, two different factors may be operating in the determination of the average hpr1Δ life span. The first may be age independent and more related to DNA damage or blocked transcription that leads to a G2 arrest-like phenotype, while the second factor seems to be age dependent and related to the wild-type aging process.
FIG. 5.
Terminal morphologies of wild-type (WT) and hpr1Δ cells. The terminal morphologies for wild-type and hpr1Δ strains were obtained from the same mother cells used to generate the life span curves in Fig. 4 (wild type; 580-10D, n = 50; hpr1Δ, HFY824-1A, n = 56). Terminal morphologies are defined as either unbudded, budded small, or budded large (see Materials and Methods). The fraction of each of these terminal phenotypes was calculated for both young (75% maximum life span) and old (>75% maximum life span) cells. The maximum life span is the average life span of the oldest 10% of cells in the experiment. The values for 75% of the maximum life span for wild-type and hpr1Δ strains were 23 and 20 generations, respectively.
While there is an increased fraction of hpr1Δ young cells that cease division as budded cells, this does not imply that growing hpr1Δ cells exhibit an obvious cell cycle defect. Indeed, the distribution of unbudded, small budded, and large budded cells in a log-phase hpr1Δ population is indistinguishable from that of the wild type and is not altered by mutations in the cell cycle checkpoint functions RAD17, RAD24, and MEC3 (H. Klein, unpublished observations).
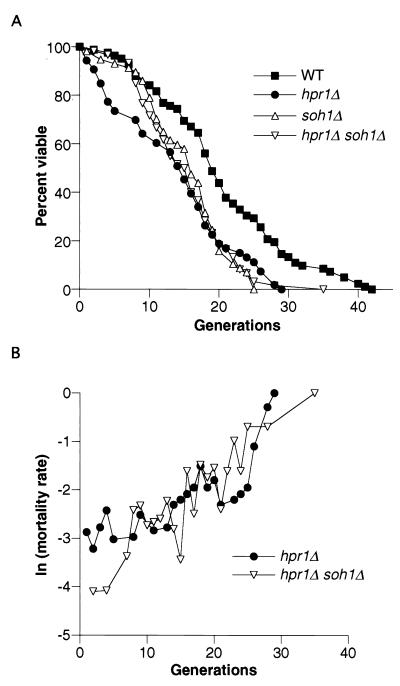
soh1Δ suppresses hpr1Δ-mediated rDNA hyperrecombination and increased mortality rate of young cells.
The soh1 mutation was identified in a selection for hpr1Δ suppressor mutants. soh1Δ suppresses 90% of hpr1Δ-mediated recombination events of non-rDNA origin (10, 11). soh1Δ also suppresses the hpr1Δ requirement for all three functional topoisomerase genes. Soh1p is connected to the RNA polymerase II machinery, and the human counterpart is part of the mediator complex (17). We wanted to determine whether the soh1Δ mutation also suppresses hpr1Δ-mediated rDNA recombination and the decrease in life span. The soh1Δ mutant has a wild-type rate of rDNA recombination (Table 2). The rDNA recombination rate of the hpr1Δ soh1Δ double mutant is indistinguishable from the wild-type and soh1Δ rates, showing that the soh1Δ mutation also suppresses the hpr1Δ-mediated increase in rDNA recombination.
Although the soh1Δ mutant has a wild-type rDNA recombination rate, the life span in this strain is slightly decreased compared to the wild type (Fig. 6A) (P < 0.01). The effect of the soh1Δ mutation is most acutely observed in the viability of older cells, a finding similar to what we observed in the cdc73Δ mutant. The soh1Δ hpr1Δ double mutant has an average life span of 15.2 generations, which is not significantly different from that of the hpr1Δ average life span (13.7 generations) (Fig. 6A and Table 3). By this measure, it cannot be said that soh1Δ suppresses the overall life span decrease of the hpr1Δ mutant. However, the shape of the curves shows that the soh1Δ mutation affects the mortality rates of young hpr1Δ cells. The mortality rate at each generation for hpr1Δ, and hpr1Δ soh1Δ cells is shown in Fig. 6B. A comparison of the mortality rates of the first six generations shows that the hpr1Δ strain has a significantly increased mortality compared to the hpr1Δ soh1Δ double mutant strain (P < 0.001). After the sixth generation, the mortality rates between hpr1Δ and hpr1Δ soh1Δ cells are not significantly different. We conclude that the soh1Δ mutation suppresses the increased early mortality rate of hpr1Δ strains but has no effect on the mortality rate during the later phase of the life span.
FIG. 6.
soh1Δ suppression of the hpr1Δ-reduced life span. (A) The life spans for soh1Δ (HFY1002-22A) and hpr1Δ soh1Δ (RMY125-7A) strains were determined. The wild-type (wt) and hpr1Δ data from Fig. 1 are reproduced here for comparison. The mean life spans were as follows: wild type, 20.3 (n = 82); hpr1Δ, 13.7 (n = 53); soh1Δ, 15.4 (n = 57); and hpr1Δ soh1Δ, 15.2 (n = 60). (B) The increased early mortality rate of hpr1Δ is suppressed by soh1Δ. Natural logarithms of age-specific mortality rates were calculated for both hpr1Δ and hpr1Δ soh1Δ strains. The log-rank test was used to determine the statistical significance between the early mortality rates (see Materials and Methods).
DISCUSSION
We have found that the hpr1Δ mutant increases recombination within the rDNA array but that this increase is not accompanied by an increase in the accumulation of ERCs, a product predicted to result from a conservative reciprocal recombination that gives two recombination products. ERC accumulation in old cells have been associated with the later stages of the yeast life span, including the onset of features characteristic of aging cells. In spite of the lack of early ERC accumulation in hpr1Δ mutants, this strain has a reduced life span, which we believe is the result of increased genomic instability. We believe that Hpr1p acts in transcription elongation to ensure unimpeded elongation through DNA regions that are prone to inducing RNA polymerase II (RNAPII) stalling. In the absence of Hpr1p, transcription is stalled, and this in turn impedes the passage of a DNA polymerase complex. Through some type of template switching or slippage event, the DNA polymerase passes through the regions where the RNAPII complex has stalled but can result in a genomic deletion event when repeated DNA sequences are close to the region of the stalled RNAPII complex. Frequent RNAPII stalling events or the accumulated genomic instability may contribute to the cell life span. The linkage of RNAPII stalling, DNA replication, and the ensuing genomic instability may explain why young hpr1Δ cells die as large budded cells, a result expected if cells die in late S phase or G2 phase. The cells do not appear to be arrested at a cell cycle arrest since DNA damage checkpoint mutants do not change the cell cycle profile of hpr1Δ cells. However, we do not yet know whether these mutants alter the terminal morphology of young hpr1Δ cells.
hpr1Δ-reduced life span and genomic instability.
In hpr1Δ mutant strains, direct repeats are unstable and deletions occur at approximately a 1,000-fold-increased rate (1, 39). This occurs through DNA homology and requires the homologous recombination repair gene RAD52 (1). Although the events occur through homologous recombination, they are not conservative and do not produce the reciprocal product which is an excised circle containing the deleted DNA (44). In terms of recombination within the rDNA this type of recombination would not give an ERC product. This is precisely what we have observed; hpr1Δ strains have an increase in rDNA recombination without an increase in ERCs in young or old cell populations. The fact that there is a reduction in life span without an increase in ERCs argues that, in the hpr1Δ mutant, life span reduction is not related to ERC production. This is further substantiated by the finding that a fob1Δ mutation eliminates all detectable ERCs and yet has no effect on the reduced life span of hpr1Δ cells. This does not mean that increased instability within the rDNA array and at other genomic sites is not related to the reduction in life span. The soh1Δ mutation suppresses hpr1Δ-induced recombination between rDNA repeats and suppresses 90% of the hpr1Δ-induced recombination events between non-rDNA direct repeats (11). soh1Δ also suppresses the increased early mortality of hpr1Δ cells. This correlation suggests that in hpr1Δ cells, reduced life span may be linked to genomic instability. The fact that the hpr1Δ soh1Δ double mutant still has a reduced average life span suggests that this reduction in life span could primarily be due to genomic instability that is not suppressed by soh1Δ.
The hpr1Δ life span curve is distinct in that young cells are particularly at risk for death. It is this increased mortality rate in young cells that is suppressed by the soh1Δ mutation. Since we have argued that soh1Δ suppresses genomic instability, this would suggest that young cells have an increased sensitivity to the rate of genomic instability. The mortality rate of young hpr1Δ cells is similar to that of the sgs1Δ mutant (Fig. 3). Both mutant strains may experience early sensitivity to spontaneous DNA damage, which is manifested as hyperrecombination and increased cell death. Recent studies on the life span of the sgs1Δ mutant have led to the conclusion that the early part of the life span curve is the result of DNA damage, while the late part of the curve is the result of age-related death (34). However, the hpr1Δ mutant has no DNA damage-sensitive phenotype (1, 2), while the sgs1Δ mutant is sensitive to DNA-damaging agents (43). This reinforces the conclusion that each mutant contributes independently to the rDNA hyperrecombination and reduced life span and shows that there are independent mechanisms that contribute to life span that are related to spontaneous DNA damage.
The terminal phenotypes of mother cells after they have ceased dividing may provide clues as to whether the majority of cell deaths are due to age-independent processes such as DNA damage or whether the deaths can be attributed to wild-type aging processes (34). hpr1Δ cells that die at a young generation tend to die as G2-arrested budded cells. This is opposite of the terminal phenotypes that are observed for young wild-type cells, which have a greater tendency to die as G1-arrested unbudded cells. This suggests that young hpr1Δ cells die from different causes than those experienced by young wild-type cells. As we have argued above, we believe that this is the genomic instability that results from DNA replication complexes passing through stalled RNAPII complexes. In contrast, the distribution of unbudded to budded cells among old dying hpr1Δ cells is more similar to the distribution observed for old wild-type cells. This indicates that young dying hpr1Δ cells may be dying from mechanisms related to DNA damage, while old dying hpr1Δ cells may be dying from mechanisms more related to wild-type aging. Therefore, at least one component of the hpr1Δ life span curve seems to be related to wild-type aging mechanisms.
Transcription and life span.
Hpr1p is part of an RNA polymerase II complex in a subcomplex with Cdc73p, Paf1p, and Ccr4p (5). Mutants in any one of the genes encoding these proteins are fully viable under normal growth conditions, but double mutants grow poorly or are inviable. We examined the cdc73Δ mutant and found that it has a reduced life span. This reduction is not readily apparent from inspection of colonies grown on rich medium. The life span curve is unusual in that the young cells have the same viability potential as wild-type cells until ca. 12 generations have passed, and then the viability of successive generations rapidly declines. We do not know the source of the inviability, but we suggest that it reflects the accumulation of some global damage to the cell.
A similar, but not so accentuated, life span curve was observed with the soh1Δ mutant. hSoh1 has been found in the human mediator complex of RNA polymerase II (17), and in yeast SOH1 has been related genetically to the RNA polymerase II complex and in particular to the CTD tail (10). Hpr1p is part of RNA polymerase II complexes, and in the mutant transcription elongation is affected (6). Our results suggests that modest transcription mutants can have a significant impact on cellular life span and that defects arising first as impairments in transcription can set off a cascade of events that leads to genomic instability and reduced generative potential of a cell. This highlights the multiple mechanisms that can compromise the cellular life span.
Transcriptional regulation through SIR2 has been linked to aging in yeast, as well as in C. elegans, and may function to couple life span to nutrient availability in these organisms (31, 53). Our results suggest that additional transcriptional components influence life span.
Acknowledgments
We thank L. Guarente for the yeast strain W303R and for the generous gift of the biotin magnetic beads. We thank E. Nudler for comments and critical reading of the manuscript.
This work was supported by grant GM30439 from the National Institutes of Health.
REFERENCES
- 1.Aguilera, A., and H. L. Klein. 1988. Genetic control of intrachromosomal recombination in Saccharomyces cerevisiae. I. Isolation and genetic characterization of hyper-recombination mutations. Genetics 119:779–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguilera, A., and H. L. Klein. 1990. HPR1, a novel yeast gene that prevents intrachromosomal excision recombination, shows carboxy-terminal homology to the Saccharomyces cerevisiae TOP1 gene. Mol. Cell. Biol. 10:1439–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115–132. [DOI] [PubMed] [Google Scholar]
- 4.Bryk, M., M. Banerjee, M. Murphy, K. E. Knudsen, D. J. Garfinkel, and M. J. Curcio. 1997. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 11:255–269. [DOI] [PubMed] [Google Scholar]
- 5.Chang, M., D. French-Cornay, H. Y. Fan, H. Klein, C. L. Denis, and J. A. Jaehning. 1999. A complex containing RNA polymerase II, Paf1p, Cdc73p, Hpr1p, and Ccr4p plays a role in protein kinase C signaling. Mol. Cell. Biol. 19:1056–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chavez, S., and A. Aguilera. 1997. The yeast HPR1 gene has a functional role in transcriptional elongation that uncovers a novel source of genome instability. Genes Dev. 11:3459–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chavez, S., T. Beilharz, A. G. Rondon, H. Erdjument-Bromage, P. Tempst, J. Q. Svejstrup, T. Lithgow, and A. Aguilera. 2000. A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J. 19:5824–5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christman, M. F., F. S. Dietrich, and G. R. Fink. 1988. Mitotic recombination in the rDNA of S. cerevisiae is suppressed by the combined action of DNA topoisomerases I and II. Cell 55:413–425. [DOI] [PubMed] [Google Scholar]
- 9.Defossez, P. A., R. Prusty, M. Kaeberlein, S. J. Lin, P. Ferrigno, P. A. Silver, R. L. Keil, and L. Guarente. 1999. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol. Cell 3:447–455. [DOI] [PubMed] [Google Scholar]
- 10.Fan, H. Y., K. K. Cheng, and H. L. Klein. 1996. Mutations in the RNA polymerase II transcription machinery suppress the hyperrecombination mutant hpr1Δ of Saccharomyces cerevisiae. Genetics 142:749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan, H. Y., and H. L. Klein. 1994. Characterization of mutations that suppress the temperature-sensitive growth of the hpr1Δ mutant of Saccharomyces cerevisiae. Genetics 137:945–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finkel, T., and N. J. Holbrook. 2000. Oxidants, oxidative stress and the biology of ageing. Nature 408:239–247. [DOI] [PubMed] [Google Scholar]
- 13.Gangloff, S., J. P. McDonald, C. Bendixen, L. Arthur, and R. Rothstein. 1994. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol. Cell. Biol. 14:8391–8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gangloff, S., H. Zou, and R. Rothstein. 1996. Gene conversion plays the major role in controlling the stability of large tandem repeats in yeast. EMBO J. 15:1715–1725. [PMC free article] [PubMed] [Google Scholar]
- 15.Gottlieb, S., and R. E. Esposito. 1989. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell 56:771–776. [DOI] [PubMed] [Google Scholar]
- 16.Gottschling, D. E., O. M. Aparicio, B. L. Billington, and V. A. Zakian. 1990. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63:751–762. [DOI] [PubMed] [Google Scholar]
- 17.Gu, W., S. Malik, M. Ito, C. X. Yuan, J. D. Fondell, X. Zhang, E. Martinez, J. Qin, and R. G. Roeder. 1999. A novel human SRB/MED-containing cofactor complex, SMCC, involved in transcription regulation. Mol. Cell 3:97–108. [DOI] [PubMed] [Google Scholar]
- 18.Guarente, L. 2000. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev. 14:1021–1026. [PubMed] [Google Scholar]
- 19.Guarente, L., and C. Kenyon. 2000. Genetic pathways that regulate aging in model organisms. Nature 408:255–262. [DOI] [PubMed] [Google Scholar]
- 20.Guthrie, C., and G. R. Fink. 1995. Guide to yeast genetics and molecular biology. Methods Enzymol. 194:3–37. [PubMed] [Google Scholar]
- 21.Heo, S. J., K. Tatebayashi, I. Ohsugi, A. Shimamoto, Y. Furuichi, and H. Ikeda. 1999. Bloom’s syndrome gene suppresses premature ageing caused by Sgs1 deficiency in yeast. Genes Cells 4:619–625. [DOI] [PubMed] [Google Scholar]
- 22.Imai, S., C. M. Armstrong, M. Kaeberlein, and L. Guarente. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403:795–800. [DOI] [PubMed] [Google Scholar]
- 23.Jazwinski, S. M. 2000. Metabolic control and ageing. Trends Genet. 16:506–511. [DOI] [PubMed] [Google Scholar]
- 24.Kaeberlein, M., M. McVey, and L. Guarente. 1999. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 13:2570–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keil, R. L., and A. D. McWilliams. 1993. A gene with specific and global effects on recombination of sequences from tandemly repeated genes in Saccharomyces cerevisiae. Genetics 135:711–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kennedy, B. K., N. R. Austriaco, Jr., and L. Guarente. 1994. Daughter cells of Saccharomyces cerevisiae from old mothers display a reduced life span. J. Cell Biol. 127:1985–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim, S., A. Benguria, C. Y. Lai, and S. M. Jazwinski. 1999. Modulation of life span by histone deacetylase genes in Saccharomyces cerevisiae. Mol. Biol. Cell 10:3125–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirchman, P. A., S. Kim, C. Y. Lai, and S. M. Jazwinski. 1999. Interorganelle signaling is a determinant of longevity in Saccharomyces cerevisiae. Genetics 152:179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kobayashi, T., D. J. Heck, M. Nomura, and T. Horiuchi. 1998. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 12:3821–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kobayashi, T., and T. Horiuchi. 1996. A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells. 1:465–474. [DOI] [PubMed] [Google Scholar]
- 31.Lin, S. J., P. A. Defossez, and L. Guarente. 2000. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289:2126–2128. [DOI] [PubMed] [Google Scholar]
- 32.Martin, G. M., and J. Oshima. 2000. Lessons from human progeroid syndromes. Nature 408:263–266. [DOI] [PubMed] [Google Scholar]
- 33.Masoro, E. J. 2000. Caloric restriction and aging: an update. Exp. Gerontol. 35:299–305. [DOI] [PubMed] [Google Scholar]
- 34.McVey, M., M. Kaeberlein, H. A. Tissenbaum, and L. Guarente. 2001. The short life span of Saccharomyces cerevisiae sgs1 and srs2 mutants is a composite of normal aging processes and mitotic arrest due to defective recombination. Genetics 157:1531–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller, R. G. 1981. Survival analysis. Wiley, New York, N.Y.
- 36.Peto, R., M. C. Pike, P. Armitage, N. E. Breslow, D. R. Cox, S. V. Howard, N. Mantel, K. McPherson, J. Peto, and P. G. Smith. 1977. Design and analysis of randomized clinical trials requiring prolonged observation of each patient. II. analysis and examples. Br. J. Cancer. 35:1–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piruat, J. I., and A. Aguilera. 1996. Mutations in the yeast SRB2 general transcription factor suppress hpr1-induced recombination and show defects in DNA repair. Genetics 143:1533–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piruat, J. I., S. Chavez, and A. Aguilera. 1997. The yeast HRS1 gene is involved in positive and negative regulation of transcription and shows genetic characteristics similar to SIN4 and GAL11. Genetics 147:1585–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prado, F., J. I. Piruat, and A. Aguilera. 1997. Recombination between DNA repeats in yeast hpr1Δ cells is linked to transcription elongation. EMBO J. 16:2826–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rine, J., and I. Herskowitz. 1987. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics 116:9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rothstein, R. J. 1983. One-step gene disruption in yeast. Methods Enzymol. 101:202–211. [DOI] [PubMed] [Google Scholar]
- 42.Roy, N., and K. W. Runge. 2000. Two paralogs involved in transcriptional silencing that antagonistically control yeast life span. Curr. Biol. 10:111–114. [DOI] [PubMed] [Google Scholar]
- 43.Saffi, J., V. R. Pereira, and J. A. Henriques. 2000. Importance of the Sgs1 helicase activity in DNA repair of Saccharomyces cerevisiae. Curr. Genet. 37:75–78. [DOI] [PubMed] [Google Scholar]
- 44.Santos-Rosa, H., and A. Aguilera. 1994. Increase in incidence of chromosome instability and non-conservative recombination between repeats in Saccharomyces cerevisiae hpr1Δ strains. Mol. Gen. Genet. 245:224–236. [DOI] [PubMed] [Google Scholar]
- 45.Santos-Rosa, H., and A. Aguilera. 1995. Isolation and genetic analysis of extragenic suppressors of the hyper-deletion phenotype of the Saccharomyces cerevisiae hpr1Δ mutation. Genetics 139:57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schneiter, R., C. E. Guerra, M. Lampl, G. Gogg, S. D. Kohlwein, and H. L. Klein. 1999. The Saccharomyces cerevisiae hyperrecombination mutant hpr1Δ is synthetically lethal with two conditional alleles of the acetyl coenzyme A carboxylase gene and causes a defect in nuclear export of polyadenylated RNA. Mol. Cell. Biol. 19:3415–3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sinclair, D. A., and L. Guarente. 1997. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell 91:1033–1042. [DOI] [PubMed] [Google Scholar]
- 48.Sinclair, D. A., K. Mills, and L. Guarente. 1997. Accelerated aging and nucleolar fragmentation in yeast sgs1 mutants. Science 277:1313–1316. [DOI] [PubMed] [Google Scholar]
- 49.Smeal, T., J. Claus, B. Kennedy, F. Cole, and L. Guarente. 1996. Loss of transcriptional silencing causes sterility in old mother cells of S. cerevisiae. Cell 84:633–642. [DOI] [PubMed] [Google Scholar]
- 50.Smith, J. S., and J. D. Boeke. 1997. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 11:241–254. [DOI] [PubMed] [Google Scholar]
- 51.Sohal, R. S., and R. Weindruch. 1996. Oxidative stress, caloric restriction, and aging. Science 273:59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Szostak, J. W., and R. Wu. 1980. Unequal crossing over in the ribosomal DNA of Saccharomyces cerevisiae. Nature 284:426–430. [DOI] [PubMed] [Google Scholar]
- 53.Tissenbaum, H. A., and L. Guarente. 2001. Increased dosage of a sir-2 gene extends life span in Caenorhabditis elegans. Nature 410:227–230. [DOI] [PubMed] [Google Scholar]
- 54.Uemura, H., S. Pandit, Y. Jigami, and R. Sternglanz. 1996. Mutations in GCR3, a gene involved in the expression of glycolytic genes in Saccharomyces cerevisiae, suppress the temperature-sensitive growth of hpr1 mutants. Genetics 142:1095–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watt, P. M., I. D. Hickson, R. H. Borts, and E. J. Louis. 1996. SGS1, a homologue of the Bloom’s and Werner’s syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics 144:935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watt, P. M., E. J. Louis, R. H. Borts, and I. D. Hickson. 1995. Sgs1: a eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell 81:253–260. [DOI] [PubMed] [Google Scholar]
- 57.Weindruch, R., and R. Walford. 1988. The retardation of aging and disease by dietary restriction. Charles C Thomas, Publisher, Springfield, Ill.