Abstract
We used mouse embryonic stem (ES) cells with systematic gene knockouts for DNA methyltransferases to delineate the roles of DNA methyltransferase 1 (Dnmt1) and Dnmt3a and -3b in maintaining methylation patterns in the mouse genome. Dnmt1 alone was able to maintain methylation of most CpG-poor regions analyzed. In contrast, both Dnmt1 and Dnmt3a and/or Dnmt3b were required for methylation of a select class of sequences which included abundant murine LINE-1 promoters. We used a novel hemimethylation assay to show that even in wild-type cells these sequences contain high levels of hemimethylated DNA, suggestive of poor maintenance methylation. We showed that Dnmt3a and/or -3b could restore methylation of these sequences to pretreatment levels following transient exposure of cells to 5-aza-CdR, whereas Dnmt1 by itself could not. We conclude that ongoing de novo methylation by Dnmt3a and/or Dnmt3b compensates for inefficient maintenance methylation by Dnmt1 of these endogenous repetitive sequences. Our results reveal a previously unrecognized degree of cooperativity among mammalian DNA methyltransferases in ES cells.
The mammalian DNA methyltransferases (DNA methyltransferase 1 [Dnmt1], Dnmt3a, and Dnmt3b) establish and maintain genomic methylation patterns which are of critical importance in various biological processes, including development, genomic imprinting, silencing of parasitic sequence elements, and tumorigenesis (3, 14, 17, 31). The individual role of each of the DNA methyltransferases in establishing and maintaining these patterns is still unclear and has been confounded by their overlapping activities with respect to their abilities to methylate unmethylated and hemimethylated DNA in the test tube (21, 30). Embryonic stem (ES) cells deficient in one or more of these enzymes can be used in one of several approaches to elucidate the roles of the individual enzymes in living cells. Earlier studies using cells deficient in the Dnmt1 enzyme showed considerable decreases in the level of genomic DNA methylation at CpG-rich repetitive elements and imprinted genes (17, 25, 27). Recent studies using cells deficient in both the Dnmt3a and -3b enzymes showed that CpG-rich retroviral and intracisternal A particle (IAP) elements became slightly demethylated, and Igf-2 and Xist became extensively demethylated, in the absence of these enzymes, implying that Dnmt1 by itself had sequence specificity in maintaining the methylation of these sequences (20).
These previous studies all focused on the methylation of CpG-rich sequences in knockout cells. However, most methylation in mammalian cells is found in non-CpG-rich regions of DNA (5), and the roles of the various enzymes in establishing and maintaining these methylation patterns have not been investigated. We have therefore used a genome-scanning approach to investigate the patterns of methylation in the various knockout cells in CpG-poor and CpG-rich regions to determine the roles of the enzymes in carrying out the bulk of methylation in mouse ES cells.
We found that methylation levels of CpG-poor sequences were, in general, uniformly reduced in Dnmt1-deficient cells. However, there was considerable variability among different regions in the efficiency with which DNA methylation was retained in Dnmt3a- and/or Dnmt3b-deficient cells indicating a sequence preference for the Dnmt1 enzyme. We further investigated one of the sequences that was poorly maintained by Dnmt1 alone and showed that it had a surprisingly high level of hemimethylation, even in wild-type cells, suggesting poor maintenance methylation balanced by a continuing high rate of de novo methylation mediated by Dnmt3a and/or Dnmt3b. This study required the development of a hemimethylation assay, which we describe in this paper. Prior to the development of this novel and straightforward method, there had been no accurate way to determine hemimethylation levels at specific CpG dinucleotides in the genome. Further evidence that Dnmt3a and/or Dnmt3b is responsible for the compensating de novo methylation is provided by the fact that these enzymes could restore methylation to pretreatment levels following transient exposure of cells to 5-aza-2′-deoxycytidine 5-aza-CdR), whereas Dnmt1 could not. We also show that Dnmt1 by itself is incapable of restoring methylation of sequences that it had been able to maintain prior to 5-aza-CdR treatment, suggesting that its de novo methylation ability is dependent on the presence of a critical level of preexisting methylation at CpG sites.
Finally, we show that methylation by Dnmt3a and/or Dnmt3b occurs close to the time of DNA replication, while Dnmt1 shows a substantial amount of delayed methylation, extending beyond 1 h post-DNA synthesis. However, this delay in maintenance methylation by Dnmt1 was not responsible for the sequence-dependent variability in methylation levels in Dnmt3a- and/or Dnmt3b-deficient cells, since both types of sequences showed this maintenance methylation delay. We conclude that the major distinction between sites that are well maintained by Dnmt1 and those that are not lies in the efficiency of postreplicative maintenance methylation efficiency by Dnmt1, rather than in a difference in de novo methylation or in delayed maintenance methylation.
MATERIALS AND METHODS
ES cell lines.
ES cell culture, transfection, and selection were carried out as described previously (18). J1 (M1/3A/3B) is a wild-type ES cell line from an inbred 129/SvJae background (18). The Dnmt1−/− (M3A/3B) and Dnmt3a−/− Dnmt3b−/− (M1) ES cells have been described previously by Okano et al. (20).
Methylation-sensitive AP-PCR.
Methylation-sensitive arbitrarily primed PCR (AP-PCR) and isolation of fragments of interest were performed as previously described (12, 19). The following CpG-poor primers were used for AP-PCR analysis: GCP1 (5′-CACATGGTTCTGC-3′), GCP2 (5′-GTCTCTATGACCC-3′), and GCP4 (5′-CTTACTGTGCCAC-3′). The following pairs were used in the AP-PCR: GCP1 and GCP2, GCP2 and GCP4, GCP1 and GCP4.
Southern blot analysis of DNA from cell lines.
Ten micrograms of DNA from cell lines was separately digested with either 50 U of RsaI, 50 U each of RsaI and HpaII, or 50 U each of RsaI and MspI (Roche) at 37°C for 16 h. Digested genomic DNA was electrophoresed on a 0.7% agarose gel and Southern transferred to Zeta-Probe (Bio-Rad) membranes overnight. Cloned DNA fragments that had previously been isolated from AP-PCR polyacrylamide gels were subsequently used as probes for hybridization to these filters. Approximately 100 ng of plasmid insert DNA was 32 labeled by random priming and used to probe the filters. All hybridizations were performed in 500 mM NaPO4 (pH 6.8)-7% sodium dodecyl sulfate (SDS)-1 mM EDTA (pH 8.0) at 65°C for 16 h. Membranes were washed twice at 65°C with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-1% SDS, followed by three washes with 0.5× SSC-1% SDS at room temperature. They were then exposed to autoradiographic film at −80°C. Quantitation of methylation levels was performed on a Molecular Dynamics PhosphorImager.
Pulse-chase experiments.
ES cells were seeded at 9 × 105 cells per 60-mm-diameter dish with feeder cells 72 h before being pulsed for 1 h in 2.5 ml of medium containing 10−4 M bromodeoxyuridine (BrdU; Sigma). The BrdU-containing medium was then removed, and cells were first washed once with regular medium at 37°C and then chased with 4 ml of medium supplemented with 10−4 M thymidine (Sigma). Genomic DNA was obtained following lysis with 100 mM NaCl-10 mM EDTA-1% SDS1 μg of proteinase K/ml and was purified by phenol and chloroform extractions and ethanol precipitation. The yield and purity of the DNA were determined by measurement of the absorbance at 260 nm. DNA (300 μg) was digested with 3,000 U of RsaI (Roche) for 16 h at 37°C. Digested DNA was purified again by phenol and chloroform extractions, ethanol precipitated, and dissolved in 500 μl of Tris-EDTA (TE) buffer (pH 7.5) with 50 μl of 10× immunoprecipitation buffer (sodium phosphate [pH 7.0], 0.14 M NaCl, and 0.05% Triton X-100). The samples were denatured at 95°C for 5 min, cooled on ice for 2 min, and immediately mixed with 2.4 μl of anti-BrdU antibody (25 μg/ml) per μg of DNA (Becton Dickinson) (23). After 30 min of incubation at room temperature with mild rocking, 0.46 μl of rabbit anti-mouse immunoglobulin G (IgG) (2.6 mg/ml) per μg of DNA (Sigma) was added and incubation was continued for another 30 min at room temperature. The precipitate was collected after 5 min of centrifugation at 13,000 rpm in a cold microcentrifuge, dissolved in 200 μl of TE (pH 7.5), and treated with proteinase K at 50°C for more than 12 h. DNA was deproteinized by phenol and chloroform extractions and ethanol precipitated.
Ms-SNuPE assays.
The mean cytosine methylation levels of CpG sites in the fragment were determined by treatment of DNA (2 μg) with sodium bisulfite according to the method of Frommer et al. (9). Methylation analysis was performed using the methylation-sensitive single-nucleotide primer extension (Ms-SNuPE) assay (11). Our PCR primers were designed specifically to amplify only bisulfite-converted DNA, and control experiments showed no amplification of unconverted DNA with these primers. Also, sequencing of PCR products after bisulfite treatment showed less than 1% residual C’s at non-CpG sites, indicating that the assays were valid for methylation status at CpG sites. The sequences of the primers used for bisulfite-treated DNA amplification were as follows: for the top strand of CI-f, the 5′ primer was 5′-AATGTGTAATATTTTTATGGTTTTTTTAGAATGG-3′ and the 3′ primer was 5′-TTACAAAAAAAATACCTCTTCCTTACTAAAC-3′; for the bottom strand of CI-f, the 5′ primer was5′-GAGTAAAGATAGAATAAATTGTTTTAATTAG-3′ and the 3′ primer was 5′-CAATACCTCTATAACCCTTCCAAA-3′; for CIId, the 5′ primer was5′-GTTTATAGGTTTAGAGGTTTT-3′ and the 3′ primer was 5′-AACACATAAACCTATTTTAAACTTA-3′; and for A-repeats, the 5′ primer was 5′-TGATTTATTTATTAGAGGTTTTAGG-3′ and the 3′ primer was 5′-ACATAAAAAAACAAACTACCC-3′.
(i) PCR conditions.
For CI-f, PCR conditions were 95°C for 2 min, 95°C for 1 min, 56°C for 50 s, and 72°C for 1 min for 35 cycles, with a final extension at 72°C for 10 min. For CII-d, PCR conditions were 95°C for 2 min, 94°C for 1 min, 50°C for 50 s, and 72°C for 1.5 min for 40 cycles, with a final extension at 72°C for 10 min. For A-repeats, PCR conditions were 95°C for 2 min, 94°C for 1 min, 50°C for 50 s, and 72°C for 1 min for 40 cycles, with a final extension at 72°C for 10 min.
PCR products were gel purified with the Qiaquick Gel Extraction Kit (Qiagen), and the template was resuspended in 30 μl of H2O.
(ii) Ms-SNuPE primers.
The CI-f primer was 5′-AATAATTTTGTTTTTTTGGATATT-3′ (HpaII site), and the CII-d primers were 5′-TTTTATTTATTGTTATTATGG-3′ (site 1), 5′-GGTATAGTTTGAGTAT-3′ (site 2), and 5′-TATTTTTTAATAGTATTATTTTTTAT-3′ (site 3, HpaII site).
Ms-SNuPE reactions were performed in a 10-μl total volume under the following conditions: 4 μl of Qiaquick product, 20 mM Tris-HCl (pH 7.5), 2.5 mM MgCl2, 100 mM KCl, 0.5 μM (final concentration) each primer, and 1 μCi of either [32P]dCTP or [32P]dTTP. Primer extension conditions for CI-f and CII-d were 95°C for 1 min, 46°C for 30 s, and 72°C for 20 s. Reaction mixtures were combined with 4 μl of stop solution before being denatured at 95°C for 5 min and loaded onto a 15% denaturing polyacrylamide gel (7 M urea). Quantitation of methylation levels was performed on a Molecular Dynamics PhosphorImager.
Bisulfite genomic sequencing by Ms-SNuPE or automated DNA sequencer.
Traditionally, patterns are determined by the cloning of individual bisulfite-treated molecules followed by DNA sequencing. We found it more convenient to subject individually cloned molecules to Ms-SNuPE analysis in order to rapidly assess how the four sites were methylated (CI-f and CII-d). The PCR products from bisulfite-converted DNA were ligated into the pCRII cloning vector (Invitrogen, San Diego, Calif.). Individual plasmid clones were amplified by M13 primers (forward and backward). The PCR product was used for sequencing by Ms-SNuPE. Conditions were the same as those given above. The primers for sequencing were as follows: for CI-f (four sites), 5′-TATGGTTTTTTTAGAATGG-3′, 5′-AATAATTTTGTTTTTTTGG ATATT-3′, 5′-AAATTTTGTTTTTGGTTGTAAA-3′, and 5′-TTGTTGTTATGTGTAATTATTTT TT-3′; for CII-d (four sites), 5′-TTTTATTTATTGTTATTATGG-3′, 5′-GGTATAGTTTGAGTAT-3′, 5′-TTTTTTAATAAGGTTATATTTTTT-3′, and 5′-TATTTTTTAATAGTATTATTTTTTAT-3′.
A-repeat clones were sequenced by automated DNA sequencer at the University of Southern California (USC)/Norris Comprehensive Cancer Center microchemical facility.
5-Aza-CdR treatments.
Cells were plated (2 × 106 cells/60-mm dish) and treated 24 h later with 3 × 10−7 M 5-aza-CdR (Sigma). The medium was changed 24 h after drug treatment and on every subsequent day. DNA was isolated at the indicated days after treatment as described previously (10).
Hemimethylation assay.
We took advantage of the fact that the enzyme HpaII will not cut the sequence CCGG in either the fully methylated or the hemimethylated configuration with bisulfite treatment to determine whether unmethylated cytosine occurred in the context of a CpG sequence in an HpaII-insensitive site. Two ∼4-μg DNAs were digested by RsaI and HpaII (10 U per μg) for 16 h at 37°C, and then HpaII was added (10 U per μg) for another 2 h. The digested DNA was then analyzed for methylation at the HpaII site by cloning of individual PCR-amplified bisulfite-treated molecules as described above.
Methylation equations.
The relative amounts of full methylation, hemimethylation, and lack of methylation of a CpG dinucleotide within an HpaII site can be derived from two measurable variables: S, which stands for the percent unmethylated cytosines found by bisulfite analysis of one of the two single DNA strands after HpaII digestion, and P, which stands for the percent methylated cytosines found by bisulfite analysis of one of the two DNA strands without prior HpaII digestion. First, the percent HpaII-resistant molecules with double-stranded (full) methylation (D) is calculated by the equation D = 1 − 2S. In this equation, it is assumed that the top and bottom strands are equal in their methylation levels and rates. We provide evidence for this in our control experiments (Fig. 3). On average, differences between the newly synthesized strand and the daughter strand after DNA replication should be distributed equally between the top and bottom strands in a large population of cells. We can then calculate the amount of hemimethylation (H) by the equation H = 2SP/(S + D) and the amount of full methylation (F) by the equation F = DP/(S + D). The basis for these two equations is the assumption that the ratio between hemimethylation and full methylation is constant for a particular DNA sample. Therefore, this ratio, which can be determined from the measurement of S (but which does not supply information on unmethylated DNA), can be applied to the measurement of all methylation within a single strand (P). The percent unmethylated CpG’s (U) can be calculated from H and F by the equation U = 100 − H − F.
FIG. 3.
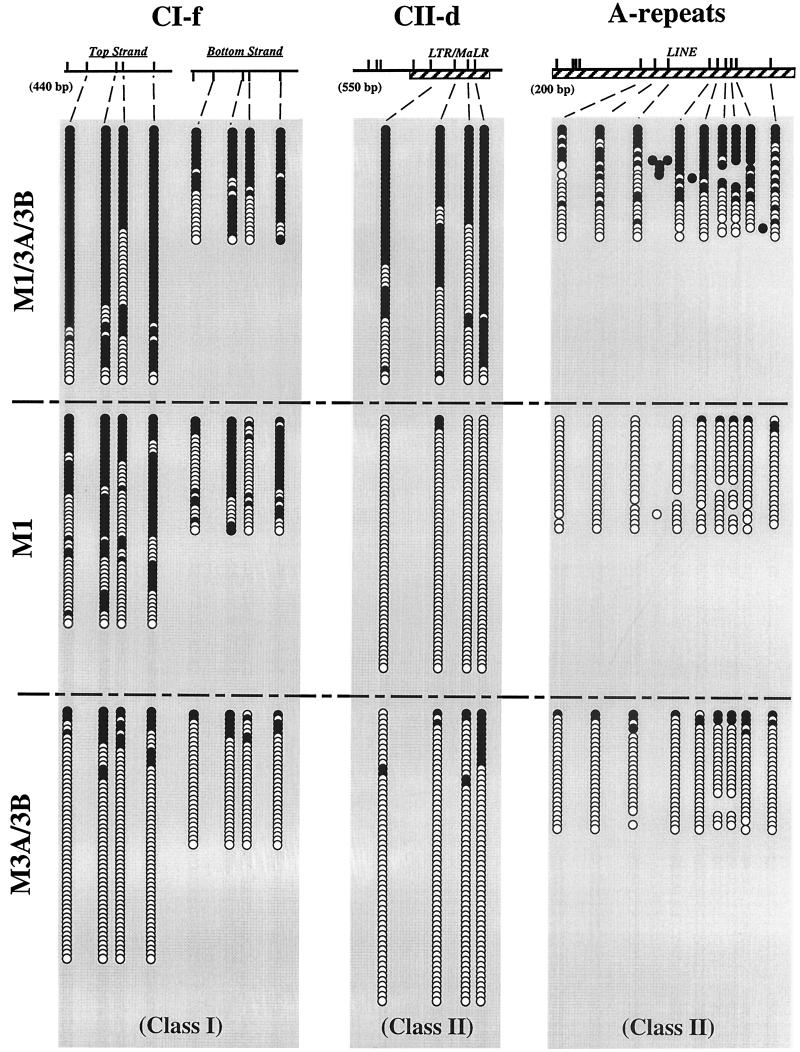
Patterns of methylation in ES cells at different regions. The methylation statuses of individual molecules of DNA from the CI-f (class I) and the CII-d and A-repeat (class II) regions were assessed by cloning of individual bisulfite-converted molecules followed by Ms-SNuPE or automated sequencing. CpG sites are represented by tick marks. Solid circles, methylated CpG dinucleotides; open circles, unmethylated CpG dinucleotides. Horizontal rows of circles indicate individual molecules that were sequenced after PCR amplification and cloning of bisulfite-treated DNA. Hatched boxes, repeat sequences.
RESULTS
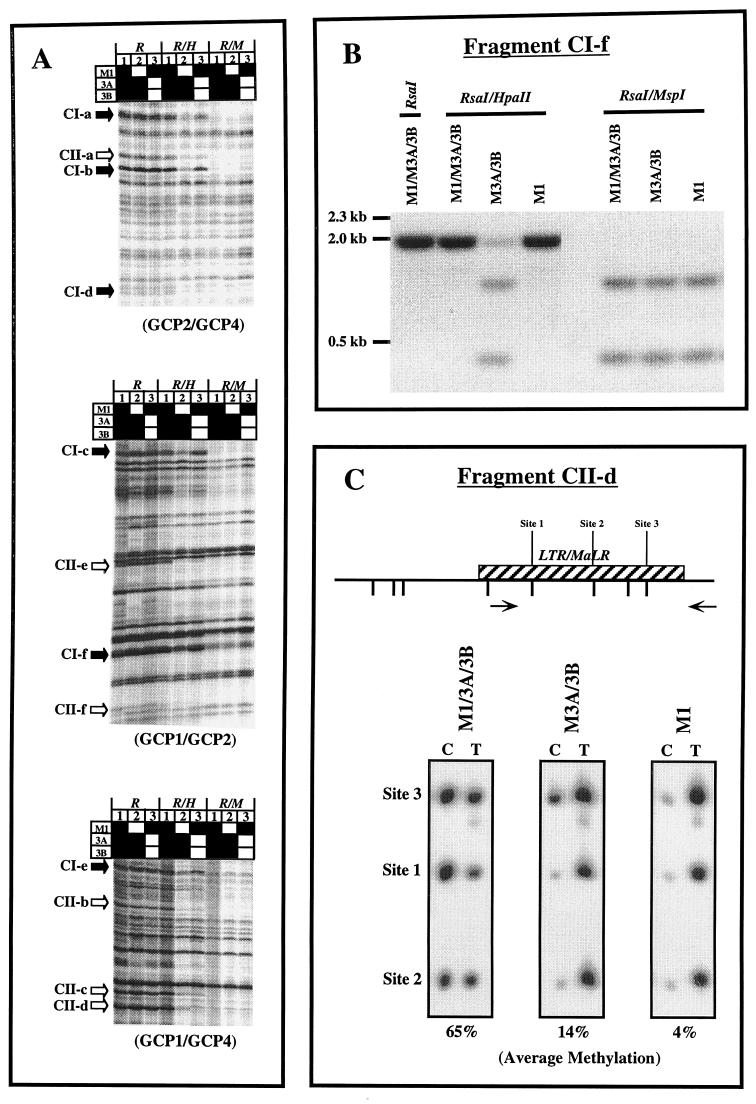
Previous studies have examined the methylation statuses of CpG islands or satellite sequences in the DNA of homozygous mutant Dnmt1−/− or Dnmt3a−/− Dnmt3b−/− ES cells (18, 20). We used methylation-sensitive (Ms) AP-PCR to fingerprint the methylation patterns in a panel of ES cells containing different combinations of DNA methyltransferases. The Ms AP-PCR method allows for a methylation pattern to be easily obtained and relies on the differential susceptibilities of unmethylated and methylated CCGG sites to cutting by the enzyme HpaII, giving a valid fingerprint of the methylation statuses of CpG islands (11, 19). The purpose and advantage of Ms AP-PCR were to perform a rapid and global screen of the genome, which would then allow us to identify representative sequences for more-detailed analysis. However, we initially focused on CpG-poor regions of DNA in the present work, since these are the regions of DNA in which the majority of 5-methylcytosine is found (5). Figure 1A shows an example of an analysis of DNA extracted from the various cell types using CpG-poor primers to target regions of DNA not located in CpG islands. The nomenclature we have used for the cells was selected to focus on the gene products that were active in the cells rather than on those that were absent. Analysis of the fingerprints showed a uniform loss of methylation at most evaluable bands in cells which contained Dnmt3a and Dnmt3b only (M3A/3B), compared to the other cell types examined (Fig. 1A; see lanes 2 and bands indicated by solid arrows). The decreased intensities in the Rsa/HpaII digests of DNA from these cells suggested a generalized decrease in methylation in cells without a functional Dnmt1 enzyme and a lack of marked sequence specificity of the retained Dnmt3a and -3b, since no bands were apparent which were completely methylated or unmethylated relative to the wild-type or mutant cell types. An important distinction was observed in cells which contained Dnmt1 only (M1 cells; Fig. 1A, lane 3). Their methylation pattern was similar to that of the wild-type cells for many of the bands. However, in 6 of 12 evaluable bands, clearly decreased methylation was apparent (indicated by open arrows). This contrasted with the situation in the M3A/3B cells, for which a generalized decrease was observed in all sequences examined. Since the AP-PCR method is only semiquantitative, we assessed the levels of methylation of some of the bands with decreased intensities on AP-PCR gels, using either Southern blot analysis or Ms-SNuPE analysis (Fig. 1B and C). Results confirmed the decreases apparent from the AP-PCR gels, thus validating the approach.
FIG. 1.
(A) Methylation-sensitive fingerprints of M1/3A/3B, M3A/3B, and M1 ES cell DNA after enzyme digestion with either RsaI, RsaI with HpaII, or RsaI with MspI for 16 h. Bands which appear to be hypomethylated only in lane 2 are indicated by solid arrows (class I). Bands which appear to be hypomethylated in both lanes 2 and 3 are indicated by open arrows (class II). Solid squares, functional Dnmt genes; open squares, inactivated genes. GCP1, -2, and -4 are GC-poor primers. (B) Southern blot analysis of genomic DNA from M1/3A/3B, M3A/3B, and M1 ES cells using isolated AP-PCR fragment CI-f as a probe. (C) Ms-SNuPE analysis for fragment CII-d from M1/3A/3B, M3A/3B, and M1 ES cell DNA. Quantitative methylation analysis of three CpG sites in CII-d was performed. The percent methylation given under each gel represents the average obtained for the three sites by using the following equation with PhosphorImager quantitation results: [C/(C + T)] × 100, where C is methylated and T is unmethylated. CpG sites are labeled; hatched boxes, repetitive sequences; arrows, PCR primers.
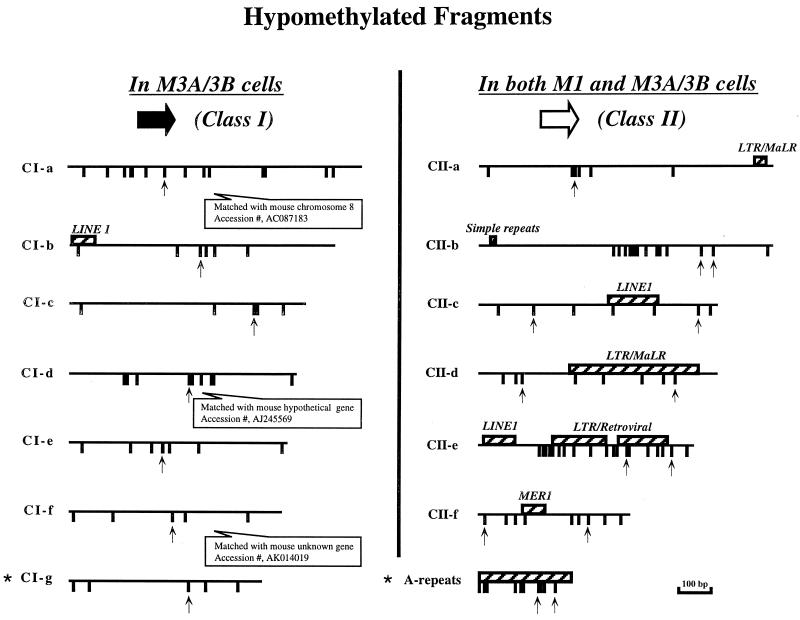
There were, therefore, two classes of fragments visible in the fingerprints. We defined class I sequences as those that had decreased methylation in M3A/3B cells (i.e., cells lacking Dnmt1) but close to normal methylation levels in M1 cells. Class II sequences, on the other hand, showed loss of methylation in both M1 and M3A/3B cells. Figure 2 shows the sequence properties of a selection of these two classes of fragments with respect to the occurrence of the CpG dinucleotides and the presence of repetitive elements. Class I sequences tended to have fewer repetitive elements, and a slightly lower CpG density, than class II sequences. Clearly, this distinction was not absolute, since there were no obvious differences with respect to these properties between fragments CI-b and CII-a, or CI-a and CII-b.
FIG. 2.
Sequence properties of hypomethylated fragments isolated from methylation-sensitive fingerprints in ES cells (see Fig. 1A). CpG sites are represented by tick marks; hatched boxes, repetitive sequences; upward-pointing arrows, HpaII sites. Stars indicate bands that are not included in Fig. 1A.
Methylation patterns on individual DNA molecules.
The analyses described above provide insight into the levels of methylation at individual CpG dinucleotides, but they do not yield information on the patterns of methylation in these regions. The sequencing of cloned PCR products of bisulfite-treated genomic DNA can reveal the patterns of methylation in individual DNA molecules. Therefore, we performed such an analysis on one of the class I sequences and two of the class II sequences. These experiments clearly illustrated the differences in methylation levels in the different knockout cells (Fig. 3). The sequence of the 440-bp fragment CI-f showed extensive methylation of all four sites sequenced in the wild-type cells and extensive methylation also in the M1 cells, but a marked decrease in methylation in M3A/3B cells on both the top and bottom strands, which had approximately equal methylation levels at the CpG sites in all cell types. Analysis of the top strands of individual molecules in the M3A/3B cells showed a strong asymmetry of patterns in that 72% of molecules were completely unmethylated, 14% had only one site methylated, but 14% had multiple sites methylated on a single molecule. The differential levels of methylation at the four sites made it difficult to distinguish between the possibility that the enzymes act processively and the possibility that methylation at one site increases the probability of methylation of adjacent sites.
Figure 3 also shows the dramatic decrease in the level of methylation of the CII-d fragment containing an LTR/MALR element in both the M1 and M3A/3B cells. There was substantial methylation in the wild-type cells but virtually no methylation in the M1 cells and low levels of methylation in M3A/3B cells. However, the data clearly showed that there was little methylation of the four sites in this fragment in the M1 cells, in contrast to the CI-f fragment, which still showed substantial levels of methylation in M1 cells. A similar situation was also observed in the A-repeats (i.e., the promoters) of LINE-1 repetitive element, which previous studies have shown to be methylated in M3A/3B cells (27). Once again, virtually no methylation occurred in M1 cells, and slightly higher levels of methylation were seen in M3A/3B cells.
Since Dnmt1 is thought to be the major cytosine-5 DNA methyltransferase in mammals, responsible for most, if not all, maintenance activity in the cell, the identification of a class of sequences that appeared to be poorly maintained by Dnmt1 alone was interesting. Dnmt3a or -3b did not seem to be independently responsible for the methylation of these sites, since these sequences were also substantially demethylated in M3A/3B cells (Fig. 1 and 3). Furthermore, the effects of Dnmt1 and Dnmt3a or -3b did not appear to be merely additive (Fig. 3), as would be the case if both Dnmt1 and Dnmt3a or -3b were independent participants in the maintenance and de novo methylation of these sequences. Rather, methylation of these sequences appears to require cooperation of both Dnmt1 and Dnmt 3a or -3b activities. The most straightforward interpretation of these data is that Dnmt3a and/or -3b is necessary for ongoing de novo methylation of these sequences to compensate for poor maintenance methylation by Dnmt1. If either of these activities is absent, the methylation of these sequences cannot be properly maintained.
A new assay for the detection of hemimethylation.
One prediction of poor maintenance methylation of class II sequences, offset by continuing de novo methylation, is that the level of hemimethylation at these sites (i.e., when only a single cytosine in the palindrome is methylated) should be substantially higher than in sequences with low levels of de novo methylation and very efficient maintenance methylation following each round of DNA replication.
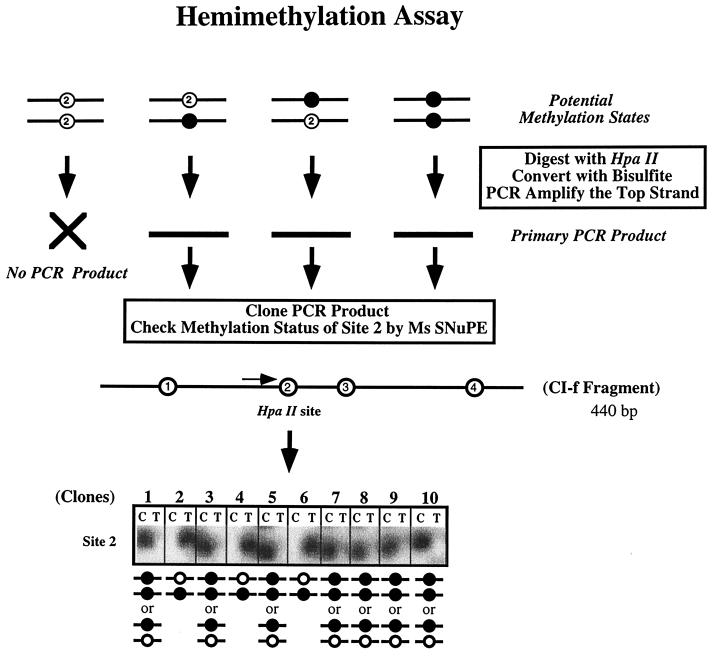
Testing of this prediction required the development of a way to measure levels of hemimethylation at individual CpG dinucleotides. Bisulfite sequencing has opened the door for the detailed analysis of methylation patterns (9), yet it has not heretofore been possible to assess the degree of hemimethylation because the parental paired DNA strands are separated before analysis. We took advantage of the fact that the enzyme HpaII will not cut the sequence CCGG in either the fully methylated or the hemimethylated configuration (13) with bisulfite treatment to determine whether the unmethylated cytosine occurred in the context of a CpG sequence in an HpaII-resistant site (see Fig. 4 for a schematic). DNA extracted from the various cell types was digested with an excess of HpaII, bisulfite treated, and then amplified by PCR with primers flanking the HpaII site. Since the HpaII site is located between the primers, molecules without full methylation or hemimethylation of the site to be interrogated would not be amplified and thus would be excluded from the analysis. The degree of hemimethylation can be accurately determined by averaging results obtained by the analysis of individual cloned PCR products for the presence of C or T at the HpaII site or directly, by Ms-SNuPE analysis.
FIG. 4.
Detection and quantitation of hemimethylation in individual cloned DNA molecules. The experimental approach used to detect hemimethylation in the ES cells consists of precutting genomic DNA with HpaII, followed by bisulfite treatment, then cloning individual PCR products, and assessing the methylation status of site 2 by Ms-SNuPE analysis. Shown are typical Ms-SNuPE results in which a signal in the T lane for a clone indicates hemimethylation, with the top strand being unmethylated. The presence of a signal in the C lane indicates that the site may be either hemimethylated (i.e., the bottom strand is unmethylated) or fully methylated. The distribution between these two scenarios is determined by calculation, assuming that there is an equal probability of hemimethylation of either the top or the bottom strand.
Figure 5 shows the results of the hemimethylation analysis of a class I sequence (CI-f) and of a class II sequence (CII-d). There was a surprisingly high level of hemimethylation of CII-d sequences in wild-type (M1/3A/3B) cells, consistent with our interpretation that the methylation of these sites is poorly maintained by Dnmt1 and requires ongoing de novo methylation mediated by Dnmt3a and/or Dnmt3b. Hemimethylation levels of CI-f were substantially lower, as would be expected for a site that is subject to efficient maintenance methylation by Dnmt1. Control experiments with these sequences in M3A/3B cells demonstrated that equal amounts of hemimethylation were present on the top and bottom strands (Fig. 5).
FIG. 5.
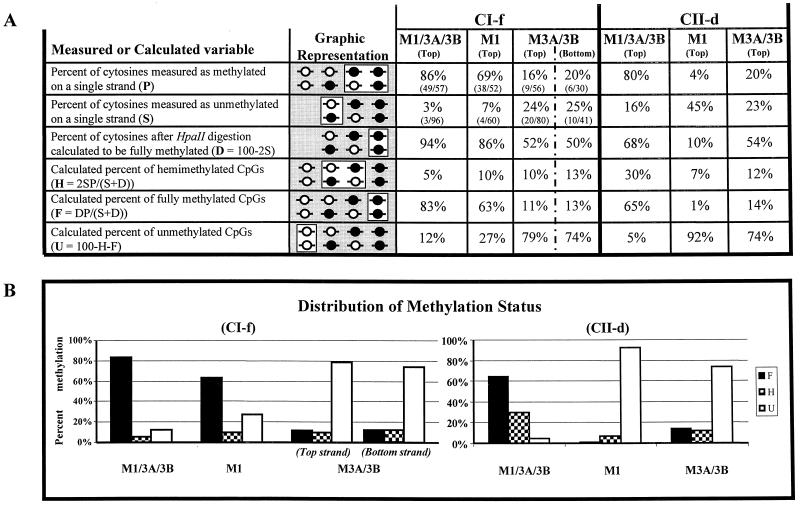
Distribution of methylation states at the CI-f and CII-d regions. (A) Overview of the procedure used to calculate distribution of methylation states (fully methylated, hemimethylated, and fully unmethylated) at a single CpG dinucleotide within an HpaII site, starting with the measurement of two experimental variables (P and S). A description of each variable is given in the leftmost column, along with the equation used to derive the calculated variables (see Materials and Methods). The second column illustrates the subset of methylation states representing that variable (open box), as well as all the other methylation states being assessed in that particular step (shaded box). The unmethylated CpG’s are absent in the cases of variables S and D, since the HpaII digestion removes these molecules from consideration at this step. The data obtained for the three cell lines for the measured variables P and S are shown in the columns on the right as percentages, followed by the absolute numbers of molecules assessed. Control experiments demonstrated that the top and bottom strands are equal in their methylation levels and rates (Fig. 3). On average, differences between the newly synthesized strand and the daughter strand after DNA replication should be distributed equally between the top and bottom strands in a large population of cells. (B) Distribution of methylation states of CI-f and CII-d. F, fully methylated; H, hemimethylated; U, fully unmethylated.
Cooperativity between enzymes after 5-aza-2′-deoxycytidine treatment.
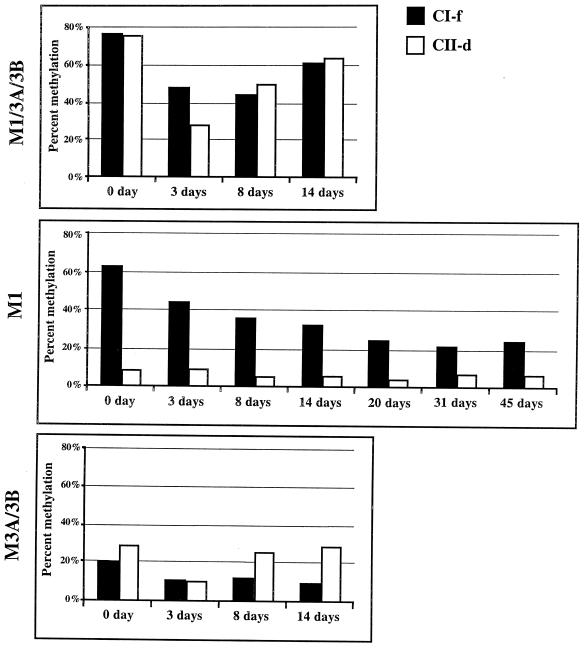
Treatment of dividing cells with the DNA methyltransferase inhibitor 5-aza-CdR results in a transient demethylation of the genome (15), followed by a rebound of methylation levels in subsequent cell divisions (4, 22). Since such rebound methylation requires de novo methylation activity, this type of analysis can provide an independent assessment of the de novo methylation capabilities of the various DNA methyltransferase enzymes at particular regions. This could lend further support to our proposition that Dnmt3a and/or Dnmt3b is responsible for ongoing de novo methylation of class II sequences. We therefore exposed the knockout cells to 5-aza-CdR and monitored the kinetics of remethylation of the CI-f and CII-d fragments to determine how the DNA methyltransferases interacted to restore methylation levels after perturbation of the equilibrium in the ES cell types (Fig. 6). The results showed that methylation levels reached their minimum 3 days after drug treatment in the wild type and that the methylation of both fragments was restored to near pretreatment levels by 14 days after treatment. Figure 6 also shows that the combination of Dnmt3a with -3b (M3A/3B) was sufficient to result in the rapid remethylation of the CII-d but not the CI-f fragment.
FIG. 6.
Recovery of methylation after 5-aza-CdR treatment. The indicated cell types were treated for 24 h with 3 × 10−7 M 5-aza-CdR; then the medium was changed, and the cells were propagated further. DNA was extracted at the times indicated after the drug was added to the cultures, and the methylation statuses of the CI-f and CII-d fragments were measured by Ms-SNuPE analysis. Results shown are average values from two separate experiments.
Perhaps the most surprising result was the failure of the Dnmt1 enzyme by itself to restore the methylation of either fragment after transient drug treatment (Fig. 6). The M1 cells showed a continued loss of methylation after treatment, suggesting that once the equilibrium had been perturbed below a certain level by 5-aza-CdR, the Dnmt1 by itself or an as-yet-unidentified methyltransferase had insufficient de novo methylation activity for the restoration of methylation to pretreatment levels. In contrast, the combined activities of Dnmt3a and Dnmt3b resulted in the full restoration of pretreatment levels of methylation at the CII-d sequence. These results suggest that the CII-d sequence is an efficient target for de novo methylation by Dnmt3a and/or Dnmt3b but not by Dnmt1. This fits our model in which the ongoing de novo methylating activity of Dnmt3a and/or 3b compensates for poor maintenance of this sequence by Dnmt1.
Kinetics of methylation of newly synthesized DNA.
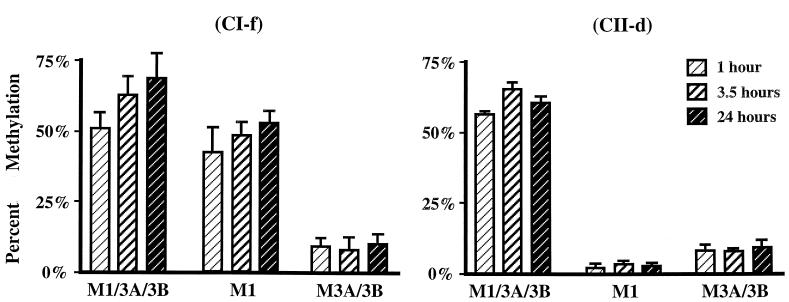
The balance of poor maintenance methylation of class II sequences, with a compensating de novo methylation by Dnmt3a and/or Dnmt3b, raises the question whether both of these methylation activities occur concurrently immediately postreplication, or whether the de novo methylation occurs at some other time in the cell cycle. We used a 1-h BrdU pulse followed by chasing for various times to investigate the timing of methylation with respect to synthesis of the two DNA sequence classes. DNA containing BrdU was isolated by immunoprecipitation (23), and the methylation statuses of CpG sites within the fragments were measured by cloning of individual DNA molecules and quantitation of methylation levels or by Ms-SNuPE analysis. Figure 7 summarizes results from such pulse-chase experiments, which show that DNA synthesized during a 1-h pulse of BrdU in wild-type (M1/3A/3B) cells was not methylated to its final level immediately after synthesis but that some further methylation occurred in the postsynthetic phase measured at 3.5 and 24 h. However, this did not appear to be de novo methylation attributable to Dnmt3a and/or Dnmt3b, since M3A/3B cells did not show additional methylation after the first hour. Indeed, the delayed increase in methylation was also evident for the CI-f sequence in M1 cells. The demonstration that Dnmt1 is localized to the replication machinery (16) in the S-phase nucleus and interacts with PCNA (7) has led to the expectation that the methylation pattern is copied immediately after synthesis (2). However, earlier studies suggested that some methylation might be delayed, with cytosine methylation occurring in a biphasic fashion (1, 26, 28). Our results are consistent with these earlier studies. We conclude that most methylation by Dnmt3a and/or Dnmt3b occurs immediately before or soon after DNA synthesis, while Dnmt1 continues to display maintenance activity more than 1 h postreplication.
FIG. 7.
Methylation kinetics of newly synthesized DNA at CI-f and CII-d. ES cells containing the genes for the indicated DNA methyltransferases were pulsed for 1 h with BrdU. The pulse was then removed, and fresh medium was added to the cells during the chase period. DNA was extracted from the cells at various times after the pulse began and was immunoprecipitated to isolate the BrdU-containing DNA. The methylation statuses of HpaII sites in CI-f and CII-d at the indicated time points were determined by quantitative Ms-SNuPE analysis. Data are mean values from two to six experiments; error bars, standard deviations.
DISCUSSION
Our experiments using ES cells with systematic knockouts of the three known active DNA methyltransferases have revealed a remarkable cooperativity in the activities of these enzymes in maintaining methylation patterns of specific sites during cell division. Studies using cell lysates (21, 30) have shown that all three enzymes are capable of methylating unmethylated and hemimethylated substrates, making it difficult to discern the relative contributions of each of the enzymes in pattern maintenance in living cells. Previous experiments using cells with disabled genes for the various enzymes have focused mainly on the methylation statuses of CpG islands in the various cell types. Since the majority of methylation occurs in CpG-depleted regions (5), we have included analysis of CpG-poor regions in addition to the CpG-rich A-repeats for this study. Our results showed that Dnmt1 by itself is capable of maintaining the methylation status in some CpG-poor class I sequences with a fairly high degree of fidelity, but not at other sequences, which we named class II. Class II sequences seem to have a higher proportion of associated repetitive elements, but this distinction is not absolute, and they resemble Igf-2 and Xist in their behavior in M1 cells (20). We performed additional studies to investigate the nature of the difference between class I and class II sequences in more detail by selecting a representative region from each of the two classes and analyzing the methylation patterns on individual molecules. In addition, we developed a novel assay to measure the level of hemimethylation at these regions and also analyzed the kinetics of methylation at these sites following DNA replication.
Methylation of the class II retroviral sequence CII-d and the type A repeats in the LINE elements was very poorly maintained by Dnmt1 alone, or by Dnmt3a and Dnmt3b without Dnmt1. Nevertheless, the CII-d sequence was highly methylated when Dnmt3a and -3b were present in addition to Dnmt1. Indeed, the level of methylation in wild-type cells (80%) far exceeded the sum of the individual contributions by Dnmt1 (4%) and Dnmt3a and -3b (20%) (see Fig. 5A). This suggests a strong cooperativity between Dnmt3a and/or -3b and Dnmt1 in ensuring the methylation of these sequences. A steady-state level of methylation of 80% can be maintained either by very efficient postreplication maintenance methylation, with relatively little de novo methylation activity necessary, or by less-efficient maintenance, balanced by substantial, continuing de novo methylation of the region. The strong synergy observed between Dnmt1 and Dnmt3a and/or Dnmt3b suggested that the latter situation may be the case for the CII-d region. One way to distinguish between efficient maintenance methylation of this region and poor maintenance combined with de novo methylation was to determine the level of hemimethylation at this region.
However, no method to analyze the levels of hemimethylation at individual regions in the genome existed. We developed such an assay for the purpose of this analysis, by taking advantage of the fact that the restriction enzyme HpaII does not cut hemimethylated DNA. Therefore, the combination of resistance to HpaII cutting, together with a lack of detectable methylation on one strand in a bisulfite analysis, is indicative of a hemimethylated state at that CpG dinucleotide in the original double-stranded DNA. An unmethylated CpG dinucleotide would be cut by HpaII, while a fully methylated CpG dinucleotide would reveal the presence of methylation in the bisulfite analysis.
This technique that we developed to measure hemimethylation was successfully applied to the analysis of both the CI-f region and the CII-d region, and it showed that the latter had considerable levels of hemimethylation in wild-type cells. As a result of this high level of hemimethylation, it appears that our original determination of 80% methylation at this region in wild-type cells, based on the analysis of a single DNA strand, was actually an underestimate. Only 5% of CpG dinucleotides at this region were completely devoid of DNA methylation (see Fig. 5A). We conclude that the high level of hemimethylation at this region (30%) must reflect poor maintenance methylation, compensated by a substantial amount of ongoing de novo methylation.
Dnmt3a and/or -3b is most likely responsible for this de novo methylation in wild-type cells. This conclusion was supported by our demethylation experiments conducted with 5-aza-CdR in which Dnmt1, by itself, was unable to restore methylation in treated cells. Indeed, the methylation level continued to decrease long after drug treatment, suggesting that once the equilibrium had been reduced below a certain level, the enzyme was incapable of sufficient de novo methylation to keep the sequences methylated. Thus, a major function of the Dnmt3a and -3b enzymes, in addition to de novo methylation of incoming retroviral sequences (20), may be to methylate endogenous repetitive DNAs so that an equilibrium level of methylation can be maintained within this sequence class. However, since some retroviral sequences and IAP sequences have been shown to be quite well maintained for methylation in M1 cells (20), Dnmt1 may have sequence specificity with respect to de novo and maintenance methylation of some repetitive DNAs. It is also possible that some unidentified DNA methyltransferases may have sequence specificity instead of Dnmt1. As Yoder and Bestor (29) have pointed out, the majority of methylation in the eukaryotic genome is found in parasitic sequences, making the complementary activities between the enzymes important in maintaining the transcriptional suppression of some of these potentially transposable elements.
Our experiments were conducted with ES cells known to express increased levels of Dnmt3a and Dnmt3b relative to their differentiated counterparts (21), so that the relevance of our findings to the maintenance of methylation in somatic cells is an issue. Since expression of Dnmt3a and -3b mRNA can easily be detected in normal fetal and adult human tissues (8, 24), in addition to mouse tissues (21), it seems likely that the two enzymes could play similar roles in somatic cells even though they appear to be downregulated during differentiation in vitro. For the above reasons, it will be important to conduct similar experiments in other suitable knockout cell types as they become available.
Our findings regarding the delayed methylation of sequences may have importance in understanding how the sequences are methylated in normal cells and cancer cells. We found that Dnmt1 acted in a biphasic mechanism with respect to the timing of methylation, with up to 10 to 20% of the methylation catalyzed by this enzyme being delayed for some time following DNA synthesis. On the other hand, the Dnmt3a and -3b enzymes appeared to act mainly at the time of replication, since little delayed methylation occurred on DNA which was synthesized in cells lacking Dnmt1. Our data show that the majority of Dnmt1-dependent methylation is concomitant with DNA replication, yet a wave of postreplication methylation is also observed. It will be of obvious importance to probe the exact timing by which these enzymes operate in the cell cycle, in order to understand in more detail how methylation patterns are copied and propagated within eukaryotic cells. Conceivably, Dnmt3a and -3b act immediately before Dnmt1, although it remains to be seen whether this occurs prior to the movement of the parental DNA through the replication machinery or whether the enzymes act on the newly synthesized DNA to cause de novo methylation of sequences unmethylated in previous cell divisions.
The fact that the methylation of sequences in CpG-poor class I DNA sequences (such as CI-f) was maintained in cells lacking Dnmt3a and -3b strongly suggests that Dnmt1 or an unidentified enzyme has some de novo methylation activity toward this sequence class. Otherwise, sites inadvertently left unmethylated in a given S phase would remain unmethylated in the next S phase, resulting in the inexorable loss of methylation with increasing numbers of cell divisions (22). However, in this regard it was of obvious interest that in the 5-aza-CdR experiments, Dnmt1 by itself could not maintain the methylation of class I sequences once the level of 5-methylcytosine had been reduced below a certain point. This resulted in the continued loss of methylation with increasing cell division. The fact that methylation was maintained at wild-type levels suggests that a significant function of the Dnmt3a and/or -3b enzyme is to restore the methylation of sites “skipped over” by Dnmt1 during the previous round of DNA replication. When these results are coupled with our observations that individual DNA strands tend to have multiple sites methylated and that preexisting methylation encourages de novo methylation by Dnmt1 (6), the most likely explanation for the maintenance failure is that the existence of methylation in sequences reinforces its propagation. For some reason Dnmt1 alone cannot keep pace with the requirement of de novo methylation of repetitive DNA sequences, indicating that a major function of Dnmt3a and/or Dnmt3b in ES cells is to ensure that this sequence class is kept modified in the genome.
Acknowledgments
Gangning Liang and Matilda F. Chan contributed equally to this work.
REFERENCES
- 1.Adams, R. L. 1971. The relationship between synthesis and methylation of DNA in mouse fibroblasts. Biochim. Biophys. Acta 254:205–212. [DOI] [PubMed] [Google Scholar]
- 2.Araujo, F. D., J. D. Knox, M. Szyf, G. B. Price, and M. Zannis-Hadjopoulos. 1998. Concurrent replication and methylation at mammalian origins of replication. Mol. Cell. Biol. 18:3475–3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baylin, S. B., and J. G. Herman. 2000. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 16:168–174. [DOI] [PubMed] [Google Scholar]
- 4.Bender, C. M., M. L. Gonzalgo, F. A. Gonzales, C. T. Nguyen, K. D. Robertson, and P. A. Jones. 1999. Roles of cell division and gene transcription in the methylation of CpG islands. Mol. Cell. Biol. 19:6690–6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bird, A., M. Taggart, M. Frommer, O. J. Miller, and D. Macleod. 1985. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell 40:91–99. [DOI] [PubMed] [Google Scholar]
- 6.Carotti, D., S. Funiciello, F. Palitti, and R. Strom. 1998. Influence of pre-existing methylation on the de novo activity of eukaryotic DNA methyltransferase. Biochemistry 37:1101–1108. [DOI] [PubMed] [Google Scholar]
- 7.Chuang, L. S., H. I. Ian, T. W. Koh, H. H. Ng, G. Xu, and B. F. Li. 1997. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science 277:1996–2000. [DOI] [PubMed] [Google Scholar]
- 8.Eads, C. A., K. D. Danenberg, K. Kawakami, L. B. Saltz, P. V. Danenberg, and P. W. Laird. 1999. CpG island hypermethylation in human colorectal tumors is not associated with DNA methyltransferase overexpression. Cancer Res. 59:2302–2306. [PubMed] [Google Scholar]
- 9.Frommer, M., L. E. McDonald, D. S. Millar, C. M. Collis, F. Watt, G. W. Grigg, P. L. Molloy, and C. L. Paul. 1992. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. USA 89:1827–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez-Zulueta, M., C. M. Bender, A. S. Yang, T. Nguyen, R. W. Beart, J. M. Van Tornout, and P. A. Jones. 1995. Methylation of the 5′ CpG island of the p16/CDKN2 tumor suppressor gene in normal and transformed human tissues correlates with gene silencing. Cancer Res. 55:4531–4535. [PubMed] [Google Scholar]
- 11.Gonzalgo, M. L., and P. A. Jones. 1997. Rapid quantitation of methylation differences at specific sites using methylation-sensitive single nucleotide primer extension (Ms-SNuPE). Nucleic Acids Res. 25:2529–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzalgo, M. L., G. Liang, C. H. Spruck III, J.-M. Zingg, W. M. Rideout III, and P. A. Jones. 1997. Identification and characterization of differentially methylated regions of genomic DNA by methylation-sensitive arbitrarily primed PCR. Cancer Res. 57:594–599. [PubMed] [Google Scholar]
- 13.Gruenbaum, Y., H. Cedar, and A. Razin. 1981. Restriction enzyme digestion of hemimethylated DNA. Nucleic Acids Res. 9:2509–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones, P. A., and P. W. Laird. 1999. Cancer epigenetics comes of age. Nat. Genet. 21:163–167. [DOI] [PubMed] [Google Scholar]
- 15.Jones, P. A., and S. M. Taylor. 1980. Cellular differentiation, cytidine analogs and DNA methylation. Cell 20:85–93. [DOI] [PubMed] [Google Scholar]
- 16.Leonhardt, H., A. W. Page, H.-U. Weier, and T. H. Bestor. 1992. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell 71:865–873. [DOI] [PubMed] [Google Scholar]
- 17.Li, E., C. Beard, and R. Jaenisch. 1993. Role for DNA methylation in genomic imprinting. Nature 366:362–365. [DOI] [PubMed] [Google Scholar]
- 18.Li, E., T. H. Bestor, and R. Jaenisch. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69:915–926. [DOI] [PubMed] [Google Scholar]
- 19.Liang, G., C. E. Salem, M. C. Yu, H. D. Nguyen, F. A. Gonzales, T.-D. T. Nguyen, P. W. Nichols, and P. A. Jones. 1998. DNA methylation differences associated with tumor tissues identified by genome scanning analysis. Genomics 53:260–268. [DOI] [PubMed] [Google Scholar]
- 20.Okano, M., D. W. Bell, D. A. Haber, and E. Li. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99:247–257. [DOI] [PubMed] [Google Scholar]
- 21.Okano, M., S. Xie, and E. Li. 1998. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 19:219–220. [DOI] [PubMed] [Google Scholar]
- 22.Pfeifer, G. P., S. D. Steigerwald, R. S. Hansen, S. M. Gartler, and A. D. Riggs. 1990. Polymerase chain reaction-aided genomic sequencing of an X chromosome-linked CpG island: methylation patterns suggest clonal inheritance, CpG site autonomy, and an explanation of activity state stability. Proc. Natl. Acad. Sci. USA 87:8252–8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rideout, W. M. III, P. Eversole-Cire, C. H. Spruck III, C. M. Hustad, G. A. Coetzee, F. A. Gonzales, and P. A. Jones. 1994. Progressive increases in the methylation status and heterochromatinization of the myoD CpG island during oncogenic transformation. Mol. Cell. Biol. 14:6143–6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robertson, K. D., E. Uzvolgyi, G. Liang, C. Talmadge, J. Sumegi, F. A. Gonzales, and P. A. Jones. 1999. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 27:2291–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warnecke, P. M., D. Biniszkiewicz, R. Jaenisch, M. Frommer, and S. J. Clark. 1998. Sequence-specific methylation of the mouse H19 gene in embryonic cells deficient in the Dnmt-1 gene. Dev. Genet. 22:111–121. [DOI] [PubMed] [Google Scholar]
- 26.Woodcock, D. M., J. K. Adams, and I. A. Cooper. 1982. Characteristics of enzymatic DNA methylation in cultured cells of human and hamster origin, and the effect of DNA replication inhibition. Biochim. Biophys. Acta 696:15–22. [DOI] [PubMed] [Google Scholar]
- 27.Woodcock, D. M., M. E. Linsenmeyer, and W. D. Warren. 1998. DNA methylation in mouse A-repeats in DNA methyltransferase-knockout ES cells and in normal cells determined by bisulfite genomic sequencing. Gene 206:63–67. [DOI] [PubMed] [Google Scholar]
- 28.Woodcock, D. M., D. L. Simmons, P. J. Crowther, I. A. Cooper, K. J. Trainor, and A. A. Morley. 1986. Delayed DNA methylation is an integral feature of DNA replication in mammalian cells. Exp. Cell. Res. 166:103–112. [DOI] [PubMed] [Google Scholar]
- 29.Yoder, J. A., and T. H. Bestor. 1998. A candidate mammalian DNA methyltransferase related to pmt1p of fission yeast. Hum. Mol. Genet. 7:279–284. [DOI] [PubMed] [Google Scholar]
- 30.Yoder, J. A., N. S. Soman, G. L. Verdine, and T. H. Bestor. 1997. DNA (cytosine-5)-methyltransferases in mouse cells and tissues. Studies with a mechanism-based probe. J. Mol. Biol. 270:385–395. [DOI] [PubMed] [Google Scholar]
- 31.Yoder, J. A., C. P. Walsh, and T. H. Bestor. 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13:335–340. [DOI] [PubMed] [Google Scholar]