Abstract
The Caenorhabditis elegans run gene encodes a Runt domain factor. Runx1, Runx2, and Runx3 are the three known mammalian homologs of run. Runx1, which plays an essential role in hematopoiesis, has been identified at the breakpoint of chromosome translocations that are responsible for human leukemia. Runx2 plays an essential role in osteogenesis, and inactivation of one allele of Runx2 is responsible for the human disease cleidocranial dysplasia. To understand the role of run in C. elegans, we used transgenic run::GFP reporter constructs and a double-stranded RNA-mediated interference method. The expression of run was detected as early as the bean stage exclusively in the nuclei of seam hypodermal cells and lasted until the L3 stage. At the larval stage, expression of run was additionally detected in intestinal cells. The regulatory elements responsible for the postembryonic hypodermal seam cells and intestinal cells were separately located within a 7.2-kb-long intron region. This is the first report demonstrating that an intron region is essential for stage-specific and cell type-specific expression of a C. elegans gene. RNA interference analysis targeting the run gene resulted in an early larva-lethal phenotype, with apparent malformation of the hypodermis and intestine. These results suggest that run is involved in the development of a functional hypodermis and gut in C. elegans. The highly conserved role of the Runt domain transcription factor in gut development during evolution from nematodes to mammals is discussed.
The PEBP2/CBF (polyomavirus enhancer-core binding protein 2/core binding factor) family of transcription factors is composed of two subunits, α and β (3, 5, 33, 34, 44). The α subunit contains an evolutionarily conserved 128-amino-acid region that harbors two different activities, the ability to bind DNA and the ability to interact with the β subunit. This conserved region has been named the Runt domain (20) because Drosophila runt (21) is the first identified gene which contains the conserved region. The function of Runt was initially characterized as one of the pair-rule genes involved in segmentation (15). Further studies have demonstrated that runt has roles in two other developmental processes in Drosophila, sex determination and neurogenesis (11, 12). Another Drosophila Runt domain-encoding gene, Lozenge, is involved in prepatterning photoreceptor precursor cells in the developing eye and is required in hematopoiesis for the specification of blood cell lineage (9, 10).
In mammals there are three PEBP2α genes, Runx1/PEBP2αB, /CBFA2/AML1 (2, 30), Runx2/PEBP2αA/CBFA1 (34), and Runx3/PEBP2αC/CBFA3 (4, 27). Recent work has identified important roles for these genes in hematopoiesis and osteogenesis in mouse and human. Runx1 has been found at the breakpoint of chromosome translocations associated with human leukemia (16, 31, 38). Targeted disruption of the Runx1 gene in mice resulted in a lack of definitive hematopoiesis in the fetal liver (35, 43). More recently, it has been demonstrated that Runx2, another Runt domain-encoding gene, plays an essential role in osteogenesis. Mice with a homozygous mutation in Runx2 died just after birth because of breathing difficulties caused by a complete lack of ossification (23, 36). Mutations in the Runx2 gene have also been demonstrated to be responsible for the human disease cleidocranial dysplasia, an autosomal dominant disorder (25, 32).
The full genomic sequencing of C. elegans is essentially complete and has recently been published (1). More than 30% of the predicted genes from the genomic sequence of C. elegans have a human homolog, many of which have been implicated in human disease (13, 17). This observation, along with increasing evidence that biological processes and molecular pathways are highly conserved, has made the nematode C. elegans a model organism for the study of developmental regulation. The C. elegans homolog of Runx, run, was recently identified in the C. elegans genome database (6).
In this paper, we used run::GFP reporter constructs to generate transgenic animals and a double-stranded RNA (dsRNA)-mediated interference (RNAi) method to determine the role of run in C. elegans. Our results show that the run gene is expressed exclusively in the nuclei of hypodermal seam cells and intestinal cells. Expression was first detected at the bean stage and the late embryonic stage for hypodermal and intestinal cells, respectively, and lasted until the L3 stage of development. The cell type specificity and expression at specific stages of development are controlled by an unusually long intron located between exons 3 and 4. Targeted inactivation of run by RNAi resulted in an early larva-lethal phenotype, with malformation of the hypodermis and intestine.
MATERIALS AND METHODS
Materials.
Reagents were purchased from the following vendors: restriction enzymes from Takara (Tokyo, Japan); T4 DNA ligase from New England Biolabs (Beverly, Mass.); and T3 and T7 RNA polymerases from Promega (Madison, Wis.). All other chemicals of the purest grade available were obtained from commercial sources.
Worm strain and culture.
Wild-type worms were the C. elegans Bristol strain N2. Worms were grown on NGM plates as described previously (8).
Construction of run::GFP reporter genes.
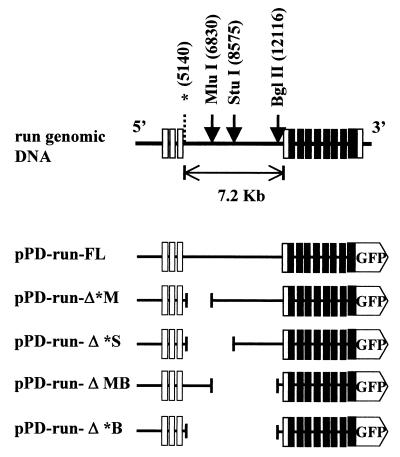
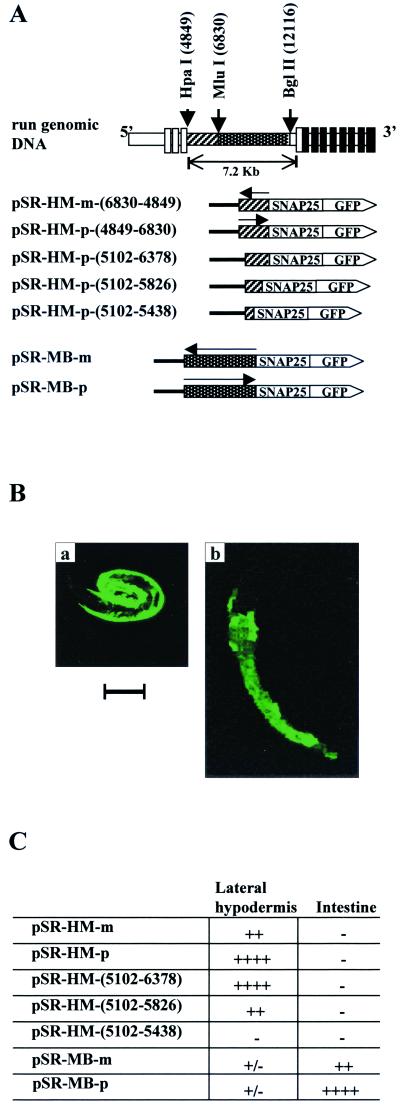
A cosmid clone, B0414, containing the entire run gene was obtained from the C. elegans genome project. To construct pPD-run-FL, a genomic DNA fragment of approximately 13 kb (nucleotides [nt] 3870 to 16791 of B0414) containing run was subcloned into the SmaI site of the reporter gene expression vector pPD95.79 (a gift from A. Fire, Carnegie Institute of Washington, Baltimore, Md.). The genomic DNA fragment included all the exons and introns of run and a 5′ region containing the adjacent 3′ noncoding region of a gene homologous to histone H1. A StuI site located six nucleotides upstream of the translation termination codon of run was used for in-frame fusion with GFP (Fig. 1).
FIG. 1.
Schematic representation of the genomic DNA structure of run and run::GFP fusion constructs. The open boxes represent noncoding exons, and the solid boxes represent coding exons (6). Restriction enzyme sites used for the deletion constructs are indicated. An asterisk indicates the deletion point at the 5′ end of the PCR-based deletion. Numbers in parentheses were obtained from the nucleotide sequence of the B0414 cosmid.
pPD-run-ΔMB was constructed by deleting an MluI/BglII fragment (nt 6830 to 12116) from pPD-FL. pPD-run-Δ*M was constructed by deleting nt 5140 to 6830 of B0414 from pPD-run-FL using a PCR-based deletion method (39). pPD-Δ*S and pPD-run-Δ*B were constructed by deleting MluI/StuI and MluI/BglII fragments, respectively, from pPD-run-Δ*M. The pSNAP25 plasmid contains the genomic region of C. elegans SNAP-25 from the TATA box to 13 amino acids in exon 1 inserted into the pPD95.77 GFP reporter vector. An HpaI/MluI fragment of B0414 cosmid DNA (corresponding to nt 4849 to 6830) was inserted into the SacI site of pSNAP25 to generate sense-oriented pSR-HM-p(4849–6830) and reverse-oriented pSR-HM-m(6830–4849) plasmids. Similarly, pSR-MB-p and pSR-MB-m plasmids were constructed by inserting an MluI/BglII fragment into the SacI site of pSNAP25.
pSR-HM(5102–6378), pSR-HM(5102–5826), and PSR-HM(5102–5438) were constructed by inserting the indicated region of B0414 amplified by PCR into pSNAP25 that had been double-digested with HindIII and XbaI. The PCR primers used for these plasmids are as follows: HM-5102 (forward primer), 5′-TTTTAGGAAGCTTTTGTGGAAGGTT-3′; HM-6378 (reverse primer), 5′-AGAATTCTAGACAATTACAGCGATT-3′; HM-5826 (reverse primer), 5′-TTCGATCTAGAGTGAAGCCCTTGCC-3′; and HM-5438 (reverse primer), 5′-CATTGTTCTAGAACAATTGTGTCA-3′. The HindIII and XbaI restriction enzyme sites in these primers used for subcloning are in italics.
Germ line transformation.
Germ line transformations were carried out by standard protocols (29). All GFP constructs were injected at a concentration of 100 ng/μl. Dominant rol-6 pRF4 DNA was coinjected at 100 ng/μl. Transgenic lines inheriting the gene of interest were selected by their rolling behaviors, and the animals were observed for GFP expression.
Fixation and microscopy.
Worms were collected from an unsaturated plate into a 1.7-ml microcentrifuge tube in M9 buffer. The worms were then centrifuged, washed with M9 buffer, and fixed overnight at 4°C by adding 20% paraformaldehyde to a final concentration of 2%. The fixed worms were then washed three times with phosphate-buffered saline (PBS) buffer and mounted on a slide glass. A Bio-Rad confocal microscope was used for observing the expression patterns and taking photos. Nomarski photos were taken with a Carl Zeiss Axoplan microscope.
Run immunostaining.
Embryos and young larvae were permeabilized by freeze-cracking and fixed in methanol (95%) and then in acetone for 5 min each at −20°C. Specimen were rehydrated in a 75, 50, and 25% methanol series (3 min at each step), then washed in PBS three times for 10 min each. The blocking reaction was carried out in 5% goat serum for 3 to 5 h at room temperature. Monoclonal antibody raised against the mammalian Runt domain (10B7G8) (19) was added (1:20), and the specimens were incubated overnight at 4°C. After washing in PBS three times for 10 min each, secondary antibody conjugated with fluorescein isothiocyanate (FITC) (Sigma) was added according to the manufacturer’s instructions.
RNAi assay.
Sense and antisense RNA of the run gene was generated by in vitro transcription of a pBluescript SK(+) vector containing run cDNA. The run cDNA fragment contained 380 bp of the 5′-UTR and 130 bp of the 3′-UTR. PCRs were carried out using T3 and T7 primers to generate a template that was then purified for transcription with T3 and T7 RNA polymerases. To make double-stranded RNA, RNA generated from both forward and reverse reactions was mixed in a 1:1 ratio in TE (Tris-EDTA) buffer at 20 or 50 ng/μl. dsRNA was injected into the syncytial gonad of young wild-type animals (pPD-run-FL) (Fig. 1). Every 12 h the P0 individuals were moved to new plates, and the number of F1 progeny eggs was counted. Each progeny was inspected for any defects and for embryonic lethality by Nomarski and fluorescent microscopy.
RESULTS
Run is expressed in the lateral hypodermis and intestine of C. elegans.
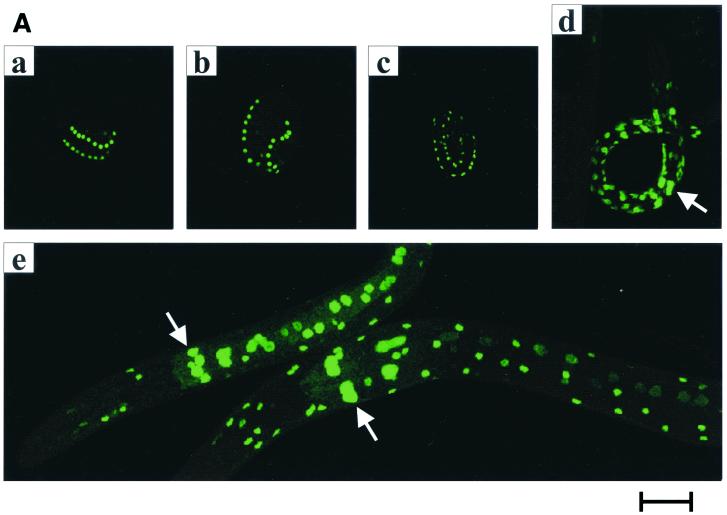
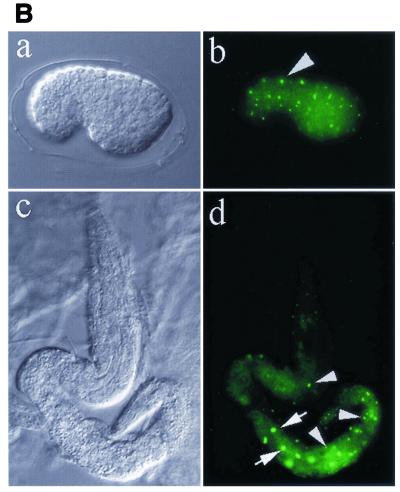
A 12,920-bp genomic DNA fragment containing the C. elegans run gene was obtained from cosmid clone B0414 and fused to a green fluorescent protein (GFP) reporter construct (pPD-run-FL) (Fig. 1). As illustrated in Fig. 2A, reporter gene expression in embryos from all transformed lines was first detected exclusively in hypodermal seam cells at the bean stage of development. Reporter gene expression was observed in the nuclei of these cells, and expression persisted throughout embryogenesis. Expression of run in seam cell nuclei was detected up until the L3 stage. Between the late embryonic stage and L3 stage, expression was also apparent in gut cell nuclei, with the anterior ring of four cells (int-1) showing the highest level of expression. Expression of run in binucleated gut cells with large nuclei became evident at the L2 stage. At the L3 stage, expression of run in gut cells was significantly decreased except in those cells in the most anterior part of the intestine. Expression in seam cells remained virtually unchanged. By the L4 stage, expression of run was undetectable (data not shown). Endogenous run expression was confirmed by immunohistochemical staining with antibody raised against the Runt domain. As shown in Fig. 2B, the pattern of endogenous run expression was essentially identical to that of run::GFP in the embryonic and larval stages, suggesting that the run::GFP reporter gene could represent the expression of endogenous run.
FIG. 2.
Expression pattern of run at various stages. (A) run::GFP reporter transgene. (a) Bean stage, (b) comma stage, (c) twofold stage, (d) L1 stage, (e) L2 (upper) and L3 (lower) stage. Arrows indicate the anterior ring of four intestinal cells (int-1). Note that run is expressed only in lateral hypodermal cells in the embryonic stage and is additionally expressed in intestine cells in the larval stage. Bar, 50 μm. (B) Endogenous expression pattern of run. (a and c) Nomarski images and (b and d) antibody staining of run visualized with FITC. (a and b) An embryo of the bean stage. Hypodermal cells express run. (c and d) An animal at the L2 stage. Intestine and hypodermal cells express run. Intestine nuclei are indicated by arrows, and hypodermal nuclei are indicated by arrowheads.
Stage- and tissue-specific expression is controlled by intronic sequence.
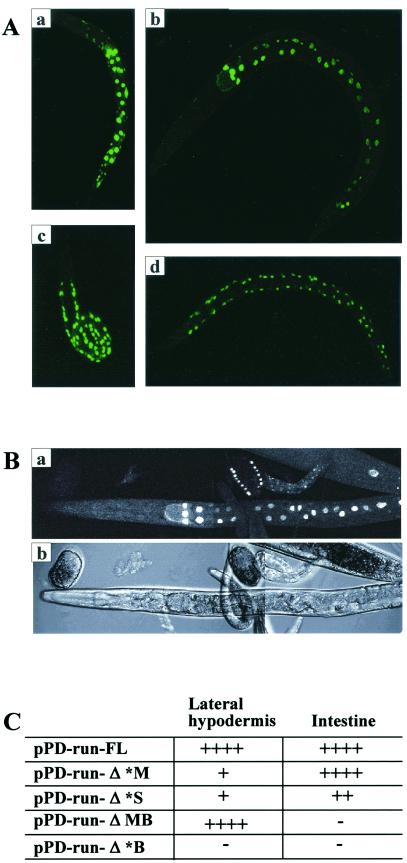
The run gene contains an exceptionally long intron (7.2 kb) between exons 3 and 4. Because of the length of this intron, we examined whether it plays roles in modulating gene expression at various stages of development and in determining expression in various cell types. For this purpose we constructed four deletion constructs from pPD-run-FL, as shown in Fig. 1. Deletion of 1.7 kb in the 5′ region of intron 3 (pPD-run-Δ*M) resulted in dramatic reduction of expression in the hypodermal seam cells at larval stage (Fig 3A). However, the reporter activity of the construct remained unchanged in the intestinal cells and embryonic seam cells (Fig. 3B). Further deletion of intron 3 up to the StuI site (pPD-run-Δ*S) showed a similar expression pattern, but the expression level was much reduced (data not shown). In contrast, deletion of 5.3 kb in the 3′ region (pPD-run-ΔMB) resulted in a complete loss of intestinal cell expression and little change in seam cell expression at the larval stage (Fig. 3A). When the majority of the intron was deleted (pPD-run-Δ*B), reporter activity was difficult to detect at any stage. Cell type-specific expression of various deletion constructs is summarized in Fig. 3C.
FIG. 3.
Expression pattern of run::GFP reporter transgenes with deletions within intron 3 at the L1 stage. (A) GFP activity of pPD-run-Δ*M (a and b) and pPD-run-ΔMB (c and d). Deletion of a 1.7-kb fragment in the 5′ region and a 5.3-kb fragment in the 3′ region of intron 3 resulted in the loss of hypodermal seam cell and intestinal cell expression of the reporter gene, respectively. (B) GFP activity of pPD-run-Δ*M. Deletion of the 1.7-kb fragment abolished postembryonic seam cell expression, but did not change the embryonic seam cell and intestinal cell expression. (a) GFP activity. (b) Nomarski photomicrograph. (C) Summary of the expression of run in deletion constructs within intron 3.
These results suggest that intron 3 is essential for stage-specific and cell type-specific expression of run. The intron can be separated into two functionally distinct regions, a 5′, 1.7-kb region and a 3′, 5.3-kb region, which are required for seam cell-specific and intestinal cell-specific expression, respectively, at the larval stage.
To examine whether the intron regions are sufficient for specifying cell type-specific expression, we examined the ability of the intronic regions to act as enhancers in a heterologous expression system. We generated pSR-HM-p(4849–6830), pSR-HM-m(6830–4849), pSR-MB-p, and pSR-MB-m by inserting the 2-kb (including the 1.7-kb region) or the 5.3-kb intron fragment into the pPD-SNAP25-GFP reporter plasmid. This plasmid contains the basal promoter of SNAP25, a neuron-specific gene (S. Hwang and J. Lee, unpublished results) (Fig. 4A). The pSNAP25 plasmid alone did not produce any expression of GFP. As shown in Fig. 4B, insertion of the 2-kb [pSR-HM-p(4849–6830)] and 5.3-kb [pSR-MB-p(6830–12116)] fragments resulted in seam cell-specific and intestinal cell-specific expression, respectively, at the larval stage. Plasmids containing the intron fragment in the reverse orientation (pSR-HM-m and pSR-MB-m) showed essentially the same results, but the activities were much lower (data not shown).
FIG. 4.
Cell type-specific enhancer activity of intron 3. (A) Heterologous reporter constructs of the run-intron::GFP fusion gene. Constructs are driven by parts of intron 3 of run, which is joined to the basal promoter of SNAP25 fused to GFP. Intron sequences essential for hypodermal seam cell expression and intestinal cell expression are indicated by hatched and shaded boxes, respectively. (B) Enhancer activity of intron 3 of run. (a) pSR-HM-p(4849–6830) and (b) pSR-MB-p. GFP activity is detected in the cytoplasm because the GFP used in this experiment does not contain a nuclear localization signal. Bar, 50 μm. (C) Summary of enhancer activities located within intron 3 of run. Note that the 0.7-kb fragment in the 5′ region and the 5.3-kb DNA fragment in the 3′ region of intron 3 are sufficient for hypodermal seam cell- and intestinal cell-specific expression, respectively, at the larval stage.
The intron region required for hypodermal seam cell expression at the larval stage was further mapped using plasmid constructs pSR-HM-p(5102–6378), pSR-HM-p(5102–5826) and pSR-HM-p(5102–5438). As summarized in Fig. 4C, the minimum region for hypodermal seam cell expression lies between nt 5102 and 5826. It was also demonstrated that the region between nt 5827 and 6378 is required for full activity. Mapping of the minimum region required for intestinal specific expression within the 5.3-kb region of the intron is the subject of a separate study.
These results strongly suggest that 0.7- and 5.3-kb fragments of intron 3 contain enhancer elements that are sufficient for seam cell and intestinal cell expression, respectively, at the larval stage.
Phenotypes of worms with RNA interference of run.
Double-stranded RNAi analysis of the run gene revealed an early larva-lethal phenotype with apparent malformation of the hypodermis and the intestine (Fig. 5). Using RNA at a concentration of 20 ng/μl, 186 animals out of 4,226 eggs (4.4%) treated with RNAi did not hatch, and 836 animals (19.8%) hatched but showed visibly abnormal phenotypes at early larval stage. The remaining animals did not have any defects. At 50 ng/ μl, 98 animals out of 1,468 eggs treated with RNAi (6.7%) failed to hatch, and 292 animals (19.9%) displayed visible abnormal phenotypes at the larval stage (Table 1).
FIG. 5.
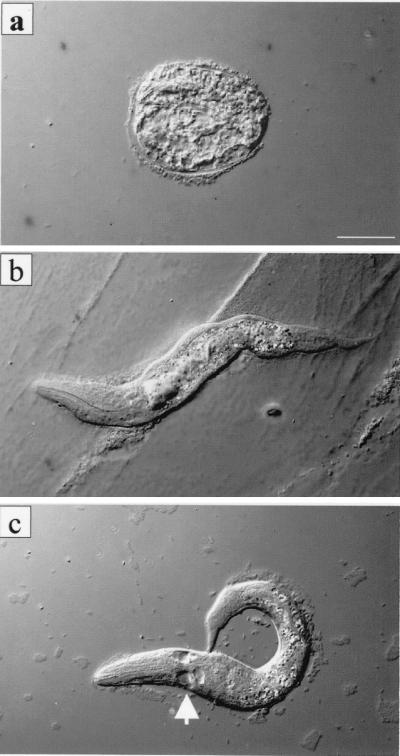
Phenotypes resulting from RNAi of run. (a) A typical egg phenotype. Although very much degraded, the egg contained a visible pharynx structure, indicating that most embryonic development had occurred. (b and c) A typical L1 phenotype. The outline of the animal is severely distorted. Arrow indicates lack of int-1 cells.
TABLE 1.
Effects of RNAi of runa
| Amt of dsRNA injected (ng/μl) | Time after injection (h) | Total no. of eggs collected | No. (%) of dead embryos (%) | No. (%) with larval defect | RNAi effect rate (%) |
|---|---|---|---|---|---|
| 0 (TE control) | 0–12 | 121 | 7 (5.8) | 6 (5.0) | 13 (10.8) |
| 12–24 | 342 | 4 (1.2) | 5 (1.5) | 9 (2.6) | |
| 24–36 | 331 | 1 (0.3) | 0 (0.0) | 1 (0.3) | |
| 36–48 | 292 | 1 (0.3) | 2 (0.7) | 3 (1.0) | |
| 48–60 | 25 | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| 20 | 0–12 | 301 | 40 (13.3) | 98 (32.6) | 138 (45.8) |
| 12–24 | 973 | 57 (5.9) | 226 (23.2) | 283 (29.1) | |
| 24–36 | 1,204 | 32 (2.7) | 180 (15.0) | 212 (17.6) | |
| 36–48 | 1,209 | 56 (4.6) | 247 (20.4) | 303 (25.1) | |
| 48–60 | 539 | 1 (0.2) | 85 (15.7) | 86 (16.0) | |
| 50 | 0–12 | 142 | 15 (10.6) | 40 (28.2) | 55 (38.7) |
| 12–24 | 313 | 13 (4.2) | 60 (19.2) | 73 (23.3) | |
| 24–36 | 371 | 24 (6.5) | 79 (21.3) | 103 (27.7) | |
| 36–48 | 401 | 29 (7.2) | 75 (18.7) | 104 (25.9) | |
| 48–60 | 241 | 17 (7.1) | 38 (15.8) | 55 (22.8) |
The numbers of dead embryos and abnormal larvae were compared to the total numbers of embryos laid at each time interval. Negative control was performed by microinjecting TE alone.
At both concentrations of dsRNAi, typical phenotypes were an abnormal morphology of the hypodermal tissue and distortion of the morphology of the intestine. This phenotype was specific to the run dsRNA interference, since we did not observe such a phenotype in the control experiment in which TE alone was microinjected (Table 1). Other organs such as the pharynx appeared to be normal. Eggs that failed to hatch apparently finished development, because fully developed organs, such as the pharynx, could be observed (Fig. 5a). We did not observe massive cell death in the hypodermis or the intestine. However, in some cases int-1 cells died at the L1 stage (Fig. 5c). In conclusion, inactivation of run by RNAi appears to cause a malformation in the development of the hypodermis and intestine. No significant effects on cell degeneration or on the formation of other organs by the embryo were observed.
DISCUSSION
The C. elegans run gene, encoding the runt domain transcription factor, is expressed in hypodermal seam cells and gut cells and is essential for the development of the corresponding tissues in early larvae. Expression of run can first be detected in hypodermal seam cells at the bean stage. Expression was observed in the nuclei of all hypodermal seam cells and remained evident up until the L3 stage. The expression pattern of run in hypodermal cells partly overlaps that of Elt-1, which has been demonstrated to play an early role in specifying epidermal cell fate. Elt-1 is expressed in hypodermal cells which appear ≈260 min postfertilization (about the 365-cell stage) along with hypodermal seam cells (37). Run expression can also be first detected at 260 min postfertilization; however, its expression is restricted to seam cells, whereas Elt-1 is expressed in all major hypodermal cells.
On each side of the newly hatched animal, there exists a single longitudinal row of specialized hypodermal cells, called seam cells. These cells are the only cells that divide after hatching. Seam cells divide following a stem cell pattern, resulting in an anterior daughter cell that fuses with the hypodermal syncytium, hyp7, and a posterior daughter cell that remains in the seam, where it is capable of further cell division (42) The majority of seam cells are blast cells that provide hypodermal nuclei and a postdeirid neuroblast (42). The male sensory rays are also derived from the seam cells. Targeted inactivation of run by RNAi resulted in abnormal morphology of hypodermal seam cells, suggesting that run is essential for the formation of normal hypodermis. Screening and characterization of run mutant C. elegans will be required to understand whether run also plays a role in the formation of postdeirid neuroblast cells and sensory rays.
The C. elegans gut is a major organ, involved not only in digestion but also in storage and macromolecule synthesis (7, 22). The intestine is derived exclusively from the founder cell E, which gives rise to no other tissue. At hatching, these cells are mononucleate, but most of them subsequently become binucleate through nuclear division (41). Furthermore, at the end of each larval stage, all the intestinal nuclei undergo endoreduplication of their DNA without accompanying nuclear division. Endoreduplication within intestinal cells results in large cell nuclei.
At the larval stage, run expression was detected in the intestine as well as in the lateral hypodermis. Expression of run in the anterior ring of four cells (int-1) and in binucleated intestinal cells can be clearly demonstrated at the L2 stage (Fig. 2). The mechanism by which the E blastomere, the clonal progenitor of the gut, becomes the functional gut is poorly understood. This is probably due to the paucity of information on transcription factors involved in the process. End-1 and elt-2 are two transcription factors that are known to play essential roles in the formation of the intestine. Both are GATA-like transcription factors that are expressed at the beginning of embryogenesis (2E cell stage) (14, 46). In contrast to the early embryonic expression of these transcription factors, run expression was only detected from the late embryonic stage and lasted until the L3 stage. This result suggests that run is involved in the establishment of a functional gut rather than in the determination of gut cell fate during embryogenesis.
This interpretation is supported by results from the RNAi experiments targeting run. Targeted disruption of run resulted in degeneration or malformation of hypodermal cells and intestinal cells. It is worth noting that targeted inactivation of run did not result in the lack of an intestine but rather in intestinal malformation. In comparison, loss-of-function phenotypes of either end-1 or elt-2 genes result in a complete lack of intestinal cells (14, 46). Runx3/PEBP2αC, a mammalian homolog of run, is expressed mainly in epithelial cells of the gastrointestinal tract. Most Runx3 mutant mice die within 24 h after birth due to a malfunctioning gastrointestinal tract that fails to absorb nutrients (Li et al., unpublished data). The major phenotype of mice with a targeted deletion of Runx3 is therefore quite similar to that of C. elegans with targeted inactivation of run. These results strongly suggest that run is required for the formation of a functional gut and that its role in gut development is evolutionarily conserved from nematodes to mammals. Although end-1 and elt-2 are also known to be gut-specific transcription factors in C. elegans, to date there has been no evidence to show that GATA family genes are involved in mammalian gut development. Therefore, run is the first gene that has been shown to be essential for both nematode and mammalian gut development.
An intimate relationship between runt domain transcription factor and transforming growth factor beta (TGF-β) superfamily signaling has been reported. Receptors for the TGF-β superfamily are composed of type I and II receptors. Following ligand binding, type I receptors associate with type II receptors and become activated through phosphorylation. The activated receptor-associated kinase phosphorylates Smads, which move into the nucleus to stimulate the transcription of a set of target genes (28). Runx2 and Runx3 are important targets of TGF-β signaling and are induced at the transcriptional level (26, 40). Furthermore, Runx proteins interact physically with activated Smads and function together (18, 26, 45). The C. elegans TGF-β-like type I receptor (sma-6), which can activate Smads of C. elegans (sma-2, sma-3, and sma-4), is expressed mainly in the intestine, and its expression overlaps that of run (24). It would therefore be of interest to study whether the functional relationship between TGF-β signaling and Runt domain transcription factor in mammals is conserved in the nematode.
An unusual characteristic of the genome of C. elegans is the short intron length. Introns in C. elegans are typically 45 to 55 nucleotides long, and more than half are shorter than 60 nucleotides. However, the third intron of the C. elegans run gene is 7.2 kb in length. All other introns in the run gene are shorter than 1 kb. Because tissue-specific enhancer elements have been identified within intron regions of fruit fly and mammalian genes, we examined whether the long intron of run plays a role in cell type-specific and stage-specific expression of the gene. Our results demonstrate that the long intron region contains enhancer elements required for stage-specific and cell type-specific expression.
The intron region could be separated into two functionally distinct fragments (a 0.7-kb fragment in the 5′ region and a 5.3-kb fragment in the 3′ region). The 0.7-kb fragment in the 5′region is sufficient for seam cell-specific expression at the larval stage. On the other hand, the 5.3-kb fragment in the 3′ region was sufficient for intestinal cell-specific expression at the larval stage. For embryonic seam cell expression, intron 3 is required but not sufficient. This is the first report demonstrating that an intron region is essential for stage-specific and cell type-specific expression of a C. elegans gene.
In this paper, we show that run is expressed in the lateral hypodermis and intestine and that cell type-specific and stage-specific expression is regulated by enhancer elements located in an intron. Our results also demonstrate that run is essential for the development of a functional hypodermis and intestine and that the role of run in C. elegans gut development has been evolutionarily conserved from nematodes to mammals.
Acknowledgments
S. Nam and Y.-H. Jin have contributed equally to this work.
This work was supported by Korean Research Foundation grant KRF-99-042-F00014 F0210.
REFERENCES
- 1.Anonymous. 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. The C. elegans Sequencing Consortium. Science 282:2012–2018. [DOI] [PubMed] [Google Scholar]
- 2.Bae, S. C., Y. Yamaguchi-Iwai, E. Ogawa, M. Maruyama, M. Inuzuka, H. Kagoshima, K. Shigesada, M. Satake, and Y. Ito. 1993. Isolation of PEBP2 alpha B cDNA representing the mouse homolog of human acute myeloid leukemia gene, AML1. Oncogene 8:809–814. [PubMed] [Google Scholar]
- 3.Bae, S. C., E. Ogawa, M. Maruyama, H. Oka, M. Satake, K. Shigesada, N. A. Jenkins, D. J. Gilbert, N. G. Copeland, and Y. Ito. 1994. PEBP2 alpha B/mouse AML1 consists of multiple isoforms that possess differential transactivation potentials. Mol. Cell. Biol. 14:3242–3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bae, S. C., E. Takahashi, Y. W. Zhang, E. Ogawa, K. Shigesada, Y. Namba, M. Satake, and Y. Ito. 1995. Cloning, mapping and expression of PEBP2 alpha C, a third gene encoding the mammalian Runt domain. Gene 159:245–248. [DOI] [PubMed] [Google Scholar]
- 5.Bae, S. C., and Y. Ito. 1999. Regulation mechanisms for the heterodimeric transcription factor, PEBP2/CBF. Histol. Histopathol. 14:1213–1221. [DOI] [PubMed] [Google Scholar]
- 6.Bae, S. C., and J. Lee. 2000. cDNA cloning of run, a Caenorhabditis elegans Runt domain encoding gene. Gene 241:255–258. [DOI] [PubMed] [Google Scholar]
- 7.Blumenthal, T., M. Squire, S. Kirtland, J. Cane, M. Donegan, J. Spieth, and W. Sharrock. 1984. Cloning of a yolk protein gene family from Caenorhabditis elegans. J. Mol. Biol. 174:1–18. [DOI] [PubMed] [Google Scholar]
- 8.Brenner, S. 1974. The Genetics of Caenorhabditis elegans. Genetics 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Canon, J., and U. Banerjee. 2000. Runt and Lozenge function in Drosophila development. Semin. Cell Dev. Biol. 11:327–336. [DOI] [PubMed] [Google Scholar]
- 10.Daga, A., C. A. Karlovich, K. Dumstrei, and U. Banerjee. 1996. Patterning of cells in the Drosophila eye by Lozenge, which shares homologous domains with AML1. Genes Dev. 10:1194–1205. [DOI] [PubMed] [Google Scholar]
- 11.Duffy, J. B., M. A. Kania, and J. P. Gergen. 1991. Expression and function of the Drosophila gene runt in early stages of neural development. Development 113:1223–1230. [DOI] [PubMed] [Google Scholar]
- 12.Duffy, J. B., and J. P. Gergen. 1991. The Drosophila segmentation gene runt acts as a position-specific numerator element necessary for the uniform expression of the sex-determining gene Sex-lethal. Genes Dev. 5:2176–2187. [DOI] [PubMed] [Google Scholar]
- 13.Fraser, A. G., R. S. Kamath, P. Zipperlen, M. Martinez-Campos, M. Sohrmann, and J. Ahringer. 2000. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408:325–330. [DOI] [PubMed] [Google Scholar]
- 14.Fukushige, T., M. G. Hawkins, and J. D. McGhee. 1998. The GATA-factor elt-2 is essential for formation of the Caenorhabditis elegans intestine. Dev. Biol. 198:286–302. [PubMed] [Google Scholar]
- 15.Gergen, J. P., and E. F. Wieschaus. 1985. The localized requirements for a gene affecting segmentation in Drosophila: analysis of larvae mosaic for runt. Dev. Biol. 109:321–335. [DOI] [PubMed] [Google Scholar]
- 16.Golub, T. R., G. F. Barker, S. K. Bohlander, S. W. Hiebert, D. C. Ward, P. Bray-Ward, E. Morgan, S. C. Raimondi, J. D. Rowley, and D. G. Gilliland. 1995. Fusion of the TEL gene on 12p13 to the AML1 gene on 21q22 in acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. USA 92:4917–4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonczy, P., G. Echeverri, K. Oegema, A. Coulson, S. J. Jones, R. R. Copley, J. Duperon, J. Oegema, M. Brehm, E. Cassin, E. Hannak, M. Kirkham, S. Pichler, K. Flohrs, A. Goessen, S. Leidel, A. M. Alleaume, C. Martin, N. Ozlu, P. Bork, and A. A. Hyman. 2000. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature 408:331–336. [DOI] [PubMed] [Google Scholar]
- 18.Hanai, J., L. F. Chen, T. Kanno, N. Ohtani-Fujita, W. Y. Kim, W. H. Guo, T. Imamura, Y. Ishidou, M. Fukuchi, M. J. Shi, J. Stavnezer, M. Kawabata, K. Miyazono, and Y. Ito. 1999. Interaction and functional cooperation of PEBP2/CBF with Smads: synergistic induction of the immunoglobulin germline Calpha promoter. J. Biol. Chem. 274:31577–31582. [DOI] [PubMed] [Google Scholar]
- 19.Huang, G., K. Shigesada, K. Ito, H. J. Wee, T. Yokomizo, and Y. Ito. 2001. Dimerization with PEBP2beta protects RUNX1/AML1 from ubiquitin-proteasome-mediated degradation. EMBO J. 20:723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kagoshima, H., K. Shigesada, M. Satake, Y. Ito, H. Miyoshi, M. Ohki, M. Pepling, and P. Gergen. 1993. The Runt domain identifies a new family of heteromeric transcriptional regulators. Trends Genet. 9:338–341. [DOI] [PubMed] [Google Scholar]
- 21.Kania, M. A., A. S. Bonner, J. B. Duffy, and J. P. Gergen. 1990. The Drosophila segmentation gene runt encodes a novel nuclear regulatory protein that is also expressed in the developing nervous system. Genes Dev. 4:1701–1713. [DOI] [PubMed] [Google Scholar]
- 22.Kimble, J., and W. J. Sharrock. 1983. Tissue-specific synthesis of yolk proteins in Caenorhabditis elegans. Dev. Biol. 96:189–196. [DOI] [PubMed] [Google Scholar]
- 23.Komori, T., H. Yagi, S. Nomura, A. Yamaguchi, K. Sasaki, K. Deguchi, Y. Shimizu, R. T. Bronson, Y. H. Gao, M. Inada, M. Sato, R. Okamoto, Y. Kitamura, S. Yoshiki, and T. Kishimoto. 1997. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89:755–764. [DOI] [PubMed] [Google Scholar]
- 24.Krishna, S., L. L. Maduzia, and R. W. Padgett. 1999. Specificity of TGFbeta signaling is conferred by distinct type I receptors and their associated SMAD proteins in Caenorhabditis elegans. Development 126:251–260. [DOI] [PubMed] [Google Scholar]
- 25.Lee, B., K. Thirunavukkarasu, L. Zhou, L. Pastore, A. Baldini, J. Hecht, V. Geoffroy, P. Ducy, and G. Karsenty. 1997. Missense mutations abolishing DNA binding of the osteoblast-specific transcription factor OSF2/CBFA1 in cleidocranial dysplasia. Nat. Genet. 16:307–310. [DOI] [PubMed] [Google Scholar]
- 26.Lee, K. S., H. J. Kim, Q. L. Li, X. Z. Chi, C. Ueta, T. Komori, J. M. Wozney, E. G. Kim, J. Y. Choi, H. M. Ryoo, and S. C. Bae. 2000. Runx2 is a common target of transforming growth factor β1 and bone morpho genetic protein 2, and cooperation between Runx2 and Smad5 induces osteoblast-specific gene expression in the pluripotent mesenchymal precursor cell line C2C12. Mol. Cell. Biol. 20:8783–8792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levanon, D., V. Negreanu, Y. Bernstein, I. Bar-Am, L. Avivi, and Y. Groner. 1994. AML1, AML2, and AML3, the human members of the runt domain gene-family: cDNA structure, expression, and chromosomal localization. Genomics 23:425–432. [DOI] [PubMed] [Google Scholar]
- 28.Massague, J. 1998. TGF-beta signal transduction. Annu. Rev. Biochem. 67:753–791. [DOI] [PubMed] [Google Scholar]
- 29.Mello, C., and A. Fire. 1995. DNA transformation. Methods Cell Biol. 48:451–482. [PubMed] [Google Scholar]
- 30.Miyoshi, H., K. Shimizu, T. Kozu, N. Maseki, Y. Kaneko, and M. Ohki. 1991. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single Gene, AML1. Proc. Natl. Acad. Sci. USA 88:10431–10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyoshi, H., T. Kozu, K. Shimizu, K. Enomoto, N. Maseki, Y. Kaneko, N. Kamada, and M. Ohki. 1993. The t(8;21) translocation in acute myeloid leukemia results in production of an AML1-MTG8 fusion transcript. EMBO J. 12:2715–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mundlos, S., F. Otto, C. Mundlos, J. B. Mulliken, A. S. Aylsworth, S. Albright, D. Lindhout, W. G. Cole, W. Henn, J. H. Knoll, M. J. Owen, R. Mertelsmann, B. U. Zabel, and B. R. Olsen. 1997. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell 89:773–779. [DOI] [PubMed] [Google Scholar]
- 33.Ogawa, E., M. Inuzuka, M. Maruyama, M. Satake, M. Naito-Fujimoto, Y. Ito, and K. Shigesada. 1993. Molecular cloning and characterization of PEBP2 beta, the heterodimeric partner of a novel Drosophila runt-related DNA binding protein PEBP2 alpha. Virology 194:314–331. [DOI] [PubMed] [Google Scholar]
- 34.Ogawa, E., M. Maruyama, H. Kagoshima, M. Inuzuka, J. Lu, M. Satake, K. Shigesada, and Y. Ito. 1993. PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proc. Natl. Acad. Sci. USA 90:6859–6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okuda, T., J. van Deursen, S. W. Hiebert, G. Grosveld, and J. R. Downing. 1996. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84:321–330. [DOI] [PubMed] [Google Scholar]
- 36.Otto, F., A. P. Thornell, T. Crompton, A. Denzel, K. C. Gilmour, I. R. Rosewell, G. W. Stamp, R. S. Beddington, S. Mundlos, B. R. Olsen, P. B. Selby, and M. J. Owen. 1997. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 89:765–771. [DOI] [PubMed] [Google Scholar]
- 37.Page, B. D., W. Zhang, K. Steward, T. Blumenthal, and J. R. Priess. 1997. ELT-1, a GATA-like transcription factor, is required for epidermal cell fates in Caenorhabditis elegans embryos. Genes Dev. 11:1651–1661. [DOI] [PubMed] [Google Scholar]
- 38.Romana, S. P., M. Mauchauffe, M. Le Coniat, I. Chumakov, D. Le Paslier, R. Berger, and O. A. Bernard. 1995. The t(12;21) of acute lymphoblastic leukemia results in a tel-AML1 gene fusion. Blood 85:3662–3670. [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1988. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 40.Shi, M. J., and J. Stavnezer. 1998. CBF alpha3 (AML2) is induced by TGF-beta1 to bind and activate the mouse germline Ig alpha promoter. J. Immunol. 161:6751–6760. [PubMed] [Google Scholar]
- 41.Sulston, J. E., and H. R. Horvitz. 1977. Postembryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56:110–156. [DOI] [PubMed] [Google Scholar]
- 42.Sulston, J. E., and J. G. White. 1980. Regulation and cell autonomy during postembryonic development of Caenorhabditis elegans. Dev. Biol. 78:577–597. [DOI] [PubMed] [Google Scholar]
- 43.Wang, Q., T. Stacy, J. D. Miller, A. F. Lewis, T. L. Gu, X. Huang, J. H. Bushweller, J. C. Bories, F. W. Alt, G. Ryan, P. P. Liu, A. Wynshaw-Boris, M. Binder, M. Marin-Padilla, A. H. Sharpe, and N. A. Speck. 1996. The CBFbeta subunit is essential for CBFalpha2 (AML1) function in vivo. Cell 87:697–708. [DOI] [PubMed] [Google Scholar]
- 44.Wang, S., Q. Wang, B. E. Crute, I. N. Melnikova, S. R. Keller, and N. A. Speck. 1993. Cloning and characterization of subunits of the T-cell receptor and murine leukemia virus enhancer core-binding factor. Mol. Cell. Biol. 13:3324–3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, Y. W., N. Yasui, K. Ito, G. Huang, M. Fujii, J. Hanai, H. Nogami, T. Ochi, K. Miyazono, and Y. Ito. 2000. A RUNX2/PEBP2alpha A/CBFA1 mutation displaying impaired transactivation and Smad interaction in cleidocranial dysplasia. Proc. Natl. Acad. Sci. USA 97:10549–10554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu, J., R. J. Hill, P. J. Heid, M. Fukuyama, A. Sugimoto, J. R. Priess, and J. H. Rothman. 1997. end-1 encodes an apparent GATA factor that specifies the endoderm precursor in Caenorhabditis elegans embryos. Genes Dev. 11:2883–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]