Abstract
Mutations in CDK4 and its key kinase inhibitor p16INK4a have been implicated in the genesis and progression of familial human melanoma. The importance of the CDK4 locus in human cancer first became evident following the identification of a germ line CDK4-Arg24Cys (R24C) mutation, which abolishes the ability of CDK4 to bind to p16INK4a. To determine the role of the Cdk4R24C germ line mutation in the genesis of other cancer types, we introduced the R24C mutation in the Cdk4 locus of mice by using Cre-loxP-mediated “knock-in” technology. Cdk4R24C/R24C mouse embryo fibroblasts (MEFs) displayed increased Cdk4 kinase activity resulting in hyperphosphorylation of all three members of the Rb family, pRb, p107, and p130. MEFs derived from Cdk4R24C/R24C mice displayed decreased doubling times, escape from replicative senescence, and escape sensitivity to contact-induced growth arrest. These MEFs also exhibited a high degree of susceptibility to oncogene-induced transformation, suggesting that the Cdk4R24C mutation can serve as a primary event in the progression towards a fully transformed phenotype. In agreement with the in vitro data, homozygous Cdk4R24C/R24C mice developed tumors of various etiology within 8 to 10 months of their life span. The majority of these tumors were found in the pancreas, pituitary, brain, mammary tissue, and skin. In addition, Cdk4R24C/R24C mice showed extraordinary susceptibility to carcinogens and developed papillomas within the first 8 to 10 weeks following cutaneous application of the carcinogens 9,10-di-methyl-1,2-benz[a]anthracene (DMBA) and 12-O-tetradecanoylphorbol-13-acetate (TPA). This report formally establishes that the activation of Cdk4 is sufficient to promote cancer in many tissues. The observation that a wide variety of tumors develop in mice harboring the Cdk4R24C mutation offers a genetic proof that Cdk4 activation may constitute a central event in the genesis of many types of cancers in addition to melanoma.
Orderly cell cycle progression of mammalian cells determines their ability to undergo regulated proliferation, differentiation, senescence, and apoptosis (43, 44). Perturbations in the expression and activities of proteins that regulate these cellular phenomena determine the emergence of a cancerous state. Two important tumor suppressor pathways, which depend on the expression and activity of two key cell cycle regulatory proteins, pRb and p53, serve as critical branch points that govern controlled cell growth (14, 17, 33, 43, 44, 49). In concordance with this, it has been observed that a majority of human tumors harbor mutations that debilitate the surveillance function of the pRb and/or p53 pathways (43, 49). Moreover, patients with inactivating mutations in both pathways have a much worse prognosis compared to patients with inactivation in just one of the two pathways. While the consequences of inactivation of p53 on cancer development has been exhaustively studied using mice that are nullizygous at the p53 loci, it has been difficult to examine the effects of simultaneous inactivation of the retinoblastoma (Rb) family proteins on cancer progression, as homozygous inactivation of both Rb alleles leads to early embryonic lethality (5, 9, 19, 27). This requirement of the Rb family proteins during embryogenesis and early development has precluded the examination of their role during adult development and on cancer predisposition by using conventional gene disruption techniques (5, 6, 19, 25–28). Since homozygous inactivation of Rb leads to early embryonic lethality, studies focused on examination of the role of Rb family proteins in development and cancer have been restricted to characterization of mouse models that either (i) harbor heterozygous inactivation of Rb family genes, (ii) harbor a heterozygous inactivation of Rb family genes in addition to disruption of genes that code for other key cell cycle regulators, such as members of the cyclin kinase inhibitor (CKI) family, or (iii) have inactivation of the Rb family proteins via expression of viral proteins, such as papillomavirus protein E7 or the simian virus 40 large T-antigen protein (14, 17). However, the unequivocal importance of the retinoblastoma proteins in regulating cell cycle progression and cell proliferation and its underlying effects on immortalization and tumorigenesis has been elucidated by studies that describe the disruption of the retinoblastoma family gene loci in fibroblasts (7, 39). Loss of expression of pRb, p107, and p130 proteins in fibroblasts resulted in increased cell proliferation, shortening of the cell cycle, and immortalization. However, these studies do not allow an extrapolation of the effects of simultaneous inactivation of the retinoblastoma proteins on adult development and cancer predisposition.
Cdk4, along with Cdk6, is the chief catalytic subunit of the regulatory cyclin D family of proteins that govern G1-to-S phase progression of mammalian cells via phosphorylation and inactivation of retinoblastoma family proteins (43, 44). Members of the INK4 family of proteins, chiefly p16Ink4a, are specific inhibitors of the cyclin D/Cdk4 complexes (43, 44). Mutations in CDK4 and its key kinase inhibitor p16INK4A have been implicated in the genesis and progression of familial human melanoma (20, 31). The importance of the CDK4 locus in human cancer was further emphasized upon identification of a germ line CDK4-Arg24Cys (R24C) mutation, which abolishes the ability of CDK4 to bind to p16INK4A, predisposing humans to hereditary melanoma (51, 53). This observation suggests that a mutant CDK4 gene can function as a dominant oncogene that is resistant to normal physiological inhibition by p16INK4A. To determine the consequence of the R24C germ line mutation on mouse development and cancer susceptibility, we introduced the R24C mutation in the Cdk4 locus of mice by using Cre-loxP-mediated “knock-in” technology (37). The presence of the Cdk4R24C mutation induced hyperphosphorylation of all three members of the Rb family, pRb, p107, and p130, and mouse embryo fibroblasts (MEFs) derived from Cdk4R24C/R24C mice escape from replicative senescence and become insensitive to contact-induced growth arrest. Moreover, the presence of the Cdk4R24C mutation resulted in transformation in vitro and spontaneous and chemical carcinogen-induced tumorigenesis in vivo, suggesting that the Cdk4R24C mutation can serve as the primary event in the progression towards a fully transformed phenotype.
MATERIALS AND METHODS
Generation of Cdk4R24C/R24C mice and tumor analysis.
Generation of the Cdk4R24C/R24C mice has been described previously (37). Cdk4+/+, Cdk4+/R24C, and Cdk4R24C/R24C mice were observed for 20 months for the appearance of detectable or palpable tumors. Tumor-bearing mice were euthanatized, and tumors were dissected and processed for histological analysis. At the end of 20 months, the remaining mice were euthanatized and grouped into tumor-prone or tumor-free categories. The number of mice that were tumor bearing or tumor-free was evaluated statistically using standard Kaplan-Meier survival analysis method. Normal and tumor tissues were fixed by immersion in either 10% neutral buffered formalin or 95% ethanol-5% glacial acetic acid overnight, dehydrated through ethanol, and embedded in paraffin. Blocks were sectioned at 5 μm and stained with hematoxylin and eosin or prepared for immunohistochemistry by mounting tissue sections on electrostatically charged slides (ProbeOn Plus; Fisher Scientific).
Protein analysis.
MEFs were lysed in buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 2.5 mM EGTA, 1 mM EDTA, 10 mM β-glycerophosphate, 0.1 mM NaVO3, 1 mM NaF, 0.1% Tween 20, 10% glycerol, 1 mM dithiothreitol) in the presence of protease inhibitor cocktail (Complete Tablets; Boehringer Mannheim). For Western blot analysis, 50 to 100 μg of total protein lysate was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride membranes (Immobilon P), followed by immunoblotting with the indicated antibodies and chemiluminescence detection. For association studies, 500 μg of total protein lysate was immunoprecipitated with the indicated antibodies. The resulting immunoprecipitates were resolved by SDS-PAGE and transferred to polyvinylidene difluoride membranes (Immobilon P) and subjected to Western blot analysis as mentioned above. For immune-complex kinase assays, 500 μg of total protein lysate was immunoprecipitated with the anti-Cdk4 antibodies (with or without the presence of competing Cdk4 peptide). The resulting immunoprecipitates were subjected to kinase assays in kinase assay buffer (500 mM HEPES [pH 7.5], 100 mM MgCl2, 25 mM EGTA, 100 mM β-glycerophosphate, 1 mM NaVO3, 10 mM NaF, 10 mM dithiothreitol, 200 μM ATP) in the presence of 20 μM [γ-32P]ATP and 0.2 to 0.5 μg of glutathione S-transferase (GST)-pRb substrate. Kinase assays were performed at 30°C for 30 min, and the reaction was stopped by addition of 25 μl of Laemmli sample buffer. The reaction mixture was resolved on SDS-10% PAGE gels and subjected to autoradiography to visualize the phosphorylated GST-pRb substrate. Antibodies used include rabbit polyclonal antibodies from Santa Cruz Biotechnology: anti-Cdk4 (C-22), anti-p16 (M-156), anti-p21Cip1 (C-19), anti-p53 (FL-393), anti-p130 (C-20), anti-p107 (C-18), and anti-pRb (G3-245; 14001A) antibodies were from BD Transduction Laboratories and anti-p19ARF (NB-200-106) antibody was from Novus Biologicals.
Growth curve and cell cycle analysis.
MEF cultures from Cdk4+/+, Cdk4+/R24C, and Cdk4R24C/R24C mouse embryos were propagated in Dulbecco minimal essential medium (DMEM) media supplemented with 10% fetal bovine serum (FBS). Early-passage MEFs (passage < 4) were used for growth curve and cell cycle phase analysis. Passage 0 refers to the stage when embryos were put in culture, and every subsequent passage is referred to as passage 1, 2, 3, etc. For the analysis of growth curves, 3 × 105 cells were cultured and the numbers of cells were counted using trypan blue exclusion analysis for 4 days. The population doubling levels were calculated according to the formula log(Nfinal/Ninitial)/log 2, where Nfinal and Ninitial are the final and initial (3 × 105cells) numbers of cells plated and counted, respectively. For the analysis of cell cycle phases, 3 × 105 cells were exponentially cultured for 24 h, upon which cells were fixed in ethanol at −20°C. The percentage of cells in G0/G1, S, and G2/M were determined by fluorescence-assorted cytometry analysis upon staining for 30 min with propidium iodide after treatment with RNase A.
Immortalization assays and colony formation assays.
MEF cultures from Cdk4+/+, Cdk4+/R24C, and Cdk4R24C/R24C mouse embryos were propagated in DMEM supplemented with 10% FBS for 25 passages according to a modified version of the 3T3 protocol. Then, 106 cells were cultured in 10-cm-diameter plates, and 3 days later, the total number of cells was counted and 106 cells were replated, which constituted one passage. This process was continued for 25 successive passages, and the cumulative increase in cell number was calculated according to the formula log(Nfinal/Ninitial)/log 2, where Nfinal and Ninitial are the final and initial numbers of cells plated and counted after 3 days, respectively. To test the ability of cells to form multiple layers (focus formation capacity), 106 cells (passage 20) representing Cdk4+/+, Cdk4+/R24C, and Cdk4R24C/R24C MEFs were plated in 10-cm-diameter dishes and grown for 2 weeks. Medium was changed every 3 days, and at the end of 2 weeks, plates were stained with Giemsa stain to determine the focus formation capacity.
Transformation assays.
For focus formation assays, early-passage (passage < 4) Cdk4+/+ and Cdk4R24C/R24C MEFs (106 cells) were seeded in plates with a 10-cm diameter and grown in DMEM plus 10% FBS overnight. The medium was changed 4 to 6 h before transfections began, and transfections were performed by standard calcium phosphate procedures with DNA mixtures containing 15 μg of the relevant plasmids plus the corresponding amount of carrier DNA plasmid, for a total of 30 μg of DNA. After 12 to 14 h of incubation with precipitates, the incubation medium was changed and the cultures were fed with fresh medium every 3 days for 14 to 21 days. At day 21 posttransfection, cells were fixed and stained with Giemsa, and foci (>2 mm in diameter) were scored visually. The plasmids used in these assays were human oncogenic H-RasV12, pE1A, and c-myc in pcDNA3 vector.
Two-stage chemical skin carcinogenesis experiments.
For chemical carcinogenesis studies, Cdk4+/+ (n = 7), Cdk4+/R24C (n = 13), and Cdk4R24C/R24C (n = 28) mice were shaved at 4 weeks of age and initiated by application of 25 μg of 9,10-di-methyl-1,2-benz[a]anthracene (DMBA) in 200 μl of acetone. Promotion was carried out after 3 days of the DMBA application by a twice-per-week application of 2 μg of 12-O-tetradecanoylphorbol-13-acetate (TPA) in 200 μl of acetone. The regimen was carried out for 20 weeks, and the mice were observed for the occurrence of skin tumors. At the end of the study period, the animals were grouped into either tumor-free or tumor-bearing categories and subjected to Kaplan-Meier survival curve analysis. At the end of 20 weeks (and in some cases earlier if the skin tumors were very aggressive and had a large burden), mice were euthanatized and the tumors were dissected and processed for histology.
RESULTS
Germ line targeting of R24C mutation in Cdk4 locus results in deregulated Cdk4R24C kinase.
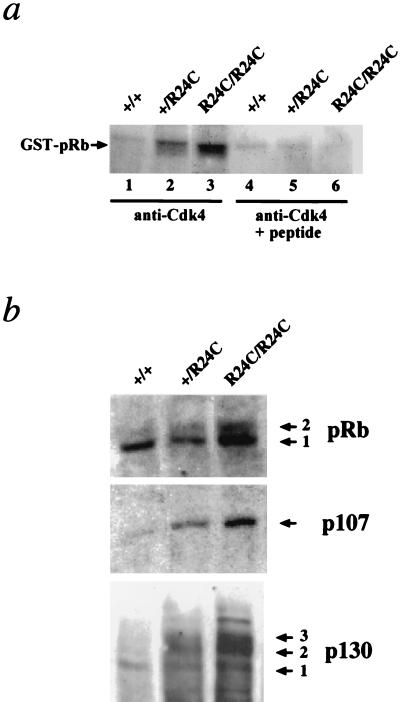
To determine the consequence of R24C germ line mutation on mouse development and cancer susceptibility, we introduced the R24C mutation in the Cdk4 locus of mice using Cre-loxP-mediated knock-in technology (37). This mutation was previously shown to result in the loss of p16INK4A binding to Cdk4 (37). To determine the effects of the Cdk4R24C mutation on kinase activity, we performed immune-complex kinase assays on protein lysates from MEFs derived from Cdk4+/+, Cdk4+/R24C, and Cdk4R24C/R24C embryos by using anti-Cdk4-specific antibodies with pRb as a substrate. To this end, anti-Cdk4 immunoprecipitates from equal amounts of cell lysates were subjected to kinase assays in the presence or absence of a competing peptide as described in Materials and Methods. Results of these kinase assays revealed that the protein expressed in Cdk4+/R24C MEFs displayed increased levels of Cdk4 kinase activity compared to the control Cdk4+/+ MEFs while the Cdk4R24C/R24C MEFs exhibited the highest levels of kinase activity (Fig. 1a).
FIG. 1.
Presence of the R24C mutation in Cdk4 results in a deregulated Cdk4R24C kinase. (a) Immune-complex kinase assays using anti-Cdk4 specific antibodies without (lanes 1 to 3) or with (lanes 4 to 6) the presence of a competing Cdk4 peptide and pRb substrate were performed with 500 μg of protein extracts isolated from Cdk4+/+, Cdk4+/R24C, and Cdk4R24C/R24C MEF cultures. The presence of the phosphorylated GST-pRb substrate is indicated. (b) Increased Cdk4R24C kinase activity results hyperphosphorylation of Rb family proteins pRb (top), p107 (middle), and p130 (bottom). Arrows indicate the phosphorylated forms of the proteins.
We next assessed the consequence of the Cdk4R24C mutation on the phosphorylation status of the three members of the Rb family proteins. To this end, the cell lysates from MEFs were subjected to Western blot analysis and probed with antibodies specific to pRb or p107 or p130. Our results demonstrate that a large percentage of the protein (pRb, p107, and p130) exists in a hyperphosphorylated state in the Cdk4R24C/R24C MEFs, as evidenced by a slower migration of phosphorylated bands (Fig. 1b). In the case of p107, it is difficult to conclusively establish the presence of a hyperphosphorylated band due to limitations of the antibody that detects both hypo- and hyperphosphorylated bands of p107.
Deregulated Cdk4R24C kinase leads to increased cell proliferation and shorter cell cycle time.
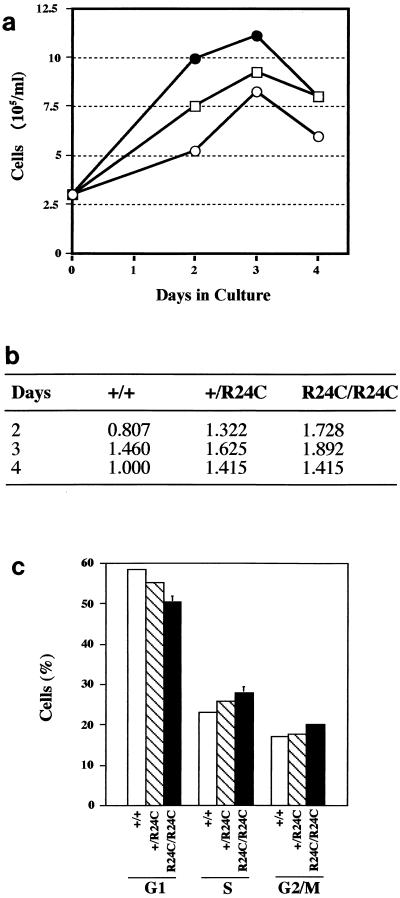
To understand the consequences of deregulated activation of the Cdk4-cyclin D pathways on growth and cell cycle progression, we examined growth and cell cycle characteristics of fibroblasts derived from Cdk4+/+, Cdk4+/R24C, and Cdk4R24C/R24C E12.5 embryos. MEFs were cultured in media with 10% FCS, and their growth characteristics in culture were determined. Cdk4R24C/R24C cells grew well in culture and displayed decreased doubling times, indicating that the expression of Cdk4R24C caused acceleration of cell proliferation (Fig. 2a). The relative increase in population doublings achieved indicates that the Cdk4R24C/R24C cells doubled at a higher rate compared to the Cdk4+/+ cells and Cdk4+/R24C cells (Fig. 2b). To analyze the effects of Cdk4R24C mutation on cell cycle progression, we performed cell cycle analysis using flow cytometry techniques. Exponentially growing Cdk4R24C/R24C MEFs exhibited a slightly higher proportion of cells in the S and G2/M phases (Fig. 2c). The consistent increase in the number of cells in the S and G2/M phases, along with a concomitant decrease in the 2n population of cells (G0/G1), together with the faster growth rate (Fig. 2a and b), suggests that the Cdk4R24C/R24C MEFs proliferate faster than the wild-type MEFs. This was in contrast to our earlier observations with Cdk4-deficient MEFs, where we observed a slight delay in S-phase entry with a concomitant increase in cells in G0/G1 phase (37). In agreement with this, we have observed a decreased cell size in Cdk4R24C/R24C MEFs compared to the Cdk4+/+ MEFs. This seems to be due to a decrease in the lengths of the G1 phase, which results in a slightly shorter doubling time. This observation is similar to that observed by Quelle et al. upon overexpression of mouse cyclin D1 in serum-stimulated mouse NIH 3T3 and rat-2 fibroblasts (36). Overexpression of cyclin D1 increased the rates of G0-to-S and G1-to-S phase transit times by several hours, leading to a proportionately reduced cell cycle time. Although such cells remained contact inhibited and anchorage dependent, similar to our observations with the Cdk4R24C/R24C MEFs, they exhibited a reduced serum requirement for growth and were also smaller in size than their normal counterparts.
FIG. 2.
Cell growth characteristics of Cdk4R24C/R24C MEFs. MEFs (passage < 4) were used for growth curve determination (a), comparison of population doublings (b), and cell cycle phase analysis (c). (a) To determine the growth curve, 3 × 105 cells from Cdk4+/+ (○), Cdk4+/R24C (□), and Cdk4R24C/R24C (•) MEF cultures were cultivated for the indicated number of days and the number of cells were counted using trypan blue exclusion analysis. The figure shown is a typical representation of the experiment that was performed two times. (b) Population doubling times between MEFs representing the three genotypes were calculated according to the formula log(Nfinal/Ninitial)/log 2, where Nfinal and Ninitial are the final and initial (3 × 105/ml) numbers of cells plated and counted at a particular time point. (c) For cell cycle phase analysis, 3 × 105 cells from Cdk4+/+ (+/+), Cdk4+/R24C (+/R24C), and Cdk4R24C/R24C (R24C/R24C) MEF cultures were exponentially cultured for 24 h and the percentage of cells in the G0/G1, S, and G2/M phases were determined by fluorescence-activated cell sorter analysis upon staining with propidium iodide.
Cdk4R24C/R24C MEFs escape replicative senescence and can be readily grown continuously in culture.
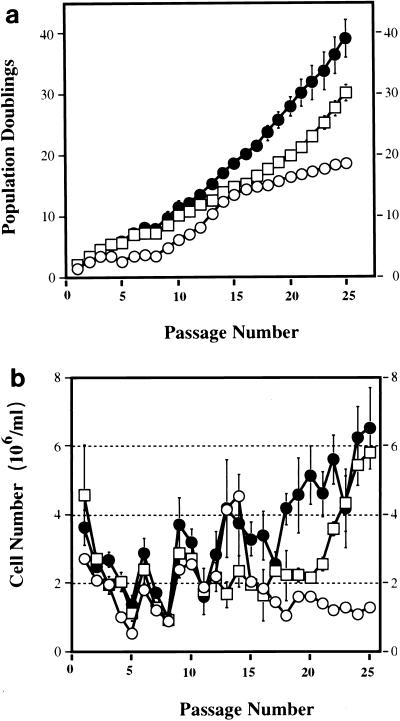
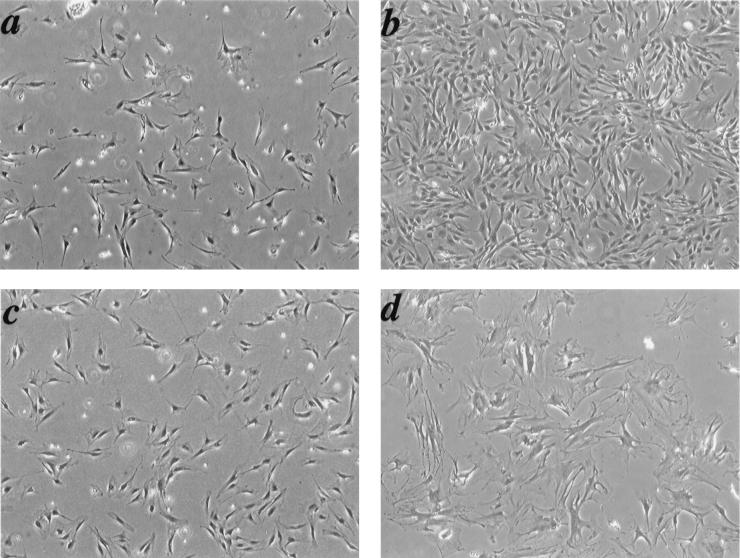
The proliferative life span of most normal cells is limited by inhibitory signals that signal cell cycle arrest after a finite number of cell divisions (1, 16, 45). Wild-type MEFs stop dividing after 15 to 30 generations and undergo replicative senescence when cultured using a 3T3 protocol (47). Recent studies demonstrate an important role played by the Rb family proteins in the regulation of an immortalized phenotype. Loss of all three known members of the Rb family, pRb, p130, and p107, in fibroblasts results in spontaneous escape from senescence and an immortal phenotype that has an increased propensity to oncogene-induced transformation (7, 39). Since the Cdk4R24C mutation leads to a partial or complete inactivation of all three members of the Rb family of proteins, it was possible that the end result could be escape from senescence, leading to increased susceptibility to transformation. To investigate this possibility, we assessed the long-term proliferative capacity and life span of the MEFs by using the 3T3 protocol (47). Like most normal cells, Cdk4+/+ cells showed a decline in the proliferation rate at about 8 to 10 passages, followed by a short growth spurt between 10 and 15 passages followed by the onset of senescence. Cdk4+/+ cells underwent approximately 18 population doublings during the first 25 passages in culture (Fig. 3a). In contrast to Cdk4+/+ cells that ceased to proliferate around passage 15, Cdk4+/R24C and Cdk4R24C/R24C cells continued to proliferate beyond passage 25 (Fig. 3b). The continued proliferation resulted in a dramatic increase in the total number of Cdk4R24C/R24C cells compared to Cdk4+/+ cells (Table 1). Morphological analysis using phase-contrast microscopy revealed a similarity between Cdk4+/+ and Cdk4R24C/R24C early-passage (passage 3) cells (Fig. 4a and c). The induction of senescence in Cdk4+/+ cells at passage 20 was confirmed by morphological changes associated with a senescence phenotype, i.e., flat, large nondividing cells (Fig. 4d). To further confirm the phenotype of replicative senescence, the activity of endogenous senescence-associated beta-galactosidase (SA-β-Gal), a specific biomarker of senescence, was examined (8). Wild-type control cells showed evidence of accumulated SA-β-Gal suggestive of senescing cells (not shown). In contrast, Cdk4R24C/R24C MEFs maintained constant proliferation rates and failed to display any morphological features of senescing cells at passage 20 (Fig. 4b). The Cdk4R24C/R24C MEFs underwent approximately 40 population doublings after 25 passages in culture (compared to 18 population doublings in control cells), indicative of a significant escape from replicative senescence (Fig. 3a). The significant increase in Cdk4R24C/R24C cell numbers as a function of passage number compared to the Cdk4+/+ cell numbers further confirms the escape of senescence observed in the Cdk4R24C/R24C cells (Table 1). The Cdk4+/R24C MEFs displayed an intermediate phenotype, with a majority of the cells maintaining a constant rate of proliferation, with approximately 30 population doublings in 25 passages (Fig. 3a). Approximately 50 to 60% of Cdk4+/R24C MEFs displayed morphological features of senescent cells (not shown). These results indicate that the expression of Cdk4R24C allows cells to escape senescence and achieve a longer life span in culture. The difference in the behavior of Cdk4+/R24C and Cdk4R24C/R24C MEFs suggests that this phenomenon may be dependent on the levels of the mutant Cdk4R24C protein with associated absence of p16INK4A binding and enhanced kinase activity, which is reflected in the degree of inactivation of the Rb family proteins.
FIG. 3.
Cdk4R24C/R24C MEFs escape from senescence. Cdk4+/+ (○), Cdk4+/R24C (n = 4) (□), and Cdk4R24C/R24C (n = 4) (•) MEF cultures were propagated in DMEM supplemented with 10% FBS for the indicated passages according to the 3T3 protocol. (a) Shown is the accumulated number of doublings that representative cultures had undergone during 25 successive passages. (b) The relative increase in cell number at successive passages is plotted. Cells were plated at 106 per plate per initial plating, and the change in the number of cells after 3 days of culture was determined. As expected, the Cdk4+/+ cells (○) stopped proliferating after passage 15, whereas the Cdk4+/R24C (□) and Cdk4R24C/R24C (•) MEF cultures continued to proliferate. Error bars indicate the standard error of the mean.
TABLE 1.
Cdk4R24C/R24C fibroblasts escape cellular senescence
| Passage no. | No. of cells
|
||
|---|---|---|---|
| +/+ | +/R24C | R24C/R24C | |
| 1 | 2.70 × 106 | 4.58 × 106 | 3.63 × 106 |
| 5 | 6.08 × 106 | 5.68 × 107 | 6.74 × 107 |
| 10 | 7.24 × 107 | 1.30 × 109 | 3.79 × 109 |
| 15 | 1.17 × 1010 | 3.91 × 1010 | 8.88 × 1011 |
| 20 | 8.44 × 1010 | 1.65 × 1012 | 7.62 × 1014 |
| 25 | 2.64 × 1011 | 2.07 × 1015 | 3.36 × 1018 |
FIG. 4.
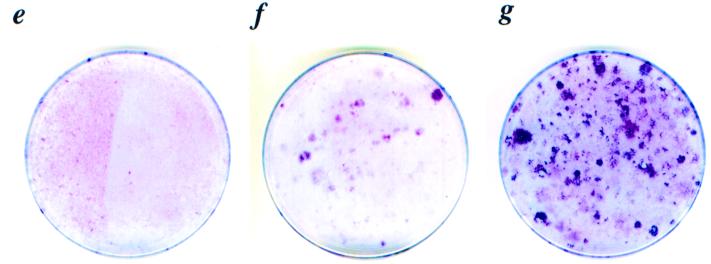
Morphological and cell growth characteristics of Cdk4R24C/R24C MEFs. (a to d) Phase-contrast pictures of Cdk4R24C/R24C (a and b) and Cdk4+/+ (c and d) cells at early passage 3 (a and c) and late passage 20 (b and d) are shown. Cdk4+/+ cells at passage 20 underwent senescence (d) while Cdk4R24C/R24C cells at passage 20 escaped senescence (b). Magnifications, ×4. (e to g) To test the ability of cells to form multiple layers (focus formation capacity), 106 cells (passage 20) representing Cdk4+/+ (e), Cdk4+/R24C (f), and Cdk4R24C/R24C (g) MEFs were plated in 10-cm-diameter dishes and grown for 2 weeks. Medium was changed every 3 days, and at the end of 2 weeks, plates were stained with Giemsa stain to determine the focus formation capacity.
To determine whether the Cdk4R24C/R24C MEFs acquired other characteristics of transformed cells, we studied their capacity to be contact inhibited after 2 weeks of confluent culturing followed by staining of colonies by Giemsa staining. After 20 passages, Cdk4+/R24C and Cdk4R24C/R24C fibroblasts became insensitive to contact-induced growth arrest and formed foci when cultured for 2 weeks (Fig. 4f and g). In contrast, the Cdk4+/+ fibroblasts (passage 20) failed to overcome contact inhibition and form foci (Fig. 4e). However, late-passage (20 passages) Cdk4R24C/R24C and Cdk4+/R24C MEFs were unable to grow in soft agar or form tumors in nude mice, suggesting that these cells were not fully transformed. Also, early-passage (passage <4) MEFs from all three genotypes were incapable of forming colonies in soft agar.
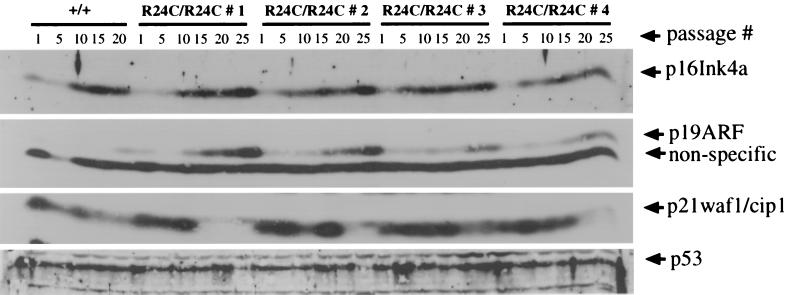
To monitor the status of other pathways that regulate the cellular senescence program, we examined the levels of proteins encoded by the INK4a-ARF locus, p16Ink4a and p19ARF, which have important roles in the induction of permanent G0/G1 arrest in cells undergoing senescence (21, 46). Levels of p16Ink4a and p19ARF increased in later-passage Cdk4R24C/R24C cells similar to Cdk4+/+ cells, indicating that the Cdk4R24C/R24C cells can resist the growth arrest signals induced by elevated levels of p16Ink4a and p19ARF (Fig. 5). This is virtually identical to that seen in Rb family triple-knockout fibroblasts (7, 39).
FIG. 5.
Expression of proteins p16Ink4a, p19ARF, p21Cip1, and p53 in Cdk4+/+ and Cdk4R24C/R24C early- and late-passage MEF cultures. Protein extracts from Cdk4+/+ and Cdk4R24C/R24C (n = 4) (R24C/R24C #1 to #4) early- and late-passage MEF cultures (passages 1, 5, 10, 15, 20, and 25) were resolved by SDS-12% PAGE and subjected to Western blot analysis using antibodies to p16Ink4a, p19ARF, p21Cip1, and p53. Not enough Cdk4+/+ cells could be harvested at passage 25 for the generation of protein extracts. The migration of proteins is identified by arrows.
Examination of the status of the p53 pathways in these cells revealed that two of the four immortal Cdk4R24C/R24C MEF clones exhibited slightly reduced levels of p53 protein at passage 25, and three of the four immortal Cdk4R24C/R24C MEF clones had a dramatic reduction in p21Cip1, a direct target of p53 (Fig. 5). These data indicate that although the loss of p53 function is not required for the immortalization of Cdk4R24C/R24C cells, p53-deficient variant cells may emerge in late-passage cells that have a selective growth advantage. These results indicate that Cdk4R24C/R24C cells can readily overcome the senescence program, establishing the importance of Cdk4-mediated phosphorylation events in determining the immortal phenotype. These results are consistent with the observation that the disruption of all three members of the Rb family proteins leads to the advent of an immortal phenotype in MEFs with a high propensity to transformation (7, 39).
Cdk4R24C/R24C MEFs can be readily transformed by activated oncogenes.
Deregulation of cell proliferation leading to neoplastic transformation in primary rodent cells requires the expression of two cooperating oncogenes (24). Therefore, the susceptibility of MEFs to neoplastic transformation can be achieved by transfection with pairs of cooperating oncogenes, such as Ha-rasV12 plus E1A, Ha-rasV12 plus myc, or Ha-rasV12 plus CDC25A. More recently, it has been shown that primary fibroblasts with disruption in all the three members of the Rb gene family can be transformed by a single oncogene, such as activated Ha-rasV12, without a need for a second cooperating oncogene (7, 39).
Based on our observations, it was important to examine whether the Cdk4R24C/R24C fibroblasts exhibit increased sensitivity to oncogene-mediated transformation since constitutive activation of Cdk4 is expected to lead to, at least, a partial inactivation of the Rb family proteins. To test this, we analyzed MEFs derived from Cdk4+/+ and Cdk4R24C/R24C embryos for susceptibility to neoplastic transformation. To this end, early-passage MEFs derived from individual embryos were grown in DMEM with 10% FBS overnight. Cells were transfected with Ha-rasV12 alone, E1A alone, myc alone, Ha-rasV12 plus myc, or Ha-rasV12 plus E1A as well as control plasmids by using standard calcium phosphate procedures. On days 14 to 21 posttransfection, cells were fixed and stained with Giemsa and foci were scored visually. Results from these experiments showed that the expression of Ha-rasV12 or E1A or v-myc genes by themselves was sufficient to induce transformation in Cdk4R24C/R24C cells, suggesting that the Cdk4R24C mutation serves as the primary event in the progression towards a fully transformed phenotype (Table 2). Furthermore, the Ha-rasV12 or E1A or v-myc-expressing Cdk4R24C/R24C cells were found to form colonies in soft agar and produce tumors in nude mice (data not shown), indicating that the Cdk4R24C mutation can cooperate with other oncogenic mutations to exacerbate the tumorigenic response. In contrast, Cdk4+/+ cells were refractory to transformation by Ha-rasV12 alone, E1A alone, or myc alone and required cooperative expression of at least two dominant oncogenes. As can be expected, Cdk4+/+ fibroblasts could be transformed by Ha-rasV12 plus myc and Ha-rasV12 plus E1A oncogenes, and moreover, the transformation potential of Cdk4R24C/R24C fibroblasts was augmented upon expression of Ha-rasV12 plus myc or Ha-rasV12 plus E1A oncogene pairs.
TABLE 2.
Neoplastic transformation of Cdk4R24C/R24C MEFs
| Oncogene | No. of cells (mean ± SD)
|
|
|---|---|---|
| Cdk4+/+ | Cdk4R24C/R24C | |
| Ras V12 | 0 | 47 (±6) |
| E1A | 0 | 29 (±7) |
| Myc | 0 | 58 (±12) |
| Ras + E1A | 34 | 124 (±23) |
| Ras + Myc | 20 | 67 (±11) |
| Control | 0 | 0 |
Germ line transmission of Cdk4R24C mutation leads to spontaneous tumorigenesis in diverse tissue types.
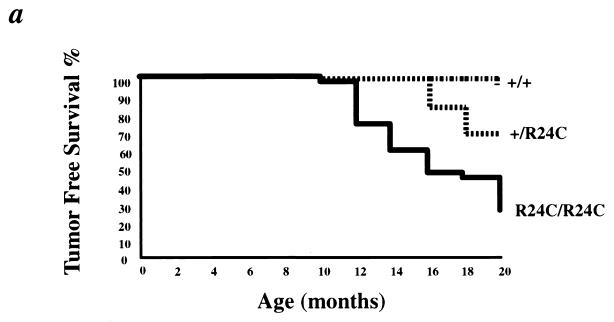
Results from our in vitro experiments suggested that the Cdk4R24C mutation might render the mice highly susceptible to spontaneous tumorigenesis. To evaluate the deleterious consequences of the germ line transmission of the R24C mutation in the Cdk4 locus on in vivo tumorigenesis, we monitored the tumor susceptibility of Cdk4R24C/R24C mice as a function of age. Cdk4R24C/R24C mice developed spontaneous tumors of various cellular origin with increasing age (Table 3). By 8 to 10 months, homozygous Cdk4R24C/R24C mice developed tumors in multiple organs, clearly demonstrating the consequence of deregulated Cdk4 expression (Fig. 6a). A summary of all the tumor types that were observed in the Cdk4R24C/R24C mice is presented (Table 3). We observed single as well as multiple primary tumors in several of the mice, and in many cases, a single primary tumor was found to have metastasized to multiple sites, such as liver, brain, and lung. Heterozygous Cdk4+/R24C mice also developed spontaneous tumors, albeit at a reduced frequency and incidence (Fig. 6a). Among the mice that were monitored for tumor development for 18 months, 74% of Cdk4R24C/R24C mice (25 out of 34) and 55% of Cdk4+/R24C mice (17 out of 31) developed tumors of various etiology. Of these, 26% of the Cdk4R24C/R24C mice (9 out of 34) developed tumors at an accelerated rate by 10 to 12 months. In contrast, none of the 21 Cdk4+/+ mice that were monitored for the same period of 18 months developed tumors. Moreover, there appears to have been no sex bias in the extent of tumor occurrence as both male and female Cdk4R24C/R24C mice were equally susceptible to increased tumorigenesis (Cdk4+/R24C mice: 7 females + 10 males = 17 out of 31 mice; Cdk4R24C/R24C mice: 14 females + 11 males = 25 out of 34 mice).
TABLE 3.
Spontaneous tumor development in Cdk4R24C/R24C mice
| Tumor pathology | Anatomic site | Occurrence (%) |
|---|---|---|
| Adenosquamous carcinoma | Mammary gland, subcutaneous | 9.5 |
| Adenocanthoma | Mammary gland | 9.5 |
| Adenoma | Pituitary | 14.3 |
| Carcinoma | Pituitary | 16.7 |
| Metastatic endocrine carcinoma | Brain | 4.8 |
| Astrocytoma | Brain | 2.4 |
| Hemagiosarcoma | Skin/subcutaneous | 16.8 |
| Hemagiosarcoma | Mesentary | 7.1 |
| Hemagiosarcoma | Liver | 31.0 |
| Hemagiosarcoma | Skeletal muscle | 2.4 |
| Hemagiosarcoma | Lungs | 2.4 |
| Hemagiosarcoma | Spleen | 7.1 |
| Rhabdomyosarcoma | Skin/subcutaneous, skeletal muscle | 4.8 |
| Spindle cell carcinoma | Kidney, liver, mesentery, pancreas, spleen | 11.9 |
| Hepatocellular adenoma | Liver | 11.9 |
| Hepatocellular carcinoma | Liver | 4.8 |
| Bronchiolar-alveolar carcinoma | Lungs | 4.8 |
| Metastatic neoplasm | Lung | 2.4 |
| Islet cell adenoma | Pancreas | 4.8 |
| Islet cell carcinoma | Pancreas | 4.8 |
| Interstitial cell tumor | Testicle | 23.8 |
| Adenoma | Seminal vesicle | 4.8 |
| Hematoma | Periovarian tissue | 2.4 |
| Fibrosarcoma | Skin, subcutaneous | 2.4 |
| Melanoma | Skin | 2.4 |
| Myxomatous mesothelioma | Skin, abdominal muscle | 2.4 |
| Keratocanthoma | Skin | 4.8 |
| Myxosarcoma | Subcutaneous | 2.4 |
| Basal cell tumor | Skin | 4.8 |
FIG. 6.
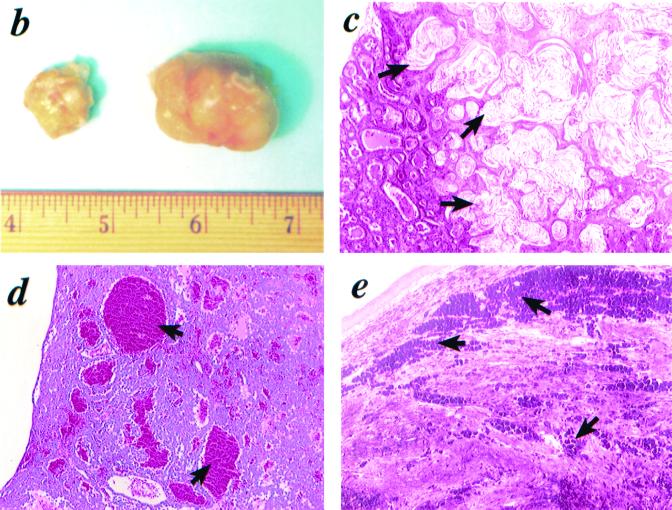
Increased spontaneous tumor incidence in Cdk4R24C/R24C mice. (a) Kaplan-Meier survival curves of Cdk4+/+ (n = 21) (+/+), Cdk4+/R24C (n = 31) (+/R24C), and Cdk4R24C/R24C (n = 34) (R24C/R24C) mice that were observed for 20 months for the appearance of detectable or palpable tumors. Tumor-prone mice were euthanized, and tumors were dissected and processed for histological analysis. (b to e) Histological analysis of spontaneous mammary and pituitary tumors and melanomas in Cdk4R24C/R24C mice. Photomicrographs of three tumor types are presented. (b) Cdk4R24C/R24C mice are susceptible to increased mammary tumorigenesis with severe tumor burden. (c) Mammary tumors are either mammary adenocarcinomas or adenocanthomas with squamous differentiation and keratinization (indicated by arrows). (d) Cdk4R24C/R24C mice are susceptible to increased incidence of pituitary tumorigenesis either in the pars distalis or pars intermedia with characteristic blood-filled lakes (indicated by arrows). (e) Cdk4R24C/R24C mice also present a low incidence of melanoma occurrence with pigment granules and cells loaded with pigment (indicated by arrows).
Cdk4R24C/R24C female mice developed severe mammary duct dilation and a high incidence of mammary tumors, which are often very aggressive with large tumor burden (Fig. 6b). A majority of the mammary tumors analyzed were adenosquamous carcinomas with papillary and cribiform elements or adenocarcinomas and adenocanthomas with squamous differentiation (Fig. 6c). The tumor cells lining the cavities underwent squamous differentiation or metaplasia with keratinization and formation of laminated horny pearls. This is consistent with the observation that mouse mammary tumor virus-cyclin D1 transgenic mice are prone to a high frequency of adenocarcinomas and adenocanthomas (50).
Cdk4R24C/R24C mice are also susceptible to an increased development of pituitary tumors arising either in the pars intermedia or the pars distalis, with characteristic angiomatous areas or dilated “blood-filled lakes” of various sizes (indicated by arrows) (Fig. 6d). In many cases, the pituitary tumors compressed adjacent nontumorous tissues, such as the hypothalamus, pons, and brain. Interestingly, mice that are heterozygous at the Rb loci and those that have disruptions in CKI genes p27Kip1 and p18Ink4c also develop pituitary tumors (11, 13, 18, 22, 30, 48).
While the germ line CDK4-Arg24Cys (R24C) mutation, which abolishes the ability of CDK4 to bind to p16INK4a, predisposes humans to hereditary melanoma, we have observed a low incidence of melanoma occurrence in the Cdk4R24C mouse model. The melanomas displayed characteristic pigment granules or cells loaded with pigment (Fig. 6e). This suggests that other carcinogenic events, such as exposure to UV radiation, could play a major role in this process. This is consistent with recent reports that melanoma genesis is dependent on the expression of H-RasV12G in a mouse melanoma model null for the tumor suppressors p16INK4a and p19ARF, due to disruption of the Ink4a-ARF locus (4). Therefore, it is likely that cooperation between other distinct genetic alterations, in addition to the Cdk4R24C mutation, may be necessary to induce melanoma in this mouse model.
Cdk4R24C mutation can cooperate with and exacerbate tumorigenic potential of activated Ras pathways.
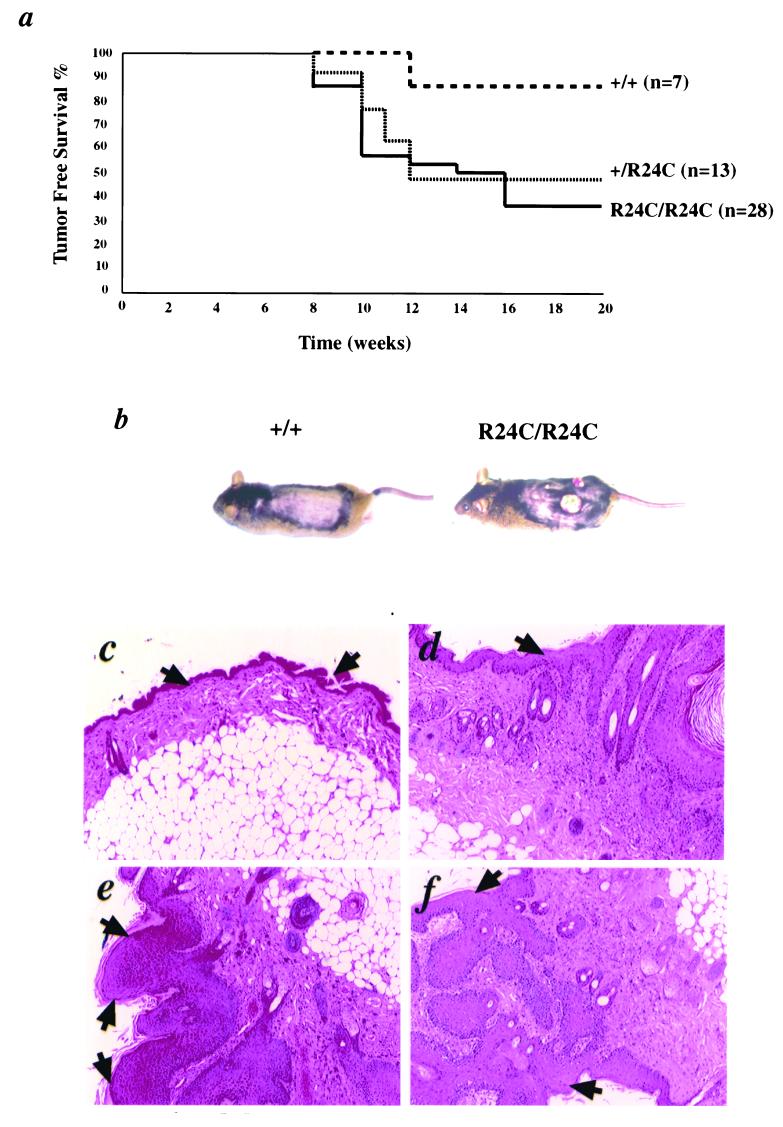
To determine the cooperation between an activated Ras pathway and the constitutively activated Cdk4-pRb pathway, we conducted chemical carcinogenesis experiments using the two-step model of skin carcinogenesis (12). This protocol involves a single application of the well-characterized mutagen DMBA followed by a twice-per-week administration of the tumor promoter TPA. Such a treatment results in the development of papillomas at the site of initiation and promotion with a characteristic oncogenic mutation in the 61st codon of the Ha-ras gene (12). Results from the DMBA/TPA skin carcinogenesis experiments indicated that the Cdk4R24C/R24C and Cdk4+/R24C mice were highly susceptible to an increased incidence of papillomas with a very short latency period compared to that of the wild-type mice (Fig. 7a and b). In contrast, topical application of DMBA alone did not lead to the development of papillomas in Cdk4+/+, Cdk4+/R24C, or Cdk4R24C/R24C mice. Histological analysis of the tumors revealed that all of the tumors were well-differentiated papillomas, with regions of hyperplasia in the epidermis and no invasion into the underlying dermis (Fig. 7c to f). In addition to papillomas, which was the predominant tumor type, we also observed a reduced incidence of benign epidermal tumors classified as keratocanthomas consisting of large keratin-filled cystic structures surrounded by a very well-differentiated squamous epithelium. These results indicate that the Cdk4R24C mutation, which deregulates the Rb pathways, can collaborate with and exacerbate the tumorigenic potential of the activated Ras pathway.
FIG. 7.
Collaboration between pRb and Ras-oncogenic pathways in Cdk4R24C/R24C mice. (a) Kaplan-Meier survival curves of DMBA-TPA-treated Cdk4+/+ (n = 7) (+/+), Cdk4+/R24C (n = 13) (+/R24C), and Cdk4R24C/R24C (n = 28) (R24C/R24C) mice that were observed for 20 weeks for appearance of detectable or palpable tumors. Tumor-prone mice were euthanatized, and tumors were dissected and processed for histological analysis. (b to f) Treatment with DMBA plus TPA leads to the development of skin tumors (b) that are histologically characterized as papillomas (d to f). The histology of a normal tumor-free skin is shown (c). The normal epidermis (c) and the epidermal hyperplasia and papillomas (d to f) are indicated by arrows.
DISCUSSION
The absolute requirement of Rb family proteins during embryogenesis and early development has hindered the examination of their role in cancer predisposition using conventional gene disruption techniques (5, 6, 19, 25–28). Most of the in vivo studies that describe the role of Rb family proteins have been restricted to characterization of mouse models that either (i) carry heterozygous disruption of Rb family genes with or without inactivation of other key cell cycle regulatory proteins or (ii) inactivate Rb protein family function via expression of viral proteins, such as the papilloma virus E7 protein or the simian virus 40 large T-antigen protein (14, 17). Recent studies that describe the disruption of the retinoblastoma family gene loci in fibroblasts have clearly elucidated the importance of the Rb family proteins in the regulation of cell cycle progression and its underlying implication in cellular immortalization and transformation (7, 39). To investigate the consequence of Rb family inactivation through Cdk4-mediated phosphorylation, we chose to introduce a germ line mutation (R24C) in the mouse Cdk4 locus that resulted in a deregulated Cdk4R24C kinase. The Cdk4R24C mutation that abolishes the ability of CDK4 to bind to and be inactivated by p16INK4a predisposes humans to hereditary melanoma (51, 53). Our results presented here show that introduction of the Cdk4R24C mutation resulted not only in the abrogation of p16Ink4a binding to Cdk4R24C but also enhanced the kinase activity of the enzyme which in turn leads to hyperphosphorylation of Rb family proteins. This technique allows the expression of the Rb family proteins throughout mouse development, ensuring the viability of the mice in contrast to studies that were based on disruption of Rb family proteins via gene targeting methods (5, 6, 19, 25–28). Expression of deregulated Cdk4R24C resulted in hyperphosphorylation of the Rb family proteins, suggestive of at least a partial inactivation of Rb protein family function.
The hyperphosphorylated state of the Rb family proteins can be regarded as a physiological inactivation of the Rb family protein function (14, 17). This mode of inactivation is distinct from that reported elsewhere, where the loci coding for all three Rb family proteins were disrupted by traditional gene disruption methods in fibroblasts (7, 39). Disruption of the retinoblastoma family gene loci in fibroblasts leads to a complete absence of Rb family proteins, whereas our studies allow expression of Rb family proteins but lead to partial or total functional inactivation of the Rb family protein function that is dependent on regulated phosphorylation by the cyclin D/Cdk4 complex.
The observation that mice with a homozygous Cdk4R24C mutation were viable suggests that (i) inactivation of Rb protein family function via phosphorylation as opposed to gene disruption (both of which may lead to deregulated E2F activity) have distinct outcomes during embryogenesis, (ii) the Cdk4R24C mutation which leads to hyperphosphorylation of the Rb family proteins may result in a partial inactivation of Rb family proteins, and (iii) functions of Rb family proteins during embryogenesis and early development are dependent on their interaction with proteins that are distinct from those that regulate cell cycle progression. It has been shown that Rb family proteins interact, in addition to their well-characterized interactions with the E2F family proteins, with proteins that are important during development, such as Myo D, myogenin, MDM2, Myc, PU.1, c-Abl, Id-2, Pax proteins, etc. (14, 17). Rb gene disruption using the more conventional gene disruption approaches may abrogate these critical interactions, resulting in embryonic lethality (5, 6, 19, 25–28). Our approach of inducing a partial or complete inactivation of Rb protein function depends on the constitutive hyperphosphorylation of Rb family proteins by the activated Cdk4R24C kinase, which allows the expression of Rb family proteins and preserves their interactions with proteins that are required during development. Our observations that expression of activated Cdk4R24C leads to (i) increased cellular proliferation, (ii) shorter cell cycle time, (iii) escape of fibroblasts from senescence, and (iv) increased susceptibility to oncogene-induced transformation are similar to those observed with fibroblasts that are nullizygous at the retinoblastoma family gene loci (7, 39). These observations highlight the importance of appropriate regulation of G1 cell cycle progression events that are dependent on timely phosphorylation of the Rb family proteins. Moreover, our observations suggest that inappropriate phosphorylation events, such as that observed in the case of Rb protein family hyperphosphorylation due to the expression of activated Cdk4R24C, can lead to the advent of a cancerous state.
The Cdk4R24C/R24C mice expressed normal levels of Cdk6, and therefore, it is likely that the observed effects are restricted to tissue and cell types in which Cdk4 is the primary D-type cyclin-associated Cdk. The diverse spectrum of tumors observed in the Cdk4R24C/R24C mice is rather surprising taking into consideration the relatively restricted tumorigenesis profile in mice with a disruption of the INK4a-ARF locus (21, 40). The INK4a-ARF locus encodes two proteins, p16INK4a and p19ARF, respectively (3, 15, 35). p16INK4a inhibits the Cdk4 and Cdk6 kinases, influencing the pRb pathways, whereas p19ARF arrests the cell cycle in a p53-dependent manner (29, 32, 52). p16INK4a:p19ARF-deficient mice develop primarily lymphomas and sarcomas that are restricted to the subcutis with invasion of the underlying musculature or a more generalized distribution that primarily involves lymphoid organs (40). Likewise, mice lacking p19ARF alone (with intact p16INK4a expression) also display increased spontaneous tumorigenesis with a predominance of lymphomas and fibrosarcomas and other rare tumor types, such as thymomas, histiocytomas, and salivary gland tumors (21). It is possible that the disparity between the tumor phenotypes displayed by the Cdk4R24C/R24C mice in comparison with mice deficient in p16INK4a and/or p19ARF is a reflection of (i) the restricted expression pattern of the two proteins p16INK4a and p19ARF, (ii) the preferential expression pattern of Cdk4 versus Cdk6 in certain tissues, and (iii) the enhanced kinase activity of the Cdk4R24C protein.
In addition, the tumor phenotype observed may also be a reflection of the importance of a specific negative regulation of cyclin D/Cdk4 complexes by p16Ink4a since the Cdk4R24C mutation is refractory to inhibition by p16Ink4a. Also, taking into consideration the established role of p16Ink4a in regulation of senescence pathways, in conjunction with our observations that Cdk4R24C/R24C MEFs fail to undergo senescence and can be readily immortalized, indicates that the efficacy of the Cdk4R24C mutation may be more pronounced with increasing age. In addition, it is likely that activated Cdk4R24C may (i) be resistant to inhibition by other members of the Ink4 family of CKIs (p15Ink4b, p18Ink4c, and p19Ink4d) and/or (ii) deregulate the downstream Cdk2-associated kinase activity due to redistribution of p21Kip1 and p27Kip2 proteins from cyclin E/A-Cdk2 complexes to the cyclin D/Cdk4R24C complexes. Recent data indicate that p21Kip1 and p27Kip2 proteins are specific inhibitors of Cdk2-associated kinase activity, whereas they are essential for stability and activity of Cdk4-associated complexes (2, 23, 44). Based on the results presented, we speculate that the tumor phenotype displayed by the Cdk4R24C/R24C mice is a reflection of the importance of Cdk4 activity in governing G1/S cell cycle progression via the regulation of Rb family activity and monitoring of senescence-associated pathways in diverse cell types. Importantly, our results present, for the first time, the deleterious consequences of simultaneous inactivation of members of the Rb family on cancer predisposition due to a naturally occurring mutation.
While the germ line Cdk4R24C mutation predisposes humans to hereditary melanoma, we have observed a low incidence of melanoma occurrence in the Cdk4R24C mouse model. This suggests that other carcinogenic events, such as exposure to UV radiation or activation of other tumor promotion pathways, could play a major role in this process. Functional collaboration between distinct tumor promotion pathways is the hallmark of an aggressive tumor phenotype (43, 44). Thus, many human tumors have alterations in multiple oncogene and/or tumor suppressor pathways. Recent evidence indicates that the Cdk4-pRb pathway is targeted by oncogenic Ras and Ras inactivation-induced cell cycle arrest is pRb dependent (10). In addition, it has been shown that cells nullizygous at the INK4 locus are permissive to Ras-induced proliferation and transformation (21, 40). However, in wild-type MEFs, oncogenic Ras promotes premature cell senescence (41). It has been shown that induction of Ha-rasV12 in rat intestinal epithelial cells leads to transformation that is associated with a sustained proliferation and accumulation of cyclin D1 (34, 42). Furthermore, cyclin D1-deficient mice have a reduced propensity to skin tumor development in response to chemical carcinogen treatments that activate oncogenic Ras pathways (38). In agreement with this, results of the two-stage skin chemical carcinogenesis experiments indicate that the R24C mutation in the Cdk4 locus can synergize with and exacerbate the tumor potential of oncogenic Ras pathways. Therefore, it is likely that cooperation between other distinct genetic alterations, in addition to the Cdk4R24C mutation, may be necessary to induce melanoma in this mouse model. Importantly, these results indicate that the Cdk4R24C mutation harbors the propensity to act in concert with distinct oncogenic or chemical carcinogenic events to further worsen the cancerous state. Therefore, the Cdk4R24C/R24C model would be a valuable tool to assess the carcinogenic activity of xenobiotic agents, aiding development of cell cycle targeted anti-neoplastic drugs or small molecules to combat cancer. Based on the wide variety of tumors that develop in mice harboring the Cdk4R24C mutation, we hypothesize that the occurrence of mutations, such as the Cdk4R24C mutation, may be the harbinger of many types of human cancers.
Acknowledgments
The work presented was supported by a Fels Foundation grant and NIH grant R24CA088261 to E.P.R. and by the Anna D. Barker Fellowship in Basic Research to S.G.R. by the American Association for Cancer Research.
We acknowledge Xavier Graña for critical reading of the manuscript. We thank Mariano Barbacid and Marcos Malumbres for helpful discussions and communication of their data.
REFERENCES
- 1.Campisi, J. 2000. Cancer, aging and cellular senescence. In Vivo 14:183–188. [PubMed] [Google Scholar]
- 2.Cheng, M. P., J. A. Olivier, M. Diehl, M. F. Fero, M. F. Roussel, J. M. Roberts, and C. J. Sherr. 1999. The p21Cip1 and p27Kip1 CDK inhibitors are essential activators of cyclin-D dependent kinases in murine fibroblasts. EMBO J. 18:1571–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chin, L., J. Pomerantz, and R. A. DePinho. 1998. The INK4a/ARF tumor suppressor: one gene—two products—two pathways. Trends Biochem. Sci. 23:291–296. [DOI] [PubMed] [Google Scholar]
- 4.Chin, L., A. Tam, J. Pomerantz, M. Wong, J. Holash, N. Bardeesy, Q. Shen, R. O’Hagan, J. Pantginis, H. Zhou, J. W. Horner II, C. Cordon-Cardo, G. D. Yancopoulos, and R. A. DePinho. 1999. Essential role for oncogenic Ras in tumour maintenance. Nature 400:468–472. [DOI] [PubMed] [Google Scholar]
- 5.Clarke, A. R., E. R. Maandag, M. van Roon, N. M. van der Lugt, M. van der Valk, M. L. Hooper, A. Berns, and H. te Riele. 1992. Requirement for a functional Rb-1 gene in murine development. Nature 359:328–330. [DOI] [PubMed] [Google Scholar]
- 6.Cobrinik, D., M. H. Lee, G. Hannon, G. Mulligan, R. T. Bronson, N. Dyson, E. Harlow, D. Beach, R. A. Weinberg, and T. Jacks. 1996. Shared role of the pRB-related p130 and p107 proteins in limb development. Genes Dev. 10:1633–1644. [DOI] [PubMed] [Google Scholar]
- 7.Dannenberg, J. H., A. van Rossum, L. Schuijff, and H. te Riele. 2000. Ablation of the retinoblastoma gene family deregulates G(1) control causing immortalization and increased cell turnover under growth-restricting conditions. Genes Dev. 14:3051–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dimri, G. P., X. Lee, G. Basile, M. Acosta, G. Scott, C. Roskelley, E. E. Medrano, M. Linskens, I. Rubelj, O. Pereira-Smith, et al. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA 92:9363–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donehower, L. A., M. Harvey, B. L. Slagle, M. J. McArthur, C. A. Montgomery, Jr., J. S. Butel, and A. Bradley. 1992. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356:215–221. [DOI] [PubMed] [Google Scholar]
- 10.Ewen, M. E. 2000. Relationship between Ras pathways and cell cycle control. Prog. Cell Cycle Res. 4:1–17. [DOI] [PubMed] [Google Scholar]
- 11.Fero, M. L., M. Rivkin, M. Tasch, P. Porter, C. E. Carow, E. Firpo, K. Polyak, L. H. Tsai, V. Broudy, R. M. Perlmutter, K. Kaushansky, and J. M. Roberts. 1996. A syndrome of multiorgan hyperplasia with features of gigantism, tumorigenesis, and female sterility in p27(Kip1)-deficient mice. Cell 85:733–744. [DOI] [PubMed] [Google Scholar]
- 12.Finch, J. S., H. E. Albino, and G. T. Bowden. 1996. Quantitation of early clonal expansion of two mutant 61st codon c-Ha-ras alleles in DMBA/TPA treated mouse skin by nested PCR/RFLP. Carcinogenesis 17:2551–2557. [DOI] [PubMed] [Google Scholar]
- 13.Franklin, D. S., V. L. Godfrey, H. Lee, G. I. Kovalev, R. Schoonhoven, S. Chen-Kiang, L. Su, and Y. Xiong. 1998. CDK inhibitors p18(INK4c) and p27(Kip1) mediate two separate pathways to collaboratively suppress pituitary tumorigenesis. Genes Dev. 12:2899–2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grana, X., J. Garriga, and X. Mayol. 1998. Role of the retinoblastoma family, pRb, p107 and p130 in the negative control of cell growth. Oncogene 17:3365–3383. [DOI] [PubMed] [Google Scholar]
- 15.Haber, D. A. 1997. Splicing into senescence: the curious case of p16 and p19ARF. Cell 91:555–558. [DOI] [PubMed] [Google Scholar]
- 16.Hayflick, L., and P. S. Moorehead. 1961. The serial cultivation of human diploid cell strains. Exp. Cell Res. 25:585–621. [DOI] [PubMed] [Google Scholar]
- 17.Herwig, S., and M. Strauss. 1997. The retinoblastoma protein: a master regulator of cell cycle, differentiation and apoptosis. Eur. J. Biochem. 246:581–601. [DOI] [PubMed] [Google Scholar]
- 18.Hu, N., A. Gutsmann, D. C. Herbert, A. Bradley, W. H. Lee, and E. Y. Lee. 1994. Heterozygous Rb-1 delta 20/+mice are predisposed to tumors of the pituitary gland with a nearly complete penetrance. Oncogene 9:1021–1027. [PubMed] [Google Scholar]
- 19.Jacks, T., A. Fazeli, E. M. Schmitt, R. T. Bronson, M. A. Goodell, and R. A. Weinberg. 1992. Effects of an Rb mutation in the mouse. Nature 359:295–300. [DOI] [PubMed] [Google Scholar]
- 20.Kamb, A., N. A. Gruis, J. Weaver-Feldhaus, Q. Liu, K. Harshman, S. V. Tavtigian, E. Stockert, R. S. Day III, B. E. Johnson, and M. H. Skolnick. 1994. A cell cycle regulator potentially involved in genesis of many tumor types. Science 264:436–440. [DOI] [PubMed] [Google Scholar]
- 21.Kamijo, T., F. Zindy, M. F. Roussel, D. E. Quelle, J. R. Downing, R. A. Ashmun, G. Grosveld, and C. J. Sherr. 1997. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell 91:649–659. [DOI] [PubMed] [Google Scholar]
- 22.Kiyokawa, H., R. D. Kineman, K. O. Manova-Todorova, V. C. Soares, E. S. Hoffman, M. Ono, D. Khanam, A. C. Hayday, L. A. Frohman, and A. Koff. 1996. Enhanced growth of mice lacking the cyclin-dependent kinase inhibitor function of p27(Kip1). Cell 85:721–732. [DOI] [PubMed] [Google Scholar]
- 23.LaBaer, J., M. D. Garrett, L. F. Stevenson, J. M. Slingerland, C. Sandhu, H. S. Chou, A. Fattaey, and E. Harlow. 1997. New functional activities for the p21 family of CDK inhibitors. Genes Dev. 11:847–862. [DOI] [PubMed] [Google Scholar]
- 24.Land, H., L. F. Parada, and R. A. Weinberg. 1983. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature 304:596–602. [DOI] [PubMed] [Google Scholar]
- 25.LeCouter, J. E., B. Kablar, W. R. Hardy, C. Ying, L. A. Megeney, L. L. May, and M. A. Rudnicki. 1998. Strain-dependent myeloid hyperplasia, growth deficiency, and accelerated cell cycle in mice lacking the Rb-related p107 gene. Mol. Cell. Biol. 18:7455–7465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LeCouter, J. E., B. Kablar, P. F. Whyte, C. Ying, and M. A. Rudnicki. 1998. Strain-dependent embryonic lethality in mice lacking the retinoblastoma-related p130 gene. Development 125:4669–4679. [DOI] [PubMed] [Google Scholar]
- 27.Lee, E. Y., C. Y. Chang, N. Hu, Y. C. Wang, C. C. Lai, K. Herrup, W. H. Lee, and A. Bradley. 1992. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature 359:288–294. [DOI] [PubMed] [Google Scholar]
- 28.Lee, M. H., B. O. Williams, G. Mulligan, S. Mukai, R. T. Bronson, N. Dyson, E. Harlow, and T. Jacks. 1996. Targeted disruption of p107: functional overlap between p107 and Rb. Genes Dev. 10:1621–1632. [DOI] [PubMed] [Google Scholar]
- 29.Lloyd, A. C. 2000. p53: only ARF the story. Nat. Cell Biol. 2:E48–E50. [DOI] [PubMed] [Google Scholar]
- 30.Nakayama, K., N. Ishida, M. Shirane, A. Inomata, T. Inoue, N. Shishido, I. Horii, and D. Y. Loh. 1996. Mice lacking p27(Kip1) display increased body size, multiple organ hyperplasia, retinal dysplasia, and pituitary tumors. Cell 85:707–720. [DOI] [PubMed] [Google Scholar]
- 31.Nobori, T., K. Miura, D. J. Wu, A. Lois, K. Takabayashi, and D. A. Carson. 1994. Deletions of the cyclin-dependent kinase-4 inhibitor gene in multiple human cancers. Nature 368:753–756. [DOI] [PubMed] [Google Scholar]
- 32.Pomerantz, J., N. Schreiber-Agus, N. J. Liegeois, A. Silverman, L. Alland, L. Chin, J. Potes, K. Chen, I. Orlow, H. W. Lee, C. Cordon-Cardo, and R. A. DePinho. 1998. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2’s inhibition of p53. Cell 92:713–723. [DOI] [PubMed] [Google Scholar]
- 33.Prives, C. 1998. Signaling to p53: breaking the MDM2–p53 circuit. Cell 95:5–8. [DOI] [PubMed] [Google Scholar]
- 34.Pruitt, K., R. G. Pestell, and C. J. Der. 2000. Ras inactivation of the retinoblastoma pathway by distinct mechanisms in NIH 3T3 fibroblast and RIE-1 epithelial cells. J. Biol. Chem. 275:40916–40924. [DOI] [PubMed] [Google Scholar]
- 35.Quelle, D. E., F. Zindy, R. A. Ashmun, and C. J. Sherr. 1995. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell 83:993–1000. [DOI] [PubMed] [Google Scholar]
- 36.Quelle, D. E., R. A. Ashmun, S. A. Shurtleff, J. Y. Kato, D. Bar-Sagi, M. F. Roussel, and C. J. Sherr. 1993. Overexpression of mouse D-type cyclins accelerates G1 phase in rodent fibroblasts. Genes Dev. 7:1559–1571. [DOI] [PubMed] [Google Scholar]
- 37.Rane, S. G., P. Dubus, R. V. Mettus, E. J. Galbreath, G. Boden, E. P. Reddy, and M. Barbacid. 1999. Loss of Cdk4 expression causes insulin-deficient diabetes and Cdk4 activation results in beta-islet cell hyperplasia. Nat. Genet. 22:44–52. [DOI] [PubMed] [Google Scholar]
- 38.Robles, A. I., M. L. Rodriguez-Puebla, A. B. Glick, C. Trempus, L. Hansen, P. Sicinski, R. W. Tennant, R. A. Weinberg, S. H. Yuspa, and C. J. Conti. 1998. Reduced skin tumor development in cyclin D1-deficient mice highlights the oncogenic ras pathway in vivo. Genes Dev. 12:2469–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sage, J., G. J. Mulligan, L. D. Attardi, A. Miller, S. Chen, B. Williams, E. Theodorou, and T. Jacks. 2000. Targeted disruption of the three Rb-related genes leads to loss of G(1) control and immortalization. Genes Dev. 14:3037–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Serrano, M., H. Lee, L. Chin, C. Cordon-Cardo, D. Beach, and R. A. DePinho. 1996. Role of the INK4a locus in tumor suppression and cell mortality. Cell 85:27–37. [DOI] [PubMed] [Google Scholar]
- 41.Serrano, M., A. W. Lin, M. E. McCurrach, D. Beach, and S. W. Lowe. 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88:593–602. [DOI] [PubMed] [Google Scholar]
- 42.Shao, J., H. Sheng, R. N. DuBois, and R. D. Beauchamp. 2000. Oncogenic Ras-mediated cell growth arrest and apoptosis are associated with increased ubiquitin-dependent cyclin D1 degradation. J. Biol. Chem. 275:22916–22924. [DOI] [PubMed] [Google Scholar]
- 43.Sherr, C. J. 2000. The Pezcoller lecture: cancer cell cycles revisited. Cancer Res. 60:3689–3695. [PubMed] [Google Scholar]
- 44.Sherr, C. J., and J. M. Roberts. 1999. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 13:1501–1512. [DOI] [PubMed] [Google Scholar]
- 45.Sherr, C. J., and R. A. DePinho. 2000. Cellular senescence: mitotic clock or culture shock? Cell 102:407–410. [DOI] [PubMed] [Google Scholar]
- 46.Stein, G. H., and V. Dulic. 1998. Molecular mechanisms for the senescent cell cycle arrest. J. Investig. Dermatol. Symp. Proc. 3:14–18. [PubMed] [Google Scholar]
- 47.Todaro, G. J., and H. Green. 1963. Quantitative studies of the growth of the mouse embryo cells in culture and their development into established cell lines. J. Cell Biol. 17:299–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vooijs, M., and A. Berns. 1999. Developmental defects and tumor predisposition in Rb mutant mice. Oncogene 18:5293–5303. [DOI] [PubMed] [Google Scholar]
- 49.Vousden, K. H. 2000. p53: death star. Cell 103:691–694. [DOI] [PubMed] [Google Scholar]
- 50.Wang, T. C., R. D. Cardiff, L. Zukerberg, E. Lees, A. Arnold, and E. V. Schmidt. 1994. Mammary hyperplasia and carcinoma in MMTV-cyclin D1 transgenic mice. Nature 369:669–671. [DOI] [PubMed] [Google Scholar]
- 51.Wolfel, T., M. Hauer, J. Schneider, M. Serrano, C. Wolfel, E. Klehmann-Hieb, E. De Plaen, T. Hankeln, K. H. Meyer zum Buschenfelde, and D. Beach. 1995. A p16INK4a-insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science 269:1281–1284. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, Y., Y. Xiong, and W. G. Yarbrough. 1998. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell 92:725–734. [DOI] [PubMed] [Google Scholar]
- 53.Zuo, L., J. Weger, Q. Yang, A. M. Goldstein, M. A. Tucker, G. J. Walker, N. Hayward, and N. C. Dracopoli. 1996. Germline mutations in the p16INK4a binding domain of CDK4 in familial melanoma. Nat. Genet. 12:97–99. [DOI] [PubMed] [Google Scholar]