Abstract
The HMG box containing protein UBF binds to the promoter of vertebrate ribosomal repeats and is required for their transcription by RNA polymerase I in vitro. UBF can also bind in vitro to a variety of sequences found across the intergenic spacer in Xenopus and mammalian ribosomal DNA (rDNA) repeats. The high abundance of UBF, its colocalization with rDNA in vivo, and its DNA binding characteristics, suggest that it plays a more generalized structural role over the rDNA repeat. Until now this view has not been supported by any in vivo data. Here, we utilize chromatin immunoprecipitation from a highly enriched nucleolar chromatin fraction to show for the first time that UBF binding in vivo is not restricted to known regulatory sequences but extends across the entire intergenic spacer and transcribed region of Xenopus, human, and mouse rDNA repeats. These results are consistent with a structural role for UBF at active nucleolar organizer regions in addition to its recognized role in stable transcription complex formation at the promoter.
In eukaryotic organisms, 18S and 28S rRNA is produced from a 35S-47S pre-rRNA that is transcribed by RNA polymerase I (Pol I). Ribosomal genes (ribosomal DNA [rDNA]) are highly repeated and organized as large tandem arrays termed nucleolar organizer regions (NORs). The Xenopus genome contains 450 copies of the rDNA repeat in a single NOR. The 400 copies of the human rDNA repeat are distributed among five NORs on the p-arms of acrocentric chromosomes 13, 14, 15, 21, and 22 (30). Interestingly, ribosomal genes appear to be the only genes found on the acrocentric p-arms. Sequences both distal and proximal to human NORs are comprised of satellite DNA packaged as heterochromatin (11, 53, 56, 60).
Within NORs, sequences carrying the pre-rRNA are separated from each other by an intergenic spacer (IGS). The IGSs in Xenopus and human rDNA repeats are 4 (8) and 30 (17) kb, respectively. IGSs typically contain an array of repetitive elements. In Xenopus and mouse, blocks of repetitive elements immediately upstream of the gene promoter function as transcriptional enhancers (47). These enhancer elements are interspersed with spacer promoters. Enhancer function has not yet been ascribed to any class of repetitive element found in the human IGS. The human IGS also contains a subset of sequences found elsewhere in the genome, such as alu repeats and a cdc27 psuedogene (17). Presumably, these latter elements are not directly involved in the regulation of human rDNA transcription.
Transcription of ribosomal genes by RNA Pol I is activated by a transcription machinery distinct from that employed by RNA Pol II and III (18). In vitro transcription studies have identified upstream binding factor (UBF), the TATA-binding protein containing complex SL1 in humans (13), TIF-1B in mice (52), and Rib 1 in Xenopus (6). Both of these factors are required for the formation of a preinitiation complex on the gene promoter. Recruitment of UBF to the ribosomal gene promoter appears to be one of the first steps in ribosomal gene activation (5). This is supported by in vivo observations of Xenopus embryogenesis, where ribosomal repeats are associated with UBF prior to the onset of transcription at the mid-blastula transition (4). In mouse cells that contain individual human chromosomes, UBF can be found associated with human rDNA despite its transcriptional silence (55). In vitro, UBF loading on promoter DNA is followed by recruitment of SL1/TIF-1B/Rib 1. The resulting preinitiation complex then recruits a specific subfraction of Pol I to the promoter. This transcriptionally competent form of Pol I is associated with Rrn3/TIF-1A and is thought to be the target of growth regulation of ribosomal gene expression in organisms ranging from yeast to humans (7, 34–36, 62.
As cells enter mitosis, ribosomal gene transcription is shut down. The Pol I transcription machinery, however, remains associated with NORs on mitotic chromosomes (54). Antibody staining of metaphase chromosomes confirms the presence of two classes of NOR in human cells, active and inactive (21, 50, 61). Active NORs have a distinct chromatin structure that is evident as a secondary constriction in metaphase (19). On inactive NORs, rDNA appears to be packaged in a form that is indistinguishable from the surrounding heterochromatin. The molecular structure of the secondary constriction is not understood. However, components of the Pol I transcription machinery, UBF in particular, are likely to play a key role in its formation.
UBF contains multiple HMG boxes, so called because of their homology to the DNA binding domain of high mobility group 1 (HMG-1) protein (20). UBF appears to bind DNA with a relaxed specificity, such that no consensus binding motif has yet been identified (15). One of the defining characteristics of UBF is its ability to bend and loop target DNA. A dimer of UBF can loop 140 bp of rDNA into a single turn (3, 44). Looping of promoter DNA appears to be required for UBF-dependent recruitment of SL1/TIF-1B/Rib 1 to the rDNA promoter (48). A significant fraction of UBF present in mammalian cells, termed UBF2, is a splice variant of full-length UBF1 (40). In UBF2, 37 amino acids from the second HMG box are deleted. This results in UBF2 being nonfunctional at the gene promoter (25) and suggests that it has an in vivo role distinct from that at the promoter.
UBF can bind in vitro to repeated elements that function as ribosomal gene-specific transcriptional enhancers (43). UBF appears to bind to these enhancer elements in a cooperative fashion (45). Experiments with in vitro systems demonstrate that UBF is required for enhancer function (33). Interestingly, UBF from heterologous species and UBF2 can support enhancer but not promoter function. Further in vitro experiments have shown that UBF can bind across the entire Xenopus IGS (37) and to sequences downstream of the transcriptional start site (29). However, it should be pointed out that UBF also binds to poly(dI-dC) and vector DNA. Consequently, in vitro binding studies do not provide reliable evidence for extensive binding of UBF to the rDNA repeat in vivo.
The abundance of UBF in nucleoli combined with the above binding characteristics suggests that UBF may bind extensively across the rDNA repeat and perform a more general structural role on active NORs. The fact that UBF can also out-compete binding of linker histones to preassembled nucleosomes in vitro provides further support of this view (23). The in vivo distribution of UBF has never previously been determined. Here, we developed a chromatin immunoprecipitation (ChIP) assay that utilizes an enriched nucleolar chromatin fraction. We used this assay to determine the distribution of UBF on the rDNA repeat in Xenopus, human, and mouse cells. We demonstrate for the first time that UBF binds in vivo to multiple sites distributed across the entire rDNA repeat (transcribed sequences and IGSs) in all three species.
MATERIALS AND METHODS
Cell culture.
HeLa and mouse A9 cells were cultured in Dulbecco’s minimal essential medium supplemented with 10% fetal calf serum (GIBCO) and penicillin-streptomycin (500 U/ml; GIBCO). The Xenopus laevis cell line XLK2 was cultured at room temperature in 50% L-15 Leibovitz medium with l-glutamine (GIBCO) supplemented with 10% fetal calf serum and penicillin and streptomycin (500 U/ml).
Antibodies.
The α-xUBF and α-human RNA Pol I (S18) antisera used here have been described elsewhere (9, 49). The α-TafI48 and α-TafI110 antibodies were provided by L. Comai (University of Southern California). Fluorescein isothiocyanate (FITC)-conjugated secondary antibodies were purchased from Sigma.
Preparation of soluble formaldehyde cross-linked nucleolar chromatin.
Nucleoli were prepared from fixed cells as described previously (55). Briefly, cells (10 175-mm dishes) were grown to approximately 70% confluence and cross-linked by formaldehyde (0.25% for 10 min). Cells were washed in phosphate-buffered saline (PBS) and harvested by being scraped into a total volume of 40 ml of PBS. After centrifugation at 200 × g for 5 min, the cell pellet was resuspended in 1.0 ml of high-magnesium buffer (10 mM HEPES [pH 7.5], 0.88 M sucrose, 12 mM MgCl2, and 1 mM dithiothreitol [DTT], plus protease inhibitors). Nucleoli were released by sonication on ice (two to three bursts of 10 s each at full power) using a Soniprep 150 (MSE) with a fine probe. The release of nucleoli was monitored microscopically. Nucleoli were pelleted by centrifugation in a microfuge (15,000 × g for 20 s), and the pellet was resuspended in 1.0 ml of low-magnesium buffer (10 mM HEPES [pH 7.5], 0.88 M sucrose, 1 mM MgCl2, and 1 mM DTT, plus protease inhibitors). Nucleoli were subject to further sonication on ice (10 s at full power) and pelleted as before. Nucleoli were resuspended in 0.2 ml of 20/2TE (20 mM Tris [pH 8.0], 2 mM EDTA) and a 1/10 volume of 20% sodium dodecyl sulfate (SDS) was added. Following incubation at 37°C for 15 min, nucleolar structures had disappeared, as determined by microscopy, and the solution had become viscous due to the high-molecular-weight nucleolar DNA. 20/2TE (0.8 ml) was added, and the solution was sonicated (three to four bursts of 5 s each at full power). The resulting sheared nucleolar chromatin was centrifuged in a microfuge (15,000 × g for 1 min), and the nucleolar chromatin supernatant was used immediately in nucleolar ChIP assays.
For determining the size range of nucleolar chromatin, 1 ml of the above supernatant was digested with RNase A (200 μg) at 37°C for 30 min, followed by proteinase K digestion (20 μl of a 14-mg/ml solution; Boehringer) at 37°C for 30 min. Formaldehyde cross-links were reversed by incubation at 65°C for 4 h. Sodium acetate (3 M; 100 μl) was added, and the mixture was extracted with phenol-chloroform. DNA was recovered by precipitation with ethanol and resuspended in 140 μl of TE (10 mM Tris [pH 8.0], 0.1 mM EDTA). Ten-microliter aliquots were subject to electrophoresis with a 1.2% Tris-acetate-EDTA (TAE) agarose gel, and signals for each lane were quantified in comparison to a marker lane with a molecular imager (Bio-Rad).
Nucleolar ChIP assays.
Nucleolar chromatin supernatant (75 μl) was combined with 1.425 ml of PBS that contained 0.1% bovine serum albumin, and 20 μl of the appropriate sera (preimmune sera, α-UBF, or α-RNA Pol I) was added. Following overnight incubation at 4°C, 50 μl of a 50% slurry of protein A-Sepharose beads preequilibrated with bovine serum albumin (0.1%) and Escherichia coli RNA (100 μg/ml) was added, and the mixture was rotated at 4°C for 1 h. Beads were recovered by centrifugation (600 × g for 15 s) and washed as follows: twice with 1 ml of 20/2TE containing 0.2% SDS, 0.5% Triton X-100, and 150 mM NaCl; twice with 1 ml of 20/2TE, 0.2% SDS, 0.5% Triton X-100, and 500 mM NaCl; and twice with 1 ml of 20/2TE. Immunoprecipitated material was eluted twice from the beads with 50 μl of 20/2TE containing 2% SDS at 37°C for 10 min each. A total of 0.3 ml of 20/2TE was added to the pooled eluates, and proteins were digested by the addition of proteinase K (10 μl of a 14-mg/ml solution; Boehringer). Following incubation at 37°C for 30 min, formaldehyde cross-links were reversed by incubation at 65°C for 4 h. Sodium acetate (3 M; 40 μl) was added, and the mixture was extracted with phenol-chloroform. Glycogen (5 μl of a 10-mg/ml solution) was added as a carrier, and DNA was recovered by ethanol precipitation and resuspended in 50 μl of TE for PCR analysis or in 9 μl of H2O for direct labeling.
Quantitative PCR.
For the sequence coordinates of human and mouse primer pairs and their resulting product sizes, see Table 1 and 2 respectively. The primers used to amplify the Xenopus ribosomal gene promoter sequence were 5′-CGATGAGGACGGATTCGCCC-3′ and 5′-TACCTTCCTGCCCGCACATG-3′, yielding a 140-bp product.
TABLE 1.
Coordinates of human primer pairsa
| Primer pair | Coordinate site
|
Product length (bp) | |
|---|---|---|---|
| Coding strand | Noncoding strand | ||
| H0 | 42952–42971 | 13–32 | 80 |
| H1 | 952–972 | 1010–1030 | 79 |
| H4 | 3990–4010 | 4072–4092 | 103 |
| H8 | 8204–8224 | 8280–8300 | 96 |
| H13 | 12855–12875 | 12950–12970 | 115 |
| H18 | 18155–18175 | 18260–18280 | 125 |
| H23/27 | 27366–27386 | 27457–27477 | 111 |
| H42 | 41982–42002 | 42055–42075 | 94 |
Sequence coordinates derived from GenBank accession no. U13369.
TABLE 2.
Coordinates of mouse primer pairs
| Primer pair | Coordinate site
|
Product length (bp) | |
|---|---|---|---|
| Coding strand | Noncoding strand | ||
| M0 | 5459–5480 | 5654–5675 | 216 |
| MEn | 3754–3774 | 3837–3857 | 103 |
| M5 | 9978–9998 | 10060–10080 | 102 |
| M9 | 14036–14056 | 14112–14132 | 96 |
| M13 | 18357–18377 | 18451–18471 | 114 |
| M16 | 21606–21626 | 21734–21754 | 148 |
Sequence coordinates derived from GenBank accession no. X82564.
PCR mixtures (50 μl) contained 10 pmol of each primer and 2.5 U of Taq DNA polymerase and were performed with the following buffer: 50 mM KCl, 10 mM Tris [pH 9.0], 0.1% Triton X-100, 2.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, and 5% dimethyl sulfoxide. Reaction mixtures also contained 0.2 pmol of the respective primer pair end labeled with T4 polynucleotide kinase and [γ-32P]ATP (3,000 Ci/mmol; Amersham). One microliter (1/50) of DNA obtained from a nucleolar ChIP assay, or the indicated amount of cloned rDNA, was used as a template. In PCRs with human DNA, 21 cycles of 0.5 min at 95°C, followed by 0.5 min at 45°C and 2.0 min at 72°C were performed. For PCRs with Xenopus and mouse cells, 23 cycles of the above were performed. Aliquots of PCR mixtures (10 μl) were subject to electrophoresis on 8% polyacrylamide gels in 1× TBE. Signals were visualized and quantified with a PhosphorImager (Bio-Rad).
Combined immunofluorescence and FISH with Xenopus metaphase spreads.
XLK2 Xenopus cells were blocked overnight with nocodazole (final concentration, 0.1 μg/ml). Cells were harvested by performing a mitotic shake-off and pelleting by centrifugation (300 × g for 3 min). Cells were washed in PBS and pelleted as above. The cells were hypotonically swollen in 17.5 mM KCl for 1 h and repelleted as above. The pellet was resuspended dropwise in fixative (a 3:1 methanol-to-acetic acid ratio) and incubated overnight. The cells were pelleted as above and refixed twice more (incubations of 20 min). Immunofluorescent in situ hybridization (immuno-FISH) was performed as described previously (55) but with Spectrum Red (Vysis)-labeled pXlr101A as a probe and α-xUBF antibodies.
Plasmids.
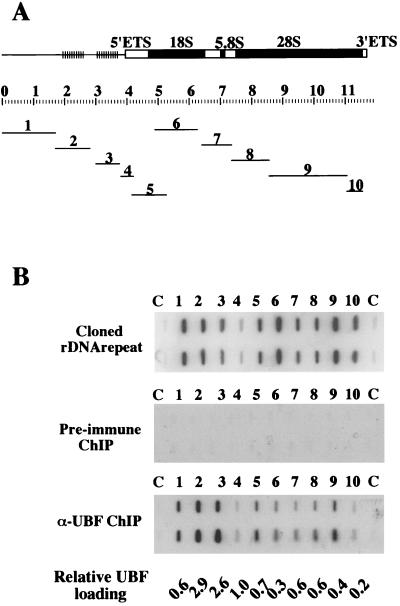
The plasmid pXlr101A contains an intact Xenopus rDNA repeat cloned as a HindIII fragment into the vector pBR322. Clones 1 to 10 were derived from pXlr101A. Clone 1 is a HindIII-BamHI restriction fragment that contains repetitive regions 0 and 1 from the IGS cloned into pBluescript SK(-) (Stratagene). Clone 2 is a BamHI restriction fragment that extends between spacer promoters and includes a single block of enhancer elements cloned into pBluescript SK(-). Clone 3 contains a single enhancer block and has been previously described as pGem14FA (26). Clone 4 contains the sequences −245 to +92 that include the gene promoter cloned into pGem3 (Promega). Clone 5 is a NotI-to-DraI restriction fragment that contains ETS and 18S sequences cloned into the NotI and SmaI sites in pBluescript SK(-). Clone 6 is a XbaI-to-EcoRI restriction fragment that contains18S sequences cloned into pBluescript SK(-). Clone 7 is a XbaI-to-BamHI restriction fragment that contains ITS and 5.8S sequences cloned into pBluescript SK(-). Clone 8 is a BamHI restriction fragment that contains 28S sequences cloned into pBluescript SK(-). Clone 9 is a BamHI-to-EcoRI restriction fragment that contains 28S sequences cloned into pBluescript SK(-). Clone 10 is an EcoRI-to-HindIII restriction fragment that contains 28S sequences cloned into pBluescript SK(-). The position of each of these inserts relative to the rDNA repeat is shown in Fig. 4A.
FIG. 4.
UBF binds extensively over the Xenopus rDNA repeat. (A) The structure of the Xenopus rDNA repeat. Solid black boxes indicate 18S, 5.8S, and 28S rRNA coding sequences. Open boxes represent 5′ and 3′ external transcribed spacers and internal transcribed spacers. Clusters of vertical bars represent blocks of enhancer elements. The scale bar below represents the length of the repeat in kilobases. The position and size of subclones 1 to 10 of the Xenopus rDNA repeat are shown below. (B) Duplicate arrays of subclones 1 to 10 (slots 1 to 10) and vector controls (slots C) were arrayed and probed with radiolabeled cloned rDNA repeat (top) and DNA extracted from both preimmune (middle) and α-xUBF (bottom) ChIP assays. Relative UBF loading values are shown below.
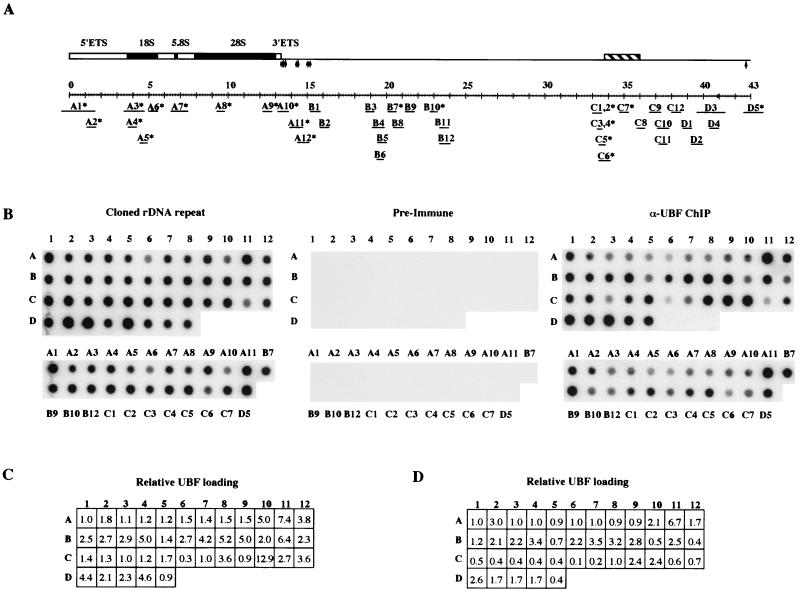
A chromosome 22 cosmid library (LL22NC03, constructed by Lawrence Livermore National Laboratory and supplied by the Human Genome Mapping Project Resource Centre, Hinxton, Cambridge, United Kingdom) was screened with 18S and 28S human rDNA sequences. The end sequence was determined for a number of positive clones. Clone N68f1 contained the entire human rDNA repeat except for sequences between nucleotides 16414 and 18112 in the IGS (see GenBank accession no. U13369). All subclones of the human ribosomal gene repeat (with the exceptioan of A1, A11, and D5) were created by shotgun cloning from N68f1. N68f1 DNA (150 μg) in a total volume of 400 μl of 10 mM Tris [pH 7.5], 50 mM NaCl, 10 mM MgCl2, and 1 mM DTT was sonicated on ice (two bursts of 15 s, each at full power). Ends were repaired using the Klenow fragment of DNA polymerase (20 U; GIBCO), 0.4 μl of 1 M DTT, and 4 μl of 10 mM deoxynucleoside triphosphates and incubation at 37°C for 30 min. Following electrophoresis on a 1.2% agarose gel in 1× TAE buffer, fragments ranging in size from 0.4 to 0.8 kb were extracted from the gel, dephosphorylated with calf intestinal alkaline phosphatase (Boehringer), and ligated into the vector pCR-Blunt II-TOPO with the Zero blunt TOPO PCR cloning kit (Invitrogen). With subclones A7, B11, D2, and D3, repaired sheared fragments were cloned into the vector pGEM-T Easy after incubation with Taq DNA polymerase and dATP (Promega). All subclones were characterized by EcoIRI digestion and sequence analysis. Subclones A1 and D5 have been previously described as pHu3 and pHuC, respectively (28). Subclone A11 was generated from pCD7, a plasmid that contains human rDNA sequences missing from cosmid N68f1 (27). The position of each of these inserts relative to the rDNA repeat is shown in Fig. 5A, and the precise coordinates of each subclone relative to the published sequence of the human rDNA repeat (GenBank accession no. U13369) can be found in Table 3.
FIG. 5.
UBF binds extensively over the human rDNA repeat. (A) The structure of the human rDNA repeat is illustrated. Solid black boxes indicate 18S, 5.8S, and 28S rRNA coding sequences. Open boxes represent 5′ and 3′ external transcribed spacers. Vertical arrows represent transcription termination elements. The cross-hatched box represents a cdc27 pseudogene. The scale bar below represents the length of the repeat, in kilobases. The position and size of subclones of the human rDNA repeat (A1 to D5) are shown below the repeat. Clones with an asterisk are devoid of alu elements. (B) Insert DNA from each of the above subclones was generated by PCR and arrayed onto nylon filters (top). Fragments derived from vector DNA (D6 to D8) provided negative controls. The identity of each insert is indicated by letters and numerals on the left and top of the array, respectively. Inserts devoid of alu elements were also arrayed independently (bottom). In panel B, the identities of each insert are shown. Arrays were hybridized with an equimolar mixture of the cosmid N68f1 and the plasmid pCD7H/B (left). In experiments shown in the middle and right panels, arrays were probed with radiolabeled DNA from α-UBF and preimmune ChIP assays, respectively. (C) The relative UBF loading values for the experiment shown above. (D) The relative UBF loading values for an independent experiment (data not shown).
TABLE 3.
Coordinates of human rDNA subclonesa
| Clone | Coordinate site
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
| A | 42486–1500 | 1199–1566 | 3542–4702 | 3689–4243 | 4449–4858 | 5093–5459 | 6507–7427 | 9280–9742 | 12,213–12,701 | 13163–13674 | 13895–14650 | 14407–15122 |
| B | 15241–15713 | 15790–16416 | 18700–19201 | 19063–19798 | 19471–20017 | 19427–19802 | 20014–20507 | 20581–21180 | 21,354–21,776 | 22795–23320 | 23357–23932 | 23569–24059 |
| C | 33022–33487 | 32998–33518 | 33246–33599 | 33284–33638 | 33429–33830 | 33372–34047 | 34697–35178 | 35771–36279 | 36,621–37,260 | 37065–37684 | 37250–37643 | 38068–38449 |
| D | 38717–39236 | 39205–39999 | 35287–41292 | 40368–41029 | 42500–700 | |||||||
Sequence coordinates derived from GenBank accession no. U13369.
Clones D6 to D8 contain sequences 659 to 1,118, 1,257 to 1,834, and 1,627 to 2,032, respectively, of the published pBluescript sequence (GenBank accession no. X52328) cloned into pCR-Blunt II-TOPO.
Array production and hybridization.
XenopusI slot blot arrays were generated with a HYBRI-SLOT filtration manifold (GIBCO). Aliquots (1.0 μg) of each plasmid to be arrayed [clones 1 to 10 and pBluescript SK(-) as a control] were diluted into 0.5 M NaOH (50 μl) and loaded onto nylon membrane (BDH) that was preequilibrated in 0.5 M NaOH. The membrane was removed from the apparatus, rinsed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 1 min, air dried, and UV cross-linked for 4 min with a Stratalinker (Stratagene).
The human dot blot arrays were generated with a Bio-Dot Microfiltration apparatus (Bio-Rad). Inserts were amplified from clones A1 to D8 by PCR. PCR mixtures (each, 100 μl) contained 10 ng of template DNA, 20 pmol of each primer (T7 and SP6), and 5 U of Taq DNA polymerase in PCR buffer (see above). A total of 35 cycles of 0.5 min at 95°C, 0.5 min at 45°C, and 3.0 min at 72°C were performed. Half of each of the purified PCR products were denatured in a final volume of 100 μl of 0.4 M NaOH and 10 mM EDTA by being heated at 100°C for 10 min. DNA was then neutralized by the addition of an equal volume of ice-cold 2 M ammonium acetate, pH 7.0. Nylon membrane, prewetted in 6× SSC, was assembled into the Bio-Dot apparatus. TE (500 μl) was filtered through each well, followed by the denatured DNA samples (200 μl) and 500 μl of 2× SSC. Loaded filters were rinsed and UV cross-linked as above.
Sequences representing the human chromosome 14/22-specific α-satellite, β-satellite, and satellite 1 were generated by PCR using primer pairs and conditions described previously (53). Chromosome 13/21-specific α-satellite sequences were prepared by restricting the 0.85-kb insert from plasmid pZ21A (1).
To probe Xenopus arrays, the appropriate DNA (9 μl from pre-UBF and α-UBF ChIP assays or 10 ng of the HindIII-excised insert from plasmid pXlr101A) was subject to random primed labeling (16) with a total volume of 20 μl. Probes for the human rDNA arrays were radiolabeled by nick translation (Roche). DNA from three ChIP assays were pooled, ethanol precipitated, resuspended in 5 μl of water, and labeled with [α-32P]dCTP (3,000 Ci/mmol; Amersham) and [α-32P]dTTP (3,000 Ci/mmol; Amersham), following the manufacturer’s protocol. The plasmid pCD7H/B is a HpaI-to-BamHI restriction fragment of pCD7 subcloned into the EcoRV and BamHI sites of pBluescript SK(-). The sequences present in pCD7H/B are restricted to those human rDNA sequences that are missing from N68f1. An equimolar mixture of N68f1 and pCD7H/B (50 ng total) was also labeled as above and used to probe human arrays. Both Xenopus and human arrays were prehybridized, hybridized, and washed as described elsewhere (12). Hybridizations took place in a volume of 50 ml. Hybridization signals were visualized and quantified with a PhosphorImager (Bio-Rad).
RESULTS
Isolation of nucleolar chromatin.
In standard ChIP protocols, proteins are cross-linked to DNA by treating cells with 1% paraformaldehyde (PFA) (41). Nuclei are isolated from fixed cells, and a soluble chromatin fraction is then prepared by sonication, followed by centrifugation to remove insoluble material. By this standard protocol, nucleolar chromatin is not solubilized (data not shown). Moreover, repeated sonication and sedimentation is an established procedure for nucleolar purification from unfixed cells (31, 38). We have previously described a protocol for isolation of nucleoli from cells in which proteins have been cross-linked to DNA by PFA treatment (55). A soluble nucleolar chromatin fraction can be prepared from these nucleoli by incubation with detergent (2% SDS), followed by sonication. We have found that cross-linking with 0.25% PFA for 10 min is optimal for subsequent nucleolar solubilization. When cells are treated with 1.0% PFA, a soluble nucleolar chromatin fraction cannot be generated. Presumably, this is a consequence of over-cross-linking. In mammalian cells, not all NORs are active. Consequently, purification of nucleolar chromatin provides the added certainty of isolating actively transcribed ribosomal gene chromatin.
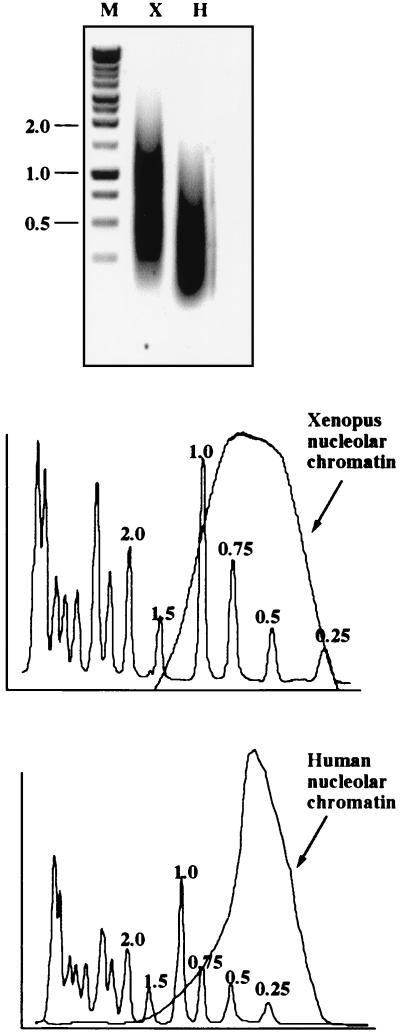
In any ChIP protocol, the resolution of the technique is limited by the size range of chromatin fragments used in the immunoprecipitation. In the results shown in Fig. 1, we demonstrate that the average sizes of Xenopus and human nucleolar chromatin fragments are ∼0.6 and ∼0.4 kb, respectively, with 90% of Xenopus fragments under 1 kb and 95% of human fragments under 1 kb.
FIG. 1.
Size range of soluble nucleolar chromatin. DNA extracted from Xenopus (X) and human (H) soluble nucleolar chromatin was electrophoresed on a 1.2% agarose gel alongside a molecular weight ladder (M). The lengths in kilobases of key bands in the marker lane are shown on the left. Ethidium bromide-stained DNA was quantified using a Molecular Imager (Bio-Rad). Profiles of DNA extracted from Xenopus and human cell nucleolar chromatin compared to the molecular weight marker are shown below the gel.
UBF binds to Xenopus and human ribosomal gene promoters in vivo.
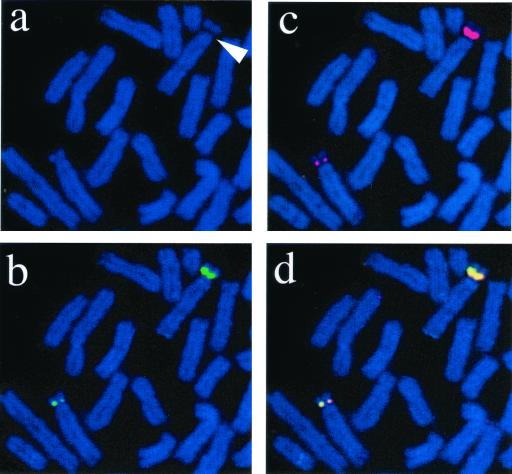
To study the distribution of UBF on Xenopus and human ribosomal gene repeats in vivo we used an antibody raised against recombinant xUBF. This antibody is highly specific and works well in Western blotting, immunoprecipitation, and immunofluorescence (4, 9, 33). UBF sequence is highly conserved between Xenopus and mammals (2, 32); consequently, the α-xUBF antibody functions across species and can recognize both mammalian UBF1 and -2. It is well documented that UBF remains associated with NORs in metaphase (10, 21, 51). To demonstrate that the α-UBF antibody used here can specifically recognize UBF bound to rDNA, we performed combined immunofluorescence and FISH analysis of metaphase chromosomes isolated from Xenopus cells (Fig. 2). UBF was visualized with a FITC-conjugated secondary antibody (Fig. 2b). To visualize Xenopus NORs, chromosomes were hybridized with Spectrum Red-labeled Xenopus rDNA (Fig. 2c). Both of these signals colocalize (Fig. 2d), and there is no detectable UBF staining elsewhere on Xenopus chromosomes. A secondary constriction is clearly associated with each NOR (Fig. 2a). The two NORs observed are of differing size as judged by rDNA hybridization signals. UBF loading appears directly proportional to the number of rDNA repeats on each NOR. Note also that the size of the secondary constriction is directly related to rDNA content and UBF loading. From this experiment, we conclude that the α-UBF antibody can specifically recognize bound UBF and is suitable for the ChIP experiments described below. Furthermore, we can conclude that UBF binding is largely restricted to NORs, since no staining is observed at other chromosomal loci and the staining observed is proportional to the rDNA content of an NOR.
FIG. 2.
α-xUBF antibodies recognize Xenopus NORs. Metaphase chromosome spreads (counterstained with 4′,6-diamidino-2-phenylindole [DAPI]) from Xenopus XLK2 cells were subject to combined immunofluorescence and FISH. UBF was visualized using α-xUBF antibody (FITC/green). FISH was performed with Spectrum Red-labeled Xenopus rDNA. Secondary constrictions on DAPI-stained chromosomes are indicated by arrows(a). Panels b and c show UBF staining and FISH signals, respectively. The merge of UBF and FISH signals is shown in panel d.
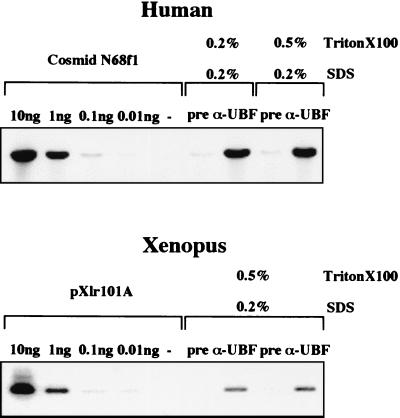
As outlined in the introduction, UBF binding to Xenopus and human promoter sequences is a key requirement of stable transcription complex formation in vitro. An examination of UBF binding to promoter sequences in vivo provides a valuable target for validating the nucleolar ChIP assay. ChIP was performed with human and Xenopus nucleolar chromatin with the α-UBF antibody (Fig. 3). After extensive optimization of immunoprecipitate wash conditions (data not shown), all immunoprecipitates were washed in a buffer containing 0.2% SDS and 0.5% Triton X-100 (unless otherwise stated). Human and Xenopus promoter sequences were detected in the immunoprecipitate by quantitative PCR with primer pairs specific to each promoter DNA (see Materials and Methods for details). In control reaction mixtures, decreasing amounts of cloned target DNA were used to validate the quantitative nature of the PCR. Strong positive signals were observed in both human and Xenopus α-UBF ChIP assays. The specificity of these signals were confirmed by the low level of product obtained with preimmune sera (60- and 40-fold-lower levels than those obtained with α-UBF anitibodies in human and Xenopus experiments, respectively). Two independent sera raised in rabbit cells against hUBF (9) and human autoantibodies that recognize UBF (55) gave the same results (data not shown). We conclude that the nucleolar ChIP protocol detects hUBF and xUBF binding in vivo to their respective ribosomal gene promoters.
FIG. 3.
UBF binds to Xenopus and human ribosomal gene promoters in vivo. Quantitative PCR was performed using promoter-specific primers and DNA extracted from α-xUBF and preimmune ChIP assays. Gels from experiments with human and Xenopus nucleolar chromatin are shown in the top and bottom panels, respectively. See Materials and Methods for details of the ChIP assay, primers, and PCR conditions. Control PCRs were performed with the human rDNA repeat cosmid N68f1 and the Xenopus plasmid pXlr101A. The amount of cloned DNA used in control reaction mixtures (10 to 0.01 ng) is shown above each lane as appropriate. Concentrations of SDS and Triton X-100 used in immunoprecipitation washes are shown above.
xUBF binds across the entire Xenopus rDNA repeat in vivo.
While PCR performed with immunoprecipitated material is a valuable technique for analyzing UBF binding to a specific target sequence, it is less suited to determining the distribution of UBF binding across the entire rDNA repeat. This is because of the large size of the rDNA repeat and the inherent difficulty in comparing signals from PCRs with different primer pairs. It would be preferable to directly label the immunoprecipitated DNA and use it as a probe against an array of subcloned fragments from the rDNA repeat. Comparison of the PCR signals obtained in ChIP assays with those obtained using cloned ribosomal DNA (Fig. 3) suggests that sufficient material may be present in ChIPs to permit its direct labeling by standard techniques. We estimate that α-UBF ChIP assays contain an amount of promoter sequence equivalent to approximately 10 ng of intact rDNA repeat. This is due to the fact that the nucleolar chromatin fraction used in our ChIP assay represents a highly enriched fraction of the genome. Furthermore, ribosomal genes are highly repeated in the Xenopus and human genomes.
To determine the in vivo distribution of UBF on the Xenopus ribosomal gene repeat, we arrayed a series of 10 plasmids that cover the entire Xenopus rDNA repeat onto a nylon filter in duplicate. The distribution of these clones is shown in Fig. 4A. This array was first hybridized with the insert of a plasmid (pXlr101A) that contains an intact Xenopus rDNA repeat (Fig. 4B). This was performed to determine the relative hybridization efficiency of each clone in the array. The signal observed over the vector control slot (Fig. 4B, slots C) is a result of contaminating vector DNA present in the probe. DNA from α-UBF ChIP assays with Xenopus nucleolar chromatin was radioactively labeled and hybridized to the array (Fig. 4B). DNA obtained from ChIP assays with the preimmune sera provided a negative control. Significant hybridization signal was observed over all 10 rDNA-containing clones when DNA from α-UBF ChIP assays was used as a probe. As we have already demonstrated UBF binding to the promoter in vivo, we calculated UBF loading on sequences represented by each subclone relative to that observed over the gene promoter (clone 4). The hybridization signal over each rDNA fragment in the array hybridized with cloned rDNA was calculated relative to that observed over clone 4. This value provided a correction factor for differences due to insert length and sequence composition across the array. Then, the hybridization signal over each rDNA fragment in arrays hybridized with α-UBF ChIP DNA was calculated relative to that observed over clone 4 and divided by the correction factor established above. This final figure is an estimate of UBF loading across the rDNA repeat relative to that observed over the gene promoter (Fig. 4B). Note that no signal is observed over the vector control slots (Fig. 4B, lanes C).
We clearly observe UBF binding over sequences represented by clones 1 to 4 that comprise the IGS (Fig. 4B). This should not be surprising, since the IGS contains regulatory elements such as the gene promoter, enhancers, and spacer promoters that are known UBF binding sites in vitro. Given the limit of resolution in this technique, one might also expect hybridization over clones that define either end of the 40S transcript (clones 5 and 10), since they are within 1 kb of known transcriptional regulatory sequences. Surprisingly, however, UBF binding is observed over the entire transcribed region, represented by clones 5 to 10 (Fig. 4B). We calculate that the amount of UBF binding over the transcribed region is, on average, 30% of that observed over the IGS. This point will be elaborated on in the discussion.
hUBF binds across the entire human rDNA repeat in vivo.
A cosmid (N68f1) containing almost the entire human rDNA repeat was obtained by screening a human chromosome 22 cosmid library with 18S and 28S rDNA sequences. Extensive restriction mapping and sequence analysis show that the structure of the rDNA repeat in this cosmid matches the published sequence (17) of the 43-kb human rDNA repeat (data not shown). Additionally, Southern blotting demonstrates that this repeat is representative of those found in HeLa cells, the human cell line used throughout this work (data not shown). A series of subcloned fragments (A1 to D5) was generated from this cosmid by shotgun cloning. The average insert size of each of these subcloned fragments is ∼0.5 kb. This insert size was chosen to reflect the limit of resolution of the ChIP assay. All inserts were fully sequenced to confirm their location relative to the intact repeat and to identify the presence of repetitive elements such as alu repeats. The position of each of these subcloned fragments is shown in Fig. 5A. Purified insert DNA from each subclone was generated by PCR and arrayed onto a nylon filter. Amplified fragments from the plasmid vector (D6 to D8) provided negative controls. Those clones that are devoid of alu repeats (Fig. 5A) were also arrayed independently. Arrays were hybridized with radiolabeled DNA from preimmune ChIP or α-UBF ChIP assays with nucleolar chromatin prepared from HeLa cells (Fig. 5B, middle and right panels, respectively). As a control, arrays were also probed with radiolabeled, cloned rDNA (an equimolar mixture of cosmid N68f1 and plasmid pCD7H/B) (Fig. 5B, left). Note the presence of hybridization signal over vector sequences (D6 to D8) in this array, due to the presence of vector DNA in the probe. The array probed with radiolabeled DNA from α-UBF ChIP (Fig. 5B, right) showed significant hybridization signal over every subcloned human rDNA fragment (A1 to D5). Specificity in this experiment was confirmed by the absence of signal over the vector controls (D6 to D8) and by the fact that no appreciable signal was observed with DNA from the preimmune ChIP assay (Fig. 5B, middle). Repetitive elements within the rDNA repeat and found elsewhere in the human genome, such as alu elements, cannot explain the presence of hybridization signal over the array, since subclones devoid of alu elements show hybridization comparable to those containing alu elements. The PCR experiments described below further underscore this point.
As with the Xenopus experiment (Fig. 4B), we have calculated the UBF loading on sequences represented by each subclone. This calculation was performed for two independent experiments (Fig. 5C and D). There is clear variation from experiment to experiment. The source of this variation could arise from the growth status of the cells or from the degree of cross-linking achieved in different experiments. This variation precludes a precise determination of the loading of UBF over the rDNA repeat. However, it would appear that UBF binding is more abundant over sequences immediately upstream of the gene promoter (C10 to D5) and over sequences immediately downstream of the 47S coding sequence (A10 and 11). From these experiments (Fig. 5 and data not shown), we can conclude that UBF is bound in substantial quantities across the entire human rDNA repeat in vivo.
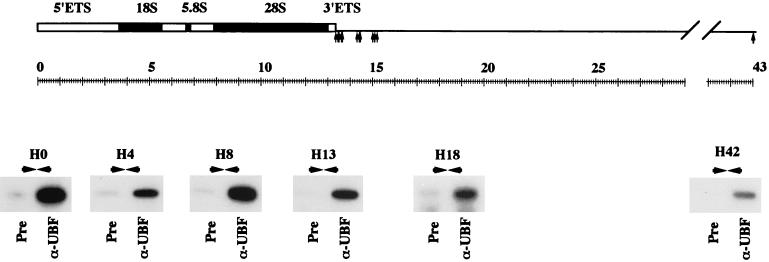
We have also analyzed DNA from preimmune and α-UBF ChIP assays by PCR with primer pairs distributed across the human rDNA repeat. In each case, primer pairs recognize a unique sequence (i.e., not duplicated elsewhere in the rDNA repeat), with the exception of the H23/27 primer pair that recognizes a duplicated element. The location of the primer pairs is shown in Fig. 6. In each case, we demonstrate the presence of target DNA, specifically in the α-UBF immunoprecipitate. This provides independent evidence for extensive UBF binding across both the IGS and transcribed sequence of the rDNA repeats in human cells. Essentially the same results (both array hybridization and PCR) (data not shown) have been obtained with an independent rabbit α-hUBF antibody (9) as well as a human autoimmune sera that recognizes hUBF (55).
FIG. 6.
PCR confirms distribution of UBF on the human rDNA repeat. PCR was performed on DNA extracted from preimmune and α-UBF ChIP assays with primer pairs from across the human rDNA repeat. The locations of the primer pairs and gels of the resulting PCRs are shown in the appropriate position below a diagram of the human rDNA repeat. The source of DNA used in each PCR is shown below the gel.
UBF does not bind to satellite DNA sequences adjacent to the NORs of human chromosomes.
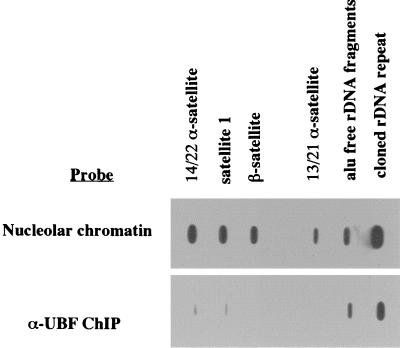
In the above sections, we demonstrated that UBF binds extensively across the rDNA repeat. As a control for the specificity of this binding, it is important to demonstrate that non-rDNA sequences present in nucleoli are not associated with UBF. The sequences surrounding the NORs on the short arms of the human acrocentric chromosomes are devoid of transcribed sequences and are composed of satellite DNA. For example, there is a 1.5-Mb contiguous array of satellites 1, 3, and β between the α-satellite at the centromere and the rDNA on human chromosome 22 (53). DNA fragments representative of α, β, and satellite 1 DNA were loaded onto slot blots alongside a cosmid clone containing the human rDNA repeat (N68f1) and a pool of rDNA sublones lacking alu repeats. When radiolabeled HeLa cell nucleolar chromatin is used as a probe, approximately equal levels of hybridization signal are observed over satellite and rDNA sequences (Fig. 7). However, when labeled DNA from α-UBF ChIP is used as a probe, the hybridization signal observed over satellite sequences is greatly reduced (approximately 10-fold) in comparison to rDNA sequences (Fig. 7). This result confirms that UBF binding is largely restricted to rDNA sequences within the nucleolus.
FIG. 7.
UBF does not bind to satellite DNA sequences adjacent to the NORs of human chromosomes. Individual slot blots were loaded with DNA fragments representing α-satellite sequences from 14/22 and 13/15 chromosome pairs, satellite 1, and β-satellite, a pool of alu-free subclones of the human rDNA repeat (see Fig.5), and an intact human rDNA repeat (cosmid N38f1). Slot blots were hybridized with radiolabeled total nucleolar chromatin (top) and DNA recovered from an α-UBF ChIP assay (bottom).
RNA Pol I selectively cross-links to transcribed sequences in the human rDNA repeat.
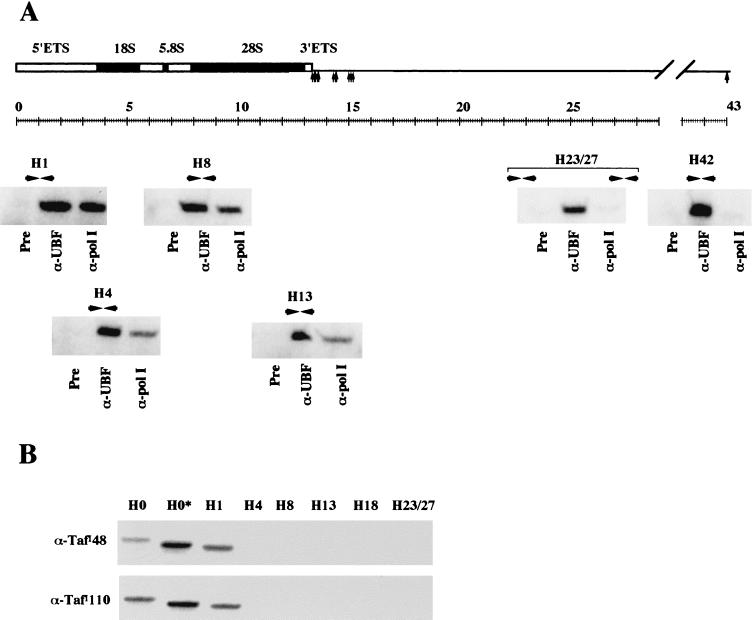
Whereas UBF binds across the entire rDNA repeat in both Xenopus and human cells, one would expect that other proteins related to ribosomal gene transcription would show a more restricted distribution. In particular, α-Pol I antibodies should only immunoprecipitate transcribed sequences within the rDNA repeat. To test this, we performed PCRs with primer pairs targeted to both transcribed and nontranscribed sequences with DNA extracted from α-Pol I ChIP assays (Fig. 8). The α-Pol I antibody used in this experiment (S18) has been extensively characterized and demonstrated to bind engaged Pol I (49).
FIG. 8.
RNA Pol I and SL1 show a resticted distribution on the human rDNA repeat. (A) PCR was performed with DNA extracted from preimmune, α-UBF, and α-Pol I ChIP assays with primer pairs from across the human rDNA repeat. The locations of the primer pairs and gels of the resulting PCRs are shown in the appropriate position below a diagram of the human rDNA repeat. The source of DNA used in each PCR is shown below the gel. (B) PCR was performed with DNA extracted from α-TafI110 and α-TafI48 ChIP assays with primer pairs from across the human rDNA repeat. The identity of the primer pairs is shown above the appropriate gel lanes, and the identity of the antibody used in ChIP is shown alongside.
Significant amounts of PCR product are recovered from both α-UBF and α-Pol I ChIP assays with primer pairs H1, H4, H8, and H13 that fall within the 47S coding sequence. Product is observed from α-UBF but not α-Pol I ChIP assays with primer pairs H23/27 and H42 that fall within the IGS (Fig. 8A). Thus, we can confirm that this assay shows the expected and restricted distribution of RNA Pol I on the human rDNA repeat. Note that the absence of Pol I over sequences targeted by the H42 primer pair (1 kb upstream of the transcriptional start) confirms the resolution of this technique.
SL1 binding is restricted to promoter sequences.
SL1 is a component of the stable transcription complex that forms at the promoter. Consequently, one would expect that SL1 should show a more-restricted distribution than either UBF or Pol I. ChIP experiments were performed with antibodies that recognize TafII48 and TafI110 components of SL1. PCR was performed with DNA recovered from each ChIP with primer pairs from across the rDNA repeat (Fig. 8B). Promoter sequences and sequences up to 1 kb downstream of the transcription start site are present in α-TafI48 and TafI110 ChIP experiments. As expected, sequences elsewhere in the transcribed region (H4, H8, and H13) and in the IGS (H18 and H23/27) are absent.
mUBF binds extensively over the mouse rDNA repeat in vivo.
To determine the UBF distribution over the rDNA repeat in mouse cells, preimmune and α-UBF antibodies were used in ChIP assays with nucleolar chromatin prepared from mouse A9 cells. PCR was performed with DNA extracted from immunoprecipitates with a series of primer pairs that recognize sequences within the transcribed region (M5, M9, and M13), downstream of the transcribed region (M16), and over the promoter and enhancer repeats (M0 and MEn, respectively) (Fig. 9). Significant amounts of PCR product were obtained from α-UBF ChIP assays with each primer pair. Note the high signal obtained with the primer pair targeted to the enhancer elements (MEn). This is presumably due to their repeated nature. The results of these PCRs clearly demonstrate that as in Xenopus and human cells, UBF binds extensively over the rDNA repeat in mouse cells.
FIG. 9.
UBF binds to sequences across the mouse rDNA repeat. PCR was performed with DNA extracted from preimmune and α-UBF ChIP assays with primer pairs from across the mouse rDNA repeat. The locations of the primer pairs and gels of the resulting PCRs are shown in the appropriate position below a diagram of the mouse rDNA repeat. The primer pair MEn is derived from the repeated enhancer elements, depicted by the cluster of vertical lines upstream of the transcribed region. The source of DNA used in each PCR is shown below the gel.
DISCUSSION
In the work presented here, we describe a method for studying the distribution of proteins bound to the rDNA repeat in vivo. In this procedure, a ChIP assay is performed with a highly enriched nucleolar chromatin fraction prepared from PFA-treated cells. We have used this technique to demonstrate for the first time that UBF is bound in vivo across the entire rDNA repeat in Xenopus and mammalian cells. This conclusion is supported by several lines of evidence. DNA, extracted from a ChIP assay and directly radiolabeled, hybridizes to an array of subcloned fragments representing the entire rDNA repeat. PCR was used to demonstrate the presence of sequences from throughout the rDNA repeat in α-UBF ChIP assays. In contrast, we observe that Pol I and SL1 cross-linking is restricted to transcribed sequences and the promoter region, respectively. This demonstrates that the assay provides an accurate description of the in vivo distribution of UBF.
Our results are further supported by the abundance and distribution of UBF in vivo. Xenopus and human cells contain approximately 106 molecules of UBF (33, 55). Indirect immunofluorescence shows that a high concentration of UBF colocalizes with rDNA at specific foci within the nucleolus (51). Furthermore, as cells double their DNA content between G1 and G2, the amount of UBF present approximately doubles (22). Thus, it would appear that the majority of UBF is engaged on DNA throughout the cell cycle. Assuming a dimer of UBF engages 100 bp of DNA, there is sufficient UBF to completely cover all the rDNA repeats on active NORs in both human and Xenopus cells. It should be pointed out that in mammalian cells, 50% of UBF is present in the form of UBF2, known to be nonfunctional at the promoter (25). Currently, we have no way of distinguishing between UBF1 and UBF2. It would be interesting to determine if they show a differential distribution.
In cells of higher eukaryotes, two distinct forms of ribosomal gene chromatin have been observed biochemically (14). A fraction of rDNA repeats (presumed to be transcriptionally active) are in an open configuration and accessible to the cross-linking agent psoralen. Inactive rDNA repeats are in a closed chromatin configuration and inaccessible. At the chromosomal level, two classes of NOR exist. Active NORs are distinguished by the presence of UBF, silver staining, and a secondary constriction (19). Inactive NORs lack these characteristics. With our α-UBF ChIP assays, we are presumably studying only those repeats that are found on active NORs. We presume that this represents the psoralen-accessible fraction.
Potorous tridactylis kidney cells have two NORs, each giving rise to a nucleolus. Within these nucleoli, three classes of rDNA repeat are observed: repeats that are transcriptionally active and associated with UBF, repeats that are not transcribed yet are associated with UBF, and repeats that are not associated with UBF (22). It seems likely that the UBF binding to transcribed sequences we have observed in this work represents the class of repeats that are not transcribed, yet are associated with UBF. This conclusion is supported by electron microscopy. Ribosomal DNA repeats that are transcriptionally active are fully packed with a Pol I molecule every 100 bp over the transcribed region (46). We envisage that this packing density is incompatible with UBF loading. It remains to be seen whether there are repeats that are entirely devoid of UBF on active NORs in human cells. As human cells can regulate the number of potentially active rDNA repeats by varying the number of active NORs, it seems more likely that all repeats on an active NOR are loaded with UBF.
UBF demonstrates low sequence specificity of DNA binding in vitro (48). This characteristic is further demonstrated by the +finding that UBF binds in vivo over the diverse collection of sequences found within the rDNA repeat. Some of these sequences are found elsewhere in the genome (e.g., alu elements and the cdc27 pseudogene in human rDNA) and clearly have not been selected for the purpose of UBF binding. Other sequences within the rDNA repeat, such as 18S and 28S coding sequences, have their own evolutionary constraints. UBF is not unique as a transcription factor in its ability to bind to nonconsensus sites across its target gene. In Drosophila melanogaster cells, the homeodomain proteins even-skipped and fushi tarazu have been shown to bind along the length of target genes in vivo (59). even-skipped and fushi tarazu also bind to genes that they are not expected to regulate, albeit to a lower degree. Also in Drosophila, the GAGA factor (designated GAF) binds to CT-GA rich sequences in vitro that are required for transcription in vivo. However, GAF also binds in vivo to genes that lack CT-rich sequences. During the heat shock response, GAF binds across the hsp70 gene. It has been proposed that the ability of GAF to disrupt nucleosomes facilitates RNA polymerase II elongation (39).
Despite UBF’s lack of sequence specificity in binding to DNA, immunofluorescent staining of interphase cells and mitotic chromosomes demonstrates that UBF binding is largely restricted to rDNA. This presents something of a paradox. How does UBF discriminate between NORs and the remainder of the genome? A clue to resolving this issue comes from in vitro binding studies with larger DNA fragments. UBF shows clear cooperative binding to a probe that comprises 10 copies of the 60-bp Xenopus enhancer repeat (45). We suggest that large blocks of enhancer elements present within an intact rDNA repeat bind UBF in a highly cooperative manner and act as nucleation centers for UBF binding across the remainder of the repeat. Thus, enhancers may be key determinants in targeting UBF to NORs.
Active NORs are visible as a secondary constriction on mitotic chromosomes. Sequences within the secondary constriction have been estimated to be 10-fold less compact than the remainder of the chromosome (19). It is interesting to speculate that extensive binding of UBF across the NOR plays a role in this phenomenon. This distinct form of chromatin is required as NORs display a number of unique characteristics. Ribosomal genes are among the most heavily transcribed genes in vertebrate genomes, and the Pol I transcription machinery remains associated with them on mitotic chromosomes. This may be required to facilitate the rapid reinitiation of transcription and nucleolar reformation as cells exit mitosis. UBF-associated NORs may be refractory to the chromosome condensation observed over the remainder of the genome during metaphase. NORs are surrounded by heterochromatin. In mouse cells, NORs are pericentromeric; in human cells, NORs are embedded in heterochromatin. It is well known that heterochromatin can have negative effects on the expression of linked sequences. UBF loading on active NORs may counteract these negative effects.
As with histones, UBF exhibits a variety of posttranslational modification including phosphorylation (24, 57, 58) and acetylation (42). In common with histones, one could imagine these modifications existing either globally over the entire NOR or locally over key regulatory elements such as enhancers and promoters. Consequently, global modification of UBF may be more relevant to overall structure of an NOR in vivo than to promoter function. In the future, it will be interesting to determine the degree to which UBF modifications vary across the rDNA repeat.
Acknowledgments
We thank Ulrich Scheer (Wurzburg, Germany) for the gift of the S18 α-RNA polymerase I antisera, Lucio Comai (University of Southern California) for α-TafI48 and −110 antibodies, Mariano Rocchi (University of Bari, Bari, Italy) for plasmid pZ21A, and Christine Mais for comments on the manuscript.
This work was supported by a program grant from the Medical Research Council to B.M. We also thank Roland Wolf (Director of the Biomedical Research Centre) for providing financial support for A.C.O.
REFERENCES
- 1.Archidiacono, N., R. Antonacci, R. Marzella, P.Finelli, A. Lonoce, and M. Rocchi. 1995. Comparative mapping of human alphoid sequences in great apes using fluorescence in situ hybridization. Genomics 25:477–484. [DOI] [PubMed] [Google Scholar]
- 2.Bachvarov, D., and T. Moss. 1991. The RNA polymerase I transcription factor xUBF contains 5 tandemly repeated HMG homology boxes. Nucleic Acids Res. 19:2331–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bazett-Jones, D. P., B. Leblanc, M. Herfort, and T. Moss. 1994. Short-range DNA looping by the Xenopus HMG-box transcription factor, xUBF. Science 264:1134–1137. [DOI] [PubMed] [Google Scholar]
- 4.Bell, P., C. Mals, B. McStay, and U. Scheer. 1997. Association of the nucleolar transcription factor UBF with the transcriptionally inactive rRNA genes of pronuclei and early Xenopus embryos. J. Cell Sci. 110:2053–2063. [DOI] [PubMed] [Google Scholar]
- 5.Bell, S. P., R. M. Learned, H. M. Jantzen, and R. Tjian. 1988. Functional cooperativity between transcription factors UBF1 and SL1 mediates human ribosomal RNA synthesis. Science 241:1192–1197. [DOI] [PubMed] [Google Scholar]
- 6.Bodeker, M., C. Cairns, and B. McStay. 1996. Upstream binding factor stabilizes Rib 1, the TATA-binding-protein-containing Xenopus laevis RNA polymerase I transcription factor, by multiple protein interactions in a DNA-independent manner. Mol. Cell. Biol. 16:5572–5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bodem, J., G. Dobreva, U. Hoffmann-Rohrer, S. Iben, H. Zentgraf, H. Delius, M. Vingron, and I. Grummt. 2000. TIF-IA, the factor mediating growth-dependent control of ribosomal RNA synthesis,is the mammalian homolog of yeast Rrn3p. EMBO Rep. 1:171–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boseley, P., T. Moss, M. Machler, R. Portmann, and M. Birnstiel. 1979. Sequence organization of the spacer DNA in a ribosomal gene unit of Xenopus laevis. Cell 17:19–31. [DOI] [PubMed] [Google Scholar]
- 9.Cairns, C., and B. McStay. 1995. HMG box 4 is the principal determinant of species specificity in the RNA polymerase I transcription factor UBF. Nucleic Acids Res. 23:4583–4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan, E. K., H. Imai, J. C. Hamel, and E. M. Tan. 1991. Human autoantibody to RNA polymerase I transcription actor hUBF. Molecular identity of nucleolus organizer region autoantigen NOR-90 and ribosomal RNA transcription upstream binding factor. J. Exp. Med. 174:1239–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choo, K. H., E. Earle, and C. McQuillan. 1990. A homologous subfamily of satellite III DNA on human chromosomes 14 and 22. Nucleic Acids Res. 18:5641–5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Church, G. M., and W. Gilbert. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 81:1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Comai, L., N. Tanese, and R. Tjian. 1992. The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor, SL1. Cell 68:965–976. [DOI] [PubMed] [Google Scholar]
- 14.Conconi, A., R. M. Widmer, T. Koller, and J. M. Sogo. 1989. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell 57:753–761. [DOI] [PubMed] [Google Scholar]
- 15.Copenhaver, G. P., C. D. Putnam, M. L. Denton, and C. S. Pikaard. 1994. The RNA polymerase I transcription factor UBF is a sequence-tolerant HMG-box protein that can recognize structured nucleic acids. Nucleic Acids Res. 22:2651–2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feinberg, A. P., and B. Vogelstein. 1983. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132:6–13. [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez, I. L., and J. E. Sylvester. 1995. Complete sequence of the 43-kb human ribosomal DNA repeat: analysis of the intergenic spacer. Genomics 27:320–328. [DOI] [PubMed] [Google Scholar]
- 18.Grummt, I. 1999. Regulation of mammalian ribosomal gene transcription by RNA polymerase I. Prog. Nucleic Acid Res. Mol. Biol. 62:109–154. [DOI] [PubMed] [Google Scholar]
- 19.Heliot, L., H. Kaplan, L. Lucas, C. Klein, A. Beorchia, M. Doco-Fenzy, M. Menager, M. Thiry, M. F. O’Donohue, and D. Ploton. 1997. Electron tomography of metaphase nucleolar organizer regions: evidence for a twisted-loop organization. Mol. Biol. Cell 8:2199–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jantzen, H. M., A. Admon, S. P. Bell, and R. Tjian. 1990. Nucleolar transcription factor hUBF contains a DNA-binding motif with homology to HMG proteins. Nature 344:830–836. [DOI] [PubMed] [Google Scholar]
- 21.Jordan, P., M. Mannervik, L. Tora, and M. Carmo-Fonseca. 1996. In vivo evidence that TATA-binding protein/SL1 colocalizes with UBF and RNA polymerase I when rRNA synthesis is either active or inactive. J. Cell Biol. 133:225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Junera, H. R., C. Masson, G. Geraud, J. Suja, and D. Hernandez-Verdun. 1997. Involvement of in situ conformation of ribosomal genes and selective distribution of upstream binding factor in rRNA transcription. Mol. Biol. Cell 8:145–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kermekchiev, M., J. L. Workman, and C. S. Pikaard. 1997. Nucleosome binding by the polymerase I transactivator upstream binding factor displaces linker histone H1. Mol. Cell. Biol. 17:5833–5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klein, J., and I. Grummt. 1999. Cell cycle-dependent regulation of RNA polymerase I transcription: the nucleolar transcription factor UBF is inactive in mitosis and early G1. Proc. Natl. Acad. Sci. USA 96:6096–6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuhn, A., R. Voit, V. Stefanovsky, R. Evers, M. Bianchi, and I. Grummt. 1994. Functional differences between the two splice variants of the nucleolar transcription factor UBF: the second HMG box determines specificity of DNA binding and transcriptional activity. EMBO J. 13:416–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Labhart, P., and R. H. Reeder. 1984. Enhancer-like properties of the 60/81 bp elements in the ribosomal gene spacer of Xenopus laevis. Cell 37:285–289. [DOI] [PubMed] [Google Scholar]
- 27.La Volpe, A., A. Simeone, M. D’Esposito, L. Scotto, V. Fidanza, A. de Falco, and E. Boncinelli. 1985. Molecular analysis of the heterogeneity region of the human ribosomal spacer. J. Mol. Biol. 183:213–223. [DOI] [PubMed] [Google Scholar]
- 28.Learned, R. M., and R. Tjian. 1982. In vitro transcription of human ribosomal RNA genes by RNA polymerase I. J. Mol. Appl. Genet. 1:575–584. [PubMed] [Google Scholar]
- 29.Leblanc, B., C. Read, and T. Moss. 1993. Recognition of the Xenopus ribosomal core promoter by the transcription factor xUBF involves multiple HMG box domains and leads to an xUBF interdomain interaction. EMBO J. 12:513–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Long, E. O., and I. B. Dawid. 1980. Repeated genes in eukaryotes. Annu. Rev. Biochem. 49:727–764. [DOI] [PubMed] [Google Scholar]
- 31.Maggio, R. 1966. Some properties of isolated nucleoli from guinea-pig liver. Biochim. Biophys. Acta 119:641–644. [DOI] [PubMed] [Google Scholar]
- 32.McStay, B., C. H. Hu, C. S. Pikaard, and R. H. Reeder. 1991. xUBF and Rib 1 are both required for formation of a stable polymerase I promoter complex in X. laevis. EMBO J. 10:2297–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McStay, B., G. J. Sullivan, and C. Cairns. 1997. The Xenopus RNA polymerase I transcription factor, UBF, has a role in transcriptional enhancement distinct from that at the promoter. EMBO J. 16:396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Milkereit, P., and H. Tschochner. 1998. A specialized form of RNA polymerase I, essential for initiation and growth-dependent regulation of rRNA synthesis, is disrupted during transcription. EMBO J. 17:3692–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller, G., K. I. Panov, J. K. Friedrich, L. Trinkle-Mulcahy, A. I. Lamond, and J. C. Zomerdijk. 2001. hRRN3 is essential in the SL1-mediated recruitment of RNA polymerase I to rRNA gene promoters. EMBO J. 20:1373–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moorefield, B., E. A. Greene, and R. H. Reeder. 2000. RNA polymerase I transcription factor Rrn3 is functionally conserved between yeast and human. Proc. Natl. Acad. Sci. USA 97:4724–4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mougey, E. B., L. K. Pape, and B. Sollner-Webb. 1996. Virtually the entire Xenopus laevis rDNA multikilobase intergenic spacer serves to stimulate polymerase I transcription. J. Biol. Chem. 271:27138–27145. [DOI] [PubMed] [Google Scholar]
- 38.Muramatsu, M., K. Smetana, and H. Busch. 1963. Quantitative aspects of isolation of nucleoli of the Walkercarcinosarcoma and liver of the rat. Cancer Res. 23:510–522. [Google Scholar]
- 39.O’Brien, T., R. C. Wilkins, C. Giardina, and J. T. Lis. 1995. Distribution of GAGA protein on Drosophila genes in vivo. Genes Dev. 9:1098–1110. [DOI] [PubMed] [Google Scholar]
- 40.O’Mahony, D. J., and L. I. Rothblum. 1991. Identification of two forms of the RNA polymerase I transcription factor UBF. Proc. Natl. Acad. Sci. USA 88:3180–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orlando, V. 2000. Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem. Sci. 25:99–104. [DOI] [PubMed] [Google Scholar]
- 42.Pelletier, G., V. Y. Stefanovsky, M. Faubladier, I. Hirschler-Laszkiewicz, J. Savard, L. I. Rothblum, J. Cote, and T. Moss. 2000. Competitive recruitment of CBP and Rb-HDAC regulates UBF acetylation and ribosomal transcription. Mol. Cell 6:1059–1066. [DOI] [PubMed] [Google Scholar]
- 43.Pikaard, C. S., B. McStay, M. C. Schultz, S. P. Bell, and R. H. Reeder. 1989. The Xenopus ribosomal gene enhancers bind an essential polymerase I transcription factor, xUBF. Genes Dev. 3:1779–1788. [DOI] [PubMed] [Google Scholar]
- 44.Putnam, C. D., G. P. Copenhaver, M. L. Denton, and C. S. Pikaard. 1994. The RNA polymerase I transactivator upstream binding factor requires its dimerization domain and high-mobility-group (HMG) box 1 to bend, wrap, and positively supercoil enhancer DNA. Mol. Cell. Biol. 14:6476–6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Putnam, C. D., and C. S. Pikaard. 1992. Cooperative binding of the Xenopus RNA polymerase Itranscription factor xUBF to repetitive ribosomal gene enhancers. Mol. Cell. Biol. 12:4970–4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Puvion-Dutilleul, F. 1983. Morphology of transcription at cellular and molecular levels. Int. Rev. Cytol. 84:57–101. [DOI] [PubMed] [Google Scholar]
- 47.Reeder, R. H. 1989. Regulatory elements of the generic ribosomal gene. Curr. Opin. Cell Biol. 1:466–474. [DOI] [PubMed] [Google Scholar]
- 48.Reeder, R. H., C. S. Pikaard, and B. McStay. 1995. UBF, an architectural element for RNA polymerase Ipromoters, p.251–263. In F. Eckstein and D. M. J. Lilley (ed.), Nucleic acids and molecular biology, vol. 9. Springer-Verlag, Berlin, Germany.
- 49.Reimer, G., K. M. Rose, U. Scheer, and E. M. Tan. 1987. Autoantibody to RNA polymerase I in scleroderma sera. J. Clin. Investig. 79:65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.ORoussel, P., C. Andre, L. Comai, and D. Hernandez-Verdun. 1996. The rDNA transcription machinery is assembled during mitosis in active NORs and absent in inactive NORs. J. Cell Biol. 133:235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roussel, P., C. Andre, C. Masson, G. Geraud, and V. D. Hernandez. 1993. Localization of the RNA polymerase I transcription factor hUBF during the cell cycle. J. Cell Sci. 104:327–337. [DOI] [PubMed] [Google Scholar]
- 52.ORudloff, U., D. Eberhard, L. Tora, H. Stunnenberg, and I. Grummt. 1994. TBP-associated factors interact with DNA and govern species specificity of RNA polymerase I transcription. EMBO J. 13:2611–2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shiels, C., C. Coutelle, and C. Huxley. 1997. Contiguous arrays of satellites 1, 3, and beta form a 1.5-Mb domain on chromosome 22p. Genomics 44:35–44. [DOI] [PubMed] [Google Scholar]
- 54.Sirri, V., P. Roussel, and D. Hernandez-Verdun. 2000. In vivo release of mitotic silencing of ribosomal gene transcription does not give rise to precursor ribosomal RNA processing. J. Cell Biol. 148:259–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sullivan, G. J., J. M. Bridger, A. P. Cuthbert, R. F. Newbold, W. A. Bickmore, and B. McStay. 2001. Human acrocentric chromosomes with transcriptionally silent nucleolar organizer regions associate with nucleoli. EMBO J. 20:2867–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tagarro, I., J. Wiegant, A. K. Raap, J. J. Gonzalez-Aguilera, and A. M. Fernandez-Peralta. 1994. Assignment of human satellite 1 DNA as revealed by fluorescent in situhybridization with oligonucleotides. Hum. Genet. 93:125–128. [DOI] [PubMed] [Google Scholar]
- 57.Voit, R., M. Hoffmann, and I. Grummt. 1999. Phosphorylation by G1-specific cdk-cyclin complexes activates the nucleolar transcription factor UBF. EMBO J. 18:1891–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Voit, R., A. Kuhn, E. E. Sander, and I. Grummt. 1995. Activation of mammalian ribosomal gene transcription requires phosphorylation of the nucleolar transcription factor UBF. Nucleic Acids Res. 23:2593–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walter, J., C. A. Dever, and M. D. Biggin. 1994. Two homeo domain proteins bind with similar specificity to a wide range of DNA sites in Drosophila embryos. Genes Dev. 8:1678–1692. [DOI] [PubMed] [Google Scholar]
- 60.Waye, J. S., and H. F. Willard. 1989. Concerted evolution of alpha satellite DNA: evidence for species specificity and a general lack of sequence conservation among alphoid sequences of higher primates. Chromosoma 98:273–279. [DOI] [PubMed] [Google Scholar]
- 61.Weisenberger, D., and U. Scheer. 1995. A possible mechanism for the inhibition of ribosomal RNA gene transcription during mitosis J. Cell Biol. 129:561–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamamoto, R. T., Y. Nogi, J. A. Dodd, and M. Nomura. 1996. RRN3 gene of Saccharomyces cerevisiae encodes an essential RNA polymerase I transcription factor which interacts with the polymerase independently of DNA template. EMBO J. 15:3964–3973. [PMC free article] [PubMed] [Google Scholar]