Abstract
Endoplasmic reticulum-associated degradation (ERAD) disposes of aberrant proteins in the secretory pathway. Protein substrates of ERAD are dislocated via the Sec61p translocon from the endoplasmic reticulum to the cytosol, where they are ubiquitinated and degraded by the proteasome. Since the Sec61p channel is also responsible for import of nascent proteins, this bidirectional passage should be coordinated, probably by molecular chaperones. Here we implicate the cytosolic chaperone AAA-ATPase p97/Cdc48p in ERAD. We show the association of mammalian p97 and its yeast homologue Cdc48p in complexes with two respective ERAD substrates, secretory immunoglobulin M in B lymphocytes and 6myc-Hmg2p in yeast. The membrane 6myc-Hmg2p as well as soluble lumenal CPY*, two short-lived ERAD substrates, are markedly stabilized in conditional cdc48 yeast mutants. The involvement of Cdc48p in dislocation is underscored by the accumulation of ERAD substrates in the endoplasmic reticulum when Cdc48p fails to function, as monitored by activation of the unfolded protein response. We propose that the role of p97/Cdc48p in ERAD, provided by its potential unfoldase activity and multiubiquitin binding capacity, is to act at the cytosolic face of the endoplasmic reticulum and to chaperone dislocation of ERAD substrates and present them to the proteasome.
Endoplasmic reticulum (ER)-associated degradation (ERAD) is a quality control process that selectively eliminates aberrant proteins in the secretory pathway. Protein substrates of ERAD are dislocated from the ER to the cytosol, where they are ubiquitinated and degraded by the proteasome (5). The Sec61p translocon is involved both in the import of nascent proteins into the ER and in dislocation of aberrant proteins from the ER. These two activities of Sec61p are mechanistically different because they involve distinct domains within Sec61p and dislocation-defective mutants of Sec61p are still proficient in protein import (40, 50, 56).
Since nascent and aberrant proteins pass through the same Sec61p translocon, this bidirectional passage requires coordination. Moreover, the Sec61p translocon is a passive conduit; thus, the driving force to move polypeptides across it should be provided by accessory proteins. Indeed, passage through the Sec61p translocon requires molecular chaperones, and their contribution further illustrates that import and dislocation must be mechanistically distinct. For example, of the two hsp70s involved in import in yeast, BiP/Kar2p in the ER lumen and Ssa1p in the cytosol, mutants of BiP/Kar2p that are defective in dislocation are still proficient in import, and mutation in SSA1 does not affect degradation of the ERAD substrates pro-α-factor and A1PiZ (9). Although the kar2 mutant initially used to link BiP to CPY* dislocation and degradation was also defective in protein import (41), the kar2 mutants that are defective only in dislocation of pro-α-factor and A1PiZ directly demonstrate the role of Kar2p in dislocation (9). Furthermore, chaperones that are required for ERAD of one protein substrate are dispensable for the degradation of another and, in particular, membrane or soluble lumenal ERAD substrates might involve different sets of chaperones. This is illustrated by BiP/Kar2p, which is required for degradation of soluble lumenal pro-α-factor, CPY* and A1PiZ, but is dispensable for the proteolysis of membrane Pdr5* and cystic fibrosis transmembrane conductance regulator (CFTR) (8, 9, 55). Also, BiP/Kar2p, as well as J domain-containing Jem1p and Scj1p, maintain unfolded lumenal proteins in a soluble form, but deletion of Jem1p and Scj1p has little effect on ERAD of a membrane protein (38).
Akin to the ratchet action of BiP during protein import, cytosolic chaperones might play a critical role in ratcheting and pulling ERAD substrates out from the ER. Indeed, cytosolic hsp70 and hsp90 are associated with CFTR in mammalian cells (34). However, Ssa1p, the major cytosolic hsc70, which facilitates ERAD of membrane CFTR expressed in yeast, is dispensable for the dislocation of the soluble lumenal pro-α-factor and A1PiZ (9, 55). Moreover, while blocking the interaction of hsp90 (the most abundant cytosolic chaperone) with the ERAD substrates apoB48 or mutant insulin receptor results in stabilization of these proteins, it accelerates the proteasomal degradation of CFTR (23, 26, 34).
In our search for accessory proteins that function in ERAD, especially cytosolic chaperones, ERAD substrates were used as bait to identify associated proteins. Here we show the pull down of the p97/Cdc48p AAA-ATPase, an abundant chaperone-like cytosolic protein, that by analogy with its archaeal homologue VAT has potential unfoldase activity (22). The mammalian p97/VCP (39) was found in a complex with μs, the heavy chain of secretory immunoglobulin M (sIgM), a soluble lumenal proteasomal substrate in B cells (2, 35). Likewise, Cdc48p, the highly conserved homologue of p97 in Saccharomyces cerevisiae (19), was found in a complex with 6myc-Hmg2p, a well-established membrane ERAD substrate in yeast (24). Moreover, the membrane 6myc-Hmg2p as well as the soluble lumenal ERAD substrate CPY* (25) were markedly stabilized in yeast strains expressing Cdc48p conditional mutants (19). These results implicate Cdc48p as a novel cytosolic ERAD component. The involvement of Cdc48p in dislocation of proteins from the ER was indicated by the accumulation of ERAD substrates in the ER and activation of the unfolded protein response (UPR) (56) in conditional cdc48 yeast mutants.
MATERIALS AND METHODS
Plasmids and strains.
Yeast strains used in this study are listed in Table 1. Strains expressing CPY* were generated by replacing wild-type CPY with CPY* (prc1-1 allele) by using the integrative plasmid bMK150 (generously provided by D. Wolf). 6myc-Hmg2p was expressed from plasmid pRH244 (generously provided by R. Hampton). The UPR reporter plasmid (56) was pMCZ2, harboring a CYC1 promoter containing the KAR2 UPR element fused to the lacZ gene (UPRE-lacZ; generously provided by R. Schekman). Strain KFY116 cdc48-1cs carries the cold-sensitive cdc48-1 mutation (19), and strains KFY188 cdc48-7ts and KFY194 cdc48-10ts are two temperature-sensitive cdc48 mutants. These were isolated independently as spontaneous revertants of cdc48-1 cold sensitivity, which acquired temperature sensitivity. Strains KFY99 and KFY100 derived from the same strain crosses, as the cdc48 conditional mutants express wild-type Cdc48p. Strains RHY696 and RHY965 (a hrd1Δ strain [24]) express integrated 6myc-Hmg2p and wild-type Cdc48p (generously provided by R. Hampton).
TABLE 1.
Yeast strains used in this study
| Name | Genotype | Reference |
|---|---|---|
| KFY99 | MATahis4-619 leu2-3,112 ura3-52 | This study |
| KFY100 | MATahis4-619 leu2-3,112 ura3-52 | This study |
| KFY116 | MATα lys2-801 leu2-3,112 ura3-52 cdc48-1cs | 19 |
| KFY188 | MATalys2-801 leu2-3,112 ura3-52 cdc48-7ts | This study |
| KFY194 | MATalys2-801 leu2-3,112 ura3-52 cdc48-10ts | This study |
| RHY696 | MATahis3Δ200 lys2-801 ade2-101 met2 hmg1::LYS2 hmg2::HIS3 trp1::hisG leu2Δ ura3-52::6MYC HMG2 | 24 |
| RHY965 | MATahis3Δ200 lys2-801 ade2-101 met2 hmg1::LYS2 hmg2::HIS3 trp1::hisG leu2Δ ura3-52::6MYC HMG2 hrd1Δ::URA3 | 24 |
Coimmunoprecipitation of cdc48p with 6myc-Hmg2p.
Cdc48p was coimmunoprecipitated with 6myc-Hmg2p from strains KFY100 (wild-type Cdc48p), KFY116 cdc48-1cs (cold-sensitive cdc48-1 mutant [19]), and KFY194 cdc48-10ts (temperature-sensitive cdc48 mutant), transfected with plasmid pRH244 encoding 6myc-Hmg2p. For comparison, mock-transfected cells that do not express 6myc-Hmg2p were used. Briefly, cells were grown at 30°C and transferred to restrictive temperatures for 3 h. The cells (4 optical densities at 600 nm [OD600]) were dissolved by 3 cycles of freeze-thaw on dry ice and vigorous vortexing in lysis buffer consisting of phosphate-buffered saline, 1% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, 100 Kallikrein inhibitor units of aprotinin/ml. 6myc-Hmg2p was immunoprecipitated with anti-myc antibodies (clone 9E10) and then with protein A-Sepharose, resolved by reducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and electroblotted onto nitrocellulose. Proteins were probed either with antibodies raised in rabbit against yeast Cdc48p or with mouse anti-myc antibodies. Blots of total protein extracts were also probed with anti-Cdc48p antibodies. The appropriate secondary antibodies, conjugated to horseradish peroxidase (HRP), were visualized by enhanced chemiluminescence (ECL).
Coimmunoprecipitation of p97 with μs.
p97 was coimmunoprecipitated with μs from the murine 38C B-lymphocyte cell line. For comparison, D2 hybridoma that expresses and secretes an abundance of the same sIgM (46) or osteosarcoma U2OS cells that do not express sIgM were used. Cells were grown and lysed, and IgM was immunoprecipitated as previously described (1). Cells (2 × 107 to 4 × 107) were dissolved in lysis buffer (phosphate-buffered saline containing 1% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, 100 kallikrein inhibitor units KIU of aprotinin/ml). Where indicated, the lysis buffer was supplemented with 1 mM of the cross-linking reagent disuccinimidyl suberate (DSS; Pierce). IgM, its cross-linked complexes, and proteins associated with IgM were precipitated with an excess of goat anti-mouse IgM antibodies followed by protein A-Sepharose. Proteins were resolved by reducing SDS-PAGE and were electroblotted onto nitrocellulose. Blots were probed either with mouse anti-p97 antibodies (clone 58.13.3; Progen) or with rabbit antiserum specific to μs. This anti-μs antibody, which was raised against the μstp peptide (the unique C-terminal 19 residues of the secretory μs heavy chain), detected only μs and not μm, the heavy chain of the membrane IgM (B. Gur and S. Bar-Nun, unpublished results). Blots of total protein extracts were also probed with anti-p97 antibodies. The appropriate secondary antibodies, conjugated to HRP, were visualized by ECL. The ∼100-kDa μs-containing complex (see Fig. 1) was isolated from preparative SDS gels (2 × 108 cells) stained with Coomassie brilliant blue and subjected to enzymatic digestion, and the resulting peptides were analyzed by matrix-assisted laser desorption ionization (MALDI)-mass spectrometry (carried out by the Keck Foundation Biotechnology Resource Laboratory, Yale University).
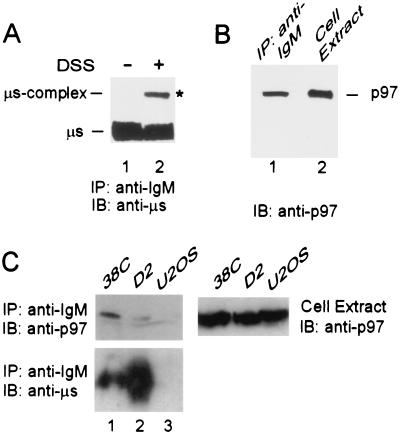
FIG. 1.
p97/VCP is coprecipitated with unstable μs in B cells. (A) 38C B cells were lysed in the absence (−) (lane 1) or presence (+) (lane 2) of DSS and IgM, and IgM-containing cross-linked complexes were immunoprecipitated with goat anti-IgM antibodies (IP: anti-IgM). Proteins were resolved by SDS-PAGE and were electroblotted onto nitrocellulose, and the blot was probed with rabbit anti-μs followed by HRP-conjugated anti-rabbit IgG (IB: anti-μs). The precipitated μs and ∼100-kDa μs complex are indicated, and the band of ∼100 kDa, marked by the asterisk, was subjected to MALDI-mass spectrometry. (B) 38C B cells were lysed, and IgM and proteins in a complex with IgM were precipitated with goat anti-IgM antibodies. The precipitated proteins (lane 1, IP: anti-IgM) and total cell extract, containing 5% of the proteins subjected to immunoprecipitation (lane 2, cell extract), were resolved by SDS-PAGE and were electroblotted onto nitrocellulose, and the blot was probed with mouse anti-p97 and then with HRP-conjugated anti-mouse IgG (IB: anti-p97). The pulled down p97 is indicated. (C) The μs and associated proteins were precipitated with anti-IgM antibodies (IP: anti-IgM) from 38C (lane 1), D2 (lane 2), and U2OS (lane 3) cells. Proteins were resolved by SDS-PAGE and were electroblotted onto nitrocellulose (left panels). For comparison, total cell extracts (10% of the proteins used for immunoprecipitation) were resolved by SDS-PAGE and were electroblotted onto nitrocellulose (right panel). Upper blots were probed with mouse anti-p97 followed by HRP-conjugated anti-mouse IgG (IB: anti-p97), and the blot in the left panel was reprobed with rabbit anti-μs followed by HRP-conjugated anti-rabbit IgG (IB: anti-μs).
Stability of CPY* and 6myc-Hmg2p.
Stability of CPY* and 6myc-Hmg2p was measured at permissive or restrictive temperatures after preincubation for 1 to 2 h. Cycloheximide (100 μg/ml) was added and cells were collected at the indicated time points. Cells (5 OD600) were dissolved in 0.5 ml of 0.2N NaOH and 36 μM β-mercaptoethanol by 20 min of incubation on ice, the pH was adjusted to 7.0, and 100 μl of total cellular proteins was resolved by reducing SDS-PAGE and electroblotted onto nitrocellulose. Blots were probed either with mouse anti-CPY antibodies (clone 10A5-B5; New Biotechnology) or with anti-myc antibodies followed by HRP-conjugated anti-mouse IgG (Jackson ImmunoResearch Laboratories), and the HRP was visualized by ECL. Blots were quantified by densitometry, and t1/2 values were calculated from slopes of the semi-logarithmic curves.
UPR induction.
UPR induction was measured with the UPRE-lacZ reporter construct (18, 56) during incubation at permissive or restrictive temperatures. Activity of β-galactosidase was detected on 5-bromo-4-chloro-3-indolyl-β-𝒹-galactoside (X-gal) reporter plates or directly measured in solubilized cells (1 OD600) with O-nitrophenyl-β-𝒟-galactopyranoside (ONPG) as a substrate (28). The maximal induction of UPR was achieved by blocking the N-linked glycosylation of glycoproteins with tunicamycin (15 μg/ml) (18, 56).
RESULTS
p97/VCP is in a complex with secretory IgM in B cells.
The secretory IgM is not an aberrant protein, but in murine B lymphocytes it undergoes developmentally regulated degradation that is blocked by proteasome inhibitors (1, 2, 35, 43, 51), suggesting that it is eliminated by an ERAD-like process. In our search for proteins that interact with secretory IgM, 38C B cells were solubilized in the presence of the cross-linking reagent DSS. When IgM was immunoprecipitated with anti-IgM and probed with antibodies specific to μs (Fig. 1A; IP: anti-IgM, IB: anti-μs), the anti-μs antibodies detected the ∼80-kDa μs polypeptide (Fig. 1A, lane 1). Yet in the immunoprecipitate from cross-linked samples these anti-μs antibodies detected an additional ∼100-kDa complex (Fig. 1A, lane 2), indicating that μs was cross-linked to a ∼20-kDa protein. When this ∼100-kDa band was isolated from preparative SDS gels and subjected to enzymatic digestion, MALDI-mass spectrometry of resulting peptides revealed the presence of p97/VCP. Clearly, p97 comigrated with the ∼100-kDa μs-containing complex. Subsequently we confirmed that p97 was actually pulled down with μs when IgM was specifically immunoprecipitated, even in samples that were not cross-linked. When IgM immunoprecipitate was probed with commercial anti-p97 antibodies (Fig. 1B, lane 1; IP: anti-IgM, IB: anti-p97), it was evident that p97 was coprecipitated via its direct or indirect association with μs. Only a small fraction of p97 cellular content coprecipitated with IgM, as shown by probing cell extract from 38C B cells (Fig. 1B, lane 2; cell extract [5% of amount used for IP], IB: anti-p97). Actually, the anti-IgM antibodies used for precipitation of μs from 38C cells (Fig. 1C, lane 1; IB: anti-μs), which evidently coprecipitated p97 (Fig. 1C, lane 1; IB: anti-p97), hardly coprecipitated p97 from the D2 hybridoma (Fig. 1C, lane 2; IB: anti-p97), although plenty of the same μs was precipitated from these cells (Fig. 1C, lane 2; IB: anti-μs). Moreover, this anti-IgM antibody did not precipitate any p97 from U2OS cells that do not express sIgM (Fig. 1C, lane 3). The amounts of coprecipitated p97 did not reflect different levels of this protein in the various cell lines (Fig. 1C, cell extract; IB: anti-p97). Taken together these results establish that p97 interacts with μs, directly or indirectly, only in cells where sIgM is unstable and subjected to an ERAD-like degradation. Conversely, p97 interacts poorly with the same sIgM in cells where this molecule is highly expressed but is stable and efficiently secreted.
Cdc48p is in a complex with the ERAD substrate 6myc-Hmg2p.
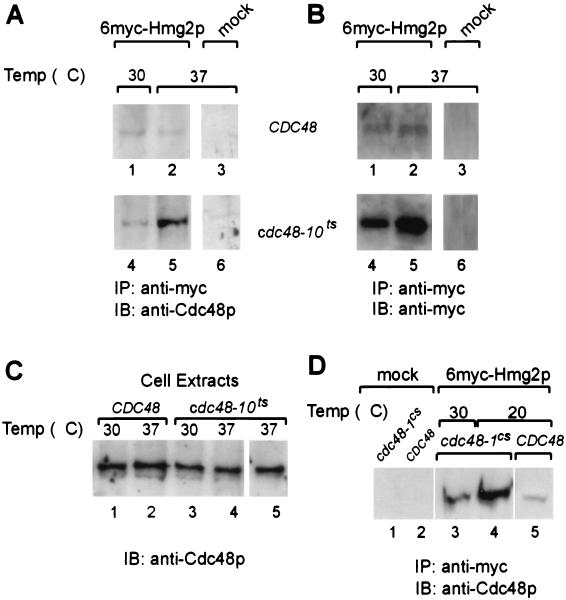
To extend our results and to examine the possible general role of p97/VCP in ERAD, we turned to study the interaction of Cdc48p with ERAD substrates in the yeast S. cerevisiae, in which conditional mutants of Cdc48p are available (19). The results shown in Fig. 2 demonstrate that Cdc48p is found in a complex with 6myc-Hmg2p, a well-established ERAD substrate (24). Immunoprecipitation of 6myc-Hmg2p with anti-myc antibodies (Fig. 2, panels A, B, and D) pulled down Cdc48p, as confirmed by positive staining of the blotted precipitates with anti-Cdc48p antibodies (Fig. 2, panels A and D). This coprecipitation was specific, and Cdc48p was pulled down via the 6myc-Hmg2p, since the anti-myc antibodies did not precipitate Cdc48p directly from cells that did not express 6myc-Hmg2p (mock; Fig. 2A and B, lanes 3 and 6; D, lanes 1 and 2). Interestingly, compared to that of the wild-type strain the amounts of Cdc48p that coprecipitated with 6myc-Hmg2p were higher in strains harboring the conditional mutations (Fig. 2A, compare lanes 4 and 5 with lanes 1 and 2; D, compare lanes 3 and 4 with lane 5), and even higher levels were pulled down at the nonpermissive temperatures (Fig. 2A, compare lane 5 with lane 4; D, compare lane 4 with lane 3). Importantly, the total amount of Cdc48p was affected neither by the mutations nor by the temperature or expression of 6myc-Hmg2p (Fig. 2C, lanes 1 to 5). These results may suggest that the interaction of mutant Cdc48p with 6myc-Hmg2p is tighter than that of the wild-type Cdc48p. Alternatively, the larger amounts of pulled down Cdc48p in the mutants may merely reflect higher steady-state levels of 6myc-Hmg2p in the mutant strains. Indeed, when the blot in Fig. 2A was reprobed with the anti-myc antibody, higher steady-state levels of 6myc-Hmg2p were detected in the cdc48 mutants (Fig. 2B, compare lanes 1 and 2 with lanes 4 and 5).
FIG. 2.
Cdc48p is coprecipitated with 6myc-Hmg2p in yeast. The indicated yeast strains KFY100 (wild-type CDC48) and KFY194 (cdc48-10ts mutation), transformed with plasmid pRH244 encoding 6myc-Hmg2p (panels A and B, lanes 1 and 2 and lanes 4 and 5; panel C, lanes 1 to 4; panel D, lanes 3 to 5) or mock-transformed (mock) (panels A and B, lanes 3 and 6; panel C, lane 5; panel D, lanes 1 and 2), were grown for 3 h at the indicated temperatures. The 6myc-Hmg2p and associated proteins were precipitated with anti-myc antibodies (IP: anti-myc). Proteins were resolved by SDS-PAGE and were electroblotted onto nitrocellulose. For comparison, total cell extracts (1% of the proteins used for immunoprecipitation) (panel C, lanes 1 to 5) were resolved by SDS-PAGE and were electroblotted onto nitrocellulose. Blots in panels A, C, and D were probed with rabbit anti-Cdc48p followed by HRP-conjugated anti-rabbit IgG (IB: anti-Cdc48p). (B) The blot in panel A was reprobed with mouse anti-myc antibody followed by HRP-conjugated anti-mouse IgG (IB: anti-myc).
ERAD is impaired in yeast cdc48 mutants.
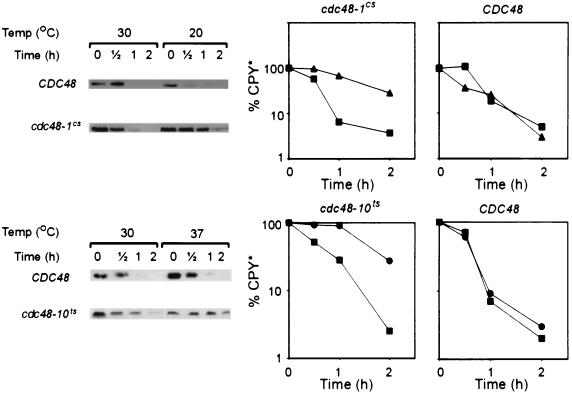
The observed increased levels of 6myc-Hmg2p (Fig. 2B, lanes 4 and 5) suggested that this ERAD substrate was stabilized in cdc48 mutants. Indeed, both membrane 6myc-Hmg2p and soluble lumenal CPY*, another short-lived protein that is eliminated by ERAD (25), were markedly stabilized in the cdc48 mutants. Employing the cycloheximide-chase assays, the half-lives of 6myc-Hmg2p (Fig. 3) and CPY* (Fig. 4) were compared in strains expressing wild-type Cdc48p and strains carrying the cold-sensitive cdc48-1 allele (19) or the temperature-sensitive mutations cdc48-7ts and cdc48-10ts. As shown in Fig. 3 and 4 and as summarized in Table 2, 6myc-Hmg2p and CPY* were rapidly degraded at all temperatures in strains expressing wild-type Cdc48p (t1/2 of 50 to 76 min and 22 to 24 min, respectively). In contrast, even at the permissive temperature (30°C) the degradation of 6myc-Hmg2p was somewhat slower in the cold-sensitive cdc48 mutant (t1/2 of 89 min) and prolonged to a t1/2 value of 2.6 to 5.2 h in the temperature-sensitive cdc48 mutants. A similar phenomenon was previously reported for the degradation of a cytosolic model substrate of the ubiquitin-dependent ubiquitin fusion degradation (UFD) pathway (27), which was impaired by the cold-sensitive cdc48-1 mutation (21). At the restrictive temperatures, the conditional cdc48 mutants exhibited a much more severe phenotype with both substrates. In the cdc48-1cs cells, the degradation of CPY* and 6myc-Hmg2p was approximately threefold slower when cells were shifted from 30 to 20°C; the half-life of CPY* was extended from 23 to 99 min, and the half-life of 6myc-Hmg2p was prolonged from 89 min to 4.1 h. Similarly, in cdc48-7ts and cdc48-10ts cells 6myc-Hmg2p was degraded at 30°C, with t1/2 values of 5.2 and 2.6 h, respectively, and these turnover rates were extended to 17.0 and 10.4 h, respectively, at 37°C. The half-life of CPY* was also extended from 22 to 83 min when cdc48-10ts cells were shifted from 30 to 37°C. The involvement of Cdc48p in disposing of ERAD substrates was further underscored when we observed a similar degree of stabilization for 6myc-Hmg2p in the cdc48 mutants and the bona fide ERAD mutant hrd1/der3 (strain RHY965 [24]). In the hrd1/der3 mutant 6myc-Hmg2p was degraded, with t1/2 values of 7.2 to 7.6 h either at 30 or 37°C, and a t1/2 value of >10 h was measured at 20°C (Fig. 3 and Table 2). The RING-H2-containing Hrd1p/Der3p forms a complex with Hrd3p to function in ERAD as an ER membrane-anchored ubiquitin ligase that collaborates with Ubc7p or Ubc1p (3, 7, 14, 20). Although Hrd1p/Der3p is not essential for degradation of every ERAD substrate (49, 55), it was specifically selected for defective ERAD of 6myc-Hmg2p (24) and is known to be required for ERAD of CPY* (6, 30).
FIG. 3.
The ERAD of 6myc-Hmg2p is impaired in yeast cdc48 mutants. The stability of 6myc-Hmg2p was measured in strains KFY100 (wild-type CDC48), KFY116 (cdc48-1cs mutation), and KFY194 (cdc48-10ts mutation) expressing 6myc-Hmg2p from plasmid pRH244. Strains RHY696 and RHY965 express integrated 6myc-Hmg2p and carry wild-type CDC48, and strain RHY965 is also hrd1Δ. Following preincubation for 2 h at the indicated permissive or restrictive temperatures, cycloheximide (100 μg/ml) was added and cells were collected at the indicated time points. Cells were dissolved and total cellular proteins were resolved by reducing SDS-PAGE and were electroblotted onto nitrocellulose. Blots were probed with mouse anti-myc antibodies followed by HRP-conjugated anti-mouse IgG, and HRP was visualized by ECL. The blots were quantified, and the semi-logarithmic plots represent the decay of 6myc-Hmg2p. The remaining 6myc-Hmg2p was calculated as the percentage of its level at the time of cycloheximide addition (100%). Upper panels, cdc48-1cs (left) and wild-type CDC48 (right) at 20°C (▴) or 30°C (▪). Lower panels, cdc48-10ts (left) and wild-type CDC48 (right) at 37°C (•) or 30°C (▪).
FIG. 4.
The ERAD of CPY* is impaired in yeast cdc48 mutants. Stability of CPY* was measured for integrated CPY* expressed in strains KFY100 (wild-type CDC48), KFY116 (cdc48-1cs mutation), and KFY194 (cdc48-10ts mutation). Following preincubation for 2 h at the indicated permissive or restrictive temperatures, cycloheximide (100 μg/ml) was added and cells were collected at the indicated time points. Cells were dissolved and total cellular proteins were resolved by reducing SDS-PAGE and were electroblotted onto nitrocellulose. Blots were probed with mouse anti-CPY antibodies followed by HRP-conjugated anti-mouse IgG, and the HRP was visualized by ECL. The blots were quantified, and the semi-logarithmic plots represent the decay of CPY*. The remaining CPY* was calculated as the percentage of its level at the time of cycloheximide addition (100%). Upper panels, cdc48-1cs (left) and wild-type CDC48 (right) at 20°C (▴) or 30°C (▪). Lower panels, cdc48-10ts (left) and wild-type CDC48 (right) at 37°C (•) or 30°C (▪).
TABLE 2.
Calculated half-lives of 6myc-Hmg2p and CPY*a
| Strain | Temp (°C) | 6myc-Hmg2p | CPY* (min) | Remark |
|---|---|---|---|---|
| KFY99 (WT CDC48) | 30 | 70 min | ND | 6myc-Hmg2p on plasmid pRH244 |
| 37 | 50 min | |||
| KFY100 (WT CDC48) | 30 | 65 ± 10 min | 23 ± 1.2 | Integrated CPY* (plasmid bMK150) 6myc-Hmg2p on plasmid pRH244 |
| 20 | 69 ± 4 min | 22 ± 1.4 | ||
| 37 | 76 ± 6 min | 24 ± 1.6 | ||
| KFY116 cdc48-1cs | 30 | 89 ± 6 min | 23 ± 1.2 | Integrated CPY* (plasmid bMK150) 6myc-Hmg2p on plasmid pRH244 |
| 20 | 4.1 ± 1 h | 99 ± 6.9 | ||
| KFY194 cdc48-10ts | 30 | 2.6 ± 0.6 h | 22 ± 1.6 | Integrated CPY* (plasmid bMK150) 6myc-Hmg2p on plasmid pRH244 |
| 37 | 10.4 ± 1.1 h | 83 ± 5.3 | ||
| KFY188 cdc48-7ts | 30 | 5.2 h | ND | 6myc-Hmg2p on plasmid pRH244 |
| 37 | 17.0 h | |||
| RHY696 (WT CDC48) | 30 | 40 min | ND | Integrated 6myc-Hmg2p |
| 20 | 75 min | |||
| RHY965 (hrd1Δ) | 30 | 7.2 h | ND | Integrated 6myc-Hmg2p |
| 37 | 7.6 h | |||
| 20 | >10.0 h |
The stability of CPY* and 6myc-Hmg2p was measured by cycloheximide-chase assays in strains expressing integrated CPY* or 6myc-Hmg2p (RHY696 and RHY965) or nonintegrated 6myc-Hmg2p on plasmid pRH244. Strains KFY99, KFY100, RHY696, and RHY965 express wild-type (WT) Cdc48p, strain KFY116 carries the cold-sensitive cdc48-1 mutation, and strains KFY188 and KFY194 are two temperature-sensitive cdc48 mutants. Strain RHY965 is hrd1Δ. Blots, represented in Fig. 3 and 4, were quantified by densitometry, and values of t½ were calculated from slopes of semi-logarithmic curves, also represented in Fig. 3 and 4. Values are the means ± standard deviations of eight independent experiments. ND, not done.
The UPR is activated in yeast cdc48 mutants.
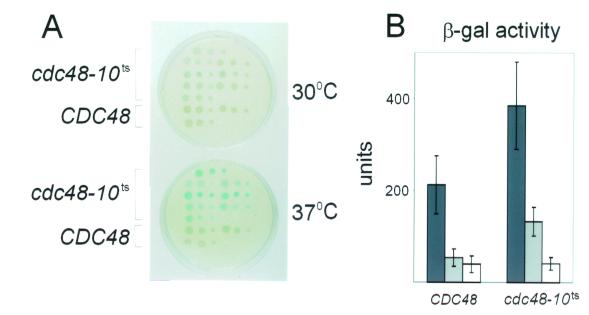
One plausible role for p97/Cdc48p in ERAD, which is supported by the known in vitro unfoldase activity of VAT (22), the archaeal homologue of p97/Cdc48p, is in the dislocation of ERAD substrates. A hallmark phenotype in dislocation-specific mutants of Sec61p is the activation of the UPR as a consequence of accumulation of unfolded proteins in the ER (56). Using the UPRE-lacZ-reporter construct (56), we showed that the UPR was markedly activated in the cdc48-10ts mutant compared to that of the wild-type strains (Fig. 5). Blue color was developed on X-gal plates by the cdc48-10ts strain even at the permissive temperature (30°C) and more evidently at the restrictive 37°C, while wild-type cells remained relatively white at both temperatures (Fig. 5A). Treatment with tunicamycin at 30°C resulted in β-galactosidase activities that were higher in the cdc48-10ts strain than in the wild-type cells (Fig. 5B). Similar phenomenon was previously reported for other ERAD-defective strains, including Δubc6, Δubc7, and Δcue1 cells (56). However, even in the absence of tunicamycin, markedly increased β-galactosidase activity was measured in cdc48-10ts at the restrictive temperature 37°C, whereas strains expressing wild-type CDC48 exhibited similar activity at either 30 or 37°C (Fig. 5B). Thus, when Cdc48p failed to function, ERAD protein substrates accumulated in the ER and consequently activated the UPR.
FIG. 5.
The UPR is activated in yeast cdc48 mutants. UPR induction was measured in strains KFY99 and KFY100 carrying wild-type CDC48 or in strain KFY194 carrying the cdc48-10ts mutation. (A) Activity of β-galactosidase (β-gal) was detected on X-gal reporter plates after a 17-h incubation at the permissive (30°C) or restrictive (37°C) temperatures. (B) β-Galactosidase activity was measured in solubilized cells with ONPG as a substrate after 2.5 to 6 h of incubation at the permissive 30°C (open bars) or restrictive 37°C (lightly shaded bars) temperature. For comparison, cells were incubated at 30°C with tunicamycin (15 μg/ml) (heavily shaded bars). Activity (units) was normalized to the OD600 units of cells used for the assay. The results (means ± standard deviations of 12 independent experiments) are collective data from strains KFY99 and KFY100 (CDC48) or from strain KFY194 (cdc48-10ts).
DISCUSSION
In this work we provide genetic evidence from yeast that implicates the cytosolic chaperone Cdc48p in the ERAD of two well-established substrates, the soluble lumenal CPY* and the membrane 6myc-Hmg2p. We also present biochemical evidence that p97/Cdc48p is in a complex with ERAD substrates; p97 is coprecipitated with the soluble lumenal μs in B cells and Cdc48p is coprecipitated with the membrane 6myc-Hmg2p in yeast. Our data are of particular interest in light of several recent reports that link p97/Cdc48 to ubiquitin-dependent processes (12, 13, 21, 31, 37, 54). In those reports, however, the role of p97/Cdc48p in degradation was demonstrated only for cytosolic or nuclear substrates. For the Ufd3p-Cdc48p complex, elimination of a cytosolic UFD model substrate was equally impaired by either the UFD3 mutation or the cold-sensitive cdc48-1 mutation (21). Likewise, multiubiquitinated proteins, including nuclear cyclins and cyclin-box-containing proteins, accumulated at 14°C in the cold-sensitive cdc48-1 mutant in vivo or stabilized upon p97/VCP depletion in vitro (13). Thus, one could argue that the involvement of p97/Cdc48p in ERAD is at a distal step common to every ubiquitin- and proteasome-dependent degradation process, merely reflecting the requirement for Cdc48p in the degradation of a general population of ubiquitinated substrates. Yet the generality of this phenomenon does not appear to hold in light of the findings that the ubiquitin- and proteasome-dependent degradation of N-end rule substrates is hardly affected in the cdc48 cold-sensitive mutant (21). Nevertheless, recent findings demonstrate that p97/VCP is a multiubiquitin binding protein (13). Moreover, p97/Cdc48p exhibits significant sequence similarities of up to six distinct ATPase subunits of the mammalian and yeast 26S proteasome, including the mammalian S7 subunit (16) and yeast Sug1p/Rpt6p (21, 45). Taken together, these observations may suggest that p97/Cdc48p is the factor that targets multiubiquitinated proteins to the proteasome.
Physical interaction of p97/Cdc48p with ubiquitinated proteins or with factors of the UFD pathway has been shown in several studies. The interaction with Ufd2 implies a role for Cdc48p in proteasomal proteolysis subsequent to the multiubiquitination step, since Ufd2p/E4 is a multiubiquitin chain assembly factor (31). The isolation of the mammalian homologues of Ufd2 and of the Ufd1/Npl4 binary complex as p97 binding proteins in rat liver cytosol connects protein ubiquitination to nuclear membrane integrity and transport (37). The copurification of p97/VCP with ubiquitinated IκBα and 26S proteasome and the identification of p97/VCP as a multiubiquitin binding protein suggest that p97 links IκBα ubiquitination to its final degradation by the proteasome (12, 13). Also, the association of cytokine-dependent tyrosine-phosphorylated p97 with complexes of engaged cytokine receptors suggests that p97 may target the ubiquitinated receptors to the proteasome for degradation (54).
p97/Cdc48p, members of the diverse family of AAA- ATPases, are implicated in a variety of cellular processes, including cell cycle regulation, vesicular transport, organelle biogenesis, homotypic membrane fusion, and proteasome-mediated degradation (39, 48). If the pleiotropic performance of p97/Cdc48p results from one basic activity in protein unfolding or disassembly (19, 22, 39, 48), specificity may be achieved by dedicated adapters or partners (37). For instance, in the homotypic fusion of mitotic Golgi fragments, p97, which unfolds t-SNARE syntaxin 5, is linked to its substrate by p47 (32, 44). By analogy, p97 may be directed to ubiquitin-related processes by association with an alternative partner(s) such as the binary complex Ufd1/Npl4, which has been shown to compete with p47 for binding to p97 (37).
The involvement of p97 in sIgM degradation, as indicated by their physical interaction (Fig. 1), could have been due to its role in homotypic membrane fusion rather than in ubiquitin-related processes. This argument may be based on our previous findings that the ERAD-like proteasomal degradation of light-chain-assembled μs requires vesicular transport (2, 43, 51). Also, requirement for vesicular transport for the degradation of soluble lumenal, but not membrane spanning, classical ERAD substrates has been recently shown in yeast (10). We and others (35, 51) have shown that free unassembled μ chains are also degraded by the proteasome. Yet, unlike the degradation of light-chain-assembled μs, we clearly demonstrated that the proteasomal degradation of unassembled μ chains does not require vesicular transport (43, 51), like most classical ERAD substrates in mammalian cells. However, three lines of evidence argue against homotypic fusion as the mechanism that underlies the involvement of p97/Cdc48p in ERAD. First, p97 is coprecipitated with light-chain-assembled μs (Fig. 1) as well as with free unassembled μs, which is degraded independently of vesicular transport (Elkabetz et al., manuscript in preparation). Formally, p97 could have contributed in different ways, i.e., to the degradation of assembled μs as a factor in homotypic fusion and to the degradation of unassembled μ as a factor in dislocation. However, our data indicate that both processes merge subsequent to the vesicular transport, and both substrates are eliminated by a common mechanism (Kerem et al., manuscript in preparation). Second, the recently described dependence of ERAD on vesicular transport applies only to lumenal proteins, while membrane-spanning ERAD substrates do not exhibit this requirement (10). This genetic evidence in yeast is in agreement with many biochemical studies in mammalian cells, showing that ERAD of membrane proteins, including 3-hydroxy-3-methylglutaryl-coenzyme A reductase (11), is brefeldin A insensitive. Nevertheless, here we show the stabilization of lumenal CPY* as well as of membrane 6myc-Hmg2p. Again, formally the pleiotropic Cdc48p could have contributed in different ways, i.e., to the degradation of lumenal CPY* as a factor in homotypic fusion and to the degradation of membrane 6myc-Hmg2p as a factor in dislocation. Yet a simpler interpretation is that Cdc48p plays a common role in ERAD in the dislocation of both lumenal and membrane substrates. An even more general role for p97/Cdc48p in targeting multiubiquitinated cytosolic, nuclear, and ERAD substrates to the proteasome is discussed below. Lastly, a screen for novel HRD mutants has revealed that Npl4 is Hrd4, showing that degradation of Hmg2p requires an active Npl4 (3a). This strongly supports our conclusion that p97/Cdc48p operates in ERAD by its interaction with the ubiquitin-related binary complex Ufd1/Npl4 rather than by association with p47, its homotypic fusion-related partner.
One cellular process unique to ERAD, which may involve p97/Cdc48p as a cytosolic molecular chaperone, is the dislocation of ERAD substrates from the ER back to the cytosol. As demonstrated when this process is directly obstructed in dislocation-specific mutants of Sec61p, the UPRE-lacZ construct faithfully reports UPR activation as a consequence of accumulation of ERAD protein substrates in the ER (56). The similar activation of the UPR reported here, when Cdc48p fails to function, suggests that the involvement of Cdc48p in ERAD is at the dislocation step. The activation of UPR upon disruption of ERAD at distal cytosolic events, including ubiquitin conjugation (18, 56), does not necessarily negate the possibility that Cdc48p is involved in dislocation, because dislocation, ubiquitination, and degradation are often coupled. For instance, dislocation of CPY* in yeast (4, 6) and of soluble truncated ribophorin I in mammalian cells (15) requires ubiquitination. Likewise, active proteasomes play a role in membrane extraction of several membrane ERAD substrates (36, 42, 52, 53). Finally, recent reports showing that multiubiquitination is required for dislocating major histocompatibility class I heavy chain from the ER into cytosol implicate multiubiquitin, alone or in conjunction with a multiubiquitin-binding protein, in a ratcheting mechanism or in active pulling out of ERAD substrates from the ER membrane (29, 47).
Taken together with our data and that of Dai and Li, identifying p97/Cdc48p as a multiubiquitin-binding protein (13), we propose a unifying model in which p97/Cdc48p recognizes multiubiquitin moieties on many proteins, including ERAD substrates, and targets them to the proteasome for degradation. In this regard it is interesting that 26S proteasomes are found in yeast in association with the ER membrane (17) and that mammalian p97 can be recruited to membranes in a phosphorylation- and/or dephosphorylation-dependent reversible fashion (33). The potential unfoldase activity (22), together with its ability to bind multiubiquitinated proteins (13), position the AAA-ATPase p97/Cdc48p as an attractive key player that acts at the cytosolic face of the ER in ratcheting and/or active pulling out of soluble lumenal and membrane ERAD substrates from the ER to the cytosol. As such, Cdc48p couples the ERAD-specific dislocation event to the distal, common recognition of multiubiquitinated proteins and their targeting to the proteasome.
Acknowledgments
We are indebted to R. Hampton for the 6myc-Hmg2p plasmid and strain and for suggesting and providing the hrd1ΔD strain. We are grateful to D. Wolf for the CPY* plasmid. We thank R. Schekman for the UPRE-lacZ plasmid. We thank members of our laboratory as well as J. Roitelman, R. Klausner, R. Kopito, and G. Kaufmann for helpful suggestions and critical reading of the manuscript.
Efrat Rabinovich and Anat Kerem contributed equally to this work.
This work was supported by grants from The Israel Science Foundation and The United States-Israel Binational Science Foundation.
REFERENCES
- 1.Amitay, R., S. Bar-Nun, J. Haimovich, E. Rabinovich, and I. Shachar. 1991. Post-translational regulation of IgM expression in B lymphocytes. J. Biol. Chem. 266:12568–12573. [PubMed] [Google Scholar]
- 2.Amitay, R., I. Shachar, E. Rabinovich, J. Haimovich, and S. Bar-Nun. 1992. Degradation of secretory IgM in B lymphocytes occurs in a post-endoplasmic reticulum compartment and is mediated by a cysteine protease. J. Biol. Chem. 267:20694–20700. [PubMed] [Google Scholar]
- 3.Bays, N. W., R. G. Gardner, L. P. Seelig, C. A. Joazeiro, and R. Y. Hampton. 2001. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat. Cell Biol. 3:24–29. [DOI] [PubMed] [Google Scholar]
- 3a.Bays, N. W., S. K. Wilhovsky, A. Goradia, K. Hodgkis-Harlow, and R. Y. Hampton. HRD4/NPL4 is required for the proteasomal porcessing of ubiquitinated ER proteins. Mol. Biol. Cell, in press. [DOI] [PMC free article] [PubMed]
- 4.Biederer, T., C. Volkwein, and T. Sommer. 1997. Role of Cue1p in ubiquitination and degradation at the ER surface. Science 278:1806–1809. [DOI] [PubMed] [Google Scholar]
- 5.Bonifacino, J. S., and A. M. Weissman. 1998. Ubiquitin and the control of protein fate in the secretory and endocytic pathways. Annu. Rev. Cell Dev. Biol. 14:19–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bordallo, J., R. K. Plemper, A. Finger, and D. H. Wolf. 1998. Der3p/Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Mol. Biol. Cell 9:209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bordallo, J., and D. H. Wolf. 1999. A RING-H2 finger motif is essential for the function of Der3/Hrd1 in endoplasmic reticulum associated protein degradation in the yeast Saccharomyces cerevisiae. FEBS Lett. 448:244–248. [DOI] [PubMed] [Google Scholar]
- 8.Brodsky, J. L., and A. A. McCracken. 1999. ER protein quality control and proteasome-mediated protein degradation. Semin. Cell Dev. Biol. 10:507–513. [DOI] [PubMed] [Google Scholar]
- 9.Brodsky, J. L., E. D. Werner, M. E. Dubas, J. L. Goeckeler, K. B. Kruse, and A. A. McCracken. 1999. The requirement for molecular chaperones during endoplasmic reticulum-associated protein degradation demonstrates that protein export and import are mechanistically distinct. J. Biol. Chem. 274:3453–3460. [DOI] [PubMed] [Google Scholar]
- 10.Caldwell, S. R., K. J. Hill, and A. A. Cooper. 2001. Degradation of endoplasmic reticulum (ER) quality control substrates requires transport between the ER and Golgi. J. Biol. Chem. 276:23296–23303. [DOI] [PubMed] [Google Scholar]
- 11.Chun, K. T., S. Bar-Nun, and R. D. Simoni. 1990. The regulated degradation of 3-hydroxy-3-methylglutaryl-CoA reductase requires a short-lived protein and occurs in the endoplasmic reticulum. J. Biol. Chem. 265:22004–22012. [PubMed] [Google Scholar]
- 12.Dai, R. M., E. Chen, D. L. Longo, C. M. Gorbea, and C. C. Li. 1998. Involvement of valosin-containing protein, an ATPase copurified with IκBα and 26S proteasome, in ubiquitin-proteasome-mediated degradation of IκBα. J. Biol. Chem. 273:3562–3573. [DOI] [PubMed] [Google Scholar]
- 13.Dai, R. M., and C. C. Li. 2001. Valosin-containing protein is a multi-ubiquitin chain-targeting factor required in ubiquitin-proteasome degradation. Nat. Cell Biol. 3:740–744. [DOI] [PubMed] [Google Scholar]
- 14.Deak, P. M., and D. H. Wolf. 2001. Membrane topology and function of Der3/Hrd1p as a ubiquitin-protein ligase (E3) involved in endoplasmic reticulum degradation. J. Biol. Chem. 276:10663–10669. [DOI] [PubMed] [Google Scholar]
- 15.de Virgilio, M., H. Weninger, and N. E. Ivessa. 1998. Ubiquitination is required for the retro-translocation of a short-lived luminal endoplasmic reticulum glycoprotein to the cytosol for degradation by the proteasome. J. Biol. Chem. 273:9734–9743. [DOI] [PubMed] [Google Scholar]
- 16.Dubiel, W., K. Ferrell, and M. Rechsteiner. 1993. Peptide sequencing identifies MSS1, a modulator of HIV Tat-mediated transactivation, as subunit 7 of the 26 S protease. FEBS Lett. 323:276–278. [DOI] [PubMed] [Google Scholar]
- 17.Enenkel, C., A. Lehmann, and P. M. Kloetzel. 1998. Subcellular distribution of proteasomes implicates a major location of protein degradation in the nuclear envelope-ER network in yeast. EMBO J. 17:6144–6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedlander, R., E. Jarosch, J. Urban, C. Volkwein, and T. Sommer. 2000. A regulatory link between ER-associated protein degradation and the unfolded-protein response. Nat. Cell Biol. 2:379–384. [DOI] [PubMed] [Google Scholar]
- 19.Fröhlich, K. U., H. W. Fries, M. Rudiger, R. Erdmann, D. Botstein, and D. Mecke. 1991. Yeast cell cycle protein CDC48p shows full-length homology to the mammalian protein VCP and is a member of a protein family involved in secretion, peroxisome formation, and gene expression. J. Cell Biol. 114:443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gardner, R. G., G. M. Swarbrick, N. W. Bays, S. R. Cronin, S. Wilhovsky, L. Seelig, C. Kim, and R. Y. Hampton. 2000. Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of hrd1p by hrd3p. J. Cell Biol. 151:69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghislain, M., R. J. Dohmen, F. Levy, and A. Varshavsky. 1996. Cdc48p interacts with Ufd3p, a WD repeat protein required for ubiquitin-mediated proteolysis in Saccharomyces cerevisiae. EMBO J. 15:4884–4899. [PMC free article] [PubMed] [Google Scholar]
- 22.Golbik, R., A. N. Lupas, K. K. Koretke, W. Baumeister, and J. Peters. 1999. The Janus face of the archaeal Cdc48/p97 homologue VAT: protein folding versus unfolding. Biol. Chem. 380:1049–1062. [DOI] [PubMed] [Google Scholar]
- 23.Gusarova, V., A. J. Caplan, J. L. Brodsky, and E. A. Fisher. 2001. Apoprotein B degradation is promoted by the molecular chaperones hsp90 and hsp70. J. Biol. Chem. 276:24891–24900. [DOI] [PubMed] [Google Scholar]
- 24.Hampton, R. Y., R. G. Gardner, and J. Rine. 1996. Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-coA reductase, an integral endoplasmic reticulum membrane protein. Mol. Biol. Cell 7:2029–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hiller, M. M., A. Finger, M. Schweiger, and D. H. Wolf. 1996. ER degradation of a misfolded luminal protein by a cytosolic ubiquitin-proteasome pathway. Science 273:1725–1728. [DOI] [PubMed] [Google Scholar]
- 26.Imamura, T., T. Haruta, Y. Takata, I. Usui, M. Iwata, H. Ishihara, M. Ishiki, O. Ishibashi, E. Ueno, T. Sasaoka, and M. Kobayashi. 1998. Involvement of heat shock protein 90 in the degradation of mutant insulin receptors by the proteasome. J. Biol. Chem. 273:11183–11188. [DOI] [PubMed] [Google Scholar]
- 27.Johnson, E. S., P. C. Ma, I. M. Ota, and A. Varshavsky. 1995. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 270:17442–17456. [DOI] [PubMed] [Google Scholar]
- 28.Kaiser, C., S. Micahaelis, and A. Mitchell. 1994. Methods in yeast genetics: a Cold Spring Harbor laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Kikkert, M., G. Hassink, M. Barel, C. Hirsch, F. J. Van der Wal, and E. Wiertz. 2001. Ubiquitination is essential for human cytomegalovirus US11-mediated dislocation of MHC class I molecules from the endoplasmic reticulum to the cytosol. Biochem. J. 358:369–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knop, M., A. Finger, T. Braun, K. Hellmuth, and D. H. Wolf. 1996. Der1, a novel protein specifically required for endoplasmic reticulum degradation in yeast. EMBO J. 15:753–763. [PMC free article] [PubMed] [Google Scholar]
- 31.Koegl, M., T. Hoppe, S. Schlenker, H. D. Ulrich, T. U. Mayer, and S. Jentsch. 1999. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 96:635–644. [DOI] [PubMed] [Google Scholar]
- 32.Kondo, H., C. Rabouille, R. Newman, T. P. Levine, D. Pappin, P. Freemont, and G. Warren. 1997. p47 is a cofactor for p97-mediated membrane fusion. Nature 388:75–78. [DOI] [PubMed] [Google Scholar]
- 33.Lavoie, C., E. Chevet, L. Roy, N. K. Tonks, A. Fazel, B. I. Posner, J. Paiement, and J. J. Bergeron. 2000. Tyrosine phosphorylation of p97 regulates transitional endoplasmic reticulum assembly in vitro. Proc. Natl. Acad. Sci. USA. 97:13637–13642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loo, M. A., T. J. Jensen, L. Cui, Y. Hou, X. B. Chang, and J. R. Riordan. 1998. Perturbation of Hsp90 interaction with nascent CFTR prevents its maturation and accelerates its degradation by the proteasome. EMBO J. 17:6879–6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mancini, R., C. Fagioli, A. M. Fra, C. Maggioni, and R. Sitia. 2000. Degradation of unassembled soluble Ig subunits by cytosolic proteasomes: evidence that retrotranslocation and degradation are coupled events. FASEB J. 14:769–778. [DOI] [PubMed] [Google Scholar]
- 36.Mayer, T. U., T. Braun, and S. Jentsch. 1998. Role of the proteasome in membrane extraction of a short-lived ER-transmembrane protein. EMBO J. 17:3251–3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meyer, H. H., J. G. Shorter, J. Seemann, D. Pappin, and G. Warren. 2000. A complex of mammalian ufd1 and npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. EMBO J. 19:2181–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishikawa, S., S. W. Fewell, Y. Kato, J. L. Brodsky, and T. Endo. 2001. Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J. Cell Biol. 153:1061–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patel, S., and M. Latterich. 1998. The AAA team: related ATPases with diverse functions. Trends. Cell Biol. 8:65–71. [PubMed] [Google Scholar]
- 40.Pilon, M., R. Schekman, and K. Romisch. 1997. Sec61p mediates export of a misfolded secretory protein from the endoplasmic reticulum to the cytosol for degradation. EMBO J. 16:4540–4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Plemper, R. K., S. Bohmler, J. Bordallo, T. Sommer, and D. H. Wolf. 1997. Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature 388:891–895. [DOI] [PubMed] [Google Scholar]
- 42.Plemper, R. K., R. Egner, K. Kuchler, and D. H. Wolf. 1998. Endoplasmic reticulum degradation of a mutated ATP-binding cassette transporter Pdr5 proceeds in a concerted action of Sec61 and the proteasome. J. Biol. Chem. 273:32848–32856. [DOI] [PubMed] [Google Scholar]
- 43.Rabinovich, E., S. Bar-Nun, R. Amitay, I. Shachar, B. Gur, M. Taya, and J. Haimovich. 1993. Different assembly species of IgM are directed to distinct degradation sites along the secretory pathway. J. Biol. Chem. 268:24145–24148. [PubMed] [Google Scholar]
- 44.Rabouille, C., H. Kondo, R. Newman, N. Hui, P. Freemont, and G. Warren. 1998. Syntaxin 5 is a common component of the NSF- and p97-mediated reassembly pathways of Golgi cisternae from mitotic Golgi fragments in vitro. Cell 92:603–610. [DOI] [PubMed] [Google Scholar]
- 45.Rubin, D. M., O. Coux, I. Wefes, C. Hengartner, R. A. Young, A. L. Goldberg, and D. Finley. 1996. Identification of the gal4 suppressor Sug1 as a subunit of the yeast 26S proteasome. Nature 379:655–657. [DOI] [PubMed] [Google Scholar]
- 46.Shachar, I., R. Amitay, E. Rabinovich, J. Haimovich, and S. Bar-Nun. 1992. Polymerization of secretory IgM in B lymphocytes is prevented by a preceding targeting to a degradation pathway. J. Biol. Chem. 267:24241–24247. [PubMed] [Google Scholar]
- 47.Shamu, C. E., D. Flierman, H. L. Ploegh, T. A. Rapoport, and V. Chau. 2001. Polyubiquitination is required for US11-dependent movement of MHC class I heavy chain from endoplasmic reticulum into cytosol. Mol. Biol. Cell 12:2546–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vale, R. D. 2000. AAA proteins. Lords of the ring. J. Cell Biol. 150:F13–F20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilhovsky, S., R. Gardner, and R. Hampton. 2000. HRD gene dependence of endoplasmic reticulum-associated degradation. Mol. Biol. Cell 11:1697–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilkinson, B. M., J. R. Tyson, P. J. Reid, and C. J. Stirling. 2000. Distinct domains within yeast Sec61p involved in post-translational translocation and protein dislocation. J. Biol. Chem. 275:521–529. [DOI] [PubMed] [Google Scholar]
- 51.Winitz, D., I. Shachar, Y. Elkabetz, R. Amitay, M. Samuelov, and S. Bar-Nun. 1996. Degradation of distinct assembly forms of IgM occurs in multiple sites in permeabilized B cells. J. Biol. Chem. 271:27645–27651. [DOI] [PubMed] [Google Scholar]
- 52.Xiong, X., E. Chong, and W. R. Skach. 1999. Evidence that endoplasmic reticulum (ER)-associated degradation of cystic fibrosis transmembrane conductance regulator is linked to retrograde translocation from the ER membrane. J. Biol. Chem. 274:2616–2624. [DOI] [PubMed] [Google Scholar]
- 53.Yang, M., S. Omura, J. S. Bonifacino, and A. M. Weissman. 1998. Novel aspects of degradation of T cell receptor subunits from the endoplasmic reticulum (ER) in T cells: importance of oligosaccharide processing, ubiquitination, and proteasome-dependent removal from ER membranes. J. Exp. Med. 187:835–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yen, C. H., Y. C. Yang, S. K. Ruscetti, R. A. Kirken, R. M. Dai, and C. C. Li. 2000. Involvement of the ubiquitin-proteasome pathway in the degradation of nontyrosine kinase-type cytokine receptors of IL-9, IL-2, and erythropoietin. J. Immunol. 165:6372–6380. [DOI] [PubMed] [Google Scholar]
- 55.Zhang, Y., G. Nijbroek, M. L. Sullivan, A. A. McCracken, S. C. Watkins, S. Michaelis, and J. L. Brodsky. 2001. Hsp70 molecular chaperone facilitates endoplasmic reticulum-associated protein degradation of cystic fibrosis transmembrane conductance regulator in yeast. Mol. Biol. Cell 12:1303–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou, M., and R. Schekman. 1999. The engagement of Sec61p in the ER dislocation process. Mol. Cell 4:925–934. [DOI] [PubMed] [Google Scholar]