Abstract
Mitogen-activated protein (MAP) kinase, extracellular-signal-regulated kinases (ERKs) play an important role in activating AP-1-dependent transcription. Studies using the JB6 mouse epidermal model and a transgenic mouse model have established a requirement for AP-1-dependent transcription in tumor promotion. Tumor promoters such as 12-O-tetradecanoylphorbol-13-acetate (TPA) and epidermal growth factor induce activator protein 1 (AP-1) activity and neoplastic transformation in JB6 transformation-sensitive (P+) cells, but not in transformation-resistant (P−) variants. The resistance in one of the P− variants can be attributed to the low levels of the MAP kinases, ERKs 1 and 2, and consequent nonresponsiveness to AP-1 activation. The resistant variant is not deficient in c-fos transcription. The purpose of these studies was to define the targets of activated ERK that lead to AP-1 transactivation. The results establish that the transactivation domain of Fra-1 can be activated, that activation of Fra-1 is ERK dependent, and that a putative ERK phosphorylation site must be intact for activation to occur. Fra-1 was activated by TPA in ERK-sufficient P+ cells but not in ERK-deficient P− cells. A similar activation pattern was seen for c-Fos but not for Fra-2. Gel shift analysis identified Fra-1 as distinguishing mitogen-activated (P+) from nonactivated (P−) AP-1 complexes. A second AP-1-nonresponsive P− variant that underexpresses Fra-1 gained AP-1 response upon introduction of a Fra-1 expression construct. These observations suggest that ERK-dependent activation of Fra-1 is required for AP-1 transactivation in JB6 cells.
The transcription factor AP-1 (activator protein 1) consists of members of the Jun and Fos family of proteins (2, 23, 56, 57, 65). AP-1 binds DNA target sites as Jun/Jun homodimers and Jun/Fos heterodimers, and activation of the AP-1 proteins can lead to an increase or decrease in transcription of AP-1 target genes. AP-1 can be activated by a variety of stimuli, including cytokines, growth factors, stress, and UV light (2, 40, 67). AP-1 regulates multiple cellular functions, including proliferation, differentiation, and cell death. Elevation of AP-1 activity can be oncogenic (11, 26–28, 47, 71).
The AP-1 family of proteins is characterized by a b-zip domain that consists of a basic region adjacent to a leucine zipper domain. The leucine zipper directs the dimerization of family members and positions these dimers for high-affinity binding to AP-1 target sites (sequence TGAC/GTCA) (32). AP-1 proteins also contain a transactivation domain. Deletion of the transactivation domain of c-Jun can lead to the formation of a dominant negative mutant, indicating that the transactivation domain is essential for AP-1 activity (11). The composition of the AP-1 dimer and the activation status of the component Jun and Fos proteins may be instrumental in determining which target genes are activated by AP-1.
Mouse epidermal JB6 cells have proven to be valuable in elucidating the mechanisms leading to activation of AP-1 and the role of AP-1 in tumor promotion, a rate-limiting step in multistage carcinogenesis. The JB6 model includes variants stably trapped in a promotable stage (7, 19–21). These variants are sensitive to AP-1-regulated neoplastic transformation by various mitogens, including 12-O-tetradecanoylphorbol-13-acetate (TPA) and epidermal growth factor (EGF). Exposure of transformation-sensitive (P+) cells to these tumor promoters produces an increase in AP-1 activity, followed by neoplastic transformation. In contrast, in resistant (P−) JB6 cells, AP-1 is not activated and cells are not transformed by TPA or EGF, although these cells do show a similar mitogenic response (21).
Neoplastic transformation requires activation of AP-1. Expression of a transactivation mutant c-jun (TAM67) or exposure to AP-1-transrepressing retinoids is sufficient to block TPA-induced AP-1 activation and transformation in culture (27, 46). AP-1 transactivation is also required for tumor promotion in vivo. Transgenic mice expressing the dominant negative c-jun in mouse skin are protected from the tumorigenic effects of 9,10-dimethyl-1,2-benzanthracene (DMBA)/TPA exposure (71). Elevated AP-1 activity is required for maintenance of the tumor phenotype in human keratinocytes (47). Thus, the observation that AP-1 is instrumental in promoting carcinogenesis made initially in JB6 cells has been validated and extended to other cell culture and in vivo models.
Mitogens like TPA and EGF activate the AP-1 transcription factor via the mitogen-activated protein kinase (MAPK) pathway (31, 37, 66). The MAPK family includes the extracellular signal-regulated protein kinase (ERK), c-Jun N-terminal kinase/stress-activated kinases (JNK/SAPK), and p38 kinase (9, 10, 24, 44, 58). ERKs (ERK 1 and ERK 2) are activated by mitogen stimulation through a cascade of kinases, including Ras, Raf, and MAPK kinase (MEK). Examination of mitogen stimulation of the MAPK pathway in JB6 cells showed that the P− variant Cl 30.7b was deficient in ERK 1 and 2 proteins compared to P+ cells (37). Restoration of ERK levels by transfecting P− cells with an ERK 2 expression vector reconstituted TPA- and EGF-induced activation of AP-1 and transformation response in these cells. Conversely, inhibition of ERK activity in JB6 P+ cells blocked TPA-induced activation of AP-1 and transformation (66). These results indicate that activation of ERK 1 and/or 2 is required for mitogen activation of AP-1 and thus for neoplastic transformation.
The MAPK cascade plays an important role in the control of cell proliferation and differentiation (3, 17, 36, 38, 39, 50, 63, 67). Once activated, ERK relocalizes to the nucleus, where it can activate transcription factors and the basal transcription complex (14, 16, 45). Among ERK’s major nuclear targets are the ternary complex factor (TCF), Elk-1, and Sap-1a (33, 39, 49, 67). Activation of TCF produces induced transcription of immediate-early genes like c-fos, although c-fos transcription is not dependent solely on ERK (41). ERK activation has also been implicated in potentiating AP-1 activity (22, 31, 37, 62, 66).
Except for the induction of c-fos transcription, little is known of the ERK-dependent events that are required for AP-1 activation. Interestingly, the AP-1-nonresponsive, ERK-deficient P− 30.7b cells are not inhibited for c-fos transcription (4). In order to better understand the pathway(s) from mitogen-activated ERK to activation of AP-1 and neoplastic transformation, we compared the activation of AP-1 proteins using Gal4 fusions in ERK-deficient P− and ERK-sufficient P+ JB6 cells. We show that the transactivation domain of Fra-1 can be activated in mouse epidermal JB6 cells, that Fra-1 activation is ERK dependent, and that Thr-231 is required for Fra-1 activation. c-Fos activation is also ERK dependent. Considering the composition of activated AP-1 as determined by electrophoretic mobility shift assays (EMSAs) and the ability of Fra-1 to restore AP-1 transactivation response in Fra-1-deficient variants, we conclude that Fra-1 is a pivotal AP-1 protein required for mitogen activation of AP-1.
MATERIALS AND METHODS
Cell culture and transfections.
JB6 mouse epidermal cell lines were cultured in monolayers at 37°C and 5% CO2 using Eagle’s minimal essential medium (EMEM) containing 4% fetal calf serum (FCS) (Life Technologies), 2 mM l-glutamine, and 25 μg of gentamicin per ml as described previously (7, 66). For transfection with Lipofectamine (Invitrogen), 5 × 104 cells were plated into each well of a 24-well dish. The following day, the cells were incubated in Optimem (Invitrogen) for 2 to 4 h. Transfections with Gal4 fusions were done in batch mixtures by adding 50 ng of Gal4 fusion construct and 12.5 ng of Gal4-luciferase reporter per well to 1 μl of Lipofectamine per well and incubating at 25°C for 15 to 45 min. The Lipofectamine-DNA mixture was then diluted in 0.3 ml of Optimem per well and added to the cells. After 6 h the cells were washed, and complete medium was added. Alternatively, 0.4 μg of pcDNA3 or pFC-MEK1 (Stratagene) DNA was added to the Gal4 fusion and reporter mix and then mixed with 1.2 μl of Fugene (Roche). The Fugene-DNA mix was added to the cells in rich medium. Following transfection, the cells were starved for 24 h in medium containing 0.2% FCS. The cells were then treated with TPA (10 ng/ml in dimethyl sulfoxide [DMSO]) for 6 h and lysed with passive lysis buffer (Promega), and luciferase activity was measured with 25 μl of cell lysate according to the manufacturer’s recommendation (Promega). Luciferase activity was read with a Dynex 96-well luminometer.
Lipofectamine-Plus was used for transfection with 4× AP-1 luciferase and CMV-fra-1. For these assays, 0.4 μg of 4× AP-1 luciferase and 0.2 to 0.8 μg of CMV-fra-1 were mixed with 3 μl of Lipofectamine and 1.2 μl of Lipofectamine-Plus according to the manufacturer’s recommendation. Total DNA was maintained at 1 μg with the addition of pcDNA3. The DNA-Lipofectamine mix was added to 104 cells seeded 24 h earlier in 24-well dishes. Cells were transfected as described above except that luciferase activity was determined 18 h after TPA treatment. For Western analysis of Gal4 fusions, 8 μg of DNA was mixed with 3.6 μl of Fugene and added to 106 cells in 150-mm dishes. Nuclear extracts were harvested as described below.
Plasmids.
The PathDetect trans-reporting system (Strategene) was used to elucidate the activation status of various AP-1 proteins in both P+ and P− cells. PathDetect vectors express a fusion protein that contains the activation domain of the protein of interest fused to the DNA-binding domain of Saccharomyces cerevisiae Gal4 (residues 1 to 147). The luciferase reporter construct contains a promoter that carries four tandem repeats of the Gal4 binding site (upstream activation sequence [UAS]). The PathDetect trans-reporting plasmids c-Fos and Elk-1 were purchased from Stratagene.
Gal4 fusions containing Jun-D (amino acids [aa] 1 to 210) and Fos-B (aa 220 to 361) were gifts from Tim Bowden (59). Gal4 fusions containing the transactivation domain from rat Fra-1 (aa 132 to 275) and rat Fra-2 (aa 148 to 326) (18, 30) were constructed by inserting DNA fragments generated by PCR of CMV-fra-1 and CMV-fra-2 into the pFA-CMV vector (Stragene). The oligonucleotides used to prime PCR synthesis for the inserts for Gal4-Fra-1 were CGCGGATCCCGCGAGCTGACAGACTTCCTGCAG and CCGGAATTCCGGTCACAAAGCCAGGAGTGTAGG, and those for Gal4-Fra-2 were CGCGGATCCCGCGAGCTGACAGAGAAGCTGCAGGCG and CCGGAATTCCGGTTACAGCCGTAGAAGTGTCGG. PCR-generated fragments were digested with BamHI and EcoRI, gel purified, and ligated in frame into pFCMV (Stratagene). Expression of the Gal4-Fra-1 and -Fra-2 fusions was confirmed by Western blot analysis. CMV-fra-1, CMV-fra-2, and antisense fra-1 were gifts from R. Bravo.
Point mutations in the Gal4-Fra-1 and the CMV-fra-1 vectors were created with the Quickchange mutagenesis kit (Stratagene) and the following oligonucleotides: S209AF (CCTTGCATCTCCCTTGCTCCAGGACCCGTAC), S209AR (GTACGGGTCCTGGAGCAAGGGAGATGCAAGG), T244AF (GTTTTCACCTATCCTAGCGCACCAGAACCTTGCTCCTCC), T244AR (GGAGGAGCAAGGTTCTGGTGCGCTAGGATAGGTGAAAAC), T231AF (CTCATGACCACACCCTCTCTGGCTCCTTTTACTCCGAGTCTG), T231AR (CAGACTCGGAGTAAAAGGAGCCAGAGAGGGTGTGGTCATGAG), S269AF (CCCTCCTCCGACCCCCTGGGCGCTCCTACACTCCTGGCTTTGTG), and S269AR (CACAAAGCCAGGAGTGTAGGAGCGCCCAGGGGGTCGGAGGAGGG). Mutated vectors were sequenced to confirm that mutagenesis was restricted to designated sites.
Preparation of nuclear extracts.
JB6 cells were collected after 24 h of starvation in 0.2% fetal bovine serum (FBS) followed by induction with TPA (10 ng/ml) for the indicated times. The collected cells were lysed with lysis buffer containing 25 mM HEPES (pH 7.7), 50 mM KCl, 2 mM phenylmethylsulfonyl fluoride (PMSF), 100 μM dithiothreitol (DTT), and 0.5% NP-40, plus 2 μg of leupeptin and 4 μg of aprotinin per ml. The resulting nuclei were washed with the above buffer minus NP-40 and subsequently disrupted with extraction buffer containing 25 mM HEPES (pH 7.7), 500 mM KCl, 1 mM PMSF, 100 μM DTT, and 10% glycerol plus 1 μg of leupeptin and 2 μg of aprotinin per ml. All of the above procedures were performed at 4°C, and aliquots of nuclear extracts were stored at −70°C. Total protein levels were determined by the bovine serum albumin (BSA) assay (Pierce).
EMSA.
Oligonucleotides containing the AP-1 consensus sequence (CGCTTGATGACTCAGCCGGAA) were purchased from Santa Cruz Biotechnology. Fifty nanograms of double-stranded oligonucleotide was end labeled with [32P]ATP and T4 polynucleotide kinase (Roche Molecular Biochemicals). Nuclear extracts (1 to 2 μg of protein) diluted in binding buffer containing 20 mM Tris-HCl (pH 7.5), 50 mM KCl, 1 μM DTT, 2 μM EDTA, and 4% glycerol were added to the EMSA reaction mixture containing 50,000 cpm of labeled and purified oligonucleotides and 1 μg of poly(dI-dC) (Roche Molecular Biochemicals) per ml. The reactions were incubated at room temperature for 30 min. The protein-DNA complexes were resolved on a 6% TBE (Tris-borate-EDTA) gel (Novex) and visualized by autoradiography. Antibody supershifts were performed by mixing nuclear extracts with labeled nucleotides and incubating for 15 min at 4°C. These extracts were then added to 1 μl of selected antibody (TransCruz Gel Supershift reagent at 2 μg/μl; Santa Cruz Biotechnology) and incubated for 15 min at room temperature. The specificity of the DNA-protein interaction was determined using 1 ng or 1 μg of unlabeled oligonucleotide.
Protein isolation and Western blot analysis.
P− 30.7b, P− SC21, and P+ clone (Cl) 41 cells (2 × 105) were seeded into six-well dishes. The following day the cells were serum starved in 0.2% FCS for 24 h. Following starvation, TPA (10 ng/ml) was used to activate the mitogen pathway for 30 min. The cells were lysed in 2% sodium dodecyl sulfate (SDS) lysis buffer, and protein concentrations were determined with a Micro-BCA protein assay (Pierce). Whole-cell extracts (4 to 6 μg) were denatured by heating at 100°C and sonicated. Proteins were fractionated on a 10% Bis-Tris polyacrylamide gel (Novex) and then transferred to nitrocellulose membranes using Novex transfer buffer and a semidry transfer apparatus. Membranes were blocked with 5% nonfat milk proteins in Tris-buffered saline (TBS)-0.5% Tween 20.
For detection of phosphorylated ERKs, the phosphospecific antibody to MAPK P42/44 (Cell Signaling Technology) was used at a dilution of 1:1,000. Anti-ERK (Promega) was used for detection of total ERK 1 and 2. For detection of AP-1-specific protein, nuclear extracts were denatured in sample buffer and fractionated as above. Antibodies from Santa Cruz were used at a dilution of 1:1,000. Gal4 fusions were detected in nuclear extracts with anti-Gal4(DBD) from Santa Cruz at a 1:1,000 dilution. Antibody-bound proteins were detected by chemiluminescence (ECL; Amersham).
RESULTS
ERK deficiency does not limit induction of c-fos mRNA.
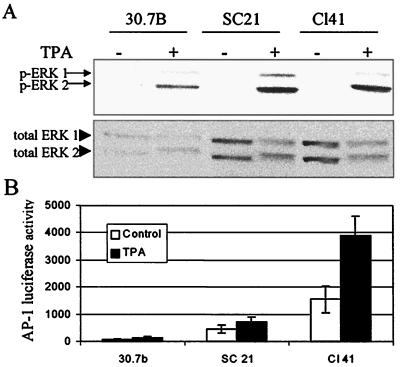
The mouse JB6 model includes variants that are sensitive (P+) and resistant (P−) to neoplastic transformation by tumor promoters. In transformation-sensitive P+ JB6 cells, exposure to tumor promoters such as TPA and EGF produces activation of the MAPK-ERK cascade, leading to transactivation of AP-1 and cellular transformation. In one of the JB6 P− variants, Cl 30.7b, resistance to AP-1 activation and thus to transformation can be attributed to a deficiency in ERK levels (37, 66). We now show in a second JB6 P− variant, Cl SC21, that the levels of ERK1/2 are similar to those in P+ Cl 41 cells (Fig. 1A), yet, like the P− Cl 30.7b variant, P− Cl SC21 cells are resistant to TPA-induced activation of AP-1 (Fig. 1B) and neoplastic transformation (15). These results suggest that some factor downstream of ERK activation is deficient in the JB6 Cl SC21 cells.
FIG. 1.
AP-1-nonresponsive JB6 P− variants include both ERK-deficient and ERK-sufficient cells. (A) Whole-cell extracts from JB6 cells serum starved for 24 h and treated with TPA (10 ng/ml) or DMSO for 30 min were analyzed by Western blot analysis with antibody specific to phosphorylated (p) ERK 1 and 2 or to total ERK. An equal amount of total protein was added to each lane. Independent experiments show similar results. (B) JB6 cells were transiently transfected with a 4× AP-1 luciferase reporter. Cells were serum starved for 24 h and then treated with TPA for an additional 16 h. Cells were lysed, and luciferase activity (relative luminescence units) was determined with a Dynex 96-well luminometer.
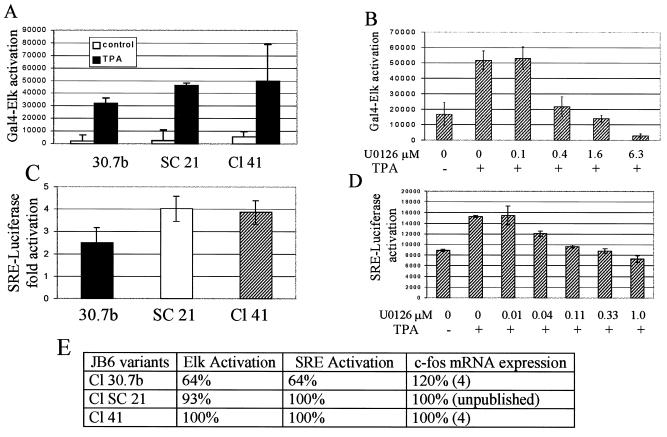
Interestingly, mitogen induction of c-fos mRNA, thought to occur through the ERK pathway, is not limited in the P− ERK-deficient Cl 30 cells, as the level of TPA-induced c-fos mRNA is slightly elevated in these cells relative to the P+ Cl 41 variants (4) (see Fig. 2E). The c-fos promoter contains a serum response element (SRE), to which the serum response factor (SRF) and the Ets transcription factors Elk-1 and SAP-1a bind (35). Activation of the MAPK cascade (either ERK or JNK) can activate Elk-1 (and SAP), which in turn induces the transcription of c-fos mRNA through the SRE (68).
FIG. 2.
MAPK-regulated serum response pathway is functional in ERK-deficient JB6 cells. (A) Activation of Elk-1 is not lacking in ERK-deficient cells. JB6 cells transiently transfected with a Gal4-Elk-1 fusion and a Gal4-luciferase reporter were serum starved for 24 h and then treated with TPA for 6 h. Cell extracts were analyzed for luciferase activity. (B) Inhibition of ERK activity blocks activation of Elk. Cl 41 cells transiently transfected with Gal4-Elk-1 and Gal4-luciferase were treated with the MEK inhibitor U0126 (Promega) or DMSO 1 h before TPA exposure. Luciferase activity was determined 6 h after TPA exposure. (C) ERK levels have little effect on activation of SRE. JB6 cells were transiently transfected with an SRE-luciferase reporter, serum starved for 24 h, and then treated with TPA for 3 h. (D) SRE activation is ERK activation dependent. Cl 41 cells transiently transfected with SRE-luciferase were treated with the MEK inhibitor U0126 or DMSO for 1 h before TPA treatment. Luciferase activity was measured 3 h later. Fold activation is defined as TPA-induced luciferase activity relative to the uninduced level. The average of three transfections is shown. Similar results were seen in multiple experiments. (E) c-fos mRNA expression is not limiting in ERK-deficient P− cells. The activation of Elk-1, SRE, and c-fos expression relative to that in the ERK-sufficient Cl 41 cells is shown. The values for Elk-1 and SRE activation are from panels A and C above. The values for c-fos expression are from Ben-Ari et al. (4) and data not shown.
In order to further analyze the ERK-dependent pathway leading to activation of c-fos transcription in JB6 cells, we measured TPA-induced activation of ERK substrate Elk-1 using a Gal4-Elk-1 fusion construct. These fusions, when activated, drive expression of luciferase from a Gal4-dependent promoter. We also measured expression of luciferase from an SRE-dependent promoter driven by a transcription factor complex containing activated Ets family proteins such as Elk-1.
Treatment of serum-starved JB6 P+ cells with TPA produced a significant induction of activated Elk-1, which was blocked by the MEK-specific inhibitor U0126 (Promega) in a dose-dependent manner (Fig. 2A and B). Elk-1 was also activated in ERK-sufficient P− SC21 cells and in the ERK-deficient 30.7b cells, although activation in 30.7b cells was not as high as that seen in Cl 41 or SC21. A similar pattern was seen for TPA induction of the SRE-driven luciferase (Fig. 2C and D). Thus, either the lower level of activated ERK present in the ERK-deficient JB6 30.7b cells is sufficient to activate Elk-1- and SRE-driven c-fos expression but is insufficient to activate AP-1, or an alternative ERK-independent pathway is mediating the mitogen-induced expression of c-fos mRNA.
The reduction of both Elk and SRE activities below basal levels in the ERK-sufficient P+ cells by the MEK inhibitor (Fig. 2B and 2D) indicates that some level of activated ERK is needed for activation of SRE-dependent transcription. Figure 2E summarizes the current and previously reported observations establishing that although the low ERK levels limit AP-1 activation in P− Cl 30.7b cells, they do not limit c-fos mRNA expression and have little effect on the activation of Elk-1- or SRE-dependent transcription.
TPA-induced AP-1 binding is enhanced in variants expressing elevated ERK.
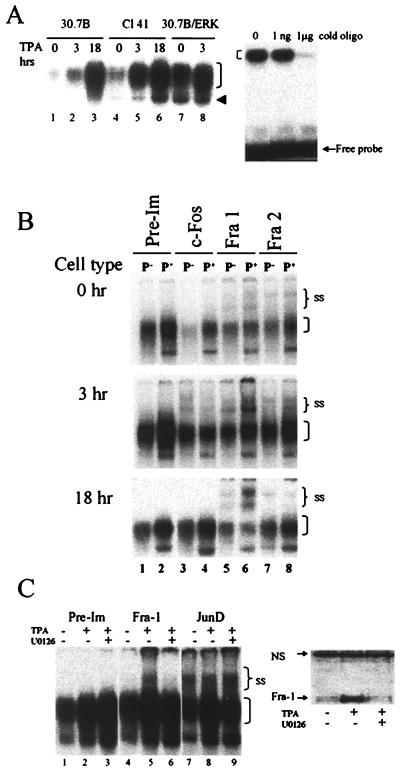
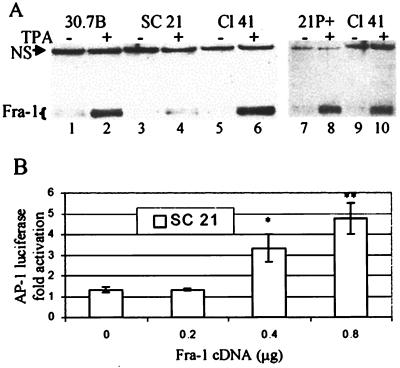
In transformation-sensitive P+ JB6 cells, exposure to tumor promoters like TPA and EGF produces transactivation of AP-1, leading to transformation. In contrast, in JB6 P− cells, exposure to TPA or EGF does not activate AP-1. It has recently been shown by EMSAs that the lack of AP-1 activation in the P− 30.7b cells could be attributed in part to a deficiency in AP-1 proteins bound to DNA (8). The results shown in Fig. 3A are consistent with these findings. P− Cl 30.7b cells have less protein bound to an AP-1 consensus site before and after TPA exposure than P+ Cl 41 cells. TPA induction of AP-1 binding is greater in the Cl 41 extracts (Fig. 3A, compare lane 2 to lane 5 and lane 3 to lane 6).
FIG. 3.
(A) AP-1 DNA binding is enhanced by ERK activation. The left panel shows 30.7B, Cl 41, and 30.7B/ERK cells that were serum starved for 24 h and then treated with TPA for 0, 3, and 18 h. Nuclear extracts were isolated and analyzed for DNA-binding activity by EMSA. The right panel shows the specificity of DNA binding. Unlabeled (cold) AP-1-binding oligonucleotide (1 ng and 1 μg) was used to compete with the labeled oligonucleotide. The bracket marks the broad (multiband) AP-1 complex. The arrowhead shows a faster-migrating band seen predominantly in ERK-sufficient cells. Electrophoresis conditions in which the free probe was visible on the gel (right) were not sufficient to separate the faster-migrating band from the major band, as seen on the left. To separate the faster-migrating band, the free probe was run off the gel. Representative gels from multiple experiments are shown. (B) TPA-activated AP-1 complex from P+ cells contains more Fra-1 than the complex from ERK-deficient P− cells. Fra-2 is also present in the AP-1 complex. Nuclear extracts were harvested from 30.7B (P−) and Cl 41 (P+) cells that had been serum starved for 24 h and then treated with TPA for 0 (top), 3 (middle), and 18 (bottom) h. Extracts were analyzed by supershift EMSA with Fos-specific antibodies as indicated. Higher antibody concentrations produced no greater shifts. It should be noted that neither these antibody concentrations nor higher concentrations (8, 42, 48) of specific AP-1 antibodies produced complete shifts in previous studies of JB6 cells in our laboratory or others. Pre-Im, preimmune; SS, supershifted bands. Unlabeled brackets mark nonsupershifted AP-1 complexes. Labeled oligonucleotide was added to the nuclear extract, and the extracts were then aliquoted to tubes containing the indicated antibodies. EMSA samples were run on two gels per sample time point. All six gels were run simultaneously. The 0-h and 3-h autoradiographs were analyzed by phosphorimaging. The 18-h autoradiograph was exposed to X-ray film and digitally rearranged for clarity. A representative autoradiogram of multiple EMSAs is shown. (C) Fra-1 recruitment to AP-1 complex is ERK dependent. JB6 cells treated for 1 h with 5 μM MEK inhibitor U0126 or DMSO were exposed to TPA for 3 h. Nuclear extracts were harvested and analyzed for AP-1 binding by supershift EMSA or for Fra-1 expression by Western blot. The left panel shows the antibody-induced supershift EMSA with preimmune (lanes 1 to 3), Fra-1 (lanes 4 to 6), and Jun-D (lanes 7 to 9) antibodies. SS, supershifted band; the bracket indicates unshifted complexes. The right panel shows the Western blot analysis of nuclear extracts detected with anti-Fra-1. NS, nonspecific cross-reactive protein to the Fra-1 antibody. Equal protein levels, as determined by the BSA assay, were loaded.
Restoration of ERK levels in the 30.7b cells (30.7b/ERK) by stable transfection elevated the basal and TPA-induced AP-1/DNA-binding levels to levels as high as or higher than those of Cl 41 (Fig. 3A, lanes 7 and 8). The increase in the basal level of activated AP-1-bound complex is likely due to the higher level of activated ERK seen in these 30.7b/ERK cells (37). The composition of the faster-migrating complex, evident in longer-run gels as shown in Fig. 3A (arrowhead), has not been identified. We and others have previously shown the specificity of AP-1 protein binding to this AP-1 oligonucleotide, as indicated by competition with unlabeled oligonucleotide and by the lack of binding to a mutant AP-1 oligonucleotide (Fig. 3A) (8, 42). These results suggest that the deficiency in ERK levels in the P− 30.7b cells causes a deficiency in DNA binding of AP-1 complex to the AP-1 consensus site compared with ERK-sufficient P+ Cl 41 cells, although other contributing factors cannot be excluded.
TPA-induced activated AP-1 complexes in P+ cells are distinguished from P− complexes by the content of Fra-1.
The AP-1 complex that binds to DNA can consist of either a Jun/Jun homodimer or a Jun/Fos heterodimer. In order to characterize the mitogen-activated AP-1 complexes seen in the EMSA, supershift assays were performed with antibodies specific to each of the Fos family proteins (Fig. 3B). In the uninduced serum-starved cells, the addition of antibodies to Fra-1 and Fra-2 produced a supershift in the AP-1-bound DNA (Fig. 3B, lanes 5 to 10), suggesting that the AP-1 complex in resting cells consists of Jun/Jun, Jun/Fra-1, or Jun/Fra-2 dimers. Fos-B is absent from AP-1 complexes in JB6 cells (8) (data not shown). Interestingly, the addition of antibodies to c-Fos produced a decrease in DNA binding (a blocked shift) in both untreated cell types without mobility shifting the bound complex (Fig. 3B, lanes 3 and 4).
Exposure to mitogens like TPA produces an increase in AP-1 DNA-binding activity and transactivation. Figure 3B shows the Fos family components of the AP-1 complexes 3 h after TPA treatment. Fra-1, Fra-2, and c-Fos were present in the AP-1 DNA-bound complexes 3 h after TPA induction. The level of Fra-2 in the AP-1 complex of both P+ and P− cells changed little in response to TPA treatment. Three hours after TPA treatment, an anti-c-Fos supershifted band was seen in both P+ and P− cells, with the level of c-Fos slightly higher in P− 30.7b cells (Fig. 3B, lanes 3 and 4). The amount of c-Fos detected in the AP-1 complex in ERK-deficient Cl 30.7b cells 1.5 h after TPA exposure was also higher than in Cl 41 cells (8). More obvious, however, was the significant increase in the amount of Fra-1 found in the AP-1 complex in Cl 41 cells compared to Cl 30.7b cells 3 h after TPA exposure (Fig. 3B, lanes 5 and 6). These results suggest that exposure to TPA produces an increase in AP-1 binding at 3 h and that the activated AP-1 seen in the ERK-sufficient P+ cell complex contains Fra-1, Fra-2, or c-Fos complexed with a Jun protein.
After 18 h of exposure to TPA, the level of Fra-1 found in the complex remained significantly higher in the P+ cells than in the P− cells (Fig. 3B, lanes 5 and 6), while c-Fos was undetectable. These results indicate that at 18 h after TPA, a time when AP-1 luciferase activity is at a peak, Fra-1 is the major Fos family protein in the activated AP-1 complex.
Fra-1 binding in the AP-1 complex is ERK dependent.
The significant difference in Fra-1 content between the activated AP-1 complex in the ERK-sufficient P+ cells and the ERK-deficient P− cells suggests that Fra-1’s role in the activated AP-1 complex is ERK dependent. To determine the ERK dependency of Fra-1 binding, P+ Cl 41 cells were treated with 5 μM MEK-1 inhibitor 1 h prior to TPA exposure. Nuclear extracts were collected 3 h after TPA treatment and analyzed by EMSA for the presence of Fra-1 in the TPA-activated AP-1 complex (Fig. 3C). Clearly, the increased level of Fra-1 in the activated complex was reduced by inhibiting ERK activation (Fig. 3C, lanes 4 to 6). In contrast, AP-1 DNA binding (Fig. 3C, lanes 2 and 3) and the level of Jun-D in the bound complex were unaffected by 5 μM MEK inhibitor treatment of TPA-treated cells (Fig. 3C, lanes 7 to 9).
The amount of MEK inhibitor (5 μM) used in these experiments was able to inhibit activation of Elk-1 as well as transcriptional activation of the SRE and AP-1 promoters (Fig. 2 and data not shown). Figure 3C also shows that inhibition of ERK activation leads to a lack of TPA-induced Fra-1 protein expression. Inhibition of ERK-dependent Fra-1 expression by MEK inhibitor has been shown previously (22, 62). Taken together, these results suggest that although different ERK activation thresholds may be operative, the synthesis of Fra-1 (Fig. 3C) and the incorporation of Fra-1 into an activated AP-1 complex (Fig. 3B) are ERK dependent.
TPA-induced activation of c-Fos protein is ERK dependent.
TPA-induced expression of c-fos mRNA and c-Fos protein is not limited by the ERK deficiency in 30.7b (4, 8), yet mitogen-induced activation of AP-1 in 30.7b cells is limited. Since mitogen-induced activation of c-Fos protein can occur through the MAPK/ERK pathway (12, 61), it is possible that c-Fos protein is not activated in 30.7b cells. Thus, the lack of TPA-induced transactivation of AP-1 might arise from an inability to activate c-Fos protein due to the deficiency of ERK in the 30.7b cells.
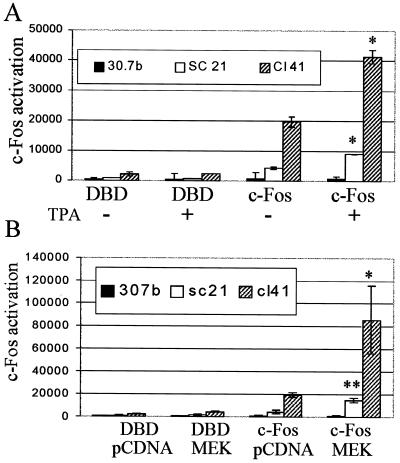
In order to determine whether c-Fos can be activated by TPA in these cells, a Gal4-c-Fos fusion construct in which the c-Fos transactivation domain is fused to the yeast Gal4 DNA-binding domain was cotransfected with a Gal4-luciferase reporter, and the c-Fos activation response to mitogen was determined. c-Fos protein was activated in the ERK-sufficient P+ Cl 41 and P− SC21 cells (though to a lesser extent), but not in the ERK-deficient P− 30.7b cells (Fig. 4A). Cl 41 and SC21 cells showed a greater than twofold increase in luciferase activity 6 h after exposure to TPA. In contrast, in the ERK-deficient P− 30.7b cells, c-Fos was not activated by TPA. Activation of the Gal4-Elk-1 fusion (see Fig. 2) was used in all assays as a control for mitogen activation.
FIG. 4.
c-Fos protein is activated by TPA or MEK-1 in ERK-sufficient but not ERK-deficient cells. (A) JB6 cells transiently transfected with a Gal4-luciferase and either a Gal4-c-Fos fusion or the empty Gal4 vector pFCMV (DBD) were serum starved for 24 h and then treated with TPA or DMSO. Luciferase activity was measured 6 h later. (B) JB6 cells transiently transfected with a Gal4-luciferase, Gal4-c-Fos, or empty pFCMV (DBD) plus activated MEK or pcDNA3 were serum starved for 24 h, and luciferase activity was measured. The average for three transfections + standard deviation is shown. Similar results were seen in multiple experiments. Activation of the Gal4-Elk-1 fusion (see Fig. 2) was used as a control for mitogen activation. * and **, statistically significant difference between TPA- or MEK-induced c-Fos-transfected cells and uninduced cells as determined by Student’s t test (*, P < 0.05; **, P < 0.01).
Expression of the constitutively activated ERK-specific MAPK kinase 1 (MEK-1) in Cl 41 and SC21 cells produced a 4-fold and a 3.5-fold activation of Gal4-c-Fos, respectively, whereas MEK-1 produced little or no activation of c-Fos in ERK-deficient 30.7b cells (Fig. 4B). These results indicate that activation of c-Fos occurs ERK dependently. Activation of the MAPK/ERK with either TPA or ERK 1- and 2-specific MEK-1 leads to the activation of c-Fos protein in the ERK-sufficient cells but not in the ERK-deficient cells. The level of ERK in the 30.7b cells, while low, may be sufficient to support the activation of Elk-1, which leads to expression of c-fos mRNA, but this level of ERK may not be sufficient to activate c-Fos protein.
Fra-1 activation occurs ERK dependently.
The elevated level of Fra-1 seen in the activated AP-1 complex in AP-1-responsive Cl 41 P+ cells and the low level of c-Fos in the P+ relative to the P− complex (Fig. 3) suggest that Fra-1 may play a more prominent role in AP-1 activation than does c-Fos. It has been reported, however, that the transactivation domain in Fra-1 is not functional (5, 69), and it has been suggested that the presence of Fra-1 in the AP-1 complex may act as a dominant negative inhibitor (70).
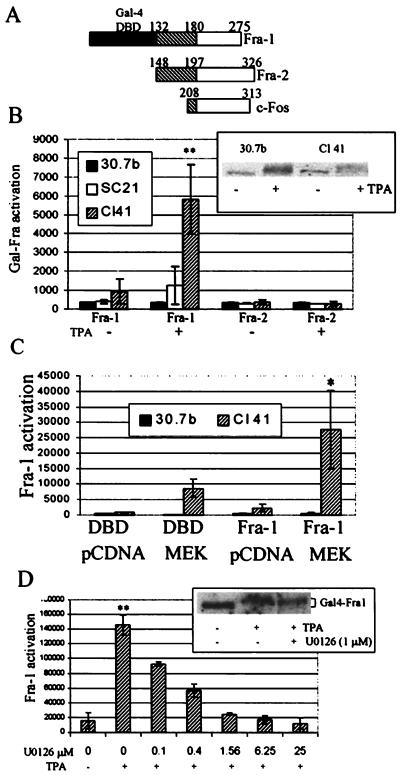
In order to determine whether the transactivation domain of Fra-1 is functional in JB6 cells, it was fused to the Gal4 binding domain (Fig. 5A). TPA treatment of Cl 41 and Cl SC21 cells cotransfected with the Gal4-Fra-1 fusion and the Gal4-luciferase reporter produced 6.3- and 3.1-fold increases in Fra-1 activity, respectively (Fig. 5B). On the other hand, Fra-1 was not activated in ERK-deficient Cl 30.7b cells after TPA treatment. Western blot analysis of P− and P+ cells transfected with the Gal4-Fra-1 fusion shows that the failure to activate Fra-1 in the Cl 30.7b P− cells is not attributable to a lack of synthesis or to an increase in degradation of the Gal4 fusion in these cells (Fig. 5B, inset). Cotransfection with the constitutively activated MEK-1 (no TPA) also activated Fra-1 in the ERK-sufficient and not in the ERK-deficient cells (Fig. 5C). Activation of Fra-1 in Cl 41 was blocked in a dose-dependent manner when ERK activation was inhibited by the MEK inhibitor U0126 (Fig. 5D). Pretreatment of TPA-treated P+ cells with 1 μM U0126, a concentration that substantially inhibits Fra-1 activation, resulted in no loss of the Gal4-Fra-1 fusion protein (Fig. 5D, inset), and 10 μM U0126 pretreatment produced complete loss of the Gal4-Fra-1 fusion protein (not shown).
FIG. 5.
Fra-1 is activated by TPA or MEK-1 in ERK-sufficient cells but not ERK-deficient cells. (A) Gal4 fusions. The Fra-1 and Fra-2 Gal4 fusions contain the yeast Gal4 DNA-binding domain (DBD) (solid box) fused to C-terminal residues of the rat Fra-1 (aa 132 to 275; NCBI accession no. NP037085) or Fra-2 protein (aa 148 to 326; accession no. P51145). Gal4-c-Fos is encoded by pFA-Fos and contains the C-terminal transactivation domain from c-Fos (aa 208 to 313) fused to the Gal4 DNA-binding domain (Stratagene). The hatched regions show the homologous domain in Fra-1, Fra-2, and c-Fos. (B) Fra-1 activation. JB6 cells transiently transfected with a Gal4-Fra-1 or Gal4-Fra-2 fusion and a Gal4-luciferase reporter were serum starved for 24 h and then treated with TPA for 6 h. Cell extracts were analyzed for luciferase activity. The inset shows a Western blot of 30.7b and Cl 41 cells transiently transfected with Gal4-Fra-1 and detected with anti-Gal4 antibody. (C) JB6 cells transiently transfected with Gal4-Fra-1 or empty pFCMV (DBD), activated MEK-1, or pcDNA3 and Gal4-luciferase were serum starved for 24 h, and luciferase activity was measured. (D) Fra-1 activation is ERK dependent. Cl 41 cells were transfected with Gal4-Fra-1 fusion and the Gal4-luciferase vector, serum starved for 24 h, and treated with MEK inhibitor U0126 or DMSO for 1 h before being exposed to TPA for 6 h. Cell extracts were analyzed for luciferase activity. Assays were done in triplicate, and a representative of multiple assays is shown. Activation of the Gal4-Elk-1 fusion (see Fig. 2) was used as a control for mitogen activation. The inset shows a Western blot of Cl 41 cells transfected with Gal4-Fra-1 and detected with anti-Gal4 antibody. * and **, statistically significant difference between TPA- or MEK-induced Gal4-Fra-1-transfected cells and uninduced cells as determined by Student’s t test (*, P < 0.05; **, P < 0.01). The P values for the inhibition of Fra-1 activation with U0126 were all <0.001.
The Gal4-Fra-2 fusion, which contains sequence (aa 148 to 197) nearly identical to that of the Fra-1 fusion (aa 132 to 180) and differs only in the C-terminal transactivation domain (Fig. 5A), was not activated by TPA or by MEK-1 in any of the JB6 variants (Fig. 5B and data not shown). Expression of the Gal4-Fra-2 fusion was confirmed by Western blot analysis (data not shown). The mitogen-dependent activation of Fra-1 can therefore not be attributed solely to a sequence shared with Fra-2. The Gal4 fusion with Jun-D, the major Jun family protein in the AP-1 complex (8), was activated by TPA in both the ERK-deficient Cl 30.7b (2.5-fold) and ERK-sufficient Cl SC21 (2.8-fold) and Cl 41 (1.8-fold) cells, suggesting that activation of Jun-D protein is not limiting for AP-1 activation in the ERK-deficient cells. c-Jun was not activated by TPA treatment in either cell type.
These results show that the Fra-1 transactivation domain, when fused to the Gal4 DNA-binding domain, can be transcriptionally activated, apparently in an ERK-dependent manner. Thus, the presence of Fra-1 in the activated AP-1 complex of ERK-sufficient cells (Fig. 3B and C), coupled with ERK-dependent activation of Fra-1’s transactivation domain (Fig. 5), suggests that activation of Fra-1 may be necessary for AP-1 transactivation in JB6 cells.
Identification of ERK-dependent activation sites in the Fra-1 transactivation domain.
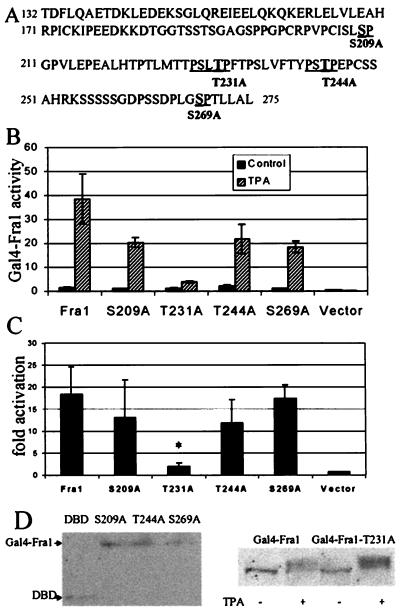
Fra-1 appears to be the predominant Fos family member in the activated AP-1 complex in ERK-sufficient P+ cells (Fig. 3B), and the transactivation domain of Fra-1 can be activated in an ERK-dependent manner (Fig. 5). To determine which site in the transactivation domain of Fra-1 is required for activation, we performed site-directed mutagenesis on the Gal4-Fra-1 fusion construct.
MAPKs catalyze the phosphorylation of substrates containing a proline in the +1 site relative to the serine or theronine being phosphorylated. A proline in the −2 or −3 position can confer specificity for ERK 1 and 2 (53, 54). The potential ERK phosphorylation sites in the transactivation domain of the Gal4-Fra-1 fusion are shown in Fig. 6A. Ser-209, Thr-231, Thr-244, and Ser-269 were mutated to Ala by site-directed mutagenesis. Mutated constructs were then cotransfected into P+ Cl 41 cells, and activation of Fra-1 was determined (Fig. 6B and C).
FIG. 6.
Thr-231 is required for activation of Fra-1. (A) Sequence of the C terminus of Fra-1 that comprises the Gal4-Fra-1 fusion. Ser/Thr-Pro sites are underlined. Mutated residues are in boldface and indicated under the sequence. (B) Fra-1 activation. Cl 41 cells cotransfected with Gal4 fusions containing wild-type or mutant Fra-1 and the Gal4-luciferase reporter were serum starved for 24 h and then untreated or treated with TPA (10 ng/ml) for 6 h. Cells were harvested, and luciferase activity was determined. A representative assay, the average of three tranfections, is shown. (C) Fold activation of Fra-1 activity was determined from the average of three individual assays. The fold activation for three separate experiments of three transfections each is shown. *, statistically significant difference between wild-type Ga14-Fra-1 and mutant Ga14-Fra-1-T231A as determined by Student’s t test (P < 0.011). (D) Expression of Gal4-Fra-1 and Gal4-Fra-1 mutants in Cl 41 cells. Cl 41 cells were transfected with Gal4-Fra-1 or Gal4-Fra-1 mutants, and nuclear extracts were harvested 48 h later (DBD) (pFCMV) (left panel). Cl 41 cells transfected with Gal4-Fra-1 or Gal4-Fra-1-T231A and serum starved overnight were treated with TPA for 6 h. Nuclear extracts were harvested and analyzed by Western blot (right panel). Gal4 fusions were detected with anti-Gal4 antibody.
Mutagenesis of Ser-209 and Thr-244 to Ala produced a slight but insignificant decrease in TPA-induced activation of Fra-1. Fra-1 activity was not affected by changing Ser-269 to Ala. Mutation of Thr-231 to Ala resulted in nearly complete loss of activity. Increasing the amount of the T231A fusion two- and fourfold failed to restore activity, suggesting that the lack of activation was not due to insufficient protein. Expression of the mutated Gal4-Fra-1 constructs was confirmed by Western blot (Fig. 6D). Exposure to TPA of Gal4-Fra-1-T231A-transfected cells produced no lack of expression of mobility-shifted Gal4-Fra-1 bands (Fig. 6D, right), similar to that seen with the wild-type Fra-1 fusion. This suggests that the unphosphorylated status of Thr-231 does not destabilize the protein. These results support the hypothesis that the transactivation domain of Fra-1 is activated by an ERK-dependent phosphorylation of Thr-231. Together with the data presented in Fig. 33 and 5, these results suggest that the ERK-dependent activation of Fra-1 in JB6 P+ cells is required for transactivation of AP-1.
Expression of endogenous Fra-1 is required for activation of AP-1 in JB6 cells.
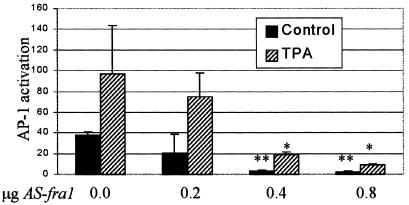
Activation of the Gal4-Fra-1 fusion in the ERK-sufficient AP-1-responsive cells and not in the ERK-deficient AP-1-nonresponsive cells leads to the expectation that Fra-1 plays an important role in the activation of AP-1 in JB6 cells. To determine whether endogenous Fra-1 is required for activation of AP-1 in JB6 P+ cells, Cl 41 cells were transiently transfected with antisense fra-1 cDNA. Figure 7 shows that expression of antisense fra-1 blocked both basal and TPA-induced activation of AP-1 in a dose-dependent manner.
FIG. 7.
Antisense (AS) fra-1 blocks AP-1 activation in P+ cells. Cl 41 cells were cotransfected with the 4× AP-1 reporter and antisense fra-1 at the indicated concentrations. pcDNA3 was used to equilibrate the DNA concentration used for transfection. At 24 h after transfection, cells were serum starved overnight and untreated or treated with TPA for 6 h. * and **, statistically significant difference between antisense fra-1- and pcDNA3-transfected cells as determined by Student’s t test (*, P < 0.05; **, P < 0.01).
P− Cl SC21 cells show levels of activated ERK similar to those of Cl 41 cells, yet they are resistant to mitogen-induced activation of AP-1 and neoplastic transformation (Fig. 1) (15). These results suggest that SC21 cells are deficient in signaling from activated ERK to activation of AP-1. As shown above, TPA induces activation of the Gal4-Elk-1 fusion and the SRE-luciferase reporter in SC21 cells (Fig. 2), suggesting that transcription from the SRE-regulated c-fos promoter is not deficient. Western blot analysis, however, showed that the TPA-induced levels of Fra-1 in the SC21 cells were lower than those seen in 30.7b and Cl 41 (Fig. 8A). These results suggest that insufficient expression of fra-1 or increased sensitivity to proteolysis in the SC21 cells may render them nonresponsive to TPA for activation of AP-1. Expression of Fra-1 in SC21 cells following transfection with a cytomegalovirus (CMV) promoter-driven fra-1 expression vector restored the MAPK cascade leading to TPA-induced activation of AP-1 in these cells (Fig. 8B). These results suggest that synthesis of Fra-1 and not Fra-1 proteolysis is the limiting factor rendering the SC21 cells resistant to AP-1 induction. Interestingly, in SC21 cells that spontaneously converted to transformation sensitivity, the level of TPA-induced Fra-1 (Fig. 8A, lane 8) and AP-1 activation (not shown) is similar to that seen in P+ Cl 41 cells.
FIG. 8.
P− SC21 cells are deficient in Fra-1. (A) Cl 30.7b, Cl SC21, and Cl 41 cells were serum starved for 24 h and then treated with TPA for 0 or 3 h. Nuclear extracts were harvested and analyzed for Fra-1 protein by Western blot. Lanes 1 to 6 show extracts from P− 30.7b, P− SC21, and P+ Cl 41 cells. Lanes 7 and 8 show extracts from SC21 cells that have spontaneously converted to P+ cells as determined by TPA-induced AP-1 activation and transformation (data not shown). NS, nonspecific cross-reactive protein to the Fra-1 antibody. Equal protein levels, as determined by BSA assay, were loaded. (B) Expression of fra-1 cDNA in SC21 cells restores AP-1 activation. SC21 cells were transiently transfected with an AP-1 luciferase reporter and increasing amounts of fra-1 cDNA. Cells were starved for 24 h and then treated with TPA for 6 h. Fold activation is defined as TPA-induced luciferase activity relative to uninduced levels. * and **, statistically significant difference between fra-1- and pcDNA3-transfected cells as determined by Student’s t test (*, P < 0.05; **, P < 0.01).
Expression of fra-1-T231A cDNA does not activate AP-1 in P+ cells.
Overexpression of Fra-1 can activate transcription of putative AP-1-regulated genes and enhance tumorigenesis (43, 52, 64). In JB6 P+ Cl 41 cells, overexpression of fra-1 cDNA results in a significant enhancement of AP-1-regulated transcription (Fig. 9) (K. Suzukawa, T. J. Weber, and N. H. Colburn, submitted for publication).
FIG. 9.
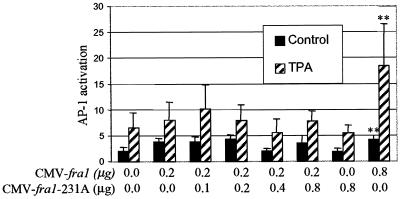
Fra-1-T231A does not activate AP-1 in JB6 cells. Cl 41 cells were cotransfected with the 4× AP-1 reporter, CMV-fra-1, and/or CMV-fra-1-T231A at the indicated concentrations. pcDNA3 was used to equalize the DNA concentration. At 24 h after transfection, cells were serum starved overnight and untreated or treated with TPA for 6 h. * and **, statistically significant difference between fra-1- and fra-1-T231A-transfected cells as determined by Student’s t test (*, P < 0.05; **, P < 0.01).
To determine if Thr-231 is required for activation of AP-1 in JB6 cells, Thr-231 in the fra-1 cDNA was mutated to Ala. CMV-fra-1 was cotransfected into P+ Cl 41 cells with increasing amounts of CMV-fra-1-T231A and the AP-1 luciferase reporter (Fig. 9). Transfection of Cl 41 cells with 0.2 μg of CMV-fra-1 produced a slight increase in both basal and TPA-induced AP-1 activation (compare lanes 1 and 2). Transfection of Cl 41 with 0.8 μg of CMV-fra-1 produced a significantly larger increase in TPA-induced activation of AP-1 (lane 8), while transfection with 0.8 μg of CMV-fra-1-T231A did not enhance either basal or TPA-induced activation of AP-1 (compare lane 7 with lane 1). Expression of T231A-mutated Fra-1 does not, however, block the enhanced AP-1 activity associated with overexpression of the wild-type cDNA (lanes 2 to 6), nor does it lower AP-1 activation in cells expressing only the endogenous wild-type fra-1 (lane 7). Thus, while CMV-fra-1-T231A does not activate AP-1, it also does not function as a dominant negative mutant blocking Fra-1 activation of AP-1. The results shown in Fig. 9 as well as those shown in Fig. 6 suggest that Thr-231 is required for ERK-dependent activation of AP-1 in JB6 cells.
DISCUSSION
These results establish that the transactivation domain of Fra-1 can be activated by mitogen stimulation in the JB6 mouse epidermal cell model, that activation of Fra-1 is ERK dependent, and that Thr-231 is required for activation. Furthermore, these observations identify Fra-1 activation as an ERK-dependent event that appears to be required for activation of AP-1-dependent transcription. This has particular significance for the cases in which another ERK-dependent event, c-fos transcription, is not a limiting factor for AP-1 transactivation.
The finding that Fra-1 is activable in JB6 P+ cells is noteworthy, as it has been reported that Fra-1 and Fra-2 lack a functional transactivation domain (5, 69). It is clear from these reports that Fra-1 and Fra-2 lack a constitutively activated transactivation domain needed for transformation of 208F cells, but these results do not address the question of whether the transactivation domain in Fra-1 and/or Fra-2 can be activated by mitogens or other stimuli. Similarly, mitogen activation of Fra-1 was not tested by Bergers et al. (5). In this report, the complete coding sequence of Fra-1 when fused to the Gal4 DNA-binding domain was unable to activate the Gal4 reporter in COS cells. Interestingly, however, Bergers et al. (5) reported that overexpression of fra-1 in rat fibroblasts produced anchorage-independent growth in vitro and tumor development in athymic mice, suggesting that Fra-1 might be activable.
In preneoplastic JB6 P+ Cl 41 cells, overexpression of fra-1 cDNA results in a significant enhancement of AP-1-regulated transcription and restores TPA-induced AP-1 activation in P− Cl SC21 cells. Others have also shown that Fra-1 expression can activate transcription of putative AP-1-regulated genes and enhance tumorigenesis (5, 43, 52, 64). Thus, although Fra-1’s function as an activator or repressor of transcription appears to be cell type dependent, it is now clear that Fra-1 is capable of being inducibly regulated as a transcription factor.
The increase in Fra-1 abundance seen in the activated AP-1 DNA-bound complex also appears to be ERK dependent. Fra-1 was recruited to AP-1 DNA complexes in TPA-treated ERK-sufficient P+ cells to a much greater degree than in ERK-deficient P− cells or in MEK inhibitor-treated P+ cells. In contrast, Rosenberger et al. (59) found that inhibition of ERK did not alter AP-1 complex composition. In the report of Rosenberger et al., the inducer (okadaic acid), the cells (308 papilloma cells), and the MEK inhibitor (PD 98059 at 50 μM) were different. Moreover, the AP-1 complex differed, consisting predominately of Fos-B and Jun-D, with little or no Fra-1. In JB6 P+ cells, AP-1 binding was reduced when the concentration of the U0126 inhibitor was raised to 20 and 40 μM (data not shown). The increase in Fra-1 abundance seen in the activated AP-1 DNA-bound complex in the AP-1-responsive JB6 P+ cells along with the ERK-dependent activation of Fra-1 in this variant is consistent with Fra-1’s role in tumor promoter-induced AP-1 transactivation. Identifying AP-1-regulated genes required for tumor promotion will be important, as will the characterization of Fra-1 target genes.
Fra-1 appears to have both unique functions and functions shared by other Fos family proteins. Fra-1 can replace certain c-Fos-dependent functions in mice (29, 51). By replacing the c-fos coding sequence with the fra-1 coding sequence in c-fos null mice, Fleischmann et al. (29) showed that Fra-1 rescues the defects in bone development seen in c-fos null mice. Fra-1 was not able, however, to rescue expression of c-Fos target genes in cultured fibroblasts isolated from the c-fos null mice. The current observations, together with those above, are compatible with the notion that Fra-1 inducibly activates AP-1-dependent transcription in a fashion that may be both tissue specific and gene specific.
Bacterially expressed Fra-1 can be phosphorylated in vitro by ERK 1, and in vitro phosphorylation by ERK increased Fra-1’s DNA-binding activity (34). However, the significance of phosphorylation relative to transactivation was not addressed. Whether Fra-1 is a direct substrate of ERK in vivo is not known. There is a potential ERK phosphorylation site (PSLTP, residues 228 to 232) in the transactivation domain of the Fra-1 protein. Mutation of Thr-231 to Ala in the Gal4-Fra-1 fusion rendered the fusion inactivable by TPA. The same mutation in fra-1 cDNA yields a Fra-1 protein that does not activate AP-1 in JB6 cells. The homologous domain in the chicken Fra-2 protein is phosphorylated on the same Thr by ERK in vitro and in response to transfection with activated MEK in culture (53).
Fra-1, like the other Fos family proteins, is also phosphorylated in vitro by other kinases, including protein kinases C and A and cdc2 (1, 34). Western blot analysis shows multiple Fra-1-specific bands in P+ and P− cells. TPA treatment also produces multiple slower-migrating bands of the Gal4-Fra-1 fusion in both P− and P+ cells and of the Gal4-Fra-1-T231A fusion in P+ cells. These results suggest that the posttranslational modification of the Gal4-Fra-1 fusions is similar to that of the endogenous Fra-1. Furthermore, the presence of both endogenous Fra-1 and Gal4-Fra-1 in ERK-deficient P− cells after TPA treatment indicates that the lack of Fra-1 activation in these cells is not attributable to an increased sensitivity to proteolysis. Interestingly, at a concentration of the MEK inhibitor U0126 (1 μM) that blocks activation of Gal4-Elk, SRE-luciferase, and Gal4-Fra-1, posttranslational modification of Gal4-Fra-1 and of endogenous Fra-1 was detected. It appears that in P− cells the level of activated ERK is sufficient to activate Elk and SRE but is not sufficient to activate Fra-1 and transactivate AP-1.
Mitogen activation of c-Fos protein was also different in the ERK-sufficient Cl 41 and Cl SC21 cells from that in the ERK-deficient 30.7b cells. These results are consistent with previous reports showing that activation of c-Fos occurs ERK dependently (12, 13). c-Fos activity is regulated by multiple mechanisms both transcriptionally and posttranslationally. Although it is clear that c-Fos is an oncogene and events that lead to enhanced expression or activation of c-Fos can be tumorigenic, a role for c-Fos in tumor promotion has not been demonstrated. In fact, c-fos null mice initiated with an activated ras gene and treated with TPA develop benign premalignant tumors at a rate similar to that of their c-fos+/+ siblings, suggesting that c-Fos does not function in tumor promotion in vivo (60). Taken together, the lack of c-fos function in the classical mouse skin tumor promotion model, the fact that c-fos transcription does not limit AP-1 transactivation in P− cells (4), and the fact that the level of c-Fos in the AP-1 complex is higher in P− than in P+ cells suggest that c-Fos does not contribute to driving tumor promotion.
It is possible that c-Fos regulates fra-1 expression in JB6 cells, as seen in RAT-1A cells (5). In NIH 3T3 cells, however, activation of ERK signaling by activated Ras increases fra-1 expression without inducing c-fos expression (62). The fact that fra-1 is expressed in the ERK-deficient P− 30.7b cells at a level similar to that seen in the P+ Cl 41 cells (4, 6) suggests that these cells contain sufficient ERK levels for c-Fos-regulated synthesis of fra-1 and that activation of the c-Fos protein is not needed to induce fra-1 mRNA synthesis. Inhibition of ERK activity by the MEK-1 inhibitor at 5 μM must reduce the threshold of activated ERK below the level required for synthesis of fra-1 (see Fig. 3C). It should nevertheless be noted that while expression of c-fos mRNA is not limiting in the P− cells and c-Fos activation is not required for fra-1 expression, the possibility that c-Fos activation may contribute to AP-1 activation in P+ cells cannot be excluded.
It is interesting that c-fos mRNA expression is not curtailed in the ERK-deficient cells. It has been well documented that activation of the MAPK/ERK cascade leads to activation of the SRE and an increase in c-fos transcription and that inhibition of ERK activation leads to an inhibition of mitogen-induced c-fos transcription (25, 55, 68). Although the 30.7b cells are deficient in ERK protein and consequently in ERK activation, they are not devoid of ERK protein. In fact, the ERK that is present in these cells is phosphorylated in response to TPA as efficiently as that seen in the P+ cells (37). Thus, it seems that a threshold level of activated ERK is needed for mitogen-induced activation of AP-1 and neoplastic transformation and that this threshold is above the level needed to activate the TCF (Elk-1) and to induce c-fos transcription.
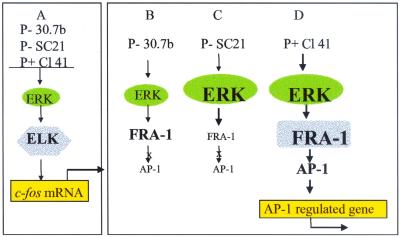
Understanding the AP-1-dependent mechanisms by which chronic exposure of initiated cells to tumor promoters leads to tumorigenesis may provide methods for prevention and/or reversal of the tumor promotion process. Fra-1 appears to be necessary to complete the signaling cascade leading to AP-1 activation, and a critical threshold level of ERK 1 and/or 2 is needed to activate Fra-1 (Fig. 10). The level of ERK in all three JB6 variants is sufficient for TPA-induced activation of Elk-1 and gene expression from genes containing an SRE promoter, such as c-fos (Fig. 10A). The level of ERK in P− Cl 30.7b is not sufficient for TPA-induced activation of Fra-1 and gene expression from AP-1-regulated genes (Fig. 10B). In the P− Cl SC21, in which the level of ERK is similar to that of the P+ Cl 41 cells, the level of Fra-1 protein rather than its activation may limit the TPA-induced activation of AP-1 (Fig. 10C). Expression of fra-1 in SC21 cells is sufficient to restore AP-1 activity. The level of both ERK and Fra-1 in the P+ Cl 41 cells is sufficient to complete the signaling cascade from ERK to AP-1 activation (Fig. 10D). Further characterization of the MAPK/ERK cascade leading to activation of Fra-1 is necessary to determine whether ERK or another kinase is phosphorylating Fra-1 directly and whether recruitment of coactivators to the AP-1 complex occurs ERK dependently.
FIG. 10.
nMAPK ERK to AP-1 pathway. (A) All three JB6 variants have sufficient ERK to drive activation of Elk-1 and transactivation of the SRE promoter, leading to c-fos expression. (B) JB6 P− 30.7b cells do not have sufficient ERK for activation of Fra-1 and/or transactivation of the AP-1 promoter. (C) P− SC21 cells have sufficient ERK for activation of exogenously added Gal4-Fra-1 but lack sufficient endogenous Fra-1, rendering them resistant to mitogen-induced transactivation of AP-1. (D) P+ Cl 41 cells have sufficient ERK and Fra-1 protein to complete the signal cascade from ERK to AP-1. Shading indicates activated proteins.
REFERENCES
- 1.Abate, C., D. R. Marshak, and T. Curran. 1991. Fos is phosphorylated by p34cdc2, cAMP-dependent protein kinase and protein kinase C at multiple sites clustered within regulatory regions. Oncogene 6:2179–2185. [PubMed] [Google Scholar]
- 2.Angel, P., and M. Karin. 1991. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim. Biophys. Acta 1072:129–157. [DOI] [PubMed] [Google Scholar]
- 3.Balmanno, K., and S. J. Cook. 1999. Sustained MAP kinase activation is required for the expression of cyclin D1, p21Cip1 and a subset of AP-1 proteins in CCL39 cells. Oncogene 18:3085–3097. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Ari, E. T., L. R. Bernstein, and N. H. Colburn. 1992. Differential c-jun expression in response to tumor promoters in JB6 cells sensitive or resistant to neoplastic transformation. Mol. Carcinog. 5:62–74. [DOI] [PubMed] [Google Scholar]
- 5.Bergers, G., P. Graninger, S. Braselmann, C. Wrighton, and M. Busslinger. 1995. Transcriptional activation of the fra-1 gene by AP-1 is mediated by regulatory sequences in the first intron. Mol. Cell. Biol. 15:3748–3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernstein, L. R., R. Bravo, and N. H. Colburn. 1992. 12-O-Tetradecanoylphorbol-13-acetateinduced levels of AP-1 proteins: a 46-kDa protein immunoprecipitated by anti-fra-1 and induced in promotion-resistant but not promotion-sensitive JB6 cells. Mol. Carcinog. 6:221–229. [DOI] [PubMed] [Google Scholar]
- 7.Bernstein, L. R., and N. H. Colburn. 1989. AP1/jun function is differentially induced in promotion-sensitive and resistant JB6 cells. Science 244:566–569. [DOI] [PubMed] [Google Scholar]
- 8.Bernstein, L. R., and S. E. Walker. 1999. Tumor promotion resistant cells are deficient in AP-1 DNA binding, JunD DNA binding and JunD expression and form different AP-1-DNA complexes than promotion sensitive cells. Biochim. Biophys. Acta 1489:263–280. [DOI] [PubMed] [Google Scholar]
- 9.Boulton, T. G., S. H. Nye, D. J. Robbins, N. Y. Ip, E. Radziejewska, S. D. Morgenbesser, R. A. DePinho, N. Panayotatos, M. H. Cobb, and G. D. Yancopoulos. 1991. ERKs: a family of protein-serine/threonine kinases that are activated and tyrosine phosphorylated in response to insulin and NGF. Cell 65:663–675. [DOI] [PubMed] [Google Scholar]
- 10.Boulton, T. G., G. D. Yancopoulos, J. S. Gregory, C. Slaughter, C. Moomaw, J. Hsu, and M. H. Cobb. 1990. An insulin-stimulated protein kinase similar to yeast kinases involved in cell cycle control. Science 249:64–67. [DOI] [PubMed] [Google Scholar]
- 11.Brown, P. H., R. Alani, L. H. Pries, E. Szabo, and M. J. Birrer. 1993. Suppression of oncogene-induced transformation by a deletion mutant of c-jun. Oncogene 8:877–886. [PubMed] [Google Scholar]
- 12.Chen, R. H., C. Abate, and J. Blenis. 1993. Phosphorylation of the c-Fos transrepression domain by mitogen-activated protein kinase and 90-kDa ribosomal S6 kinase. Proc. Natl. Acad. Sci. USA 90:10952–10956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, R. H., P. C. Juo, T. Curran, and J. Blenis. 1996. Phosphorylation of c-Fos at the C-terminus enhances its transforming activity. Oncogene 12:1493–1502. [PubMed] [Google Scholar]
- 14.Chen, R.-H., C. Sarnecki, and J. Blenis. 1992. Nuclear localization and regulation of erk- and rsk-encoded protein kinases. Mol. Cell. Biol. 12:915–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cmarik, J. L., H. Min, G. Hegamyer, S. Zhan, M. Kulesz-Martin, H. Yoshinaga, S. Matsuhashi, and N. H. Colburn. 1999. Differentially expressed protein Pdcd4 inhibits tumor promoter-induced neoplastic transformation. Proc. Natl. Acad. Sci. USA 96:14037–14042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cobb, M. H. 1999. MAP kinase pathways. Prog. Biophys. Mol. Biol. 71:479–500. [DOI] [PubMed] [Google Scholar]
- 17.Cobb, M. H., S. Xu, J. E. Hepler, M. Hutchison, J. Frost, and D. J. Robbins. 1994. Regulation of the MAP kinase cascade. Cell. Mol. Biol. Res. 40:253–256. [PubMed] [Google Scholar]
- 18.Cohen, D. R., and T. Curran. 1988. fra-1: a serum-inducible, cellular immediate-early gene that encodes a Fos-related antigen. Mol. Cell. Biol. 8:2063–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colburn, N. H., B. F. Former, K. A. Nelson, and S. H. Yuspa. 1979. Tumour promoter induces anchorage independence irreversibly. Nature 281:589–591. [DOI] [PubMed] [Google Scholar]
- 20.Colburn, N. H., T. D. Gindhart, G. A. Hegamyer, P. M. Blumberg, K. B. Delclos, B. E. Magun, and J. Lockyer. 1982. Phorbol diester and epidermal growth factor receptors in 12-O-tetradecanoylphorbol-13-acetate-resistant and -sensitive mouse epidermal cells. Cancer Res. 42:3093–3097. [PubMed] [Google Scholar]
- 21.Colburn, N. H., E. J. Wendel, and G. Abruzzo. 1981. Dissociation of mitogenesis and late-stage promotion of tumor cell phenotype by phorbol esters: mitogen-resistant variants are sensitive to promotion. Proc. Natl. Acad. Sci. USA 78:6912–6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cook, S. J., N. Aziz, and M. McMahon. 1999. The repertoire of Fos and Jun proteins expressed during the G1 phase of the cell cycle is determined by the duration of mitogen-activated protein kinase activation. Mol. Cell. Biol. 19:330–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Curran, T., and B. R. Franza, Jr. 1988. Fos and Jun: the AP-1 connection. Cell 55:395–397. [DOI] [PubMed] [Google Scholar]
- 24.Davis, R. J. 1994. MAPKs: new JNK expands the group. Trends Biochem. Sci. 19:470–473. [DOI] [PubMed] [Google Scholar]
- 25.Deng, T., and M. Karin. 1994. c-Fos transcriptional activity stimulated by H-Ras-activated protein kinase distinct from JNK and ERK. Nature 371:171–175. [DOI] [PubMed] [Google Scholar]
- 26.Domann, F. E., J. P. Levy, M. J. Birrer, and G. T. Bowden. 1994. Stable expression of a c-JUN deletion mutant in two malignant mouse epidermal cell lines blocks tumor formation in nude mice. Cell Growth Differ. 5:9–16. [PubMed] [Google Scholar]
- 27.Dong, Z., M. J. Birrer, R. G. Watts, L. M. Matrisian, and N. H. Colburn. 1994. Blocking of tumor promoter-induced AP-1 activity inhibits induced transformation in JB6 mouse epidermal cells. Proc. Natl. Acad. Sci. USA 91:609–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong, Z., H. C. Crawford, V. Lavrovsky, D., Taub, R. Watts, L. M. Matrisian, and N. H. Colburn. 1997. A dominant negative mutant of Jun blocking 12-O-tetradecanoylphorbol-13-acetate-induced invasion in mouse keratinocytes. Mol. Carcinog. 19:204–212. [DOI] [PubMed] [Google Scholar]
- 29.Fleischmann, A., F. Hafezi, C. Elliott, C. E. Reme, U. Ruther, and E. F. Wagner. 2000. Fra-1 replaces c-fos-dependent functions in mice. Genes Dev. 14:2695–2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Foletta, V. C., M. H. Sonobe, T. Suzuki, T. Endo, H. Iba, and D. R. Cohen. 1994. Cloning and characterisation of the mouse fra-2 gene. Oncogene 9:3305–3311. [PubMed] [Google Scholar]
- 31.Frost, J. A., T. D. Geppert, M. H. Cobb, and J. R. Feramisco. 1994. A requirement for extracellular signal-regulated kinase (ERK) function in the activation of AP-1 by Ha-Ras, phorbol 12-myristate 13-acetate, and serum. Proc. Natl. Acad. Sci. USA 91:3844–3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gentz, R., F. J. Rauscher III, C. Abate, and T. Curran. 1989. Parallel association of Fos and Jun leucine zippers juxtaposes DNA binding domains. Science 243:1695–1699. [DOI] [PubMed] [Google Scholar]
- 33.Gille, H., A. D. Sharrocks, and P. E. Shaw. 1992. Phosphorylation of transcription factor p62TCF by MAP kinase stimulates ternary complex formation at c-fos promoter. Nature 358:414–417. [DOI] [PubMed] [Google Scholar]
- 34.Gruda, M. C., K. Kovary, R. Metz, and R. Bravo. 1994. Regulation of Fra-1 and Fra-2 phosphorylation differs during the cell cycle of fibroblasts and phosphorylation in vitro by MAP kinase affects DNA binding activity. Oncogene 9:2537–2547. [PubMed] [Google Scholar]
- 35.Hayes, T. E., A. M. Kitchen, and B. H. Cochran. 1987. Inducible binding of a factor to the c-fos regulatory region. Proc. Natl. Acad. Sci. USA 84:1272–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hill, C. S., and R. Treisman. 1995. Transcriptional regulation by extracellular signals: mechanisms and specificity. Cell 80:199–211. [DOI] [PubMed] [Google Scholar]
- 37.Huang, C., W. Y. Ma, M. R. Young, N. Colburn, and Z. Dong. 1998. Shortage of mitogen-activated protein kinase is responsible for resistance to AP-1 transactivation and transformation in mouse JB6 cells. Proc. Natl. Acad. Sci. USA 95:156–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson, G. L., and R. R. Vaillancourt. 1994. Sequential protein kinase reactions controlling cell growth and differentiation. Curr. Opin. Cell Biol. 6:230–238. [DOI] [PubMed] [Google Scholar]
- 39.Karin, M., and T. Hunter. 1995. Transcriptional control by protein phosphorylation: signal transmission from the cell surface to the nucleus. Curr. Biol. 5:747–757. [DOI] [PubMed] [Google Scholar]
- 40.Karin, M., Z. Liu, and E. Zandi. 1997. AP-1 function and regulation. Curr. Opin. Cell Biol. 9:240–246. [DOI] [PubMed] [Google Scholar]
- 41.Kim, D. W., V. Cheriyath, A. L. Roy, and B. H. Cochran. 1998. TFII-I enhances activation of the c-fos promoter through interactions with upstream elements. Mol. Cell. Biol. 18:3310–3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim, H., W. D. Pennie, Y. Sun, and N. H. Colburn. 1997. Differential functional significance of AP-1 binding sites in the promoter of the gene encoding mouse tissue inhibitor of metalloproteinases-3. Biochem. J. 324:547–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kustikova, O., D. Kramerov, M. Grigorian, V. Berezin, E. Bock, E. Lukanidin, and E. Tulchinsky. 1998. Fra-1 induces morphological transformation and increases in vitro invasiveness and motility of epithelioid adenocarcinoma cells. Mol. Cell. Biol. 18:7095–7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kyriakis, J. M., P. Banerjee, E. Nikolakaki, T. Dai, E. A. Rubie, M. F. Ahmad, J. Avruch, and J. R. Woodgett. 1994. The stress-activated protein kinase subfamily of c-Jun kinases. Nature 369:156–160. [DOI] [PubMed] [Google Scholar]
- 45.Lenormand, P., C. Sardet, G. Pages, G. L’Allemain, A. Brunet, and J. Pouyssegur. 1993. Growth factors induce nuclear translocation of MAP kinases (p42mapk and p44mapk) but not of their activator MAP kinase kinase (p45mapkk) in fibroblasts. J. Cell Biol. 122:1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li, J. J., Z. Dong, M. I. Dawson, and N. H. Colburn. 1996. Inhibition of tumor promoter-induced transformation by retinoids that transrepress AP-1 without transactivating retinoic acid response element. Cancer Res. 56:483–489. [PubMed] [Google Scholar]
- 47.Li, J. J., J. S. Rhim, R. Schlegel, K. H. Vousden, and N. H. Colburn. 1998. Expression of dominant negative Jun inhibits elevated AP-1 and NF-kappaB transactivation and suppresses anchorage independent growth of HPV immortalized human keratinocytes. Oncogene 16:2711–2721. [DOI] [PubMed] [Google Scholar]
- 48.Li, J. J., C. Westergaard, P. Ghosh, and N. H. Colburn. 1997. Inhibitors of both nuclear factor-kappaB and activator protein-1 activation block the neoplastic transformation response. Cancer Res. 57:3569–3576. [PubMed] [Google Scholar]
- 49.Marais, R., J. Wynne, and R. Treisman. 1993. The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell 73:381–393. [DOI] [PubMed] [Google Scholar]
- 50.Marshall, M. S. 1995. Ras target proteins in eukaryotic cells. FASEB J. 9:1311–1318. [DOI] [PubMed] [Google Scholar]
- 51.Matsuo, K., J. M. Owens, M. Tonko, C. Elliott, T. J. Chambers, and E. F. Wagner. 2000. Fosl1 is a transcriptional target of c-Fos during osteoclast differentiation. Nat. Genet. 24:184–187. [DOI] [PubMed] [Google Scholar]
- 52.Mechta, F., D. Lallemand, C. M. Pfarr, and M. Yaniv. 1997. Transformation by ras modifies AP1 composition and activity. Oncogene 14:837–847. [DOI] [PubMed] [Google Scholar]
- 53.Murakami, M., M. Ui, and H. Iba. 1999. Fra-2-positive autoregulatory loop triggered by mitogen-activated protein kinase (MAPK) and Fra-2 phosphorylation sites by MAPK. Cell Growth Differ. 10:333–342. [PubMed] [Google Scholar]
- 54.Pearson, G., F. Robinson, T. Beers Gibson, B. E. Xu, M. Karandikar, K. Berman, and M. H. Cobb. 2001. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr. Rev. 22:153–183. [DOI] [PubMed] [Google Scholar]
- 55.Rao, V. N., and E. S. Reddy. 1994. elk-1 proteins interact with MAP kinases. Oncogene 9:1855–1860. [PubMed] [Google Scholar]
- 56.Rauscher, F. J. D., D. R. Cohen, T. Curran, T. J. Bos, P. K. Vogt, D. Bohmann, R. Tjian, and B. R. Franza, Jr. 1988. Fos-associated protein p39 is the product of the jun proto-oncogene. Science 240:1010–1016. [DOI] [PubMed] [Google Scholar]
- 57.Rauscher, F. J. D., L. C. Sambucetti, T. Curran, R. J. Distel, and B. M. Spiegelman. 1988. Common DNA binding site for Fos protein complexes and transcription factor AP-1. Cell 52:471–480. [DOI] [PubMed] [Google Scholar]
- 58.Robbins, D. J., E. Zhen, M. Cheng, S. Xu, C. A. Vanderbilt, D. Ebert, C. Garcia, A. Dang, and M. H. Cobb. 1993. Regulation and properties of extracellular signal-regulated protein kinases 1, 2, and 3. J. Am. Soc. Nephrol. 4:1104–1110. [DOI] [PubMed] [Google Scholar]
- 59.Rosenberger, S. F., J. S. Finch, A. Gupta, and G. T. Bowden. 1999. Extracellular signal-regulated kinase 1/2-mediated phosphorylation of JunD and FosB is required for okadaic acid-induced activator protein 1 activation. J. Biol. Chem. 274:1124–1130. [DOI] [PubMed] [Google Scholar]
- 60.Saez, E., S. E. Rutberg, E. Mueller, H. Oppenheim, J. Smoluk, S. H. Yuspa, and B. M. Spiegelman. 1995. c-fos is required for malignant progression of skin tumors. Cell 82:721–732. [DOI] [PubMed] [Google Scholar]
- 61.Swanson, K. D., L. K. Taylor, L. Haung, A. L. Burlingame, and G. E. Landreth. 1999. Transcription factor phosphorylation by pp90(rsk2): identification of Fos kinase and NGFI-B kinase I as pp90(rsk2). J. Biol. Chem. 274:3385–3395. [DOI] [PubMed] [Google Scholar]
- 62.Treinies, I., H. F. Paterson, S. Hooper, R. Wilson, and C. J. Marshall. 1999. Activated MEK stimulates expression of AP-1 components independently of phosphatidylinositol 3-kinase (PI3-kinase) but requires a PI3-kinase signal to stimulate DNA synthesis. Mol. Cell. Biol. 19:321–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Treisman, R. 1996. Regulation of transcription by MAP kinase cascades. Curr. Opin. Cell Biol. 8:205–215. [DOI] [PubMed] [Google Scholar]
- 64.Vallone, D., S. Battista, G. M. Pierantoni, M. Fedele, L. Casalino, M. Santoro, G. Viglietto, A. Fusco, and P. Verde. 1997. Neoplastic transformation of rat thyroid cells requires the junB and fra-1 gene induction which is dependent on the HMGI-C gene product. EMBO J. 16:5310–5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wagner, E. F. 2001. AP-1 articles. Oncogene 20:2333–2497. [Google Scholar]
- 66.Watts, R. G., C. Huang, M. R. Young, J. J. Li, Z. Dong, W. D. Pennie, and N. H. Colburn. 1998. Expression of dominant negative Erk2 inhibits AP-1 transactivation and neoplastic transformation. Oncogene 17:3493–3498. [DOI] [PubMed] [Google Scholar]
- 67.Whitmarsh, A. J., and R. J. Davis. 1996. Transcription factor AP-1 regulation by mitogen-activated protein kinase signal transduction pathways. J. Mol. Med. 74:589–607. [DOI] [PubMed] [Google Scholar]
- 68.Whitmarsh, A. J., P. Shore, A. D. Sharrocks, and R. J. Davis. 1995. Integration of MAP kinase signal transduction pathways at the serum response element. Science 269:403–407. [DOI] [PubMed] [Google Scholar]
- 69.Wisdom, R., and I. M. Verma. 1993. Transformation by Fos proteins requires a C-terminal transactivation domain. Mol. Cell. Biol. 13:7429–7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yoshioka, K., T. Deng, M. Cavigelli, and M. Karin. 1995. Antitumor promotion by phenolic antioxidants: inhibition of AP-1 activity through induction of Fra expression. Proc. Natl. Acad. Sci. USA 92:4972–4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Young, M. R., J. J. Li, M. Rincon, R. A. Flavell, B. K. Sathyanarayana, R. Hunziker, and N. Colburn. 1999. Transgenic mice demonstrate AP-1 (activator protein-1) transactivation is required for tumor promotion. Proc. Natl. Acad. Sci. USA 96:9827–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]