Abstract
The replication initiation pattern of the murine β-globin locus was analyzed in totipotent embryonic stem cells and in differentiated cell lines. Initiation events in the murine β-globin locus were detected in a region extending from the embryonic Ey gene to the adult βminor gene, unlike the restricted initiation observed in the human locus. Totipotent and differentiated cells exhibited similar initiation patterns. Deletion of the region between the adult globin genes did not prevent initiation in the remainder of the locus, suggesting that the potential to initiate DNA replication was not contained exclusively within the primary sequence of the deleted region. In addition, a deletion encompassing the six identified 5′ hypersensitive sites in the mouse locus control region had no effect on initiation from within the locus. As this deletion also did not affect the chromatin structure of the locus, we propose that the sequences determining both chromatin structure and replication initiation lie outside the hypersensitive sites removed by the deletion.
The initiation of DNA replication is a critical step in normal cell cycle progression, yet little is known about the biochemical pathways that control this event. The replicon model (24) proposed that cells regulate DNA replication through a bipartite mechanism involving an interaction between an initiator and a replicator. Initiators are trans-acting factors that initiate replication in response to cell cycle signals. Replicators are DNA sequences that enable initiation of DNA replication in cis, presumably by binding to initiators. Replication origins, or initiation regions (IR), are the DNA sequences within which replication synthesis begins. As initiation within an origin may depend on the presence of distant auxiliary sequences (3, 17, 25), a functional replicator may require multiple sequence elements separated within the genome. Understanding the mechanisms governing the initiation process requires the identification of origins, replicators, and the initiators with which they interact.
A small number of replication origins and even fewer replicators have been identified in mammalian cells. The search for these elements had been difficult due to the lack of reliable genetic methods that assess the initiation competence of specific sequences. Moreover, different biochemical strategies designed to identify IR in mammalian cells have generated controversy over whether discrete elements initiate replication (11). Nevertheless, several lines of evidence suggest that defined cis-acting elements are required for DNA replication in mammalian cells. First, replication initiates from identical regions in wild-type chromosomes and in rearranged loci resulting from naturally occurring processes, such as gene amplification (8, 22, 27). Second, an 8-kb deletion of the single biochemically defined IR in the human β-globin locus (Lepore deletion) prevents initiation within this locus (28), suggesting that the deleted sequence contained information essential for initiation. Third, replication initiates from identical regions in cosmid or yeast artificial chromosomes inserted into ectopic chromosomal regions and in the native chromosomal loci (7, 20, 27, 33). Finally, transfer of the 8-kb IR from the human β-globin locus to an ectopic site induces initiation of DNA replication from the new ectopic location (1). These data demonstrate that the potential to initiate DNA replication is encoded in the primary sequence of cis-acting elements.
The chromosomal environment plays an important role in determining replicator function. For example, as assayed in a naturally occurring deletion, initiation from the human β-globin replication origin in its native site requires a region located 50 kb upstream (3). This deletion spans 35 kb and includes the locus control region (LCR) and additional upstream sequences. This distal region also has a role in controlling globin gene expression and in establishing tissue-specific and developmentally regulated changes in chromatin structure within the locus (16). The involvement of distal elements in replication implies that replication initiation requires interactions between the biochemically defined origin and sequences far upstream. Recent data demonstrating the requirement for distant sequences for initiation in the Chinese hamster dihydrofolate reductase (DHFR) locus (25) imply that interactions between multiple dispersed elements may be commonly required for initiation in metazoan replicons. These observations suggest that the capacity to initiate replication from a particular origin might depend on its location relative to distant auxiliary elements. As replication initiation from the human β-globin origin introduced into an ectopic site does not require these distal upstream elements (1), it seems likely that auxiliary elements may substitute for each other.
The multipartite nature of replicators underlines the importance of mapping replication origins within their native loci to reveal how interactions between origins and auxiliary elements lead to initiation. In loco analyses are essential for assessing potential roles for gene expression and chromatin remodeling in the formation of prereplication complexes. The ability to modify complex loci at the native site using homologous recombination in totipotent embryonic stem (ES) cells makes the murine model particularly attractive for replication studies. Chromosomes modified in ES cells can be analyzed during development to assess the impact of replicator mutations on chromosome structure, gene expression, and replication initiation. However, it has not been determined whether DNA replication in ES cells initiates from fixed origins, as in somatic cells, or from apparently random sites, as in the rapidly dividing embryonic cells of other species (23). Whether the pattern of origin distribution is conserved throughout evolution has also not been evaluated. This information will be necessary for identifying conserved regulatory elements.
We addressed these issues by determining the replication initiation profile in the murine β-globin locus. Previous work with the human β-globin locus identified a single 8-kb IR. This region fulfills the genetic criteria for a replicator, as initiation is prevented when it is deleted from the native locus (28), and it is able to confer initiation competence to ectopic chromosomal sites (1). The data presented here reveal a different picture of replication initiation in the murine locus. Biochemical analyses using several methods, confirmed by deletion studies, showed that initiation occurred at multiple sites in both ES and differentiated somatic cells. Furthermore, a deletion of the known DNase I-hypersensitive sites in the LCR in ES cells did not alter the initiation pattern in the mouse locus. Since the LCR deletion-containing chromosome still exhibited developmentally programmed chromatin reorganization and transcriptional regulation (14), we infer that additional regulatory sequences outside the deleted region contribute to the regulation of both gene expression and replication initiation.
MATERIALS AND METHODS
Cells and culture conditions.
Murine erythroleukemia (MEL) cells were grown in Dulbecco modified Eagle’s medium supplemented with 10% heat-inactivated fetal calf serum and ℓ-glutamine (Gibco-BRL) as described previously (3). Strain AK7 ES cells were grown on gelatinized plates in Dulbecco modified Eagle’s medium supplemented with 15% dialyzed fetal calf serum, nonessential amino acids (BRL), 50 μM β-mercaptoethanol, and l-glutamine (BRL). Cells were supplemented with a 1:100 dilution of medium from Chinese hamster ovary cells producing leukemia-inhibiting factor as described previously (4).
Replication initiation assays. (i) Nascent-strand abundance analyses.
Nascent replicating strands were isolated based either on incorporation of bromodeoxyuridine (BrdU) (35, 38) or on their resistance to λ-exonuclease (5). BrdU substitution-containing nascent strands were isolated as described previously (1) and size fractionated on alkaline agarose gels. Specific PCR primers spanning the loci of interest were used to amplify gel fractions with DNA strands ranging in size from 0.6 to 8 kb. λ-exonuclease-resistant nascent strands were prepared from an exponentially growing population as originally described for replication initiation point mapping (5) and modified for mammalian cell nascent-strand analysis (29). Small strands were isolated after size fractionation on alkaline agarose gels as described previously (1). PCRs comparing various DNA preparations included three controls. First, the amount of DNA in each sample was normalized by using mitochondrial DNA primers. Second, primer pairs from the murine adenosine deaminase locus (ADA) (36) were used to assess the selectivity of the nascent-strand preparation (see below). For semiquantitative PCR, constant amounts of nascent strands were amplified with serial dilutions of PCR competitors as described previously (1) or subjected to real-time PCR analysis as described below, to estimate the abundance of specific sequences within a nascent-strand population. Finally, amplification of genomic DNA showed that all the primers used in the assay can efficiently amplify genomic DNA. Genomic DNA was prepared according to the protocol used to prepare nascent strands, with the omission of the exonuclease step. Such genomic DNA was used as a standard in the quantitative real-time PCR analyses.
(ii) Leading-strand direction analyses.
Leading strands were isolated as BrdU substitution-containing DNA strands from emetine-treated cells as described previously (2, 7, 20). BrdU substitution-containing DNA strands from cells that were not treated with emetine were used as total genomic DNA controls. Slot blots containing 3 μg of leading strands or control DNA were hybridized to single-stranded DNA probes to estimate the relative intensity of hybridization to leading strands versus total genomic DNA (bias ratio; see reference 2 for details). The hybridization results were quantitated with a Molecular Dynamics PhosphorImager and the ImageQuant software package. Preferential hybridization of a probe to leading strands suggests that the majority of the leading strands are complementary to the probe. The hybridization bias was calculated as follows: (leading-strand hybridization signal for the 5′-3′ probe/signal for the 3′-5′ probe)/(genomic standard hybridization signal for the 5′-3′ probe/signal for the 3′-5′ probe). Hybridization biases are considered significant if the values are consistently over 1.2 or below 0.8 (2).
PCR.
Nascent DNA strands were amplified with primers encompassing the murine globin locus. The primer sequences, listed from the 5′ to the 3′ end of the locus, are as follows. Primer pair 5′HS includes GGACTGTGCTCTCAGCTATTG (forward) and ACAGGCAAGGCAGCTTCCTC (reverse). Primer pair Ey includes TGGAGGTGAAGCCTTGGG (forward) and AGTCAGCACCTTCTTGCC (reverse). Primer pair βh1 includes CACTTGAGATCATCTCCAAGC (forward) and TAACCCCCAAGCCCAAGGATG (reverse). Primer pair βmajor includes CAGTAAGCCACAGATCCTATTG (forward) and CCCCATAGTGACTATTGACTGTG (reverse). Primer pair βminor includes CAAGTAGAAGCTGGGTGCTTGGAGA (forward) and TTAAAGGCAGTTATCACCTTTTTGCC (reverse).
The following two primers were used to verify the results obtained for the Ey and βmajor regions. Primer pair 5′Ey includes GGCATTTCTGTGTCCACAGC forward) and CAGGTCATGGGAGGGTCAG (reverse); primer pair 3′βmajor includes CCCTGGCTATTCTGCTCAACCTTC (forward) and CCCCTGTCACCCTGGCATAAAA (reverse). Primers from the murine mitochondrial genome, used to calibrate the amount of nascent strands in the preparations, were GACATCTGGTTCTTACTTCA (forward) and GTTTTTGGGGTTTGGCATTA (reverse). Primers A, B, and C from the murine ADA locus were exactly as described previously (36).
Amplification conditions were optimized for each primer pair on a genomic DNA template using the conditions specified in the Invitrogen PCR optimization kit.
For competitive PCR analysis, the PCRs included competitors synthesized as described previously (19) with the following modifications. Genomic DNA was used to amplify a PCR product using the forward and reverse primers. These products were cloned into a PCR2.TOPO vector (Invitrogen), and a 75-bp oligonucleotide was inserted into convenient restriction sites. The final competitors were thus similar in sequence to the amplified genomic sequences except for the insertion of 75 bp. This procedure allowed separation of the competitor from the genomic sequence on 1.5% agarose gels.
For real-time quantitative PCR, samples were prepared as described above and amplified in an ABI 7700 sequence detector. Nascent strands were amplified with a series of primer-probe sets for fluorescent resonance energy transfer (FRET)-based detection. This detection system relies on the 5′ exonuclease activity of Taq polymerase. A probe homologous to an internal sequence in the amplified product contains a 5′ fluorescent dye and a 3′ quencher. The probe does not emit detectable fluorescence due to FRET from the dye to the quencher. During PCR, probes that specifically hybridize to the amplification product are digested by Taq, releasing the dye from its proximity to the quencher and allowing detection of the fluorescence. The quantity of the template DNA directly correlates with the earliest cycle in which significant fluorescence can be detected. The abundance of the sequence was calculated based on a comparison with a standard curve of genomic DNA containing a single copy of the target per haploid genome. We designed a series of FRET-based probes, listed below, to quantify the abundance of specific targets in nascent DNA. In addition, we have used the primer pairs designed for the gel-based competitive PCR to scan for initiation sites within the locus in the presence of SYBR Green. SYBR Green detection was followed by agarose gel electrophoresis to ensure that a single fragment was amplified during the reaction. The primer pairs βmajor and βminor, listed above, could not amplify products using the manufacturer’s recommended amplification protocols. These regions were analyzed using FRET-based primer-probe sets.
FRET-based fluorescent primer-probe combinations were as follows. Primer set M5′HS included TCATCAGTCTTGTGTGAAAGTGCTT (forward), AGTAGAGTGGAGAATCAAAAACACATTT (reverse), and FAM-ATCTAGTGAACCACATTAACTGGCCCTGGC-TAMRA (probe) (FAM is 6-carboxy-fluorescein and TAMRA is 6-carboxy-tetramethyrhodamine). Primer set m5′β1 included TGGATTGTTCCTCATGGCTTT (forward), GTGCCACCCCTCATCAGG (reverse), and FAM-CGGTCTGCTTTCTTATCGCATCATGAAGC-TAMRA (probe). Primer set mβ1 included CCAGCCTCAGTGAGCTCCA(forward), CCCATCAGACTCACCCTGAAG (reverse), and FAM-TGTGACAAGCTGCATGTGGATCCTGA-TAMRA (probe). Primer set m5′β2 included TGATTGGTGCCCAGTTCAGTT (forward), TGGGTCTCTGTGTCTGCTCCT (reverse), and FAM-TTGGAGAGTCATCATCCAGCAACGGA-TAMRA (probe). Primer set m3′β included AGGAGTTCTTGAGCCCTGGTC (forward), CCTGTTCTCTAGTTCAAGTTGTGAGTATG (reverse), and FAM-CCAGCCTCTCCCATTAACAATGATAATGAAACTT-TAMRA (probe).
Genomic DNA preparations ranging in quantity from 10 pg to 10 ng were used as standards. Assays were performed in duplicate using two different concentrations of DNA per primer pair. Similar results were obtained from two independent preparations of λ-exonuclease-resistant nascent strands.
Replication timing analysis.
Approximately 107 log-phase cells were labeled with 50 μM BrdU (Sigma) for 90 min. Cells were harvested, washed, fixed in phosphate-buffered saline-70% ethanol, and stored at −20°C overnight. For fluorescence-activated cell sorting, fixed cells were rehydrated, permeabilized with 0.1% NP-40, and stained with propidium iodide (50 μg/ml) in the presence of RNase A. Using a Becton Dickinson Vantage fluorescence-activated cell sorter, cells were separated into six compartments of the cell cycle based on propidium iodide staining intensity (DNA content), corresponding roughly to G1, S1 to S4, and G2. Equal numbers of cells from each compartment (6,000 to 10,000, depending on the experiment) were sorted into tubes containing a sodium dodecyl sulfate-protease K lysis buffer. DNA was then purified, sonicated to a size range of 0.5 to 2 kb, and denatured by boiling. Immunoprecipitation of BrdU-DNA was performed with a mouse monoclonal primary antibody (Becton Dickinson) and a rabbit anti-mouse immunoglobulin G secondary antibody (Sigma). Pellets were washed thoroughly, and DNA was purified following sodium dodecyl sulfate-protease K digestion. From each cell cycle fraction, DNA equivalent to 500 sorted cells was used as a template for standard 23-cycle PCRs. The products were immobilized on nylon membranes and probed with radiolabeled fragments amplified from genomic DNA with the primers used to amplify the BrdU-DNA.
Two primers from the murine globin locus (5′Ey and 5′βmajor) were used to determine the replication timing of the murine globin locus. In addition, the following loci were examined as markers for early and late replication: for murine pancreatic amylase 2.1y (accession number M16540), amylase-1 (5′-AGCACTGAGGATTCAGTCTATG) and Amyl −2 (5′-CCCGTACAAGGAGAATTACAAC), and for murine cyclin D1 (accession number S78355), mcyD1-1 (5′-CAGACCTCTTAACCTTATAGATG) and mcyD1-2 (5′-ACACTGCCTCCAGCTAGCTG).
RESULTS
Initiation of DNA replication within the murine β-globin locus.
We mapped the replication initiation profile within the murine β-globin locus by using three different biochemical approaches that rely on nonoverlapping assumptions. The first two methods identify specific sequences in nascent replicated DNA strands. Both methods isolate nascent replicated DNA from exponentially growing cell populations without drug treatment to reveal where replication initiates. The first protocol uses short, BrdU substitution-containing DNA strands obtained after pulse-labeling with this thymidine analog (35). The second protocol relies on resistance to λ-exonuclease to isolate short DNA strands that contain an RNA primer (5, 18, 29). Sequences that are adjacent to or within replication origins are expected to be present in the nascent-strand preparations, while sequences that are located far from replication origins should not be found in these preparations. The third method measures the direction of the majority of replication forks traversing a genomic region and identifies replication origins as sites in which the leading-strand templates diverge from each other. This is accomplished by the isolation of denatured, sonicated, BrdU substitution-containing DNA strands under conditions that allow the synthesis of leading strands but not Okazaki fragments (7). If a genomic locus contains a single replication origin, all three methods should yield identical results. However, if initiation occurs at several different sites within a specific locus, the leading-strand analysis may identify the center of the region within which the most frequent initiation events occur, while the nascent-strand abundance methods should detect additional lower-frequency initiation events (38).
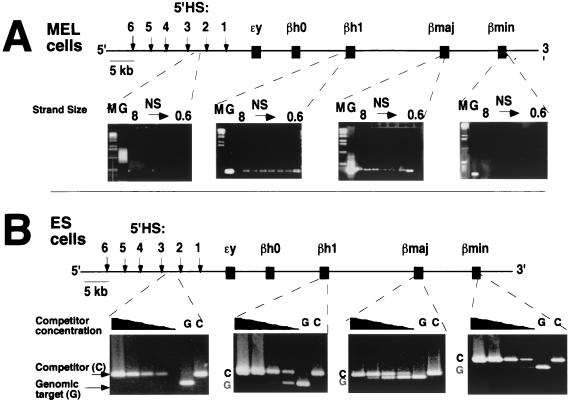
We first used the BrdU-labeled nascent-strand assay to analyze the replication pattern in the murine β-globin locus in MEL and ES cells. In the experiment illustrated in Fig. 1A, short nascent strands were size fractionated on an agarose gel and each fraction was amplified with primers spanning the locus. We observed that primers in the region encompassing the βh1 and βmajor globin genes yielded amplification products in all nascent-strand fractions. The presence of primer sequences in the lowest-molecular-size fraction (0.6 to 0.8 kb) indicates close proximity of the primers to the site of replication initiation. By contrast, primers 3′ to the βminor gene and in the murine 5′ hypersensitive site region (5′HS) did not yield amplification products from the low-molecular-weight fractions. Since primer pairs spaced 10 kb apart detected low-molecular-weight nascent DNA, these results are consistent with DNA replication initiating from multiple sites within the murine locus.
FIG. 1.
Analysis of BrdU-labeled-nascent-strand abundance within the murine β-globin locus. The abundance of origin-specific DNA sequences in BrdU substitution-containing nascent strands from differentiated and totipotent cells was determined. (A) Nascent strands derived from MEL cells, a differentiated cell line in the hematopoietic lineage, were size fractionated on an agarose gel. Each size fraction was used as a template for amplification with the 5′HS, βh1, βmajor, and βminor primers (see Materials and Methods for sequences). PCR with the βh1 and βmajor primers yielded products from both large and small nascent strands, indicating that these primers are near sites of replication initiation. In contrast, primers within 5′HS and βminor failed to yield products from small nascent strands, as expected if they are not near an IR. (B) Nascent strands from totipotent ES cells. Nascent strands were isolated from gel fractions containing fragments ranging from 0.8 to 1.5 kb. These strands were amplified with the 5′HS, βh1, βmajor, and βminor primers in the presence of variable quantities of competitor templates (1, 0.1, 0.01, and 0.001 pg per reaction, from left to right in each panel) to control for variations in amplification efficiency. PCR primers from the βh1 and βmajor regions amplified products from small nascent strands, whereas primers from the 5′HS and the βminor regions did not, indicating that initiation in ES cells occurred in the region similar to the IR in MEL cells. M, molecular weight markers; G, total genomic DNA; C, competitor; NS, nascent strands.
We next determined whether the differentiation status of the cells affected the replication initiation pattern by analyzing the composition of nascent strands in totipotent ES cells. These nascent DNA strands were amplified in the presence of specific competitor templates that yielded products slightly larger than the product expected from each endogenous template (19). Amplification using a constant amount of nascent-strand template in the presence of variable quantities of competitor yields an estimate of the relative abundance of nascent strands in each preparation. As shown in Fig. 1B, primers from the βh1 and βmajor gene region were able to amplify products from nascent strands, whereas primers from the 5′HS and in the βminor region did not. Thus, the 5′HS and the βminor gene delineate the boundaries of the IR. These results revealed that, similar to the replication pattern observed in MEL cells, replication in ES cells initiated within the protein-coding region of the murine β-globin locus.
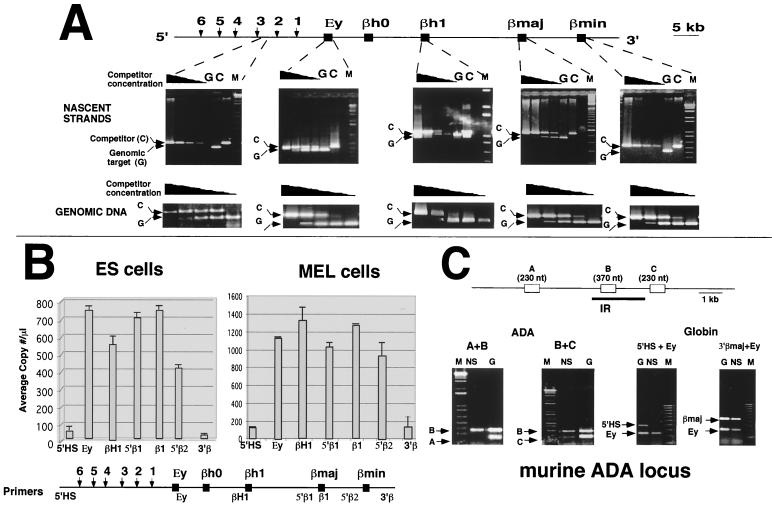
We have confirmed and extended these results by using a second strategy that employed λ-exonuclease to digest all DNA except strands primed by RNA (5). This strategy should yield only newly synthesized DNA strands. Size fractionation was used to separate Okazaki fragments from longer, origin-proximal nascent strands. Figure 2A shows the results of a nascent-strand abundance assay using λ-exonuclease resistant strands from ES cells. PCR primers from the 5′βmajor, βh1, and Ey globin regions amplified nascent DNA. No products were detected with primer pairs situated within the 5′HS or the βminor regions. By contrast, all the primers amplified products from genomic DNA, suggesting that the differences in amplification reflect differences in abundance of the amplified sequences in nascent strands rather than differences in replication efficiency. These results were consistent with the data from analyses employing BrdU substitution-containing nascent strands (Fig. 1) and suggested that replication initiated at multiple sites within the murine globin locus. Similar results were obtained with MEL cells (not shown), confirming that the differentiation status did not alter the initiation profile in the murine β-globin locus.
FIG. 2.
Analysis of exonuclease-resistant-nascent-strand abundance within the murine β-globin locus. The abundance of origin-specific DNA sequences in λ-exonuclease-resistant nascent strands from ES cells was determined by PCR. (A) DNA strands isolated from a gel fraction containing strands ranging from 1 to 1.8 kb were amplified with the primer pairs 5′HS, Ey, βh1, βmajor, and βminor (see Materials and Methods for primer locations and sequences). Amplification reactions were performed in the presence of a series of 10-fold dilutions of PCR competitors as outlined in the legend to Fig. 1B. Ey, βh1, and βmajor primer pairs could amplify products from nascent strands, while 5′HS and βminor primers could not. All primers amplified product from genomic DNA (lower panels). (B) Short, λ-exonuclease-resistant, newly replicated DNA strands were prepared as described above and amplified by real-time quantitative PCR using the indicated primer pairs. Detection of amplification with the 5′HS, 5′β1, β1, 5′β2, and 3′β primers sets was based on FAM fluorescence using FRET-based probing (ABI TaqMan chemistry). Detection of amplification with the Ey and βh1 primers was based on SYBR Green fluorescence followed by gel electrophoresis to verify single-fragment amplification. Data are averages from four measurements for each primer pair from a single preparation of nascent strands. Similar results were obtained from two independent preparations of λ-exonuclease-resistant nascent strands for each cell line. (C) Verification of nascent-strand selectivity by examination of nascent strands abundance in the previously characterized murine ADA locus. λ-exonuclease-resistant DNA strands ranging from 0.6 to 8 kb from the preparation used for panel A were amplified with primer pairs A, B, and C from the murine ADA locus (36). Duplex PCR was performed either with pairs A and B or with pairs B and C. The origin-proximal sequences from the ADA locus (primer pair B) were preferentially amplified, demonstrating that the nascent-strand preparation was enriched for origin-proximal sequences. Although the original analysis (36) suggested that primer pair C is closer to the origin that primer pair B, both primers were able to amplify nascent strands from the smallest fraction used in this analysis at equal frequencies. M, molecular weight markers; G, total genomic DNA; C, competitor; NS, nascent strands.
The extent of amplification of newly replicated λ-exonuclease-resistant DNA was also measured using real-time quantitative PCR (Fig. 2B). This analysis used specific primer-probe combinations that allowed FRET-based detection of real-time amplification. To allow more complete coverage of the locus, we used primers developed for gel-based analysis and followed the amplification by SYBR Green detection (see Materials and Methods). The real-time quantitative PCR data (Fig. 2B) demonstrated that nascent strands are amplified by primers from the region from the Ey gene to the interval between the two adult β-globin genes, whereas primers from outside this region failed to amplify products in nascent strands. These analyses confirmed that replication of the murine β-globin locus initiated in the structural gene region in ES and MEL cells.
To verify whether the λ-exonuclease resistant short strands detected above represented bona fide initiation events and did not result from nonspecific degradation of longer strands, we analyzed the nascent-strand synthesis pattern at another, previously characterized murine replication origin. To that end, we used the preparations described above as substrates for amplification with primers located at various distances from an origin near the murine ADA gene (36). If our DNA preparations consisted mainly of newly replicated DNA, we expected that ADA primers near the origin should amplify products in short nascent-strand preparations while primers located farther from the ADA IR should exhibit little or no amplification from the same substrate (36). On the other hand, if the nascent-strand preparations contained primarily nonorigin sequences resulting from nonspecific DNA breakage, all primers should exhibit similar amplification efficiencies. Figure 2C shows that primers from origin-proximal sequences in the ADA locus (primer pairs B and C) amplified nascent strands with a higher efficiency than origin-distal primers (primer pair A), indicating that the nascent-strand preparation contained primarily origin-proximal sequences. These controls support the interpretation that replication initiated in the structural gene region of the β-globin locus but not in the 5′HS or the downstream regions. Similar results were obtained with MEL cells (data not shown), confirming that a similar initiation pattern was maintained in differentiated and totipotent cells.
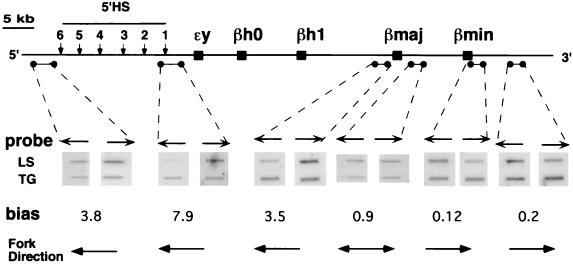
We used the leading-strand polarity assay (20) to investigate whether the multiple initiation sites could be detected as multiple conversion points between leading and lagging strands. Interestingly, hybridization with single-stranded probes within the murine globin locus showed that the majority of leading strands traveled towards the LCR in the region 5′ to the βmajor gene (leftward in Fig. 3), while leading strands 3′ of the βminor gene traveled in the opposite direction (rightward in Fig. 3). There was no significant bias in leading-strand signals in the region just downstream of the βmajor gene (0.9 in the example in Fig. 3), as was expected if this region contained an origin of bidirectional replication. Similar results were obtained with leading strands from MEL cells (data not shown). These data were similar to those obtained for the replication profile in the human β-globin locus, where replication initiates from a short IR that acts as an origin of bidirectional replication. The similar results obtained in ES and MEL cells confirmed that the initiation pattern in the murine β-globin was similar in somatic and ES cells.
FIG. 3.
Replication fork direction within the murine β-globin locus. Replication fork direction was determined by leading-strand analysis as described previously (3). BrdU substitution-containing leading strands were isolated from emetine-treated ES cells. Hybridization of these strands with single-stranded probes, normalized against the hybridization of total genomic DNA to the same probes, yielded a bias which indicated the direction of replication forks at a specific site through the murine globin locus. Hybridization with 5′HS, 5′Ey, and 5′βmajor probes, located 5′ to the βmajor gene, showed a strong bias towards strands traveling towards the 5′ end of the locus. Hybridization to 3′βminor and 3′HS probes, located upstream of the βminor gene, showed a strong bias towards strands traveling towards the 3′ end of the locus. Hybridization with the 3′βmajor probe, located in the interval between the two adult genes, exhibited no reproducible hybridization bias (biases between 0.8 and 1.2 are considered insignificant; see Materials and Methods for calculation). These results were confirmed with additional probes and leading-strand preparations. Arrows above the panels show the direction of the probes used in the analyses, while arrows below the panels show the direction of leading-strand progression. Preferential hybridization of a probe to leading strands suggests that the majority of the leading strands are complementary to the probe. LS, leading strands; TG, total genomic DNA.
Deletion of the region between the two murine adult genes still allows initiation of DNA replication in the murine β-globin locus.
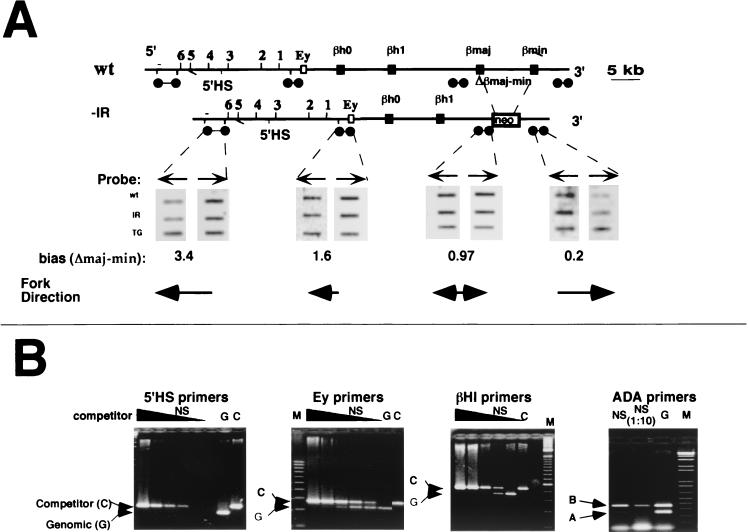
We next determined whether a deletion of the βmajor region perturbed the replication initiation pattern in the remainder of the locus. Figure 4 illustrates the replication pattern in a chromosome from which the region between the adult βmajor and βminor genes was removed by homologous recombination in ES cells (9). As in the wild-type cells, leading-strand polarity analysis (Fig. 4A) showed that replication forks moved in the 3′-to-5′ direction upstream of the Ey gene, while downstream of βminor they moved in the opposite direction. Immediately upstream of βmajor, there was no significant bias, suggesting that this probe lies in the vicinity of an IR. These data implied that the majority of initiation events occurred in the region immediately 5′ to the initiation site observed in the cells that did not harbor the deletion. Analysis of λ-exonuclease-resistant nascent strands (Fig. 4B) showed that initiation events occurred within the undeleted structural gene region but did not extend through the boundaries established in the undeleted region (i.e., 5′ of Ey and 3′ of βminor). Quantitative real-time PCR analysis confirmed that initiation occurred within the region delineated by the competitive PCR data (not shown). Thus, deletion of a region encompassing at least one initiation site still allowed initiation of DNA replication to occur in the remainder of the region, and the majority of the loci initiated DNA replication immediately 5′ to the initiation site observed in unmodified loci.
FIG. 4.
Initiation of DNA replication in the absence of the region between the two adult genes. The replication pattern in the β-globin locus was analyzed in an ES cell line homozygous for a deletion of the region encompassing the adult globin genes. These cells harbor an active neomycin resistance gene replacing the deleted region. (A) Leading-strand analysis, as outlined in the legend to Fig. 3, showed that replication forks far 5′ to the deleted region traveled towards the 5′ end of the locus, while replication forks 3′ to the βminor gene traveled towards the 3′ end of the locus. We did not observe a significant hybridization bias in the 5′ region proximal to the deletion (5′ to the deleted βmajor gene). Probes used were identical to the probes used in Fig. 3 except for the omission of probes 3′βmajor and 3′βminor, which hybridize with sequences deleted in the cell line lacking βmajor and βminor. The hybridization biases and fork directions for the IR deletion-containing locus are indicated below the panels. The biases obtained for the wild-type locus in the panels were (5′ to 3′) 3.7, 2.4, and 0.17. WT, leading strands from wild-type ES cells; −IR, leading strands from ES cells carrying the deletion; TG, total genomic DNA from ES cells carrying the deletion. (B) Nascent-strand abundance analysis in λ-exonuclease-resistant nascent strands from an ES line lacking βmajor and βminor. The relative abundance of the globin sequences in nascent strands was measured by performing the amplification reaction in the presence of a series of 10-fold dilutions of competitor DNA as outlined in the legend to Fig. 1B. Primers from the Ey and βh1 regions amplified DNA sequences from λ-exonuclease-resistant DNA strands, suggesting that DNA replication initiated in this region, while primers from the 5′HS region did not. As a control for the selectivity of the isolation procedure, we verified that origin-proximal primers from the murine ADA region were preferentially amplified from the λ-exonuclease-resistant-nascent-strand preparation, as outlined in the legend to Fig. 2B. Similar results were obtained with other nascent-strand preparations and additional primers and with BrdU substitution-containing nascent strands (not shown). M, molecular weight markers; NS, nascent strands; G, genomic DNA; C, competitor only.
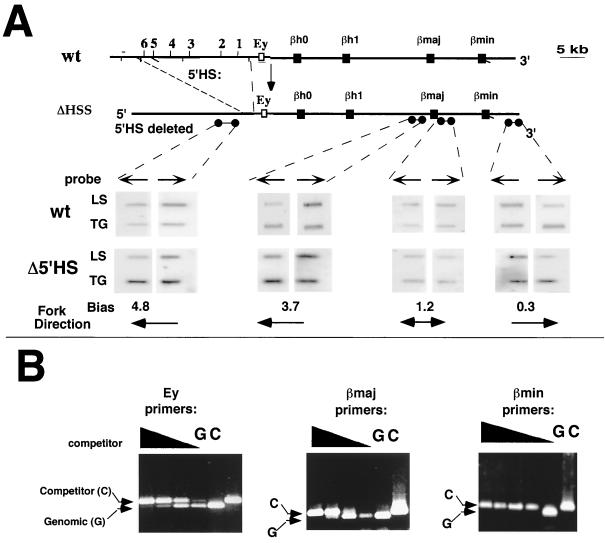
Deletion of the 5′HSs does not alter the replication profile in the murine β-globin locus.
Initiation of DNA replication in the human β-globin locus depends on the presence of sequences within the 35 kb removed in the Hispanic deletion (3). This deletion includes the LCR, a 5′ genetic element containing five DNase-hypersensitive regions located 50 kb upstream of the IR, as well as sequences upstream of the LCR. Chromosomes harboring the Hispanic deletion adopt a closed chromatin conformation in erythroid cells, do not express globin, and replicate the locus late in S phase (15). The murine globin locus has six DNase-hypersensitive sites located 5′ to the embryonic globin gene, Ey (14), but a deletion of these sequences does not alter chromatin conformation and influences the magnitude but not the developmental control of gene expression. We analyzed replication in ES cells harboring a homozygous deletion of all six hypersensitive sites to determine the effect of the deletion on DNA replication (Fig. 5). The leading-strand analysis in Fig. 5A revealed no alterations in replication fork patterns following the deletion of the 5′HSs. Analyses of exonuclease-resistant nascent strands (Fig. 5B) and BrdU substitution-containing nascent strands (data not shown) demonstrated that initiation occurred within the globin locus in a manner similar to that observed in the parental locus.
FIG. 5.
Initiation of DNA replication in the absence of the 5′HSs. The replication pattern in the β-globin locus was analyzed in an ES cell strain that harbors a deletion of all the six hypersensitive sites located 5′ to the Ey gene (ΔHSS) (14). (A) Leading-strand analysis, as outlined in the legend to Fig. 3. Probes used were 5′HS, 5′βmajor, 3′βmajor, and 3′HS. Replication forks 5′ to the βmajor gene travel towards the 5′ end of the locus, while replication forks 3′ to the βminor gene travel towards the 3′ end of the locus. We did not observe a significant hybridization bias in the region between the two adult genes. The bias ratios were similar in wild-type and 5′HS deletion chromosomes. (B) Analysis of nascent-strand abundance in the 5′HS deletion-containing cells. The relative abundance of the globin sequences in nascent strands was measured by performing the amplification reaction in the presence of a series of 10-fold dilutions of competitor DNA as outlined in the legend to Fig. 1B. Primers from the Ey, βmajor, and βminor regions were used to amplify DNA sequences from λ-exonuclease-resistant DNA strands ranging from 1 to 1.8 kb. Primers from the Ey and βmajor regions amplified products in the nascent-strand preparations, while primers from the βminor region did not. Similar results were obtained with real-time quantitative PCR, with other primers throughout the locus, and with BrdU substitution-containing nascent strands (not shown), suggesting that the deletion of the 5′HSs did not alter the initiation pattern within the murine β-globin locus. G, genomic DNA control; C, competitor only.
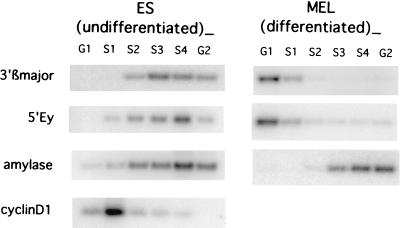
Replication timing of the murine β-globin locus during S phase.
In human cells, the wild-type globin locus replicates early in erythroid cells, in which globin is expressed, and late in nonerythroid cells, in which the globin locus is silent (13). Replication initiates from the same origin in both cell types (28). As the replication initiation pattern differed between the human and murine cells, we asked whether timing control was also different. We determined the replication timing of the globin locus in both the erythroid differentiated MEL cells and the totipotent ES cells (Fig. 6) to learn if it is affected by the differentiation status of the cells. Asynchronous cells were labeled briefly with BrdU, fixed, and then fractionated by fluorescence-activated cell sorting according to DNA content. BrdU-labeled DNA from cells at various stages of the cell cycle was amplified with primers from the murine β-globin locus as well as from the cyclin D1 and amylase loci (21). The cyclin D1 and amylase loci served as markers for early and late replication, respectively. The sorting windows were set to include early-S-phase cells in the G1 fraction. We found that sequences within the murine β-globin locus replicated in early S phase in erythroid cells and in late S phase in ES cells. Thus, the replication timing patterns in the human and mouse globin loci were consistent and reflected the pattern of globin gene expression (14). This characteristic is, therefore, independent of the pattern of replication initiation.
FIG. 6.
Timing of β-globin locus DNA replication in undifferentiated (ES) and erythroid (MEL) cells. For each cell line, cells were labeled with BrdU for 90 min and then fractionated with a fluorescence-activated cell sorter to cell cycle compartments corresponding to G1, S1 to S4, and G2 (the G1 and G2 fractions contained some early and late S-phase cells, respectively). BrdU substitution-containing DNA from the cell cycle fractions was isolated by immunoprecipitations as described previously (21). PCR primers corresponding to globin, cyclin D1, and amylase genes were used to amplify specific sequences in the BrdU substitution-containing DNA preparation. Cyclin D1 and amylase were used as early- and late-replicating controls, respectively. In ES cells, sequences within the β-globin locus (3′β major and 5′Ey) replicate late in the cell cycle, at roughly the same time as the late-replicating amylase locus and much later than the early-replicating cyclin D1 locus. In contrast, the β-globin locus in MEL cells displays early replication timing relative to the late-replicating amylase locus.
DISCUSSION
We used three different methods to analyze the initiation pattern in the murine β-globin locus. In the human locus, the results of both directional and nascent-strand analyses indicate initiation from a single origin, while in the murine locus, results from different methods yield apparently conflicting interpretations. The leading-strand analysis showed no bias in leading-strand signals in the region just downstream of the βmajor gene, as was expected if this region contained an origin of bidirectional replication. The observed bias in the region 5′ and 3′ to the interval between the adult genes suggested that the majority of the replication forks traversed this region in the upstream and downstream directions, respectively. These results suggested that the murine β-globin locus replicated by DNA strands originating at the interval between the adult genes. On the other hand, both nascent-strand assays allowed amplification of short, newly replicated DNA by primers throughout the gene coding region, suggesting that initiation events occurred within an extended region encompassing all the structural genes. These results were similar to those obtained by two-dimensional gel analyses, where replication forks traveling in two directions were observed throughout the structural gene region in the murine β-globin locus (C. Schildkraut, personal communication). The nascent-strand analyses as well as the two-dimensional gel analyses confirmed that the 5′HS and the 3′βminor regions replicated passively by replication forks originating at the structural gene region. The presence of IR 5′ to the adult genes was confirmed by removal of the interval between the two adult genes. This deletion still allowed initiation from the origins located 5′ to the deletion region, suggesting that these sequences contained genetic information required for initiation.
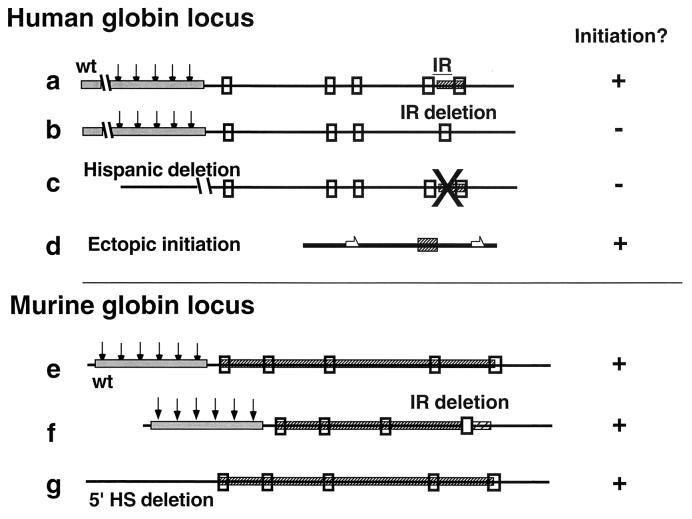
The combined results of our genetic and biochemical mapping in the human and murine β-globin loci (Fig. 7) suggest that although these two orthologous loci exhibit a highly conserved order of structural genes and regulatory elements, the replication patterns within these loci are not identical. In the human locus, initiation is confined to a region between the two adult globin genes. This region was identified as an origin of bidirectional replication by leading-strand analyses (3, 28), nascent-strand abundance assays (3, 10), and in situ hybridization (28). A report of replication initiation 5′ of the human adult globin genes (26) was not consistent with these observations or with a recent study using the same method, a λ-exonuclease-resistant-nascent-strand abundance assay (10). This apparent discrepancy may be due to methodological differences. The IR in the human β-globin locus fulfills the genetic criteria for a replicator, because its removal by the naturally occurring Lepore hemoglobin deletion results in lack of replication initiation within the locus (28) and its insertion in an ectopic site confers replication initiation (1). This region is recognized as an IR by the murine replication machinery, as human chromosomes transferred to murine cells in murine-human somatic cell hybrids conserve the initiation pattern observed in the human chromosome (3). This ability to recognize replicators across species extends to chicken-human somatic cell hybrids (3). By contrast, the data presented here show that replication in the murine locus initiates from multiple sites, and the removal of the interval between the adult genes did not prevent initiation from the locus. While we did not enumerate the replicators in the murine locus, these analyses demonstrated that the genetic information required for initiation was not confined to the region between the adult murine β-like globin genes. It should be noted, however, that unlike the natural Lepore deletion, which fuses the two structural adult genes, deletion of the region between the two adult murine genes replaced the deleted sequence with an antibiotic marker. It is therefore formally possible that the artificial deletion did not recapitulate the Lepore deletion because the neomycin gene or the plasmid sequences that replaced the native sequences in the deletion mutant contained replicator sequences. However, gene transfer experiments using vectors containing the neomycin expression cassette did not detect replicator activity in the neomycin gene or in the associated control elements (25; M. I. Aladjem, J. L. Kolman, and G. M. Wahl, unpublished data).
FIG. 7.
Comparison of the replication features in murine versus human β-globin loci. In the human locus, replication from unaltered loci (a) initiates at the IR (hatched box), located between the two adult globin genes (genes are depicted as small empty boxes). A deletion of the IR (b) shows no initiation within the locus. The dependence of initiation on 5′ sequences (gray box) is manifested in the Hispanic thalassemia chromosome (c), where the presence of the IR is not sufficient to direct initiation in the absence of the LCR. The IR fulfills the genetic requirements for a replicator because it is capable of directing initiation at ectopic loci (d). In the murine locus, replication initiates from an extended origin or from multiple initiation points within an extended region (e). Deletion of the sequences between the two adult genes does not abolish initiation in the locus (f), and the initiation pattern does not change when the 5′HS region is deleted (g).
The replication pattern in the murine β-globin locus strongly resembles the pattern observed in the well-characterized Chinese hamster DHFR locus. In both loci, nascent-strand abundance assays and methods directed at isolation of replication intermediates demonstrate that replication initiates at multiple origins over an extended region of 50 kb, while directional methods identify preferred origins of bidirectional replication (see reference 11 for a review). These apparently conflicting data may be reconciled by a model (38) suggesting that leading-strand analyses point towards the center of the most frequent IR but do not delineate the boundaries of the IR. Similar to our observations with the murine β-globin locus, deletion of the most abundant DHFR replication origin, as defined by directional methods, still allows initiation within the locus (25). These observations suggest that multiple initiation sites can function in both the DHFR and the murine globin loci and that in both loci the replication origin detected by the leading-strand method is not the sole origin.
The cellular factors determining which potential initiation site is used in a particular cell remain to be determined. In yeast, origin recognition complexes (ORC) were shown to bind potential initiation sites regardless of whether these sites actively initiate DNA replication or replicated passively from adjacent origins that initiated earlier during S phase. Removal of the earlier replicating origins allows replication from the “dormant” origins (34). These data suggest that ORC binding correlates with the potential to act as replication origins, while other factors determine which ORC binding site will fulfill the potential and initiate DNA replication. Our data suggest that a similar dynamic situation occurs in mammalian cells. Murine cells, which initiate DNA replication from an expanded region in the murine globin locus, are capable of initiating DNA replication from the relatively confined IR in the human globin locus (3). These observations imply that the determinants of differential origin usage between the murine and human loci reside in the DNA sequence, not in the cellular environment. On the other hand, replication timing patterns are conserved and correlate with chromatin decondensation in both species. The conservation of replication timing patterns, but not replication sites, suggests that specific DNA sequences determine potential initiation sites, while the cellular environment, and chromatin organization in particular, dictates whether this potential will translate into actual initiation. Studies with isolated nuclei transferred to Xenopus egg extracts suggest that the timing of replication and the initiation sites are both determined afresh during the G1 phase in each cell cycle; replication timing domains are established before replication initiation sites (12). It is plausible to assume that the DNA sequence sets the stage for potential replication initiation sites while epigenetic factors dictate the temporal order of replication initiation.
In the human β-globin locus, deletion of the 5′HSs and upstream sequences by Hispanic thalassemia prevents replication initiation from the normal origin (3). The involvement of distal sequences in initiation was also demonstrated in the Chinese hamster DHFR locus (25). These observations suggest that proper initiation of DNA replication requires interaction between the origin and distant sequences. The human LCR contains five DNase-hypersensitive sites, whose presence contributes to gene expression but is not required for establishing or maintaining an open chromatin conformation or for initiation of DNA replication (10). Similarly, the murine 5′HS deletion (5′HSs 1 to 6) did not recapitulate the Hispanic thalassemia phenotype, suggesting that the 5′HS region is not required to establish an open chromatin structure and developmentally regulated globin gene expression (14). The results presented here show that this deletion still allowed initiation of DNA replication from within the murine globin locus. These data are consistent with the observations with the human globin locus (3), demonstrating that the ability to initiate DNA replication at the normal origin correlates with the potential to decondense chromatin in the erythroid environment, not with the expression status of individual genes within the locus. It is possible that the murine IR does not require interaction at a distance with a 5′ sequence. However, it is also possible that the deletion of the hypersensitive sites left intact the crucial 5′ elements required for initiation of DNA replication. The 5′ sequences deleted in Hispanic thalassemia may serve to insulate the globin locus from an inactivating effect of the odorant receptor array in which both human and murine loci are embedded (6). This insulation may be similar to that observed in Drosophila chorion gene amplification, where initiation capacity can be protected from position effects by transcriptional insulators (31). These observations suggest that initiation of DNA replication in metazoans may require specific alterations in chromatin structure, the nature of which has yet to be determined.
Our results reveal that undifferentiated wild-type murine ES cells initiate DNA replication within a region similar to the IR observed in differentiated somatic cells. We previously found that undifferentiated ES cells appear to oscillate between S phase and M phase, like embryonic cells in other species, and that ES cells do not activate p53-mediated controls that arrest the cell cycle in response to DNA damage (4). The p53-mediated checkpoint responses become functional upon differentiation of ES cells. Taken with the results presented here, the data indicate that the establishment of origin usage in the murine system is independent of, and occurs prior to, the establishment of the prolonged G1 phase and some of the cell cycle controls that typify differentiated somatic cells.
Although DNA replication initiates within a similar region in ES and somatic cells, evidence from other metazoans suggests that these sites do change in early development. DNA replication initiates without regard to specific sequences in the early cell divisions in Xenopus and Drosophila (23). Before the midblastula transition in Xenopus, interphase chromosomes are organized in independent units called karyomeres (30) within which chromosomes replicate autonomously. Replication initiation becomes restricted to preferred sites at the midblastula transition, which coincides with the onset of transcription and chromatin (23). Murine embryos undergo chromatin remodeling and initiation of transcription before the four-cell stage, accompanied by changes in histone synthesis (32, 37). ES cells are collected from the inner cell mass of the murine blastula after chromatin remodeling has occurred and, as shown here, initiate replication at the region used in somatic cells. While all previous studies concerning replication in mammalian cells involved permanent lines derived from tumor cells, the present study demonstrates the usefulness of ES cells as a source of normal cells for such analyses. The facility with which ES cells can be manipulated genetically and the ability to study the effects of such changes on the replication program during differentiation suggest the general utility of this system for future replication studies.
Acknowledgments
We gratefully acknowledge D. J. Ciavatta and T. M. Townes for the ES cells containing the adult β-globin deletion, C. Schildkraut and N. Lam for sharing data prior to publication and helpful comments, and Kurt Kohn, Yves Pommier, and Fred Indig for insightful discussions.
This work was supported by grants from the NIH to G.M.W. (CA48405 and GM51104) and to M.G. (DK44746), a grant from the G. Harold and Leila Y. Mathers Charitable Foundation to G.M.W., and a fellowship from the American Cancer Society to D.M.C. M.I.A. was a Special Fellow of the Leukemia Society of America.
REFERENCES
- 1.Aladjem, M., L.-W. Rodewald, J. L. Kolman, and G. M. Wahl. 1998. Genetic dissection of a mammalian replicator in the human beta-globin locus. Science 281:1005–1009. [DOI] [PubMed] [Google Scholar]
- 2.Aladjem, M., and G. M. Wahl. 1997. Mapping replication origins by leading strand analysis in the absence of protein synthesis. Methods Companion Methods Enzymol. 13:281–292. [DOI] [PubMed] [Google Scholar]
- 3.Aladjem, M. I., M. Groudine, L. L. Brody, E. S. Dieken, R. E. K. Fournier, G. M. Wahl, and E. M. Epner. 1995. Participation of the human beta globin locus control region in initiation of DNA replication. Science 270:815–819. [DOI] [PubMed] [Google Scholar]
- 4.Aladjem, M. I., B. T. Spike, L. W. Rodewald, T. J. Hope, M. Klemm, R. Jaenisch, and G. M. Wahl. 1998. ES cells do not activate p53-dependent stress responses and undergo p53-independent apoptosis in response to DNA damage. Curr. Biol. 8:145–155. [DOI] [PubMed] [Google Scholar]
- 5.Bielinsky, A. K., and S. A. Gerbi. 1998. Discrete start sites for DNA synthesis in the yeast ARS1 origin. Science 279:95–98. [DOI] [PubMed] [Google Scholar]
- 6.Bulger, M., J. H. von Doorninck, N. Saitoh, A. Telling, C. Farrell, M. A. Bender, G. Felsenfeld, R. Axel, and M. Groudine. 1999. Conservation of sequence and structure flanking the mouse and human beta-globin loci: the beta-globin genes are embedded within an array of odorant receptor genes. Proc. Natl. Acad. Sci. USA 96:5129–5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burhans, W. C., L. T. Vassilev, J. Wu, J. M. Sogo, F. S. Nallaseth, and M. L. DePamphilis. 1991. Emetine allows identification of origins of mammalian DNA replication by imbalanced DNA synthesis, not through conservative nucleosome segregation. EMBO J. 10:4351–4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carroll, S. M., M. L. DeRose, J. L. Kolman, G. H. Nonet, R. E. Kelly, and G. M. Wahl. 1993. Localization of a bidirectional DNA replication origin in the native locus and in episomally amplified murine adenosine deaminase loci. Mol. Cell. Biol. 13:2971–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciavatta, D. J., T. M. Ryan, S. C. Farmer, and T. M. Townes. 1995. Mouse model of human beta zero thalassemia: targeted deletion of the mouse beta maj- and beta min-globin genes in embryonic stem cells. Proc. Natl. Acad. Sci. USA 92:9259–9263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cimbora, D. M., D. Schubeler, A. Reik, J. Hamilton, C. Francastel, E. M. Epner, and M. Groudine. 2000. Long-distance control of origin choice and replication timing in the human beta-globin locus are independent of the locus control region. Mol. Cell. Biol. 20:5581–5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DePamphilis, M. L. 1997. The search for origins of DNA replication. Methods 13:211–219. [DOI] [PubMed] [Google Scholar]
- 12.Dimitrova, D. S., and D. M. Gilbert. 1999. The spatial position and replication timing of chromosomal domains are both established in early G1 phase. Mol. Cell 4:983–993. [DOI] [PubMed] [Google Scholar]
- 13.Epner, E., W. C. Forrester, and, G. M. 1988. Asynchronous DNA replication within the human beta-globin gene locus. Proc. Natl. Acad. Sci. USA 85:8081–8085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Epner, E., A. Reik, D. Cimbora, A. Telling, M. A. Bender, S. Fiering, T. Enver, D. I. Martin, M. Kennedy, G. Keller, and M. Groudine. 1998. The beta-globin LCR is not necessary for an open chromatin structure or developmentally regulated transcription of the native mouse beta-globin locus. Mol. Cell 2:447–455. [DOI] [PubMed] [Google Scholar]
- 15.Forrester, W. C., E. Epner, M. C. Driscoll, T. Enver, M. Brice, T. Papayannopoulou, and M. Groudine. 1990. A deletion of the human beta-globin locus activation region causes a major alteration in chromatin structure and replication across the entire beta-globin locus. Genes Dev 4:1637–1649. [DOI] [PubMed] [Google Scholar]
- 16.Forrester, W. C., S. Takegawa, T. Papayannopoulou, G. Stamatoyannopoulos, and M. Groudine. 1987. Evidence for a locus activation region: the formation of developmentally stable hypersensitive sites in globin-expressing hybrids. Nucleic Acids Res 15:10159–10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman, K. L., J. D. Diller, B. M. Ferguson, S. V. M. Nyland, B. J. Brewer, and W. L. Fangman. 1996. Multiple determinants controlling activation of yeast replication origins late in S phase. Genes Dev. 10:1565–1607. [DOI] [PubMed] [Google Scholar]
- 18.Gerbi, S. A., and A. K. Bielinsky. 1997. Replication initiation point mapping. Methods 13:271–280. [DOI] [PubMed] [Google Scholar]
- 19.Giacca, M., L. Zentilin, P. Norio, S. Diviacco, D. Dimitriva, G. Contreas, G. Biamonti, G. Perini, F. Weighardt, S. Riva, and A. Falaschi. 1994. Fine mapping of a replication origin of human DNA. Proc. Natl. Acad. Sci. USA 91:7119–7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Handeli, S., A. Klar, M. Meuth, and H. Cedar. 1989. Mapping replication units in animal cells. Cell 57:909–920. [DOI] [PubMed] [Google Scholar]
- 21.Hansen, R. S., T. K. Canfield, M. M. Lamb, S. M. Gartler, and C. D. Laird. 1993. Association of fragile X syndrome with delayed replication of the FMR1 gene. Cell 73:1403–1409. [DOI] [PubMed] [Google Scholar]
- 22.Heintz, N., and J. Hamlin. 1982. An amplified chromosomal sequence that includes the gene for dihydrofolate reductase initiates replication within specific restriction fragments. Proc. Natl. Acad. Sci. 79:4083–4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hyrien, O., C. Maric, and M. Mechali. 1995. Transition in specification of embryonic metazoan DNA replication origins. Science 270:994–997. [DOI] [PubMed] [Google Scholar]
- 24.Jacob, F., J. Brenner, and F. Cuzin. 1964. On the regulation of DNA replication in bacteria. Cold Spring Harbor Symp. Quant. Biol. 288:329. [Google Scholar]
- 25.Kalejta, R. F., X. Li, L. D. Mesner, P. A. Dijkwel, H. B. Lin, and J. L. Hamlin. 1998. Distal sequences, but not ori-beta/OBR-1, are essential for initiation of DNA replication in the Chinese hamster DHFR origin. Mol. Cell 2:797–806. [DOI] [PubMed] [Google Scholar]
- 26.Kamath, S., and M. Leffak. 2001. Multiple sites of replication initiation in the human beta-globin gene locus. Nucleic Acids Res 29:809–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelly, R. E., M. L. DeRose, B. W. Draper, and G. M. Wahl. 1995. Identification of an origin of bidirectional DNA replication in the ubiquitously expressed mammalian CAD gene. Mol. Cell. Biol. 15:4136–4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kitsberg, D., S. Selig, I. Keshet, and H. Cedar. 1993. Replication structure of the human beta-globin gene domain. Nature 366:588–590. [DOI] [PubMed] [Google Scholar]
- 29.Kobayashi, T., T. Rein, and M. L. DePamphilis. 1998. Identification of primary initiation sites for DNA replication in the hamster dihydrofolate reductase gene initiation zone. Mol. Cell. Biol. 18:3266–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lemaitre, J. M., G. Geraud, and M. Mechali. 1998. Dynamics of the genome during early Xenopus laevis development: karyomeres as independent units of replication. J. Cell Biol. 142:1159–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu, L., and J. Tower. 1997. A transcriptional insulator element, the su(Hw) binding site, protects a chromosomal DNA replication origin from position effects. Mol. Cell. Biol. 17:2202–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Majumder, S., Z. Zhao, K. Kaneko, and M. L. DePamphilis. 1997. Developmental acquisition of enhancer function requires a unique coactivator activity. EMBO J. 16:1721–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nonet, G. H., S. M. Carroll, M. L. DeRose, and G. M. Wahl. 1993. Molecular dissection of an extrachromosomal amplicon reveals a circular structure consisting of an imperfect inverted duplication. Genomics 15:543–558. [DOI] [PubMed] [Google Scholar]
- 34.Santocanale, C., K. Sharma, and J. F. Diffley. 1999. Activation of dormant origins of DNA replication in budding yeast. Genes Dev. 13:2360–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vassilev, L., and E. M. Johnson. 1989. Mapping initiation sites of DNA replication in vivo using polymerase chain reaction amplification of nascent strand segments. Nucleic Acids Res. 17:7693–7705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Virta-Pearlman, V., P. H. Gunaratne, and A. C. Chinault. 1993. Analysis of a replication initiation sequence from the adenosine deaminase region of the mouse genome. Mol. Cell. Biol. 13:5931–5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiekowski, M., M. Miranda, J. Y. Nothias, and M. L. DePamphilis. 1997. Changes in histone synthesis and modification at the beginning of mouse development correlate with the establishment of chromatin mediated repression of transcription. J. Cell Sci. 110:1147–1158. [DOI] [PubMed] [Google Scholar]
- 38.Yoon, Y., J. A. Sanchez, C. Brun, and J. A. Huberman. 1995. Mapping of replication initiation sites in human ribosomal DNA by nascent-strand abundance analysis. Mol. Cell. Biol. 15:2482–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]