Abstract
Pancreatic β-cell-type-specific expression of the insulin gene requires both ubiquitous and cell-enriched activators, which are organized within the enhancer region into a network of protein-protein and protein-DNA interactions to promote transcriptional synergy. Protein-protein-mediated communication between DNA-bound activators and the RNA polymerase II transcriptional machinery is inhibited by the adenovirus E1A protein as a result of E1A’s binding to the p300 coactivator. E1A disrupts signaling between the non-DNA-binding p300 protein and the basic helix-loop-helix DNA-binding factors of insulin’s E-element activator (i.e., the islet-enriched BETA2 and generally distributed E47 proteins), as well as a distinct but unidentified enhancer factor. In the present report, we show that E1A binding to p300 prevents activation by insulin’s β-cell-enriched PDX-1 activator. p300 interacts directly with the N-terminal region of the PDX-1 homeodomain protein, which contains conserved amino acid sequences essential for activation. The unique combination of PDX-1, BETA2, E47, and p300 was shown to promote synergistic activation from a transfected insulin enhancer-driven reporter construct in non-β cells, a process inhibited by E1A. In addition, E1A inhibited the level of PDX-1 and BETA2 complex formation in β cells. These results indicate that E1A inhibits insulin gene transcription by preventing communication between the p300 coactivator and key DNA-bound activators, like PDX-1 and BETA2:E47.
Pancreatic islet β-cell-specific expression of the insulin gene is due to a unique combination of factors that stimulate transcription through an enhancer located between nucleotides −340 and −90 relative to the transcription start site (2, 5, 55, 62). These sequences can replicate the tissue-restricted and metabolically regulated expression pattern of the endogenous insulin gene when linked to a reporter construct in transgenic mice (3, 6, 20, 66) and cell lines (14, 25, 32, 56, 57, 59). C2 (−317 to −311 bp) (15, 25, 54), A3 (−201 to −196 bp) (15, 49, 50), C1 (−118 to −107 bp) (59, 73), and E1 (−100 to −91 bp) (9, 25, 71) are the cis-acting enhancer elements within the mammalian insulin gene that are essential for activation. (These insulin elements are labeled in accordance with the nomenclature proposed by German et al. [16].) Because each of these activator-binding motifs is found within the transcription unit of all characterized mammalian insulin genes (65), a common regulator strategy is likely to be involved in control.
The PAX6 paired-homeodomain transcription factor activates insulin C2 element-stimulated transcription (64), and the PDX-1 homeodomain protein mediates A3 element-activated expression (42, 47, 49, 50). PDX-1 is the name (i.e., pancreatic and duodenal homeobox-1) given by the International Committee on Standardized Genetic Nomenclature for Mice, but it has also been referred to as IPF-1 (42), IDX-1 (33), and STF-1 (28). The activator of E element-directed expression is a heterodimer of proteins in the basic helix-loop-helix (bHLH) family that are islet enriched, BETA2 (36) (also known as NeuroD1 [26]), and ubiquitously distributed, E47 (3, 8, 17, 59) and HEB (51). The C1 activator, RIPE3b1 (56, 59, 73), has not been isolated. In addition, PDX-1 (29, 31, 44, 50), RIPE3b1 (44, 56, 73), and BETA2:E47 (14, 39, 56) activation is controlled by glucose, the principal physiological regulator of β-cell gene expression.
Gene targeting experiments performed on PAX6, PDX-1, and BETA2 have also demonstrated their contribution to pancreas formation. Thus, PDX-1 expression in a common progenitor cell population appears to be essential for development of both the endocrine and exocrine compartments of the pancreas, by permitting proliferation, branching, and differentiation of the pancreatic epithelium (1, 24, 40). In contrast, PAX6 (54, 69) and BETA2 (37) act downstream of PDX-1 in the islet endocrine cell differentiation pathway. Collectively, these results have established a central role for each of the isolated insulin gene transcription factors in islet cell development and function.
Preventing PDX-1, BETA2:E47, PAX-6, or RIPE3b1 binding drastically reduces insulin enhancer-driven transcription as well as promoter activation from a number of other islet-enriched genes, including glucokinase, GLUT2, somatostatin, and glucagon (reviewed in references 5, 55, and 62). The data suggest that these activators are components of regulatory circuits that respond in a synergistic fashion to mediate transcription. Although the exact mechanisms involved in control are unclear, experiments performed in other systems imply that transactivation is dependent upon the arrangement of activator recognition sites and a precise complement of bound activators that together form a unique network of protein-protein and protein-DNA interactions.
The non-DNA-binding p300 coactivator and its paralogue CBP are involved in transducing transcriptional signals between RNA polymerase II and a number of the essential DNA-bound activators (reviewed in references 12 and 60), including the insulin gene BETA2:E47 activator (35, 52). In this study, p300 is also shown to enhance PDX-1 activation and to act in conjunction with the BETA2:E47 activator to synergistically stimulate insulin gene transcription. Our results further suggest that transcriptional stimulation requires direct interactions between the insulin enhancer-bound activators and the p300 coactivator, presumably enabling each to communicate signals through the coactivator to the basal RNA polymerase II transcriptional complex.
MATERIALS AND METHODS
DNA constructs.
The GAL4:PDX-1(1–79)ΔABC mutant was constructed from full-length GAL4:PDX-1ΔABC (48) by restriction enzyme digestion. The ΔABC mutation removes evolutionarily conserved amino acids (aa) 13 to 22 (A), 32 to 38 (B), and 60 to 73 (C) from the transactivation domain region (aa 1 to 79) of the rat PDX-1 protein (48). The GST:PDX-1 chimeras were constructed by subcloning PCR-amplified rat PDX-1 fragments into pGEX2T (Pharmacia) to create in-frame glutathione S-transferase (GST) fusion proteins. The correctness of each plasmid was verified by restriction enzyme digestion and partial DNA sequence analysis.
The construction of the cytomegalovirus (CMV)-driven wild-type and mutant PDX-1 expression vectors was described previously (48). The adenovirus type 5 E1A expression plasmids encode the wild-type 243 aa protein and mutants deficient in p300 binding (E1A Δ2–36; internal deletion of aa 2 to 36 [63]) and Rb pocket protein binding (E1A 928; cysteine 124 to glycine [34]). The CMV-driven wild-type (27) and p300 dl10 mutant (deletion of aa 1680 to 1811) (27), as well as the wild-type (27), Np300 (aa 1 to 1257) (27), Cp300 (aa 1258 to 2414) (27), and Cp300ΔQ (deletion of aa 1945 to 2377) (52) in vitro translation mutants of p300 have been described.
E47 is expressed from the simian virus 40 (SV40) enhancer/promoter in pJ3Ω (48). The CMV BETA2 expression vectors contain either the complete coding sequence of the hamster protein or lack aa 156 to 355 (BETA2 1–155) (52); this mutant contains the bHLH region (i.e., aa 100 to 155) and dimerizes with E47. Myc epitope sequences were fused in frame to the BETA2 sequences (52, 58). GST:TAL1 (22) and GST:BETA2 (52) are in-frame GST fusions of the full-length mouse and hamster proteins, respectively. The −238 insulin-chloramphenicol acetyltransferase (CAT) expression plasmids contain wild-type rat insulin II gene sequences from bp −238 to +2 and E1 or A3 element mutants (53). The construction of the rat insulin I minienhancer (−247 to −197 bp; FF CAT) constructs in tkCAT (pTE2ΔS/N) has been described previously (15).
Cell culture and transfections.
The β (HIT T-15 2.2.2 and βTC3) and non-β (HeLa) cell lines were maintained as described previously (53). The insulin-CAT constructs were transiently transfected by the calcium phosphate coprecipitation procedure (70). The SV40 enhancer-driven luciferase (LUC) expression plasmid SV2 LUC (11) was used as a recovery marker. Extracts were prepared 40 to 48 h after transfection, and LUC and CAT enzymatic assays were performed as described by DeWet et al. (11) and Nordeen et al. (38), respectively. The CAT activity from the reporter construct was normalized to the LUC activity of the cotransfected internal control plasmid. Each experiment was repeated several times with at least two different plasmid preparations.
Electrophoretic mobility shift assays.
HIT T-15 cells were transiently transfected with GAL4:PDX-1(1–79) or GAL4:PDX-1(1–79)ΔABC and (GAL4)5E1bCAT and assayed for CAT and gel shift activity. The nuclear extract was prepared as described previously (48). Double-stranded oligonucleotides to the GAL4 (5′-GGCGGAAGACTCTCCTCCG-3′) DNA-binding element were end labeled using [γ-32P]dATP and T4 polynucleotide kinase. The reactions were performed using 2.5 μg of nuclear extract in binding buffer (25 mM HEPES [pH 7.9], 0.1 mM EDTA, 10 mM MgCl2, 0.1 mM dithiothreitol [DTT], 10% glycerol, 0.5 μg of salmon sperm DNA), and labeled probe; each sample was incubated for 15 min at room temperature. A polyclonal antibody raised to the GAL4 DNA-binding domain (Santa Cruz Biotech, Santa Cruz, Calif.) was used to localize the GAL4:PDX-1(1–79) complexes. The samples were resolved on a 6% nondenaturing polyacrylamide gel (acrylamide-bisacrylamide ratio, 29:1) and run in TGE buffer (50 mM Tris, 380 mM glycine, 2 mM EDTA, pH 8.5). The gel was dried and subjected to autoradiography.
In vitro translation and GST binding assay.
Interactions between PDX-1 and p300 were assessed by examining binding of in vitro-translated p300 with GST:PDX-1. Wild-type and mutant GST:PDX-1 proteins were prepared using the manufacturer’s conditions (Pharmacia), and [35S]methionine-labeled p300 was synthesized using the TNT Kit (Promega). Labeled p300 protein was incubated for 1 h in binding buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.1% NP-40, 2 mM EDTA, 10 mM MgCl2, 20 μM ZnCl2) with GST:PDX-1 coupled to glutathione-Sepharose beads (Pharmacia). The beads were washed three times with binding buffer, and the bound protein complexes were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography.
Immunoprecipitation and Western blotting.
HIT T-15 and HeLa cells were transfected with PDX-1, BETA2-Myc, E1A, and p300 expression plasmids and harvested after 24 h for immunoprecipitation and Western blot analyses. The cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris-HCl, pH 8.0, 140 mM NaCl, 0.025% NaN3, 0.5% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 1 mM phenylmethylsulfonyl fluoride, 2 μg of aprotinin per ml) and incubated overnight at 4°C with antiserum to PDX-1, p300 (RW128 (Upstate Biotechnology, Lake Placid, N.Y.), MN11 (10)), FLAG (M2; Tony Weil, Department of Molecular Physiology and Biophysics, Vanderbilt University), Myc (9E10; Sigma, St. Louis, Mo.), or normal immunoglobulin G (IgG).
PDX-1 polyclonal antiserum were developed to N- (aa 1 to 75 [48]) or C-terminal (aa 271 to 283 [68]) epitopes. The monoclonal antibodies to p300 (MN11 and RW128) were raised to distinct peptide domains. The proteins were immunoprecipitated with either protein A- or protein G-Sepharose beads (Sigma, St. Louis, Mo.), washed three times with RIPA buffer, subjected to SDS-PAGE, and then electrotransferred to Immobilon polyvinylidene difluoride membrane (Millipore, Bedford, Mass.). The blot was incubated for 1 h at 4°C in blocking buffer (10 mM Tris, pH 8.0, 150 mM NaCl, 0.05% Tween 20, and 5% nonfat dry milk) and then at 4°C overnight with either PDX-1, p300, or Myc antiserum. Detection was performed using enhanced chemiluminescence (Pierce, Rockford, Ill.) after incubation with a horseradish peroxidase-conjugated secondary antibody.
RESULTS
Adenovirus E1A inhibits A3/A4 element activation.
The observation that adenovirus E1A inhibited activation directed by the bp −238 to −101 region of the rat insulin II enhancer demonstrated that a control element(s), distinct from the E1 element at −100/−91, was a target of repression (64). The PDX-1 and RIPE3bl activator binding sites at −201 to −196 bp (i.e., A3) and −115 to −107 bp (i.e., C1) are the principal conserved control elements within this area, although previous studies have shown that E1A does not influence C1-mediated activation (52).
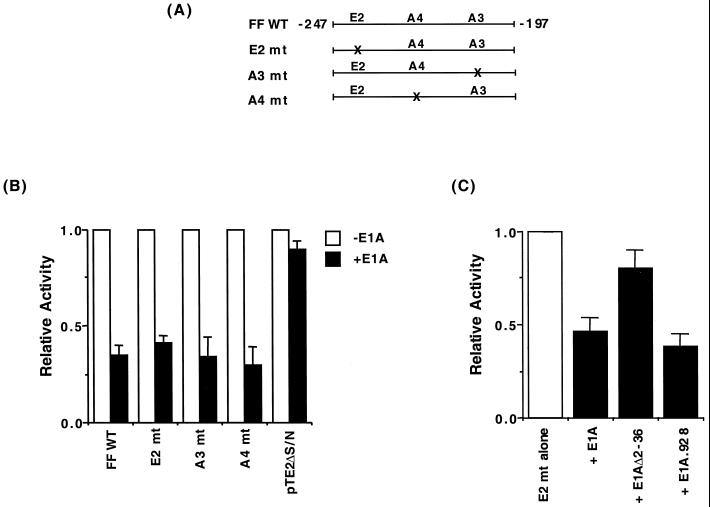
To test the effect of E1A on A3 element activation, regulation from an insulin enhancer-driven CAT expression construct spanning nucleotides −247 to −197 of the rat insulin I gene was examined in HIT T-15 β cells. Stimulation by this minienhancer region (termed FF) is mediated by the A3, A4, and E2 elements (15) (Fig. 1A. Upon cotransfection of E1A, the level of FF CAT activation was reduced markedly (Fig. 1B). In contrast, E1A had no effect on the activity of the parent tkCAT cloning vector pTE2ΔS/N. The level of repression was also unaffected in the E element binding site mutant (E2 mt in Fig. 1B), suggesting that stimulation by A3 and/or A4 was altered by E1A.
FIG. 1.
Repression of A3/A4 element-driven activity by the p300 binding region of adenovirus E1A. (A) Schematic representation of the FF region in the rat insulin I gene, illustrating the E2 (−239/−228 bp), A4 (−221/−217 bp), and A3 (−212/−208 bp) elements. The nonallelic rat insulin I gene, unlike the rat II or human insulin gene, contains a second upstream E-element (termed E2) (65). The mutant element is denoted mt. FF CAT contains three copies of the −247 to −197 region (15). (B) HIT T-15 cells were transfected with wild-type (WT) FFCAT, mutant (mt) FFCAT, or pTE2ΔS/N (2.5 μg) in the presence or absence of E1A (2.5 μg), and a recovery marker (1.0 μg) for transfection efficiency, pSV2 LUC. (C) E2 mt FF CAT (2.5 μg) was transfected into HIT T-15 cells in the presence of wild-type E1A (2.5 μg) or a p300 (E1AΔ2–36)- or Rb (E1A.928) binding-defective mutant. The normalized activity ± standard error of the mean (SEM) is presented relative to FFCAT alone (B and C), which in the absence of E1A corresponded to 1.36 × 106 cpm/reaction, 0.72 × 106 cpm/reaction, 6.10 × 106 cpm/reaction, 0.27 × 106 cpm/reaction, and 0.08 × 106 cpm/reaction for the FF WT, FF A4 mt, FF A3 mt, FF E2 mt, and pTE2ΔS/N, respectively. The relative activity of the various FF CAT constructs in HIT T-15 cells is similar to that described previously (15). FF E2mt activity was not statistically changed in the presence of E1AΔ2–36, but was with wild-type or 928 mt E1A (P < 0.05).
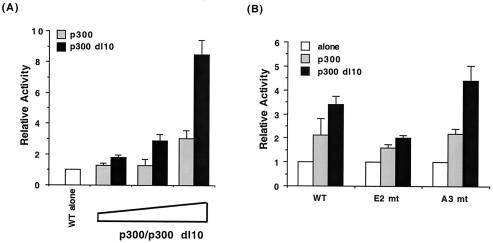
E1A influences cellular gene transcription by binding to coregulatory factors (7) like the retinoblastoma (Rb) pocket-binding proteins (i.e., p105, p107, and p110 [19, 45]) and p300 (12, 60). To investigate whether these factors were involved in E1A-mediated inhibition of A3/A4-directed transcription, specific mutants that fail to bind either p300 (E1AΔ2–36 [63]) or Rb (E1A.928 [34]) were used. In contrast to wild-type E1A, the E1AΔ2–36 mutant was unable to inhibit A3/A4-dependent transcription effectively, whereas E1A.928 fully repressed activity (Fig. 1C). In addition, wild-type p300 and a mutant that lacks the E1A binding domain, p300 dl10, increased FF CAT activity (Fig. 2A).
FIG. 2.
p300 coactivator potentiates A3/A4-mediated activation. (A) p300 or p300 dl10 (5, 10, or 20 μg) was transfected with FFCAT (2.5 μg) into HIT T-15 cells. The sequences in p300 required for E1A association (aa 1680 to 1811) are missing in p300 dl10 (26). The ability of p300 dl10 to stimulate more effectively than p300 has also been found with other insulin-driven reporter genes (52). (B) WT, E2 mt, and A3 mt FF CAT (2.5 μg) were cotransfected into HIT T-15 cells in the presence (10 μg) or absence of p300 or p300 dl10 (10 μg). Relative activity ± SEM is calculated as the normalized activity of FFCAT plus E1A divided by that of FFCAT alone.
p300 also stimulated both A3/A4-and E2-dependent activity (Fig. 2B). These data strongly implicate p300 in both A3/A4 and E element-dependent transcription. Because the A3 site and not the A4 site is conserved among the mammalian insulin genes (13, 15), the following experiments were designed to determine if stimulation by its activator, PDX-1, was affected by p300.
p300 interacts with PDX-1.
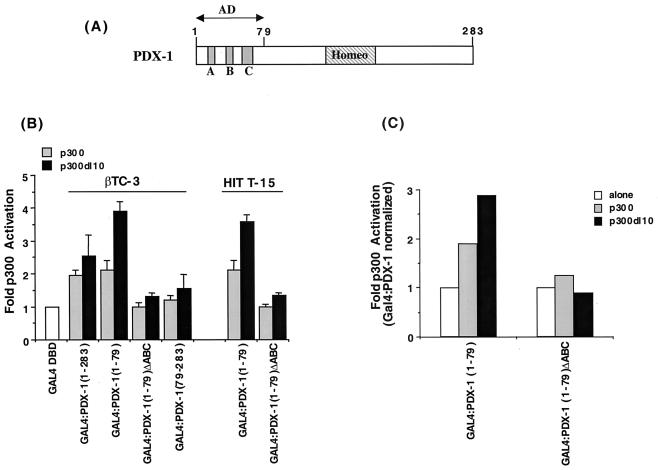
To test if stimulation by PDX-1 was influenced by p300, β cells were cotransfected with expression plasmids producing p300 and either wild-type PDX-1 or N- and C-terminal deletion mutants fused in frame to the DNA-binding domain of the Saccharomyces cerevisiae GAL4 transcription factor. The GAL4:PDX-1 fusions represented regions of PDX-1 important in activation (aa 1 to 79) and DNA binding (homeodomain, aa 146 to 206) (Fig. 3A). Because p300 contains two distinct activation domain regions (72), we reasoned that functional interactions between PDX-1 and p300 would enhance transcription from the GAL4 DNA binding site-driven reporter, (GAL4)5E1bCAT, as observed with the insulin gene activators BETA2 and E47 (52) and other p300-regulated transcription factors (12, 60).
FIG. 3.
p300 activates PDX-1 dependent transcription. (A) Diagrammatic representation of PDX-1 illustrating the location of the homeodomain region (Homeo; aa 146 to 206) and conserved amino acid segments (A, 13 to 22; B, 32 to 38; C, 60 to 73) within the aa 1 to 79 activation domain (AD) (48). (B) The transcriptional activity of the wild-type and mutant GAL4:PDX-1 fusion proteins was examined in βTC-3 and HIT T-15 cells. The PDX-1 sequences within the GAL4 chimeras are noted in parentheses. Each transfection contained GAL4:PDX-1 (2.5 μg), p300 or p300 dl10 (10.0 μg), (GAL4)5E1bCAT (2.5 μg), and pSV2 LUC (1.0 μg). The normalized fold p300 activation ± SEM is the ratio of GAL4:PDX-1 and p300 to GAL4:PDX-1 alone. (C) HIT T-15 cells were transfected with GAL4:PDX-1(1–79) or GAL4:PDX-1(1–79)ΔABC (2.5 μg), p300 or p300 dll0 (10.0 μg), and (GAL4)5E1bCAT (2.5 μg). The extracts were assayed for both CAT and GAL4 DNA-binding activity, and the CAT activity from each GAL4:PDX-1 construct was normalized to the level of GAL:PDX-1 binding activity in the gel shift assay. The ratio of the normalized GAL4:PDX-1 activity with p300 to GAL4:PDX-1 alone from a representative experiment is shown.
Indeed, p300 stimulated GAL4:PDX-1 activity from the wild-type and aa 1 to 79 activation domain-spanning constructs in βTC-3 and HIT T-15 cells (Fig. 3B). In contrast, p300 did not affect activation from the homeodomain and C-terminal region GAL4:PDX-1 construct [i.e., GAL4:PDX-1(79–283)] or an activation-defective mutant of the aa 1 to 79 region [GAL4:PDX-1(1–79)ΔABC in Fig. 3B]. Gel shift and (GAL4)5E1bCAT reporter assays performed on transfected HIT T-15 cells demonstrated that p300 affected GAL4:PDX-1(1–79) activation, and not protein expression (Fig. 3C).
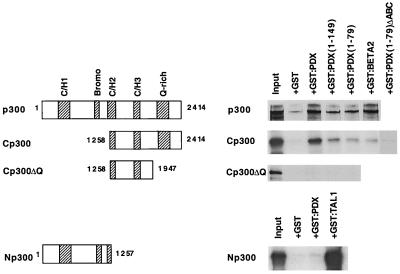
To determine if p300 physically interacted with PDX-1, affinity chromatography methods were developed using regions of PDX-1 fused in frame to the GST gene. GST:PDX protein was expressed in bacteria and immobilized on glutathione beads. In vitro-labeled p300 translation products were incubated separately with GST:PDX and GST control resin, and the bound material was eluted and analyzed by SDS-PAGE. Little if any of the radiolabeled p300 proteins bound to the GST resin alone (Fig. 4). In contrast, p300 bound to the GST:PDX fusions spanning the activation domain (full length, aa 1 to 149, and aa 1 to 79). The level of PDX-1 binding to p300 was comparable to that of BETA2 (Fig. 4).
FIG. 4.
In vitro interactions between p300 and PDX-1. Schematic representation of the 35S-labeled in vitro-synthesized human p300 proteins. The cysteine/histidine-rich (CH/1, CH/2, and CH/3), bromo-rich, and glutamine-rich (Q-rich) domains that are involved in protein-protein interactions are shown (12, 60). Radiolabeled p300 was incubated with purified GST, GST:PDX, GST:BETA2 or GST:TAL1 protein bound to glutathione-Sepharose beads. Bound p300 was eluted, separated by SDS-10% PAGE, and detected by autoradiography. The input lane represents 5% of the total volume used in the binding assay. GST:TAL1 (22) and GST:BETA2 (52) served as positive controls for Np300 and Cp300 binding, respectively.
To localize the PDX-1 interaction domain in p300, various p300 mutant proteins were analyzed for binding to GST:PDX resins. GST:TAL1 (22) and GST:BETA2 (52) binding to the N- or C-terminal p300 regions served as controls. PDX-1 binding was only detected with the C-terminal p300 expression construct (i.e., aa 1258 to 2414), an interaction lost in the activation domain dysfunctional mutant GST:PDX(1–79)ΔABC (Fig. 4). Furthermore, removing the glutamine-rich sequences from the C-terminal region of p300 prevented both PDX-1 (Cp300ΔQ in Fig. 4) and BETA2 (52) binding. The results demonstrate that the N-terminal activation domain region of PDX-1 interacts with C-terminal sequences of p300.
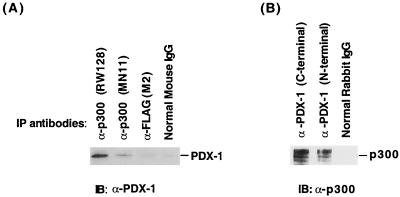
Having established that PDX-1 and p300 directly bind in vitro, we asked whether this complex could be detected in vivo. Immunoprecipitation assays were performed in HIT T-15 cell extracts with antibodies to p300 (RW128 and MN11) and PDX-1 (N- and C-terminal). Western blotting revealed that PDX-1 was coprecipitated with p300 antibodies (Fig. 5A), and p300 was coprecipitated with PDX-1 antibodies (Fig. 5B). These complexes appear to be specific, as neither the unrelated anti-FLAG monoclonal antibody (M2), mouse IgG, nor rabbit IgG precipitated p300 or PDX-1 (Fig. 5). These results demonstrated that p300 and PDX-1 also associate in vivo.
FIG. 5.
p300 and PDX-1 associate in vivo. Immunoprecipitation analysis was performed with HIT T-15 nuclear extract using (A) p300 (RW128 and MN11) antiserum, FLAG (M2) antiserum, and normal mouse IgG or (B) PDX-1 antiserum (N-terminal and C-terminal) and normal rabbit IgG. The precipitate was washed with RIPA buffer, and the released proteins were separated by SDS-10% PAGE and immunoblotted (IB) with (A) PDX-1 (N terminal) or (B) p300 (RW128) antiserum.
p300 cooperates with PDX-1 and BETA2:E47 to induce insulin gene transcription.
The results presented above and elsewhere have shown that p300 interacts directly with PDX-1 and BETA2:E47 (35, 52) to stimulate transcription. Because insulin enhancer activation is drastically reduced when the binding site for either factor is mutated, we considered that p300 might mediate the synergy between PDX-1 and BETA2:E47.
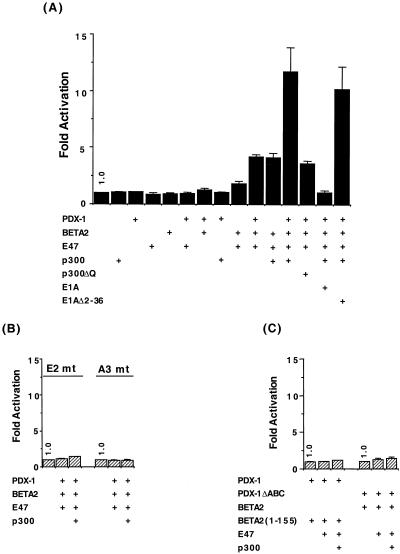
To test this possibility, p300, PDX-1, E47, and BETA2 expression constructs were cotransfected with an insulin enhancer/promoter-driven reporter construct (−238 CAT) into a cell line (HeLa) that does not produce insulin or the islet-enriched transcription factors BETA2 or PDX-1. As expected, this reporter does not respond to p300 or any of the individual DNA-binding activators (Fig. 6A). p300 also has little or no influence on activation by BETA2 plus E47 or PDX-1 alone. However, transactivation by PDX-1, E47, and BETA2 was potentiated when these factors were combined, an activity induced to a much higher level in the presence of p300 and not by the coactivator binding-defective mutant p300ΔQ. In addition, wild-type E1A but not the p300 binding-defective E1A mutant E1AΔ2–36 prevented activation (Fig. 6A).
FIG. 6.
PDX-1, BETA2: E47 and p300 synergistically stimulate insulin enhancer-driven reporter expression. The rat insulin II (A and C) WT, (B) E1 mt, and (B) A3 mt −238 CAT reporter (1.0 μg), and expression vectors encoding E47 (2.5 μg), p300 (10 μg), BETA2 (2.5 μg), PDX-1 (2.5 μg), and E1A (2.5 μg) were transfected into HeLa cells, as indicated. Values are expressed as the normalized fold induction ± SEM relative to the reporter gene alone.
Stimulation of −238 CAT activity by p300, PDX-1, E47, and BETA2 was also dependent upon activator binding, as the PDX-1 (A3 mt) and BETA2:E47 (E1 mt) binding site mutants were inactive, even in the presence of p300 (Fig. 6B). The p300 binding sequences within each activator were necessary for stimulation, as neither PDX-1ΔABC nor BETA2(1–155) could support p300-mediated synergistic activation (Fig. 6C). BETA2(1–155) contains an intact bHLH region, which is necessary for DNA binding and dimerization with E47, but lacks the C-terminal p300 binding and transactivation domains (52, 58). Together, these results strongly indicate that communication between p300, PDX-1, E47, and BETA2 plays a key role in regulating insulin gene transcription.
PDX-1 and BETA2:E47 complex formation in vivo is mediated by p300.
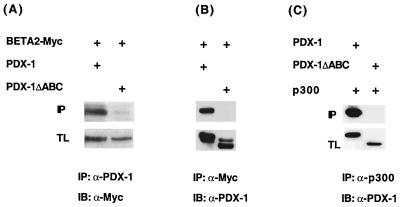
To further investigate the basis for cooperation between the insulin activator factors, we tested whether physical interactions between PDX-1 and BETA2 were detected in vivo. HIT T-15 cells were transfected with PDX-1 and BETA2, and an immunoprecipitation analysis was performed with antibodies that recognized the introduced PDX-1 and BETA2 proteins. BETA2 was detected in a complex with PDX-1 and not PDX-1ΔABC (Fig. 7A and B), although similar amounts of protein were made.
FIG. 7.
The p300 binding sequences in PDX-1 are important for complex formation with BETA2 in vivo. Expression plasmids (2.5 μg each) encoding Myc epitope-tagged BETA2 (BETA2-Myc), wild-type PDX-1, PDX-1ΔABC, and p300 were transfected into (A and B) HIT T-15 and (C) HeLa cells, as indicated. The extracts were immunoprecipitated (IP) with (A) α-PDX-1 (N terminal), (B) α-Myc, or (C) α-p300 (RW128) antibody. BETA2-Myc and PDX-1 in the precipitates were analyzed by immunoblotting (IB) with Myc and PDX-1 (N-terminal)-specific antisera. The total lysate (TL; approximately 5% of total) shows the expression level of the transfected BETA2-Myc and PDX-1 proteins.
To examine if p300 was involved in formation of the PDX-1:BETA2 complex, an α-p300 immunoprecipitation was conducted with cells transfected with p300 binding-defective mutants of PDX-1 (PDX-1ΔABC) and BETA2 [BETA2(1–155)]. As expected, p300-insulin activator complex formation was not detected with either of these mutants [PDX-1ΔABC, Fig. 7C; BETA2(1–155), data not shown].
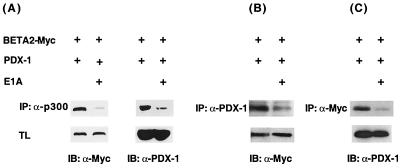
Collectively, our results suggested that the PDX-1 and BETA2 sequences required in physical (Fig. 3 and 4) and functional interactions (Fig. 6) with p300 were also necessary for formation of a BETA2 and PDX-1 precipitation complex in cells. If so, we predicted that E1A should inhibit the formation of the PDX-1:BETA2 complex. To test this proposal, HIT T-15 cells were transfected with BETA2, PDX-1, and E1A, and an immunoprecipitation analysis was performed with p300, BETA2 (i.e., Myc tag), and PDX-1-specific antisera. The precipitated proteins were probed for the presence of BETA2 and PDX-1. The addition of E1A profoundly reduced the amount of BETA2 and PDX-1 coprecipitating with either p300 (Fig. 8A), PDX-1 (Fig. 8B), or BETA2 (Fig. 8C). In contrast, E1A did not affect PDX-1 or BETA2 protein expression (see TL in Fig. 8).
FIG. 8.
Adenoviral E1A reduces PDX-1:p300:BETA2 complex levels. HIT T-15 cells were transfected with the E1A (5 μg), PDX-1 (2.5 μg), and BETA2-Myc (2.5 μg) expression plasmids, as indicated. Nuclear extracts were immunoprecipitated (IP) with (A) α-p300 (RW128), (B) α-PDX-1 (N terminal), or (C) α-Myc antibody and immunoblotted (IB) with (A and B) Myc and (A and C) PDX-1 (N terminal)-specific antisera. (A) BETA2 and PDX-1 were reduced to 0.03 and 0.09 of the no-E1A-transfected control, respectively; (B) 0.16; (C) 0.15. BETA2 and PDX-1 are expressed at similar levels under these conditions (5% of total lysate [TL]).
As the BETA2:PDX-1 complex levels were reduced by E1A to a similar low in the p300, PDX-1, and BETA immunoprecipitations (Fig. 8), we propose that interactions between these DNA-binding factors are principally, if not exclusively, mediated through p300. Most significantly, when these results are considered together with the transfection and biochemical experiments performed with p300, our data strongly indicate that the recruitment of this coactivator by PDX-1 and BETA2:E47 plays an essential role in controlling insulin gene transcription in vivo.
DISCUSSION
PDX-1 and BETA2:E47 act upon the insulin enhancer to control β-cell-specific insulin gene transcription. This unique combination of activators promotes the RNA polymerase II machinery to stimulate insulin gene transcription synergistically. The greater-than-additive effect results from multiple protein-protein interactions between activators and the general transcriptional machinery, as a consequence of activator binding directly to the RNA polymerase II transcriptional apparatus and/or through a bridging coactivator(s).
Previously we had shown that adenovirus E1A protein binding to the p300 coactivator blocked insulin gene transcription in islet β cells (63), at least in part due to actions on BETA2 and E47 (52). In this study, we found that E1A also inhibits PDX-1 activation through binding to p300. PDX-1 and BETA2:E47 were shown to act together with p300 to stimulate insulin gene transcription, with p300 complex formation involving activation domain sequences within each factor. Our results suggest that p300 provides a docking and recruitment interface between PDX-1, BETA2:E47, and the general transcriptional machinery.
PDX-1 became a candidate for p300 regulation due to the ability of E1A to influence insulin A3/A4 element activation (Fig. 1). As the A3 site and not the A4 site is conserved among the mammalian insulin genes (13, 15), our experiments focused on determining if E1A specifically affected activation by the A3 activator, PDX-1. Interestingly, the HNF1α transcription factor, which can bind to the mouse (13) and human (43) A4 elements in gel shift assays, also appears to be regulated by p300 (61). However, chromatin immunoprecipitation assays performed with HNF1α antiserum strongly suggest that this factor does not bind to the A4 element in vivo (46). In contrast, a similar analysis performed with BETA2- and PDX-1-specific antisera confirmed that each bound within the enhancer region of the endogenous insulin gene (M. Cissell and R. Stein, data not shown).
Stimulation by PDX-1 and BETA2 was shown to be mediated by the p300 coactivator in both immunoprecipitation and insulin gene reporter transfection experiments performed in β and non-β cells. The p300 binding surface within PDX-1 [aa 13 to 22 (A), 32 to 38 (B), and 60 to 73 (C), Fig. 4 and 7] was mapped by in vitro GST pulldown and in vivo immunoprecipitation assays to sequences essential in activation domain function. Ashara et al. (4) also found that N-terminal trans-activation domain-spanning sequences of PDX-1 (aa 1 to 140) were bound to CBP, the paralog of p300. The activation domain region of BETA2 was also necessary in p300 binding (i.e., within aa 156 to 355) (52). Both PDX-1 and BETA2 associate with p300 through the C-terminal glutamine-rich domain (Fig. 4), whereas E47 interacts with the N-terminal cysteine/histidine-rich domain of p300 (52).
PDX-1 has also been shown to bind directly to BETA2 and E47 by in vitro GST pulldown analysis (41). The sequences involved in BETA2 (aa 94 to 162) and PDX-1 (aa 138 to 213) binding were distinct from those involved with p300. Interestingly, we found that the reduction by E1A of PDX-1-, BETA2-, and E47-mediated activation (Fig. 1 and 6) (52) closely paralleled the loss in PDX-1:p300 and BETA2:p300 complex formation in β cells (Fig. 8). Glick et al. (18) have also shown that PDX-1, BETA2, and E47 act cooperatively to activate insulin enhancer-directed expression. Although potentiation by p300 was not investigated, these investigators found that the C-terminal p300-binding region of BETA2 was involved in activation. p300 also enhances transactivation by PAX6 on the glucagon gene (23). Collectively, the data strongly indicate that insulin gene transcription is mediated by p300 association with the key β-cell-enriched activators.
Synergistic transcription results from multiple interactions between activators and the general transcriptional machinery. Activator-activator interactions between PDX-1, BETA2, and E47 have been shown to promote cooperative DNA binding on insulin enhancer DNA in vitro (≈3.5-fold between E47 and PDX-1) (41). These results imply that cooperative binding of activators to DNA, and possibly direct contacts between activator and the basal transcriptional machinery, are involved in insulin gene activation. However, as p300 binding to E1A resulted in a profound and parallel loss in the capacity of PDX-1 and BETA2 to interact in vivo and activate insulin gene transcription, we propose that functional interactions between these DNA-binding factors are principally, if not exclusively, mediated through p300.
Binding between p300 and basal factors, like the TATA-binding protein (TBP) and TFIIB (12, 60), are presumably essential in this process. In addition, the intrinsic acetyltransferase activity of p300 and the p300-associated factor P/CAF may contribute to activation through modifications of histone proteins (12, 60) and/or the insulin activators directly. The adenovirus E1A protein appears to inhibit insulin transcription by either preventing assembly of or destabilizing the p300 complex formed with the insulin enhancer factors PAX6 (23), PDX-1, and BETA2 and the general transcriptional machinery (Fig. 8).
The ability of the β cell to provide insulin in sufficient amounts to meet the body’s needs is compromised in type 2 diabetes mellitus patients with mutations in BETA2 (30) and PDX-1 (21, 67). As mutations within the p300 binding domain of each factor, Q59L and D76N in PDX-1 (21) and 206+C in BETA2 (30), have been identified in diabetic families, a defect in p300:activator complex formation could be the cause of β-cell dysfunction. Thus, p300-mediated activation of the insulin gene would be inhibited in these mutants if BETA2:E47 or PDX-1 could not engage in combinatorial interactions with p300. In addition, as both BETA2:E47 (14, 39, 56) and PDX-1 (29, 30, 44, 50) control glucose-regulated transcription of the insulin gene, metabolic signaling may also be disrupted in these mutants. Experiments are in progress to test whether p300 provides PDX-1 and BETA2 the transactivation capacity to support metabolic and differentiation control.
Acknowledgments
We thank Kevin Gerrish, Michelle Cissell, and Chris Wright for helpful discussions, E. Henderson for technical assistance, and Michael German for providing the FF CAT plasmids.
This work was supported by grants from the National Institutes of Health (NIH RO1 DK-55091 and DK-50203 to R.S.) and partial support from the Vanderbilt University Diabetes Research and Training Center Molecular Biology Core Laboratory (Public Health Service grant P60 DK20593 from the National Institutes of Health). The FLAG (termed M2) and C-terminal PDX-1-specific antisera were generously provided by Tony Weil and Joel Habener, respectively.
REFERENCES
- 1.Ahlgren, U., J. Jonsson, L. Jonsson, K. Simu, H. Edlund. 1998. Beta-cell-specific inactivation of the mouse Ipf1/Pdx1 gene results in loss of the beta-cell phenotype and maturity onset diabetes. Genes Dev. 12:1763–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alpert, S., D. Hanahan, and G. Teitelman. 1988. Hybrid insulin genes reveal a developmental lineage for pancreatic endocrine cells and imply a relationship with neurons. Cell 53:295–308. [DOI] [PubMed] [Google Scholar]
- 3.Aronheim, A., R. Shiran, A. Rosen, and M. D. Walker. 1993. The E2A gene product contains two separable and functionally distinct transcription activation domains. Proc. Natl. Acad. Sci. USA 90:8063–8067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asahara, H., S. Dutta, H.-Y. Kao, R. M. Evans, and M. Montminy. 1999. Pbx-Hox heterodimers recruit coactivator-corepressor complexes in an isoform-specific manner. Mol. Cell. Biol. 12:8219–8225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bramblett, D. E., H.-P. Huang, and M.-J. Tsai. 2000. Pancreatic islet development. Adv. Pharmacol. 47:255–315. [DOI] [PubMed] [Google Scholar]
- 6.Bucchini, D., O. Madsen, M.-A. Stinnakre, P. Desbois, P. Lores, E. Monthioux, J. Absil, J.-A. Lepesant, R. Pictet, R., and J. Jami. 1986. Pancreatic expression of the human insulin gene in transgenic mice. Proc. Natl. Acad. Sci. USA 83:2511–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Condorelli, G., and A. Giordano. 1997. Synergistic role of E1A-binding proteins and tissue-specific transcription factor in differentiation. J. Cell. Biochem. 67:423–431. [DOI] [PubMed] [Google Scholar]
- 8.Cordle, S. R., E. Henderson, H. Masuoka, P. A. Weil, and R. Stein. 1991. Pancreatic β-cell-type-specific transcription of the insulin gene is mediated by basic helix-loop-helix DNA-binding proteins. Mol. Cell. Biol. 11:1734–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crowe, D. T., and M.-J. Tsai. 1989. Mutagenesis of the rat insulin II 5′-flanking region defines sequences important for expression in HIT cells. Mol. Cell. Biol. 9:1784–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dallas, P. B., P. Yaciuk, and E. Moran. 1997. Characterization of monoclonal antibodies raised against p300: both p300 and CBP are present in intracellular TBP complexes. J. Virol. 71:1726–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Wet, J. R., K. V. Wood, M. DeLuca, D. R. Helinski, and S. Subramani. 1987. Firefly luciferase gene: structure and expression in mammalian cells. Mol. Cell. Biol. 7:725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eckner, R. 1997. p300 and CBP as transcriptional regulators and targets of oncogenes. Biol. Chem. 377:685–688. [PubMed] [Google Scholar]
- 13.Emens, L. A., D. W. Landers, and L. G. Moss. 1992. Hepatocyte nuclear factor 1α is expressed in a hamster insulinoma line and transactivates the rat insulin I gene. Proc. Natl. Acad. Sci. USA 89:7300–7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.German, M. S., and J. Wang. 1994. The insulin gene contains multiple transcriptional elements that respond to glucose. Mol. Cell. Biol. 14:4067–4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.German, M. S., L. G. Moss, J. Wang, and W. J. Rutter. 1992. The insulin and islet amyloid polypeptide genes contain similar cell-specific promoter elements that bind identical β-cell nuclear complexes. Mol. Cell. Biol. 12:1777–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.German, M. S., S. Ashcroft, K. Docherty, H. Edlund, T. Edlund, S. Goodison, H. Imura, G. Kennedy, O. Madsen, D. Melloul, L. Moss, K. Olson, M. A. Permutt, J. Philippe, R. P. Robertson, W. J. Rutter, P. Serup, R. Stein, D. Steiner, M.-J. Tsai, and M. D. Walker. 1995. The insulin promoter: a simplified nomenclature. Diabetes 44:1002–1004. [DOI] [PubMed] [Google Scholar]
- 17.German, M. S., M. A. Blanar, C. Nelson, L. G. Moss, and W. J. Rutter. 1991. Two related helix-loop-helix proteins participate in separate cell-specific complexes that bind the insulin enhancer. Mol. Endocrinol. 5:292–299. [DOI] [PubMed] [Google Scholar]
- 18.Glick, E., D. Leshkowitz, and M. D. Walker. 2000. Transcription factor BETA2 acts cooperatively with E2A and PDX1 to activate the insulin gene promoter. J. Biol. Chem. 275:2199–2204. [DOI] [PubMed] [Google Scholar]
- 19.Granna, X., J. Garriga, and X. Mayol. 1998. Role of the retinoblastoma protein family, pRB, p107 and p130 in the negative control of growth. Oncogene 17:3365–3383. [DOI] [PubMed] [Google Scholar]
- 20.Hanahan, D. 1985. Heritable formation of pancreatic β-cell tumours in transgenic mice expressing recombinant insulin/simian virus 40 oncogenes. Nature 315:115–122. [DOI] [PubMed] [Google Scholar]
- 21.Hani, E. H., D. A. Stoffers, J. C. Chevre, E. Durand, V. Stanojevic, C. Dina, J. F. Habener, and P. Froguel. 1999. Defective mutations in the insulin promoter factor-1 (IPF-1) gene in late-onset type 2 diabetes mellitus. J. Clin. Investig. 104:R41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, S., Y. Qiu, R. Stein, and S. J. Brandt. 1999. p300 functions as a transcriptional coactivator for the TAL1/SCL oncoprotein. Oncogene 18:4958–4967. [DOI] [PubMed] [Google Scholar]
- 23.Hussain, M.A., and J. F. Habener. 1999. Glucagon gene transcription activation mediated by synergistic interactions of PAX-6 and Cdx-2 with the p300 coactivator. J. Biol. Chem. 274:28950–28957. [DOI] [PubMed] [Google Scholar]
- 24.Jonsson, J., L. Carlsson, T. Edlund, and H. Edlund. 1994. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature 371:606–609. [DOI] [PubMed] [Google Scholar]
- 25.Karlsson, O., T. Edlund, J. B. Moss, W. J. Rutter, and M. D. Walker. 1987. A mutational analysis of the insulin gene transcription control region: expression in beta cells is dependent on two related sequences within the enhancer. Proc. Natl. Acad. Sci. USA 84:8819–8823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, J. E., S. M. Hollenberg, L. Snider, D. L. Turner, N. Lipnick, and H. Weintraub. 1995. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science 268:836–844. [DOI] [PubMed] [Google Scholar]
- 27.Lee, J.-S., K. M. Galvin, R. H. See, R. Eckner, D. Livingston, E. Moran, and Y. Shi. 1995. Relief of YY1 transcriptional repression by adenovirus E1A is mediated by E1A-associated protein p300. Genes Dev. 9:1188–1198. [DOI] [PubMed] [Google Scholar]
- 28.Leonard, J., B. Peers, T. Johnson, K. Ferreri, S. Lee, and M. Montminy. 1992. Characterization of Somatostatin Transactivating Factor-1, a novel homeobox factor that stimulates somatostatin expression in pancreatic islet cells. Mol. Endocrinol. 7:1275–1283. [DOI] [PubMed] [Google Scholar]
- 29.Macfarlane, W. M., S. B. Smith, R. F. James, A. D. Clifton, Y. N. Doza, P. Cohen, K. Docherty. 1997. The p38/reactivating kinase mitogen-activated protein kinase cascade mediates the activation of the transcription factor insulin upstream factor 1 and insulin gene transcription by high glucose in pancreatic beta-cells. J. Biol. Chem. 272:20936–20944. [DOI] [PubMed] [Google Scholar]
- 30.Malecki, M. T., U. S. Jhala, A. Antonellis, L. Fields, A. Doria, T. Orban, M. Saad, J. H. Warram, M. Montminy, and A. S. Krolewski. 1999. Mutations in NeuroD1 are associated with the development of type 2 diabetes mellitus. Nat. Genet. 23:323–328. [DOI] [PubMed] [Google Scholar]
- 31.Marshak, S., H. Totary, E. Cerasi, and D. Melloul. 1996. Purification of the beta-cell glucose-sensitive factor that transactivates the insulin gene differentially in normal and transformed islet cells. Proc. Natl. Acad. Sci. USA 93:15057–15062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Melloul, D., Y. Ben-Neriah, and E. Cerasi. 1993. Glucose modulates the binding of an islet-specific factor to a conserved sequence within the rat I and the human insulin promoters. Proc. Natl. Acad. Sci. USA 90:3865–3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller, C. P., R. E. McGehee Jr., and J. F. Habener. 1994. IDX-1: a new homeodomain transcription factor expressed in rat pancreatic islets and duodenum that transactivates the somatostatin gene. EMBO J. 13:1145–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moran, E., B. Zerler, T. M. Harrison, and M. B. Mathews. 1986. Identification of separate domains in the adenovirus E1A gene for immortalization activity and the activation of virus early genes. Mol. Cell. Biol. 6:3470–3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mutoh, H., F. J. Naya, M.-J. Tsai, and A. B. Leiter. 1998. The basic helix-loop-helix protein BETA2 interacts with p300 to coordinate differentiation of secretin-expressing enteroendocrine cells. Genes Dev. 12:820–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naya, F. J., C. M. M. Stellrecht, and M.-J. Tsai. 1995. Tissue-specific regulation of the insulin gene by a novel basic helix-loop-helix transcription factor. Genes Dev. 9:1009–1019. [DOI] [PubMed] [Google Scholar]
- 37.Naya, F. J., H.-P. Huang, Y. Qiu, H. Mouth, F. J. DeMayo, A. B. Leiter, and M.-J. Tsai. 1997. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/NeuroD-deficient mice. Genes Dev. 11:2323–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nordeen, S. K., P. P. Green, I. I. I., and D. M. Fowles. 1987. Laboratory methods: a rapid, sensitive, and inexpensive assay for chloramphenicol acetyltransferase. DNA 6:173–178. [DOI] [PubMed] [Google Scholar]
- 39.Odagari, H., J. Wang, and M. S. German. 1996. Function of the human insulin promoter in primary cultured islet cells. J. Biol. Chem. 271:1909–1915. [DOI] [PubMed] [Google Scholar]
- 40.Offield, M. F., T. L. Jetton, R. Stein, T. Labosky, M. Ray, M. Magnuson, B. Hogan, and C. V. E. Wright. 1996. PDX-1 is required for development of the pancreas and differentiation of the rostral duodenum. Development. 122:983–995. [DOI] [PubMed] [Google Scholar]
- 41.Ohneda, K., R. G. Mirmira, J. Wang, J. D. Johnson, and M. S. German. 2000. The homeodomain of PDX-1 mediates multiple protein-protein interactions in the formation of a transcriptional activation complex on the insulin promoter. Mol. Cell. Biol. 20:900–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohlsson, H., K. Karlsson, and T. Edlund. 1993. IPF-1, a homeodomain-containing transactivator of the insulin gene. EMBO J. 12:4251–4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okita, K., Q. Yang, K. Yamagata, K. A. Hangenfeldt, J. Miyagawa, Y. Kajimoto, H. Nakajima, M. Namba, Wollheim, C. B., Hanafusa, T., and Matsuzawa, Y. 1999. Human insulin gene is a target of hepatocyte nuclear factor-1 alpha (HNF1α) and HNF1β. Biochem. Biophys. Res. Commun. 263:566–569. [DOI] [PubMed] [Google Scholar]
- 44.Olson, L. K., A. Sharma, C. V. E. Wright, H. C. Towle, R. P. Robertson, and R. Stein. 1995. Reduction of insulin gene transcription in HIT-T15 β cells chronically exposed to a supraphysiological glucose concentration is associated with loss of STF-1 transcription factor expression. Proc. Natl. Acad. Sci. USA 92:9127–9131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paggi, M. G., A. Baldi, F. Bonetto, and A. Giordano. 1996. Retinoblastoma protein family in cell cycle and cancer: a review. J. Cell. Biochem. 62:418–430. [DOI] [PubMed] [Google Scholar]
- 46.Parrazas, M. M. A. Maestro, S. F. Boj, A. Paniagua, R. Casamitjana, R. Gomis, F. Rivera, and J. Ferrer. 2001. Hepatic nuclear factor 1-alpha directs nucleosomal hyperacetylation to its tissue-specific targets. Mol. Cell. Biol. 21:3234–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peers, B., J. Leonard, S. Sharma, G. Teitelman, and M. R. Montminy. 1994. Insulin expression in pancreatic islet cells relies on cooperative interactions between the helix loop helix factor E47 and the homeobox factor STF-1. Mol. Endocrinol. 8:1798–1806. [DOI] [PubMed] [Google Scholar]
- 48.Peshavaria, M., E. Henderson, A. Sharma, C. V. E. Wright, and R. Stein. 1997. Functional characterization of the transactivation properties of the PDX-1 homeodomain protein. Mol. Cell. Biol. 17:3987–3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peshavaria, M., L. Gamer, E. Henderson, G. Teitelman, C. V. E. Wright, and R. Stein. 1994. XlHbox 8, an endoderm-specific Xenopus homeodomain protein, is closely related to a mammalian insulin gene transcription factor. Mol. Endocrinol. 8:806–816. [DOI] [PubMed] [Google Scholar]
- 50.Petersen, H. V., P. Serup, J. Leonard, B. K. Michelsen, and O. D. Madsen. 1994. Transcriptional regulation of the human insulin gene is dependent of the homeodomain proteins STF1/IPF1 acting through the CT boxes. Proc. Natl. Acad. Sci. USA 91:10465–10469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peyton, M., L. Moss, and M.-J. Tsai. 1994. Two distinct class A helix-loop-helix transcription factors, E2A and BETA1, form separate DNA-binding complexes on the insulin E-box. J. Biol. Chem. 269:25936–25941. [PubMed] [Google Scholar]
- 52.Qiu, Y., A. Sharma, and R. Stein. 1998. p300 mediates transcriptional stimulation by the basic helix-loop-helix activators of the insulin gene. Mol. Cell. Biol. 18:2957–2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson, G. L. W. G., M. Peshavaria, E. Henderson, S.-Y. Shieh, M.-J. Tsai, G. Teitelman, R. Stein. 1994. Analysis of transcription regulatory signals of the insulin gene: expression of the trans-active factor that stimulates insulin control element mediated expression precedes insulin gene transcription. J. Biol. Chem. 269:2452–2460. [PubMed] [Google Scholar]
- 54.Sander, M., A. Neubuser, J. Kalamaras, H. C. Ee, G. R. Martin, and M. S. German. 1997. Genetic analysis reveals that PAX6 is required for normal transcription of pancreatic hormone genes and islet development. Genes Dev. 11:1662–1673. [DOI] [PubMed] [Google Scholar]
- 55.Sander, M., and M. S. German. 1997. The beta cell transcription factors and development of the pancreas. J. Mol. Med. 75:327–340. [DOI] [PubMed] [Google Scholar]
- 56.Sharma, A., and R. Stein. 1994. Glucose-induced transcription of the insulin gene is mediated by factors required for β-cell-type-specific expression. Mol. Cell. Biol. 14:871–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sharma, A., L. K. Olson, R. P. Robertson, and R. Stein. 1995. Inhibition of insulin gene transcription in HIT T-15 β Cells by high glucose is mediated by the RIPE3b1 and STF-1 activators. Mol. Endocrinol. 9:1127–1134. [DOI] [PubMed] [Google Scholar]
- 58.Sharma, A., M. Moore, E. Marcora, J. E. Lee, Y. Qiu, S. Samaras, S., and R. Stein. 1999. The NeuroD1/BETA2 sequences essential for insulin gene transcription colocalize with those necessary for neurogenesis and p300/CREB binding protein binding. Mol. Cell. Biol. 18:704–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shieh, S.-Y., and M.-J. Tsai. 1991. Cell-specific and ubiquitous factors are responsible for the enhancer activity of the rat insulin II gene. J. Biol. Chem. 266:16708–16714. [PubMed] [Google Scholar]
- 60.Shikama, N., J. Lyon, and N. B. La Thangue. 1997. The p300/CBP family: integrating signals with transcription factors and chromatin. Trends Cell Biol. 7:230–236. [DOI] [PubMed] [Google Scholar]
- 61.Soutoglou, E., G. Papafotiou, N. Katrakili, and I. Talianidis. 2000. Transcriptional activation by hepatocyte nuclear factor-1 requires synergism between multiple coactivator proteins. J. Biol. Chem. 275:12515–12520. [DOI] [PubMed] [Google Scholar]
- 62.Stein, R. 2001. Insulin gene transcription: the factors involved in cell-type-specific and glucose-regulated expression in islet β cells are also essential during pancreatic development, p. 25–78. In A. Cherrington and J. Jefferson (ed.), Handbook of physiology, section 7. The endocrine system, vol. II. American Physiology Society, Washington, D.C.
- 63.Stein, R. W., M. Corrigan, P. Yaciuk, J. Whelan, and E. Moran. 1990. Analysis of E1A-mediated growth regulation functions: binding of the 300-kilodalton cellular product correlates with E1A enhancer repression function and DNA synthesis-inducing activity. J. Virol. 64:4421–4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stein, R. W., and J. Whelan. 1989. Insulin gene enhancer activity is inhibited by adenovirus 5 E1a gene products. Mol. Cell. Biol. 9:4531–4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Steiner, D. F., S. J. Chan, J. M. Welsh, and S. C. K. Kwok. 1985. Structure and evolution of the insulin gene. Annu. Rev. Genet. 19:463–484. [DOI] [PubMed] [Google Scholar]
- 66.Stellrecht, M. M., F. J. DeMayo, M. J. Finegold, and M.-J. Tsai. 1997. Tissue-specific and developmental regulation of the rat insulin II gene enhancer, RIPE3, in transgenic mice. J. Biol. Chem. 270:21503–21508. [DOI] [PubMed] [Google Scholar]
- 67.Stoffers, D. A., J. Ferrer, W. L. Clarke, and J. F. Habener. 1997. Early onset type II diabetes (MODY4) linked to IPF-1. Nat. Genet. 17:138–139. [DOI] [PubMed] [Google Scholar]
- 68.Stoffers, D. A., N. T. Zinkin, V. Stanojevec, W. L. Clarke, and J. F. Habener. 1997. Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nat. Genet. 15:107–110. [DOI] [PubMed] [Google Scholar]
- 69.St-Onge, L., B. Sosa-Pineda, K. Chowdhury, A. Mansouri, P. Gruss. 1997. Pax6 is required for differentiation of glucagon-producing α-cells in mouse pancreas. Nature 387:406–409. [DOI] [PubMed] [Google Scholar]
- 70.Whelan, J., D. Poon, P. A. Weil, and R. Stein. 1989. Pancreatic β-cell-type-specific expression of the rat insulin II gene is controlled by positive and negative transcriptional elements. Mol. Cell. Biol. 9:3253–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Whelan, J., S. R. Cordle, E. Henderson, P. A. Weil, and R. Stein. 1990. Identification of a pancreatic β-cell insulin gene transcription factor that binds to and appears to activate cell-type-specific expression: its possible relationship to other cellular factors that bind to a common insulin gene sequence. Mol. Cell. Biol. 10:1564–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yuan, W., G. Condorelli, M. Caruso, A. Felsani, and A. Giordano. 1996. Human p300 protein is a coactivator for the transcription factor MyoD. J. Biol. Chem. 271:9009–9013. [DOI] [PubMed] [Google Scholar]
- 73.Zhao, L., M. A. Cissell, E. Henderson, R. Colbran, and R. Stein. 2000. The RIPE3b1 activator of the insulin gene is composed of a protein(s) of approximately 43 kDa, whose DNA binding activity is inhibited by protein phosphatase treatment. J. Biol. Chem. 275:10532–10537. [DOI] [PubMed] [Google Scholar]