Abstract
BRCA1 carboxyl-terminal (BRCT) motifs are present in a number of proteins involved in DNA repair and/or DNA damage-signaling pathways. Human DNA topoisomerase II binding protein 1 (TopBP1) contains eight BRCT motifs and shares sequence similarity with the fission yeast Rad4/Cut5 protein and the budding yeast DPB11 protein, both of which are required for DNA damage and/or replication checkpoint controls. We report here that TopBP1 is phosphorylated in response to DNA double-strand breaks and replication blocks. TopBP1 forms nuclear foci and localizes to the sites of DNA damage or the arrested replication forks. In response to DNA strand breaks, TopBP1 phosphorylation depends on the ataxia telangiectasia mutated protein (ATM) in vivo. However, ATM-dependent phosphorylation of TopBP1 does not appear to be required for focus formation following DNA damage. Instead, focus formation relies on one of the BRCT motifs, BRCT5, in TopBP1. Antisense Morpholino oligomers against TopBP1 greatly reduced TopBP1 expression in vivo. Similar to that of ataxia telangiectasia-related protein (ATR), Chk1, or Hus1, downregulation of TopBP1 leads to reduced cell survival, probably due to increased apoptosis. Taken together, the data presented here suggest that, like its putative counterparts in yeast species, TopBP1 may be involved in DNA damage and replication checkpoint controls.
Cell cycle checkpoints induced by DNA damage are essential for maintaining genetic integrity. Signals of DNA damage lead to cell cycle arrest and allow time for the repair of damaged DNA (for recent reviews, see references41, 45, and 72). Failure of checkpoint responses results in genetic instability, frequently leading to cancer development.
In mammals, ataxia telangiectasia mutated protein (ATM) and ataxia telangiectasia-related protein (ATR), two phosphatidylinositol-3 kinase (PI3K)-related protein kinases, are essential components in DNA damage-signaling pathways. In response to DNA damage and/or replication blocks, ATM and ATR activate the downstream checkpoint kinases Chk1 and Chk2/Cds1 (see references 41, 45, and 72 for details). Together, these four DNA damage-activated kinases phosphorylate and regulate a number of proteins, including Cdc25C (4, 7, 13, 35, 39, 51), Cdc25A (21, 36), NBS1 (24, 34, 65, 70), p53 (3, 11, 14, 28, 31, 55, 58), BRCA1 (15, 17, 23, 25, 32, 59), and CtIP (33). By regulating the functions of these proteins and other unidentified substrates, these kinases play essential roles in coordinating DNA repair, cell cycle progression, transcriptional regulation, and apoptosis in response to various DNA-damaging events.
In order to understand in detail the mammalian DNA damage-signaling pathway, one has to identify the physiological substrates of ATM and ATR. It is interesting that several ATM and/or ATR substrates, including BRCA1 and NBS1, contain BRCA1 carboxyl-terminal (BRCT) motifs. BRCT motifs were originally identified in the breast cancer tumor suppressor protein BRCA1 (30) and have since been identified in a number of proteins involved in DNA repair (e.g., XRCC1 and DNA ligases III and IV) and cell cycle checkpoints (e.g., Cut5/Rad4, Crb2, and Saccharomyces cerevisiae Rad9 [scRad9]) (6, 10). At least for BRCA1, the BRCT motifs appear to be critical for its tumor suppression function, since these motifs are frequently lost or mutated in tumor-associated BRCA1 mutants.
DNA topoisomerase II binding protein 1 (TopBP1), a protein containing eight BRCT motifs, was cloned through its association with topoisomerase IIβ in a yeast two-hybrid screen (68). While the biological significance of TopBP1-topoisomerase II interaction remains to be resolved, TopBP1 shares sequence and structural similarities with the fission yeast Rad4/Cut5 protein. Rad4/Cut5 is a checkpoint Rad protein involved in cellular responses to DNA damage and replication blocks (22, 40, 47–50, 60). Genetic and biochemical studies suggest that Schizosaccharomyces pombe Rad4/Cut5 (pRad4/Cut5) and its associated protein spCrb2 interact with the checkpoint kinase spChk1 and act upstream of spChk1 in the checkpoint signaling pathway (47). Thus, eight checkpoint Rad proteins (Rad3, Rad17, Rad9, Rad1, Hus1, Cut5/Rad4, Crb2, and Rad26) are required to activate the downstream checkpoint protein kinases Chk1 and/or Cds1/Chk2 in fission yeast (for reviews, see references 41, 45, and 72).
The S. cerevisiae homologue of spRad4/Cut5 is DPB11, a protein that interacts with DNA polymerase and is required for S-phase progression as well as DNA damage and S-phase checkpoint controls (2, 62). DPB11 is required for the proper activation of the checkpoint kinase RAD53, the budding yeast homologue of spCds1/human Chk2 (hChk2), following DNA damage and replication blocks (62), suggesting that DPB11 acts upstream of RAD53 in the DNA damage-signaling pathway.
In Drosophila melanogaster, the mutagen-sensitive 101 protein (Mus101) is the closest homologue of spRad4/Cut5 and scDPB11 (67). Mus101 contains seven BRCT motifs distributed throughout its primary sequence. Some mus101 mutant phenotypes include hypersensitivity to DNA-damaging agents and ionizing radiation, defects in DNA synthesis, and chromosome instability, suggesting that, like spRad4/Cut5 and scDPB11, Mus101 also plays a role in DNA repair, replication, and checkpoint controls.
Because TopBP1 shares sequence similarity with spRad4/Cut5, scDPB11, and the Drosophila Mus101 protein (dMus101), we examined whether TopBP1 would be regulated in response to DNA damage. Here we report that TopBP1 is phosphorylated and localizes to the sites of DNA damage in response to DNA double-strand breaks and replication blocks. TopBP1 expression peaks in S-phase cells. Similar to what occurs with other proteins (ATR, Chk1, or hHus1) involved in S-phase checkpoints, downregulation of TopBP1 results in reduced cell survival due to increased apoptosis. Taken together, these results suggest that TopBP1 participates in the mammalian DNA damage- and/or replication block-signaling pathways.
MATERIALS AND METHODS
Cell culture and ionizing radiation.
Cells were grown in RPMI 1640 medium supplemented with 10% fetal calf serum in a humidified atmosphere of 5% CO2 and 95% air at 37°C. The cells were irradiated in a JL Shepherd 137Cs radiation source at a rate of 10 Gy/min. The cells were then returned to the culture conditions and maintained for the times indicated in the figure legends.
Immunoprecipitation, immunoblotting, and antibodies.
Immunoprecipitation and immunoblotting were performed as described previously (54). TopBP1 was extracted from cells with NETN buffer (0.5% NP-40, 1 mM EDTA, 20 mM Tris-HCl [pH 8.0], 100 mM NaCl) (5 volumes of cell pellet) on a slowly moving shaker for 10 min. The sample was centrifuged to isolate the supernatant. Immunoprecipitation experiments were carried out in NETN buffer. Rabbit polyclonal anti-TopBP1 serum was purified by using glutathione S-transferase (GST)-B6-78 (69) immobilized on aminolink-plus columns according to the manufacturer’s instructions (Pierce). All experiments described here were performed with this affinity-purified anti-TopBP1 antibody.
In phosphatase treatment experiments, TopBP1 immunoprecipitates (30 μl of a mixture containing a 50% slurry of protein A and 7 μg of antibody in NETN buffer) were treated with 400 U of lambda protein phosphatase by incubation in the accompanying buffer (New England Biolabs) for 30 min at 30°C. Polyacrylamide gels (5 or 4%) were used for the separation of unphosphorylated and phosphorylated TopBP1 proteins.
Anti-BRCA1 monoclonal antibody SD118 and anti-NBS1 monoclonal antibody EE15 were kindly provided by David M. Livingston and Xiaohua Wu (Dana-Farber Cancer Institute, Boston, Mass.). Anti-PCNA monoclonal antibody PC10 was purchased from Santa Cruz Biotechnology, Inc. The generation of anti-53BP1 antibodies was described previously (44).
Anti-phospho-TopBP1 antibodies were raised against a peptide (CDEDLLS405QYENGS) containing phosphorylated Ser405 of TopBP1. The antisera were precleared with a column containing nonphosphorylated peptides and then affinity purified with a column containing phosphorylated peptides. Anti-phospho-Tyr antibody was purchased from Upstate Biotechnology, Inc. Anti-Myc epitope antibody 9E10 was purchased from Berkeley Antibody Company.
Immunostaining.
Cells grown on coverslips were washed with phosphate-buffered saline and fixed with 50% acetone-50% methanol (vol/vol) for 10 min unless otherwise stated. After being washed with TBST (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.2% Tween 20), the coverslips were incubated for 1 h with primary antibodies in 10% goat serum in TBST at room temperature. The coverslips were washed three times with TBST and incubated with secondary antibodies, fluorescein-conjugated goat anti-mouse immunoglobulin G (IgG) and rhodamine-conjugated anti-rabbit IgG (1:200 dilution; Jackson Immunoresearch Lab), in 10% goat serum in TBST at room temperature for 20 min. Hoechst dye was added for nuclear staining. After repeated washing, the coverslips were mounted on glass slides with 2 drops of a mixture containing 1 mg of p-phenylenediamine/ml and 90% glycerol in phosphate-buffered saline. The samples were observed with a fluorescent microscope (Nikon E800) at a magnification of ×600. Images were captured with a Metamorph imaging system and processed with Adobe Photoshop software.
ATM kinase assay.
ATM was immunoprecipitated from K562 cells using anti-ATM antibody 132 raised against a peptide (CKSLASFIKKPFDRGEVESMEDDTNG). The kinase assay was performed as described previously (29), except that the immunoprecipitates were washed three times with buffer containing 1.0 M LiCl.
DNA fragments coding for the SQ or TQ sites of TopBP1 were cloned into the BamHI and XhoI sites of pGEX-5X-1 (Amersham) by a procedure similar to that described previously (29). GST fusion proteins (2 μg) were purified as described previously (69) and used as substrates in the ATM kinase assay.
Transient expression of Myc-tagged TopBP1 and its derivatives.
Myc epitope-tagged TopBP1 in a modified pcDNA3 (Invitrogen) vector was introduced into SaOS2 cells by using Fugene 6 (Roche) transfection reagent. After 2 days, the cells were irradiated, fixed, and immunostained as described above.
Colony formation assay and fluorescence-activated cell sorter (FACS)-apoptosis analysis.
A pcDNA3.1 derivative with a neomycin resistance gene (26 μg) was cotransfected with antisense oligomers (500 μM, 12.5 μl) containing artificial backbone structures (Gene Tools; Morpholino). The Morpholino oligomers used were As1 (5′-TTGGGACACATCGCTGGTGGTGCAT) (at the translational initiation site) and As2 (5′-AAACGGTTCTTTGTCATTTCTGGAC) (at the 5′ untranslated region). The control Morpholino oligonucleotide (5′-CCTCTTACCTCAGTTACAATTTATA) was a standard control from Gene Tools. The mixture containing 160 μl of cells was subjected to electroporation at 344 V for 10 ms in a 0.4-cm gapped cuvette. The cells were maintained for 3 days before being seeded on plates containing RPMI 1640 medium and 1 mg of neomycin/ml. Colonies were stained and counted 15 days later. Following formaldehyde fixation, terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assays were performed as suggested by the manufacturer (Apoalert; Clontech).
RESULTS
TopBP1 is phosphorylated in response to ionizing radiation.
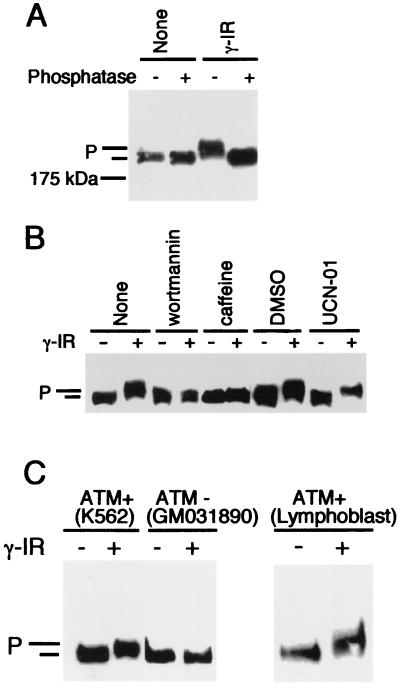
To examine whether TopBP1 is phosphorylated in response to DNA damage, K562 cells were treated with γ irradiation. One hour later, whole-cell lysates were prepared, immunoprecipitated, and analyzed by Western blotting with anti-TopBP1 antibody. As shown in Fig. 1A, TopBP1 migrated slower in a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel following DNA damage. This slower mobility of TopBP1 was reversed when the immunoprecipitates were treated with lambda protein phosphatase (Fig. 1A), suggesting that TopBP1 is phosphorylated in response to DNA damage.
FIG. 1.
TopBP1 is phosphorylated in response to DNA damage. (A) K562 cells were treated with γ irradiation (γ-IR) (10 Gy) or left untreated (None). After 1 h, whole-cell lysates were immunoprecipitated with anti-TopBP1 antibody. Duplicate samples were treated with lambda phosphatase (+) or left untreated (−). Western blotting was performed with anti-TopBP1 antibody. (B) Treatments of cells with wortmannin (100 μM), caffeine (10 mM), or UCN-01 (1 μM) were performed 30 min before γ irradiation. Dimethyl sulfoxide (DMSO) was the solvent for wortmannin and was used here as a negative control. Duplicate samples were treated with γ irradiation (+) (10 Gy) or left untreated (−) and analyzed by Western blotting. (C) Cells expressing wild-type ATM (K562 and normal lymphoblast) and ATM-deficient cells (GM031890) were treated with γ irradiation (+) (10 Gy) or left untreated (−) and collected 1 h later. Western blotting was performed using anti-TopBP1 antibody. Phosphorylated (P) and unphosphorylated (unlabeled bar) TopBP1 bands are indicated for all panels.
ATM is required for the ionizing radiation-induced phosphorylation of TopBP1.
ATM plays a central role in cell cycle checkpoint controls. To examine whether ATM contributes to the phosphorylation of TopBP1 following DNA damage, we pretreated cells with wortmannin or caffeine, two agents that can inhibit PI3K-related protein kinases, including ATM (for examples, see references 5 and 52). As shown in Fig. 1B, pretreatment of cells with wortmannin or caffeine inhibited the γ radiation-induced phosphorylation of TopBP1, suggesting that ATM or other PI3K-related kinases participate in TopBP1 phosphorylation. UCN-01 (7-hydroxystaurosporine), an inhibitor of hChk1 (9, 26), did not inhibit TopBP1 phosphorylation, indicating that the downstream checkpoint kinase hChk1 may not be required for this phosphorylation event.
To examine directly whether ATM is required for TopBP1 phosphorylation following DNA damage, we used an ATM-deficient lymphoblast cell line, GM03189D. As shown in Fig. 1C, damage-induced phosphorylation of TopBP1 was absent in the ATM-deficient cells but was readily observed 1 h after radiation of K562 cells or normal lymphoblasts that express wild-type ATM, suggesting that ATM is required for damage-induced phosphorylation of TopBP1. We also examined the time course of TopBP1 phosphorylation in ATM wild-type and ATM-deficient cells. While TopBP1 phosphorylation persisted for at least 6 h following radiation in wild-type cells, no detectable phosphorylation of TopBP1 was observed in ATM-deficient cells (data not shown). Taken together, these data suggest that ATM is required for radiation-induced phosphorylation of TopBP1.
ATM phosphorylates TopBP1 in vitro and in vivo.
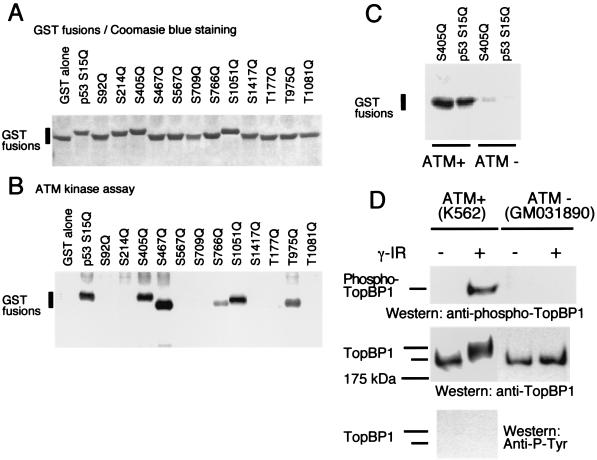
To examine whether ATM directly phosphorylates TopBP1, we generated GST fusion proteins containing potential ATM phosphorylation sites (serine-/threonine-glutamine sites) of TopBP1 (Fig. 2A). The ATM kinase assay was performed with these GST peptides as the substrates and with immunoprecipitated ATM. As shown in Fig. 2B, ATM efficiently phosphorylated peptides containing the S405Q, S467Q, and S1051Q sites of TopBP1 in vitro. The extent of phosphorylation at these three positions was similar to that at S15Q of p53, a known ATM phosphorylation site. ATM also phosphorylated TopBP1, to a lesser extent, at two additional sites, S766Q and T975Q. As a negative control, GST alone was not phosphorylated by ATM in these experiments. While ATM immunoprecipitates prepared from K562 cells (containing wild-type ATM) efficiently phosphorylated Ser405 of TopBP1 and Ser15 (a known ATM phosphorylation site) of p53, immunoprecipitates using the same anti-ATM antibody prepared from ATM-deficient cells did not lead to efficient phosphorylation of these two substrates (Fig. 2C). Taken together, these results suggest that TopBP1 is a substrate of ATM in vitro.
FIG. 2.
ATM is required for phosphorylation of TopBP1. (A) GST fusion proteins with corresponding SQ or TQ mutation sites of TopBP1 were expressed in Escherichia coli and purified on glutathione-Sepharose beads. Proteins were eluted with SDS loading buffer, separated by gel electrophoresis, and visualized with Coomassie blue staining. (B) ATM kinase assays were performed with the purified GST fusion proteins as substrates, which were prepared as described for panel A, and ATM kinase immunoprecipitated from K562 lysates. (C) ATM kinase assays were performed with anti-ATM immunoprecipitates prepared from K562 cells (containing wild-type ATM) (ATM+) or from ATM-deficient GM031890 cells (ATM−). (D) Anti-TopBP1 immunoprecipitates were prepared as described in the legend to Fig. 1A. A phosphospecific antibody raised against the phospho-Ser405 of TopBP1 (anti-phospho-TopBP1 antibody) was used for Western blotting to detect phosphorylated TopBP1 following ionizing radiation. Anti-TopBP1 and anti-phosphotyrosine antibodies were used here as the positive and negative controls, respectively. +, γ irradiated; −, untreated; unlabeled bar, unphosphorylated TopBP1.
To confirm that TopBP1 is phosphorylated by ATM in vivo, we generated a phosphospecific antibody raised against phospho-Ser405 of TopBP1. This antibody recognized phosphorylated TopBP1 isolated from irradiated K562 cells but failed to recognize TopBP1 isolated from control cells or from irradiated ATM-deficient cells (Fig. 2D). When used as a control, an anti-phospho-Tyr-specific antibody did not recognize phosphorylated TopBP1 in the same experiment. Thus, TopBP1 is phosphorylated by ATM following ionizing radiation.
TopBP1 forms foci and colocalizes with BRCA1 and NBS1 following ionizing radiation.
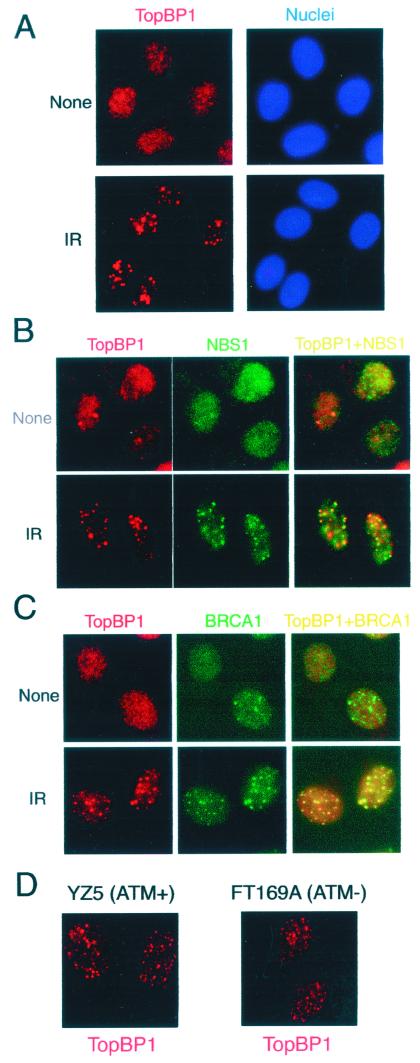
To examine the cellular localization of TopBP1, we performed immunostaining experiments with U2OS cells by using anti-TopBP1 antibodies. As shown in Fig. 3A, TopBP1 normally localized in nuclei, as demonstrated by granular nuclear staining. Following γ irradiation, TopBP1 became concentrated in nuclear foci (Fig. 3A), suggesting that the subnuclear localization of TopBP1 is regulated in response to DNA damage.
FIG. 3.
TopBP1 forms foci and colocalizes with NBS1 and BRCA1 after ionizing irradiation. U2OS cells were treated with γ irradiation (IR) (10 Gy) or left untreated (None) and incubated for 6 (A and B) or 20 (C) h. Cells were fixed and stained with anti-TopBP1 (red, TopBP1) antibody and Hoechst dye (blue, Nuclei) (A), anti-TopBP1 (red, TopBP1) and anti-NBS1 (green, NBS1) antibodies (B), or anti-TopBP1 (red, TopBP1) and anti-BRCA1 (green, BRCA1) antibodies (C). (B and C) Two images were merged on the unlabeled right panels, and yellow dots indicate colocalization of TopBP1 with NBS1 or BRCA1. (D) YZ5 cells containing wild-type ATM and ATM-deficient FT169 cells were treated with γ irradiation (10 Gy) and incubated for 6 h before being fixed and stained with anti-TopBP1 antibody (red).
These TopBP1-containing nuclear foci are reminiscent of the ionizing radiation-induced foci (IRIF) that were reported previously. NBS1/Mre11/Rad50 and BRCA1 localize to these IRIF following γ irradiation (12, 38, 64, 71). Thus, we examined whether TopBP1 colocalizes with NBS1 and BRCA1 following DNA damage. Six hours after γ irradiation, U2OS cells were fixed and stained with anti-TopBP1 and anti-NBS1 antibodies (Fig. 3B). As shown in a merged image, TopBP1 foci colocalized with NBS1 foci following DNA damage (Fig. 3B) (∼43% of TopBP1 foci colocalized with those of NBS1). Similarly, coimmunostaining experiments were performed with anti-BRCA1 and anti-TopBP1 antibodies to examine whether TopBP1 colocalizes with BRCA1 before and after DNA damage. In agreement with published observations (16, 54), BRCA1 localized to foci in normal S- and G2-phase cells (Fig. 3C). TopBP1 did not colocalize with BRCA1 in these S-phase foci (Fig. 3C). However, TopBP1 colocalized with BRCA1 in IRIF following γ irradiation (Fig. 3C) (65% of TopBP1 foci colocalized with those of BRCA1). Taken together, these data suggest that TopBP1 colocalizes with NBS1 and BRCA1 in IRIF following DNA damage. Identical results were obtained when MCF-7, HBL100, IMR90, and SaOS2 cells were used.
To examine whether the focus formation of TopBP1 depends on its phosphorylation by ATM following DNA damage, we used ATM-deficient FT169 cells and wild-type ATM-reconstituted YZ5 cells. While TopBP1 foci formed readily in YZ5 cells, we also detected TopBP1 foci in ATM-deficient FT169A cells (Fig. 3D). Thus, phosphorylation of TopBP1 by ATM is not required for its focus formation.
IRIF formation requires BRCT5 of TopBP1.
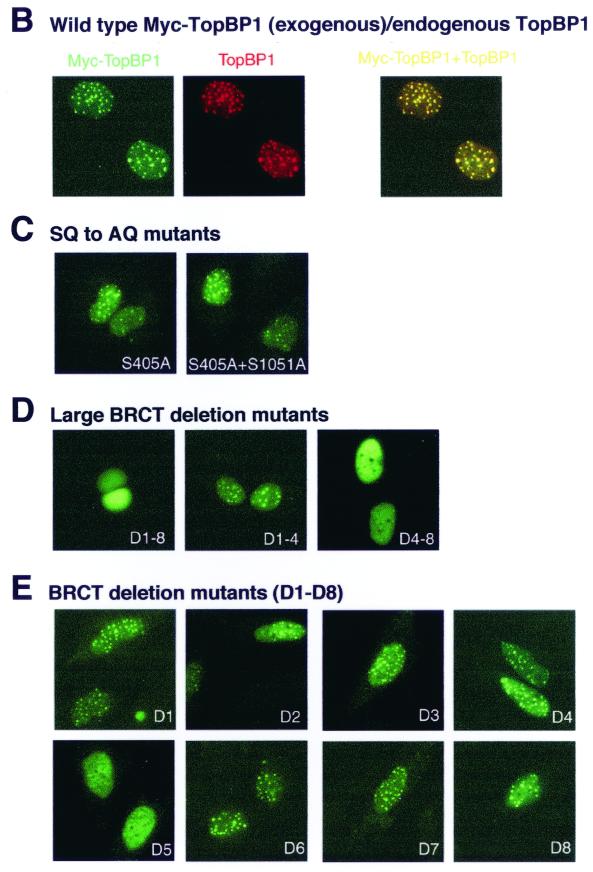
To explore the regulation of TopBP1 focus formation, we generated expression constructs of Myc-epitope-tagged wild-type TopBP1 and a series of TopBP1 deletion mutants (Fig. 4A). These constructs were transiently transfected into SaOS2 cells. Neither myc-tagged wild-type TopBP1 nor any of these deletion mutants formed foci in unirradiated cells (data not shown), but Myc-tagged wild-type TopBP1 readily localized to foci following radiation (Fig. 4B).
FIG. 4.
BRCT5 is required for IRIF localization of TopBP1. (A) Schematic diagram of wild-type TopBP1 and its derivatives. All mutations were generated with the QuickChange mutagenesis protocol (Stratagene). Thick lines indicate remaining regions in TopBP1. Results of focus formation as described below are summarized. (B to E) Plasmids carrying Myc-epitope-tagged wild-type or mutant TopBP1 were transfected into SaOS2 cells. The cells were treated with γ irradiation (10 Gy), incubated for 6 h, and fixed and stained with anti-Myc (green, Myc-TopBP1) and anti-TopBP1 (red, TopBP1) antibodies. Only transfected cells showed moderate or strong positive signals for anti-Myc antibody, whereas untransfected cells showed weak, uniform nuclear staining. (B) Two images were merged on the unlabeled right panel, and yellow dots indicate colocalization of TopBP1 with Myc-TopBP1.
To determine the regions of TopBP1 required for the formation of its damage-induced foci, immunostaining experiments were performed in cells transfected with various TopBP1 mutants. In agreement with the hypothesis that TopBP1 focus formation is independent of ATM, two phosphorylation mutants (S405A and S405A+S1051A) also localized to foci following DNA damage (Fig. 4C). TopBP1 contains eight BRCT motifs. Deletion of all eight BRCT motifs or C-terminal BRCT motifs (Fig. 4D, panel D-8) abolished or reduced damage-induced focus formation of TopBP1. However, deletion of the four N-terminal BRCT motifs (Fig. 4D, panel D1-8) did not affect TopBP1 focus formation, suggesting that the C terminus of TopBP1 controls its localization following DNA damage. Indeed, deletion of BRCT5 (Fig. 4E, panel D5), but not any other BRCT motifs, diminished damage-induced focus formation of TopBP1. Thus, BRCT5 is required for TopBP1 focus formation.
TopBP1 colocalizes and associates with 53BP1.
The tumor suppressor p53 binding protein 53BP1 participates early in the DNA damage-signaling pathway (1, 44, 53, 66). 53BP1 contains tandem BRCT motifs at the C terminus (27) and shares sequence homology with spCrb2, a protein that interacts with spRad4/Cut5 (47). Because TopBP1 may be the mammalian homologue of spRad4/Cut5, we examined whether TopBP1 associates with 53BP1. As shown in Fig. 5A, TopBP1 coimmunoprecipitated with 53BP1 and vice versa. In addition, TopBP1, synthesized by in vitro transcription and translation, associated with a GST fusion protein containing residues 956 to 1354 of 53BP1 (Fig. 5B), suggesting that TopBP1 may directly interact with 53BP1 in vivo.
FIG. 5.
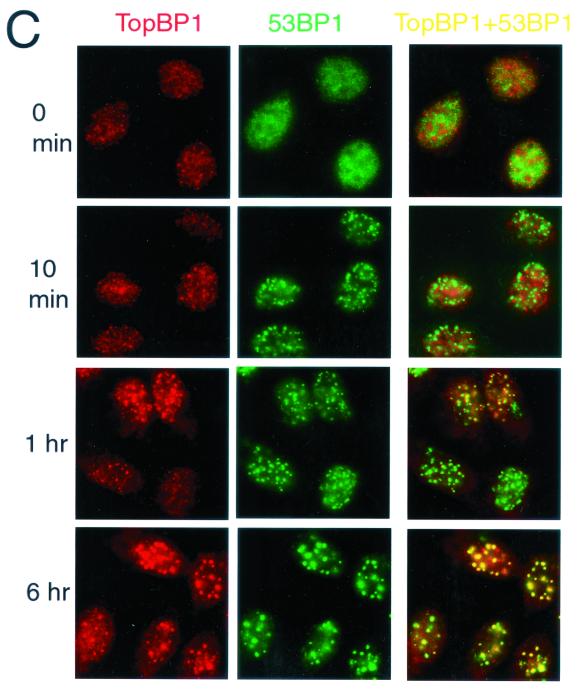
TopBP1 associates with 53BP1 and colocalizes with 53BP1 following DNA damage. (A) Cell lysates prepared from HBL100 cells were immunoprecipitated with the indicated antibodies, and Western blotting was performed with anti-53BP1 or anti-TopBP1 antibodies. In each panel, 10% of the immunoprecipitates was applied in the two left lanes. (B) TopBP1 was translated and 35S labeled in vitro by using the T7 quick-coupled transcription-translation system (Promega). The products were incubated with GST-53BP1 fusion proteins immobilized on beads. The beads were washed, and the proteins bound to the beads were eluted in SDS sample buffer, separated by SDS-PAGE, and visualized by autoradiography. Fragments 1, 2, 3, 4, 5, and 6 correspond to 53BP1 residues 1 to 346, 339 to 671, 628 to 962, 956 to 1354, 1331 to 1664, and 1657 to 1972, respectively. (C) U2OS cells were left untreated (0 min) or treated with γ irradiation at 10 Gy, incubated for the indicated times, and fixed and immunostained with anti-TopBP1 (red, TopBP1) and anti-53BP1 (green, 53BP1) antibodies. Two images were merged on the unlabeled right panels, and yellow dots indicate colocalization of TopBP1 with 53BP1.
53BP1 forms nuclear foci and colocalizes with the phosphorylated histone H2AX at the sites of DNA double-strand breaks within minutes after irradiation (1, 43, 44, 46, 53, 66). Thus, we examined the time course of TopBP1 focus formation in U2OS cells. As shown in Fig. 5C, while 53BP1 readily localized to foci within 10 min after irradiation, TopBP1 foci were not apparent at this early time point. After 1 h, approximately 50% of TopBP1 foci colocalized with 53BP1 foci. After 6 h, more than 90% of TopBP1 foci colocalized with 53BP1. Thus, TopBP1 appears to enter these foci after 53BP1 does.
Regulation of TopBP1 following replication blocks.
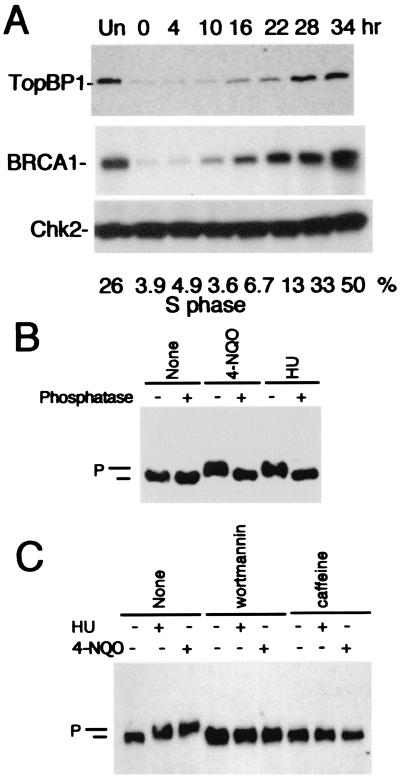
TopBP1 shares sequence and structural homology with the fission yeast protein Rad4/Cut5 and the budding yeast protein DPB11. Both proteins are involved not only in DNA damage but also in replication checkpoint controls. We have observed that, similar to that of BRCA1 (16), the protein level of TopBP1 peaks in S phase (Fig. 6A and data not shown), suggesting that TopBP1 may participate in certain S-phase functions. Similar findings regarding cell cycle regulation of TopBP1 were published recently (37).
FIG. 6.
Regulation of TopBP1 in S-phase cells. (A) MCF7 cells were serum starved for 2 days. The cells were then sequentially released from serum starvation for the indicated times. Western blotting was performed with anti-TopBP1, anti-BRCA1 (MS110), and anti-Chk2 (monoclonal antibody no. 7) antibodies, respectively. The percentages of the cell populations in the S phase were analyzed by FACS and are indicated at the bottom of the panel. Un, unsynchronized cells. (B) K562 cells were left untreated (None) or treated with HU (1 mM) or 4-NQO (2 μg/ml). After 1 h, whole-cell lysates were prepared and immunoprecipitated with anti-TopBP1 antibody. Portions of samples were treated with lambda phosphatase. Western blotting was performed with anti-TopBP1 antibody. Phosphorylated (P) and unphosphorylated (unlabeled bar) TopBP1 bands are indicated. (C) K562 cells were treated with wortmannin (100 μM) or caffeine (10 mM) for 30 min before the procedures described above were performed. (D) U2OS cells were treated with 4-NQO or HU, fixed 1 h later, and stained with anti-TopBP1 (red, TopBP1) and anti-PCNA (green, PCNA) antibodies. Two images were merged on the unlabeled right panels, and yellow dots indicate colocalization of TopBP1 with PCNA.
To explore whether TopBP1 is involved in the replication checkpoints, we treated cells with hydroxyurea (HU) or 4-nitroquinoline-1-oxide (4-NQO), a UV-mimetic agent, both of which lead to the inhibition of DNA synthesis. As shown in Fig. 6B, inhibition of DNA replication leads to the phosphorylation of TopBP1. As with ionizing radiation, replication block-induced TopBP1 phosphorylation is inhibited by wortmannin or caffeine (Fig. 6C), suggesting that ATM and/or ATR are involved in these phosphorylation events. To examine the localization of TopBP1 following replication blocks, the control and treated cells were fixed with methanol-acetone. As shown in Fig. 6D, HU or 4-NQO treatments led to relocalization of TopBP1 to the arrested replication forks (indicated by anti-PCNA staining), a finding similar to that reported by Makiniemi and colleagues (37). Thus, TopBP1 is likely involved in the replication checkpoint controls.
TopBP1 is required for cell survival.
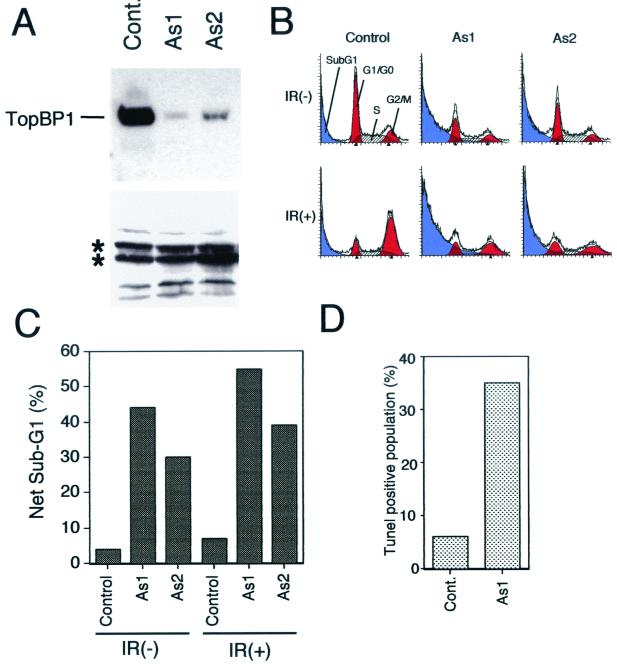
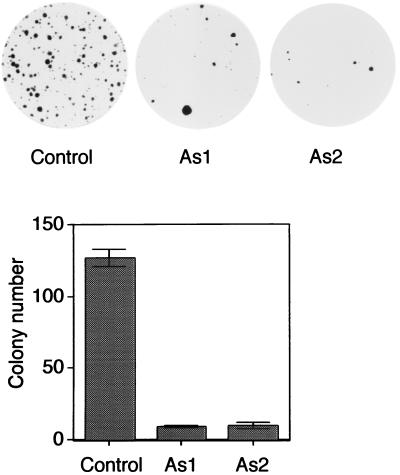
ATR, Chk1, and Hus1 are required for checkpoint controls in mammals. ATR-, Chk1-, and Hus1-deficient cells all show reduced cell viability, probably due to increased apoptosis (8, 18, 35, 57, 63), suggesting that checkpoint function is critical for cell survival. Using a control and two antisense Morpholino oligomers, we examined the consequences of abolishing TopBP1 expression in vivo. Two antisense Morpholino oligomers of TopBP1 (AS1 and AS2) strongly inhibited TopBP1 expression in the cell (Fig. 7A). These two antisense Morpholino oligomers also induced apoptosis in the transfected cells, as measured by sub-G1 (i.e., apoptotic) populations and also by TUNEL assays (Fig. 7B to D). These increases in apoptosis are independent of exogenous DNA damage (Fig. 7B and C), suggesting that TopBP1 is required for normal cell survival. Indeed, the two antisense Morpholino oligomers strongly inhibited cell survival in colony formation assays, confirming that TopBP1 is essential for cell viability (Fig. 8).
FIG. 7.
Antisense Morpholino oligomers for TopBP1 mRNA strongly inhibit TopBP1 expression and induce apoptosis. Two different antisense Morpholino oligomers, As1 and As2, for the TopBP1 mRNA 5′ region were transfected into HeLa cells by electroporation. More than 95% of the cells were transfected as suggested, with fluorescein-conjugated Morpholino oligomers being used as a control (data not shown). After 3 days, cells were collected and equal amounts of whole-cell lysates were analyzed by Western blotting using anti-TopBP1 antibody. (A) Although TopBP1 levels were reduced in the cells transfected with As1 or As2, the intensities of several cross-reacting bands (indicated by stars) in the lower portion of the panel were not altered. (B) Cells transfected with control or As1 or As2 Morpholino oligomers were fixed with ethanol. Apoptosis and cell cycle distribution were analyzed by FACS analysis. (C) The percentages of the net sub-G1 populations are shown; from each of these values, the value representing the sub-G1 population derived from electroporation (∼20%) has been subtracted. IR(−), unirradiated; IR(+), irradiated. (D) Cells transfected with control or As1 Morpholino oligomers were fixed with formaldehyde, double-stained with propidium iodide, and analyzed by the TUNEL method using fluorescein-labeled dUTP. Percentages of TUNEL-positive cell populations are shown. Cont, control.
FIG. 8.
Reduction of TopBP1 expression leads to reduced cell survival. Colony formation assays were performed with HeLa cells cotransfected with a vector carrying a neomycin-resistant gene and the control, As1, or As2 Morpholino oligomers. Transfected cells were selected with neomycin-containing medium. Colonies were stained and counted. Representative plates are shown in the top panel. Averages of results from three independent experiments (plus or minus standard errors of the mean) are summarized in the bottom panel.
DISCUSSION
We report here that TopBP1 is phosphorylated after γ irradiation. Several lines of evidence suggest that ATM is required for this damage-induced phosphorylation of TopBP1. First, the inhibitors of the ATM kinase, wortmannin and caffeine, inhibit γ irradiation-induced TopBP1 phosphorylation. Second, TopBP1 phosphorylation is absent in the ATM-deficient cells following DNA damage. Third, ATM efficiently phosphorylates several residues of TopBP1 in vitro. Fourth, using a phosphospecific anti-TopBP1 antibody, we have shown that TopBP1 is phosphorylated at the S405Q site following DNA damage in an ATM-dependent manner. Taken together, these data suggest that TopBP1 is phosphorylated by ATM following ionizing radiation.
The subnuclear localization of TopBP1 is regulated by DNA damage. Following exposure to ionizing radiation, TopBP1 colocalizes with NBS1, BRCA1, and 53BP1 at the sites of DNA breaks. Although both phosphorylation and localization of TopBP1 are regulated by DNA damage, the phosphorylation of TopBP1 does not appear to be essential for its relocalization following DNA damage. Damage-induced focus formation of TopBP1 still occurs in ATM-deficient cells (Fig. 3D) and is not blocked by wortmannin or caffeine at concentrations sufficient to inhibit TopBP1 phosphorylation (data not shown). Phosphorylation-deficient mutants of TopBP1 localize normally to foci following DNA damage (Fig. 5C). Similar damage-dependent but ATM-independent focus localization has been reported for other ATM substrates, including BRCA1 and NBS1. For example, phosphorylation-deficient mutants of BRCA1 localize normally to foci (17). Relocalization of BRCA1 following DNA damage normally occurs in ATM-deficient cells (unpublished observations). In addition, there are reports that ATM-dependent phosphorylation events are not required for NBS1 focus formation following DNA damage (24, 65). Mirzoeva and Petrini have also shown that ATM does not influence the relocalization of the NBS1 complex following DNA double-strand breaks (40a). Thus, it seems that ATM-dependent phosphorylation events may regulate other functions of these substrates but not their subnuclear localization. What, then, determines the damage-induced focus formation of TopBP1? Using a series of TopBP1 deletion mutants, we have shown that only one of the eight BRCT motifs, BRCT5, is required for its damage-induced focus formation. It would be interesting to identify any protein specifically interacting with TopBP1 through the BRCT5 motif. Such a protein may be required for recruiting TopBP1 to the sites of DNA damage.
As shown by Makiniemi and colleagues (37), the TopBP1 protein level peaks in the S phase (see also Fig. 6). TopBP1 is phosphorylated and relocalizes to the stalled replication fork in response to HU or 4-NQO treatment. Furthermore, Makiniemi and colleagues have also shown that TopBP1 associates with the checkpoint protein hRad9 and DNA polymerase ɛ (37), suggesting that TopBP1 is involved in the S-phase checkpoints. Here, we have demonstrated that TopBP1 is required for cell survival. Inhibition of TopBP1 expression in cells leads to reduced cell viability due to increased apoptosis, a phenomenon similar to that in ATR-, Chk1-, or hHus1-deficient cells. Taken together, these data imply that in addition to its role in DNA damage-signaling pathways, TopBP1 participates in the replication checkpoint controls.
TopBP1 shares sequence and structural homology with the fission yeast Rad4/Cut5 protein, the budding yeast DPB11 protein, and the Drosophila Mus101 protein. All these proteins contain multiple BRCT motifs distributed throughout their corresponding primary sequences. Similar to the roles of spRad4/Cut5 and scDPB11 in DNA damage and replication checkpoint controls in yeast species, some Drosophila mus101 mutants are hypersensitive to DNA-damaging agents and ionizing radiation. Here, we have shown that TopBP1 may participate in the DNA damage-signaling pathway as well as in replication checkpoint controls. While spRad4/Cut5 interacts with spCrb2, a protein containing C-terminal tandem BRCT motifs (47), TopBP1, associates with 53BP1, also a protein with tandem BRCT motifs at the C terminus (27). These similarities suggest that TopBP1 may be the functional homologue of spRad4/Cut5 and scDPB11 in mammals. spRad4/Cut5 is required for Chk1 activation following DNA damage (47). In budding yeast, both scDPB11 and scRAD9 (the homologue of spCrb2) are required for RAD53 activation (19, 20, 42, 56, 61, 62). If TopBP1 were the spRad4/Cut5 and scDPB11 homologue, we would expect TopBP1 to be required for Chk1 and/or Chk2 activation following DNA damage. Given that TopBP1 binds to DNA breaks directly in vitro (69) and localizes to DNA damage sites in vivo, it is possible that TopBP1 participates in the recognition of DNA damage and cooperates with ATM to activate downstream checkpoint kinases (Chk1 and Chk2). Because TopBP1 is essential for cell survival, we have not yet been able to examine directly the roles of TopBP1 in mammalian DNA damage-signaling pathways. The development of conditional TopBP1 knockout cells will be useful in addressing these issues.
Acknowledgments
We thank Takashi Tsuruo for TopBP1 reagents, David M. Livingston and Xiaohua Wu for anti-BRCA1 and anti-NBS1 antibodies, Irene M. Ward for materials related to 53BP1, and Yossi Shiloh for YZ5 and FT169 cells. We also thank Scott Kaufmann, Larry Karnitz, and Jann Sarkaria for stimulating conversations and members of the J.C. laboratory for helpful discussions.
This study was supported by the Mayo Cancer Center and by a grant from the Charlotte Geyer Foundation to J. Chen.
REFERENCES
- 1.Anderson, L., C. Henderson, and Y. Adachi. 2001. Phosphorylation and rapid relocalization of 53BP1 to nuclear foci upon DNA damage. Mol. Cell. Biol. 21:1719–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Araki, H., S. H. Leem, A. Phongdara, and A. Sugino. 1995. Dpb11, which interacts with DNA polymerase II(epsilon) in Saccharomyces cerevisiae, has a dual role in S-phase progression and at a cell cycle checkpoint. Proc. Natl. Acad. Sci. USA 92:11791–11795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banin, S., L. Moyal, S. Shieh, Y. Taya, C. W. Anderson, L. Chessa, N. I. Smorodinsky, C. Prives, Y. Reiss, Y. Shiloh, and Y. Ziv. 1998. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 281:1674–1677. [DOI] [PubMed] [Google Scholar]
- 4.Blasina, A., I. V. de Weyer, M. C. Laus, W. H. Luyten, A. E. Parker, and C. H. McGowan. 1999. A human homologue of the checkpoint kinase Cds1 directly inhibits Cdc25 phosphatase. Curr. Biol. 9:1–10. [DOI] [PubMed] [Google Scholar]
- 5.Blasina, A., B. D. Price, G. A. Turenne, and C. H. McGowan. 1999. Caffeine inhibits the checkpoint kinase ATM. Curr. Biol. 9:1135–1138. [DOI] [PubMed] [Google Scholar]
- 6.Bork, P., K. Hofmann, P. Bucher, A. F. Neuwald, S. F. Altschul, and E. V. Koonin. 1997. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 11:68–76. [PubMed] [Google Scholar]
- 7.Brown, A. L., C. H. Lee, J. K. Schwarz, N. Mitiku, H. Piwnica-Worms, and J. H. Chung. 1999. A human Cds1-related kinase that functions downstream of ATM protein in the cellular response to DNA damage. Proc. Natl. Acad. Sci. USA 96:3745–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, E. J., and D. Baltimore. 2000. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 14:397–402. [PMC free article] [PubMed] [Google Scholar]
- 9.Busby, E. C., D. F. Leistritz, R. T. Abraham, L. M. Karnitz, and J. N. Sarkaria. 2000. The radiosensitizing agent 7-hydroxystaurosporine (UCN-01) inhibits the DNA damage checkpoint kinase hChk1. Cancer Res. 60:2108–2112. [PubMed] [Google Scholar]
- 10.Callebaut, I., and J. P. Mornon. 1997. From BRCA1 to RAP1: a widespread BRCT module closely associated with DNA repair. FEBS Lett. 400:25–30. [DOI] [PubMed] [Google Scholar]
- 11.Canman, C. E., D. S. Lim, K. A. Cimprich, Y. Taya, K. Tamai, K. Sakaguchi, E. Appella, M. B. Kastan, and J. D. Siliciano. 1998. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 281:1677–1679. [DOI] [PubMed] [Google Scholar]
- 12.Carney, J. P., R. S. Maser, H. Olivares, E. M. Davis, M. Le Beau, J. R. Yates III, L. Hays, W. F. Morgan, and J. H. Petrini. 1998. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 93:477–486. [DOI] [PubMed] [Google Scholar]
- 13.Chaturvedi, P., W. K. Eng, Y. Zhu, M. R. Mattern, R. Mishra, M. R. Hurle, X. Zhang, R. S. Annan, Q. Lu, L. F. Faucette, G. F. Scott, X. Li, S. A. Carr, R. K. Johnson, J. D. Winkler, and B. B. Zhou. 1999. Mammalian Chk2 is a downstream effector of the ATM-dependent DNA damage checkpoint pathway. Oncogene 18:4047–4054. [DOI] [PubMed] [Google Scholar]
- 14.Chehab, N. H., A. Malikzay, M. Appel, and T. D. Halazonetis. 2000. Chk2/hCds1 functions as a DNA damage checkpoint in G(1) by stabilizing p53. Genes Dev. 14:278–288. [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, J. 2000. Ataxia telangiectasia-related protein is involved in the phosphorylation of BRCA1 following deoxyribonucleic acid damage. Cancer Res. 60:5037–5039. [PubMed] [Google Scholar]
- 16.Chen, Y., A. A. Farmer, C. F. Chen, D. C. Jones, P. L. Chen, and W. H. Lee. 1996. BRCA1 is a 220-kDa nuclear phosphoprotein that is expressed and phosphorylated in a cell cycle-dependent manner. Cancer Res. 56:3168–3172. [PubMed] [Google Scholar]
- 17.Cortez, D., Y. Wang, J. Qin, and S. J. Elledge. 1999. Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science 286:1162–1166. [DOI] [PubMed] [Google Scholar]
- 18.de Klein, A., M. Muijtjens, R. van Os, Y. Verhoeven, B. Smit, A. M. Carr, A. R. Lehmann, and J. H. Hoeijmakers. 2000. Targeted disruption of the cell-cycle checkpoint gene ATR leads to early embryonic lethality in mice. Curr. Biol. 10:479–482. [DOI] [PubMed] [Google Scholar]
- 19.de la Torre-Ruiz, M. A., C. M. Green, and N. F. Lowndes. 1998. RAD9 and RAD24 define two additive, interacting branches of the DNA damage checkpoint pathway in budding yeast normally required for Rad53 modification and activation. EMBO J. 17:2687–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emili, A. 1998. MEC1-dependent phosphorylation of Rad9p in response to DNA damage. Mol. Cell 2:183–189. [DOI] [PubMed] [Google Scholar]
- 21.Falck, J., N. Mailand, R. G. Syljuasen, J. Bartek, and J. Lukas. 2001. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature 410:842–847. [DOI] [PubMed] [Google Scholar]
- 22.Fenech, M., A. M. Carr, J. Murray, F. Z. Watts, and A. R. Lehmann. 1991. Cloning and characterization of the rad4 gene of Schizosaccharomyces pombe; a gene showing short regions of sequence similarity to the human XRCC1 gene. Nucleic Acids Res. 19:6737–6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gatei, M., S. P. Scott, I. Filippovitch, N. Soronika, M. F. Lavin, B. Weber, and K. K. Khanna. 2000. Role for ATM in DNA damage-induced phosphorylation of BRCA1. Cancer Res. 60:3299–3304. [PubMed] [Google Scholar]
- 24.Gatei, M., D. Young, K. M. Cerosaletti, A. Desai-Mehta, K. Spring, S. Kozlov, M. F. Lavin, R. A. Gatti, P. Concannon, and K. Khanna. 2000. ATM-dependent phosphorylation of nibrin in response to radiation exposure. Nat. Genet. 25:115–119. [DOI] [PubMed] [Google Scholar]
- 25.Gatei, M., B. B. Zhou, K. Hobson, S. Scott, D. Young, and K. K. Khanna. 2001. Ataxia telangiectasia mutated (ATM) kinase and ATM and Rad3 related kinase mediate phosphorylation of Brca1 at distinct and overlapping sites. In vivo assessment using phospho-specific antibodies. J. Biol. Chem. 276:17276–17280. [DOI] [PubMed] [Google Scholar]
- 26.Graves, P. R., L. Yu, J. K. Schwarz, J. Gales, E. A. Sausville, P. M. O’Connor, and H. Piwnica-Worms. 2000. The chk1 protein kinase and the Cdc25C regulatory pathways are targets of the anticancer agent UCN-01 J. Biol. Chem. 275:5600–5605. [DOI] [PubMed] [Google Scholar]
- 27.Iwabuchi, K., B. Li, H. F. Massa, B. J. Trask, T. Date, and S. Fields. 1998. Stimulation of p53-mediated transcriptional activation by the p53-binding proteins, 53BP1 and 53BP2. J. Biol. Chem. 273:26061–26068. [DOI] [PubMed] [Google Scholar]
- 28.Khanna, K. K., K. E. Keating, S. Kozlov, S. Scott, M. Gatei, K. Hobson, Y. Taya, B. Gabrielli, D. Chan, S. P. Lees-Miller, and M. F. Lavin. 1998. ATM associates with and phosphorylates p53: mapping the region of interaction. Nat. Genet. 20:398–400. [DOI] [PubMed] [Google Scholar]
- 29.Kim, S. T., D. S. Lim, C. E. Canman, and M. B. Kastan. 1999. Substrate specificities and identification of putative substrates of ATM kinase family members. J. Biol. Chem. 274:37538–37543. [DOI] [PubMed] [Google Scholar]
- 30.Koonin, E. V., S. F. Altschul, and P. Bork. 1996. BRCA1 protein products, functional motifs. Nat. Genet. 13:266–268. [DOI] [PubMed] [Google Scholar]
- 31.Lakin, N. D., B. C. Hann, and S. P. Jackson. 1999. The ataxia-telangiectasia related protein ATR mediates DNA-dependent phosphorylation of p53. Oncogene 18:3989–3995. [DOI] [PubMed] [Google Scholar]
- 32.Lee, J. S., K. M. Collins, A. L. Brown, C. H. Lee, and J. H. Chung. 2000. hCds1-mediated phosphorylation of BRCA1 regulates the DNA damage response. Nature 404:201–204. [DOI] [PubMed] [Google Scholar]
- 33.Li, S., N. S. Ting, L. Zheng, P. L. Chen, Y. Ziv, Y. Shiloh, E. Y. Lee, and W. H. Lee. 2000. Functional link of BRCA1 and ataxia telangiectasia gene product in DNA damage response. Nature 406:210–215. [DOI] [PubMed] [Google Scholar]
- 34.Lim, D. S., S. T. Kim, B. Xu, R. S. Maser, J. Lin, J. H. Petrini, and M. B. Kastan. 2000. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature 404:613–617. [DOI] [PubMed] [Google Scholar]
- 35.Liu, Q., S. Guntuku, X. S. Cui, S. Matsuoka, D. Cortez, K. Tamai, G. Luo, S. Carattini-Rivera, F. DeMayo, A. Bradley, L. A. Donehower, and S. J. Elledge. 2000. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 36.Mailand, N., J. Falck, C. Lukas, R. G. Syljuasen, M. Welcker, J. Bartek, and J. Lukas. 2000. Rapid destruction of human Cdc25A in response to DNA damage. Science 288:1425–1429. [DOI] [PubMed] [Google Scholar]
- 37.Makiniemi, M., T. Hillukkala, J. Tuusa, K. Reini, M. Vaara, D. Huang, H. Pospiech, I. Majuri, T. Westerling, T. P. Makela, and J. E. Syvaoja. 2001. BRCT domain-containing protein TopBP1 functions in DNA replication and damage response. J. Biol. Chem. 276:30399–30406. [DOI] [PubMed] [Google Scholar]
- 38.Maser, R. S., K. J. Monsen, B. E. Nelms, and J. H. J. Petrini. 1997. hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol. Cell. Biol. 17:6087–6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsuoka, S., M. Huang, and S. J. Elledge. 1998. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science 282:1893–1897. [DOI] [PubMed] [Google Scholar]
- 40.McFarlane, R. J., A. M. Carr, and C. Price. 1997. Characterisation of the Schizosaccharomyces pombe rad4/cut5 mutant phenotypes: dissection of DNA replication and G2 checkpoint control function. Mol. Gen. Genet. 255:332–340. [DOI] [PubMed] [Google Scholar]
- 40a.Mirzoeva, O. K., and J. H. Petrini. 2001. DNA damage-dependent nuclear dynamics of the Mre11 complex. Mol. Cell. Biol. 21:281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Molinari, M. 2000. Cell cycle checkpoints and their inactivation in human cancer. Cell Prolif. 33:261–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Navas, T. A., Y. Sanchez, and S. J. Elledge. 1996. RAD9 and DNA polymerase epsilon form parallel sensory branches for transducing the DNA damage checkpoint signal in Saccharomyces cerevisiae. Genes Dev. 10:2632–2643. [DOI] [PubMed] [Google Scholar]
- 43.Paull, T. T., E. P. Rogakou, V. Yamazaki, C. U. Kirchgessner, M. Gellert, and W. M. Bonner. 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10:886–895. [DOI] [PubMed] [Google Scholar]
- 44.Rappold, I., K. Iwabuchi, T. Date, and J. Chen. 2001. Tumor suppressor p53 binding protein 1 (53BP1) is involved in DNA damage-signaling pathways. J. Cell Biol. 153:613–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rhind, N., and P. Russell. 2000. Checkpoints: it takes more than time to heal some wounds. Curr. Biol. 10:908–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rogakou, E. P., C. Boon, C. Redon, and W. M. Bonner. 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146:905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saka, Y., F. Esashi, T. Matsusaka, S. Mochida, and M. Yanagida. 1997. Damage and replication checkpoint control in fission yeast is ensured by interactions of Crb2, a protein with BRCT motif, with Cut5 and Chk1. Genes Dev. 11:3387–3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saka, Y., P. Fantes, T. Sutani, C. McInerny, J. Creanor, and M. Yanagida. 1994. Fission yeast cut5 links nuclear chromatin and M phase regulator in the replication checkpoint control. EMBO J. 13:5319–5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saka, Y., P. Fantes, and M. Yanagida. 1994. Coupling of DNA replication and mitosis by fission yeast rad4/cut5. J. Cell Sci. 18(Suppl.):57–61. [DOI] [PubMed] [Google Scholar]
- 50.Saka, Y., and M. Yanagida. 1993. Fission yeast cut5+, required for S phase onset and M phase restraint, is identical to the radiation-damage repair gene rad4+. Cell 74:383–393. [DOI] [PubMed] [Google Scholar]
- 51.Sanchez, Y., C. Wong, R. S. Thoma, R. Richman, Z. Wu, H. Piwnica-Worms, and S. J. Elledge. 1997. Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science 277:1497–1501. [DOI] [PubMed] [Google Scholar]
- 52.Sarkaria, J. N., E. C. Busby, R. S. Tibbetts, P. Roos, Y. Taya, L. M. Karnitz, and R. T. Abraham. 1999. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 59:4375–4382. [PubMed] [Google Scholar]
- 53.Schultz, L. B., N. H. Chehab, A. Malikzay, and T. D. Halazonetis. 2000. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell Biol. 151:1381–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scully, R., J. Chen, A. Plug, Y. Xiao, D. Weaver, J. Feunteun, T. Ashley, and D. M. Livingston. 1997. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell 88:265–275. [DOI] [PubMed] [Google Scholar]
- 55.Shieh, S. Y., J. Ahn, K. Tamai, Y. Taya, and C. Prives. 2000. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 14:289–300. [PMC free article] [PubMed] [Google Scholar]
- 56.Sun, Z., J. Hsiao, D. S. Fay, and D. F. Stern. 1998. Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science 281:272–274. [DOI] [PubMed] [Google Scholar]
- 57.Takai, H., K. Tominaga, N. Motoyama, Y. A. Minamishima, H. Nagahama, T. Tsukiyama, K. Ikeda, K. Nakayama, M. Nakanishi, and K. Nakayama. 2000. Aberrant cell cycle checkpoint function and early embryonic death in Chk1(—/—) mice. Genes Dev. 14:1439–1447. [PMC free article] [PubMed] [Google Scholar]
- 58.Tibbetts, R. S., K. M. Brumbaugh, J. M. Williams, J. N. Sarkaria, W. A. Cliby, S. Y. Shieh, Y. Taya, C. Prives, and R. T. Abraham. 1999. A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev. 13:152–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tibbetts, R. S., D. Cortez, K. M. Brumbaugh, R. Scully, D. Livingston, S. J. Elledge, and R. T. Abraham. 2000. Functional interactions between BRCA1 and the checkpoint kinase ATR during genotoxic stress. Genes Dev. 14:2989–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Verkade, H. M., and M. J. O’Connell. 1998. Cut5 is a component of the UV-responsive DNA damage checkpoint in fission yeast. Mol. Gen. Genet. 260:426–433. [DOI] [PubMed] [Google Scholar]
- 61.Vialard, J. E., C. S. Gilbert, C. M. Green, and N. F. Lowndes. 1998. The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tel1-dependent hyperphosphorylation and interacts with Rad53 after DNA damage. EMBO J. 17:5679–5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang, H., and S. J. Elledge. 1999. DRC1, DNA replication and checkpoint protein 1, functions with DPB11 to control DNA replication and the S-phase checkpoint in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 96:3824–3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weiss, R. S., T. Enoch, and P. Leder. 2000. Inactivation of mouse Hus1 results in genomic instability and impaired responses to genotoxic stress. Genes Dev. 14:1886–1898. [PMC free article] [PubMed] [Google Scholar]
- 64.Wu, X., J. H. Petrini, W. F. Heine, D. T. Weaver, D. M. Livingston, and J. Chen. 2000. Independence of R/M/N focus formation and the presence of intact BRCA1. Science 289:11. [DOI] [PubMed] [Google Scholar]
- 65.Wu, X., V. Ranganathan, D. S. Weisman, W. F. Heine, D. N. Ciccone, T. B. O’Neill, K. E. Crick, K. A. Pierce, W. S. Lane, G. Rathbun, D. M. Livingston, and D. T. Weaver. 2000. ATM phosphorylation of Nijmegen breakage syndrome protein is required in a DNA damage response. Nature 405:477–482. [DOI] [PubMed] [Google Scholar]
- 66.Xia, Z., J. C. Morales, W. G. Dunphy, and P. B. Carpenter. 2001. Negative cell cycle regulation and DNA damage-inducible phosphorylation of the BRCT protein 53BP1. J. Biol. Chem. 276:2708–2718. [DOI] [PubMed] [Google Scholar]
- 67.Yamamoto, R. R., J. M. Axton, Y. Yamamoto, R. D. Saunders, D. M. Glover, and D. S. Henderson. 2000. The Drosophila mus101 gene, which links DNA repair, replication and condensation of heterochromatin in mitosis, encodes a protein with seven BRCA1 C-terminus domains. Genetics 156:711–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yamane, K., M. Kawabata, and T. Tsuruo. 1997. A DNA-topoisomerase-II-binding protein with eight repeating regions similar to DNA-repair enzymes and to a cell-cycle regulator. Eur. J. Biochem. 250:794–799. [DOI] [PubMed] [Google Scholar]
- 69.Yamane, K., and T. Tsuruo. 1999. Conserved BRCT regions of TopBP1 and of the tumor suppressor BRCA1 bind strand breaks and termini of DNA. Oncogene 18:5194–5203. [DOI] [PubMed] [Google Scholar]
- 70.Zhao, S., Y. C. Weng, S. S. Yuan, Y. T. Lin, H. C. Hsu, S. C. Lin, E. Gerbino, M. H. Song, M. Z. Zdzienicka, R. A. Gatti, J. W. Shay, Y. Ziv, Y. Shiloh, and E. Y. Lee. 2000. Functional link between ataxia-telangiectasia and Nijmegen breakage syndrome gene products. Nature 405:473–477. [DOI] [PubMed] [Google Scholar]
- 71.Zhong, Q., C. F. Chen, S. Li, Y. Chen, C. C. Wang, J. Xiao, P. L. Chen, Z. D. Sharp, and W. H. Lee. 1999. Association of BRCA1 with the hRad50-hMre11–p95 complex and the DNA damage response. Science 285:747–750. [DOI] [PubMed] [Google Scholar]
- 72.Zhou, B. B., and S. J. Elledge. 2000. The DNA damage response: putting checkpoints in perspective. Nature 408:433–439. [DOI] [PubMed] [Google Scholar]