Abstract
The chlorate-resistant mutant cr88 is defective in photomorphogenesis, as shown by the phenotypes of long hypocotyls in red light and yellow cotyledons under all light conditions. A subset of light-regulated genes is expressed at subnormal levels in cr88. To analyze further the role that CR88 plays in photomorphogenesis, we investigated the genetic interactions between cr88 and mutants of two other loci affecting photomorphogenesis, CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) and LONG HYPOCOTYL5 (HY5). COP1 represses the expression of light-regulated genes in the dark, and HY5 inhibits hypocotyl elongation in the light. Using morphological, cellular, and gene expression criteria for epistasis analyses to position CR88 in the genetic hierarchy of the photomorphogenesis pathway, we determined that CR88 acts downstream of COP1 but in a branch separate from HY5. In the course of our analysis, we discovered that light causes extensive destruction of plastids in dark-grown cop1 seedlings and that cr88 prevents this destruction.
INTRODUCTION
Light signals are essential environmental cues for seedlings to elaborate their developmental program during the transition from heterotrophy to autotrophy. The effects of light on seedling development can be observed at many levels, including gene expression, cell differentiation, and plant morphology. During seedling development, higher plants, such as Arabidopsis, can adopt two contrasting development schemes: skotomorphogenesis, which takes place in the dark, and photomorphogenesis, which occurs in the light (Kendrick and Kronenberg, 1994). Dark-grown seedlings undergoing skotomorphogenesis exhibit etiolated morphologies, including long hypocotyls, apical hooks, and closed and undeveloped cotyledons. The genes required for chloroplast development and photosynthesis are expressed at low or undetectable levels. When exposed to light, seedlings undergo photomorphogenesis and exhibit deetiolated morphologies, such as short hypocotyls and open and developed cotyledons. Expression of genes encoding photosynthetic proteins is strongly induced, and chloroplasts develop from etioplasts (Mullet, 1988; Susek et al., 1993; Deng, 1994; Reiter et al., 1994).
Genetic analyses have identified components in the photomorphogenesis pathway. In addition to mutants defective in the phytochrome photoreceptors PhyA (Dehesh et al., 1993; Parks and Quail, 1993; Whitelam et al., 1993) and PhyB (Koornneef et al., 1980; Reed et al., 1993), and the blue light photoreceptor cryptochrome (CRY1) (Koornneef et al., 1980; Ahmad and Cashmore, 1993), two classes of regulatory mutants have been identified. One class consists of mutants that exhibit an etiolated morphology when germinated in the light, suggesting a positive role for the wild-type gene products in photomorphogenesis. These mutants define loci, such as LONG HYPOCOTYL5 (HY5) (Koornneef et al., 1980); PHYTOCHROME SIGNALING–EARLY FLOWERING (PEF1, PEF2, and PEF3) (Ahmad and Cashmore, 1996); RED ELONGATED1 (red1) (Wagner et al., 1997); and ELONGATED HYPOCOTYL IN FAR-RED LIGHT (FHY1 and FHY3) (Whitelam et al., 1993; Johnson et al., 1994). The hy5 mutant displays long hypocotyls under red, far-red, and blue light conditions (Koornneef et al., 1980), suggesting that HY5 encodes a component that functions after convergence of the pathways that transduce signals triggered by red, far-red, and blue light (Koornneef et al., 1980). The predicted protein sequence encoded by HY5 resembles a basic leucine zipper transcription activator (Oyama et al., 1997). HY5, which is required for light-induced expression of the gene encoding chalcone synthase (CHS), binds to the CHS promoter (Ang et al., 1998). The remaining mutants in this class are defective in the photomorphogenic response specific to either or both of the PhyA and PhyB signaling pathways (Whitelam et al., 1993; Johnson et al., 1994; Ahmad and Cashmore, 1996; Wagner et al., 1997). Except for hy5, all mutants in this class appear to define early steps of light signaling (Ahmad and Cashmore, 1996; Wagner et al., 1997). Recently, these loci, together with SPA1 (Hoecker et al., 1998, 1999) and PHYTOCHROME SIGNALING2 (PSI2) (Genoud et al., 1998), which cause a hypersensitive photomorphogenic response when mutated, have been proposed to act downstream of photoreceptors and upstream of the protein encoded by CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) (Ni et al., 1998).
Another class of mutants displays deetiolated morphologies when grown in the dark. The recessive nature of the mutants suggests that the gene products are required to repress photomorphogenesis in the dark. These mutants include at least 16 cop/deetiolated/fusca loci (Chory et al., 1989; Deng et al., 1991; Wei and Deng, 1992; Misera et al., 1994; Wei et al., 1994; Kwok et al., 1996). Genetic analyses suggest that these genes act at or after the convergence of the PhyA, PhyB, and CRY1 signaling pathways (Chory, 1993; Ang and Deng, 1994). Among these loci, COP1 encodes a key repressor that is hypothesized to interact with specific transcription factors in the nucleus to repress their activities in the dark. One of these transcription factors is HY5, which has been shown to interact directly with the COP1 WD-40 repeats (Ang et al., 1998; Chattopadhyay et al., 1998). Exposure to light causes COP1 to be excluded from the nucleus, thereby allowing the expression of downstream genes (reviewed in Osterlund et al., 1999).
While selecting chlorate-resistant mutants that were defective in the expression of genes encoding nitrate reductase (NR), we identified a novel chlorate-resistant mutant, cr88, that displays long hypocotyls in red light but not in far-red or blue light. Cotyledons and young leaves of light-grown cr88 are yellow-green, correlating with delayed chloroplast development. The expression of light-induced genes encoding the chlorophyll a/b binding protein (CAB), the small subunit of ribulose bisphophate carboxylase (RBCS), and NR2—but not the expression of CHS and NR1—was altered.
On the basis of these observations, we hypothesized that CR88 acts in the pathways for transduction of light signals to regulate a subset of photomorphogenesis responses that are repressed by COP1 and, moreover, that CR88 plays both a major role in controlling the greening process and a minor role in controlling hypocotyl elongation (Lin and Cheng, 1997). By analyzing epistatic relationships between mutants cr88 and cop1 and between mutants cr88 and hy5, we found that CR88 is likely to act downstream of COP1 but in a branch separate from that affected by HY5. During the course of the analysis, we discovered that the loss of greening ability of cop1 mutants after prolonged growth in the dark (Ang and Deng, 1994) is caused by the extensive destruction of plastids and other cellular structures when exposed to light. However, the cr88 allele is able to prevent the destruction caused by cop1. In addition, we show that the long hypocotyls in red light and the impaired light induction of gene expression in cr88 are unlikely to be secondary effects caused by a delay in the greening process.
RESULTS
Inhibition of Photosynthesis Neither Increases the Hypocotyl Length nor Decreases the Expression of Photosynthetic Genes in Wild-Type and cr88 Seedlings
We showed previously that cr88 exhibits reduced expression of NR2, CAB, and RBCS genes as well as impaired deetiolation in red light. In addition, the chloroplasts of cr88 cotyledons and young leaves are not as well developed as those of the wild type (Lin and Cheng, 1997). These observations raised the possibility that the lack of inhibition of hypocotyl elongation and reduced expression of NR2, CAB, and RBCS mRNA in cr88 might be caused by a decrease in the photosynthetic activity of the underdeveloped chloroplasts. Therefore, we tested whether inhibiting photosynthesis in wild-type seedlings would result in an increase in hypocotyl length in red light and a decrease of the expression of NR2, CAB, and RBCS genes. To determine whether inhibiting photosynthesis would result in longer hypocotyls, we grew the wild-type and cr88 seedlings in continuous red light for 4 days on media containing 0, 0.1, 1, 5, or 10 μM 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU), a photosynthesis inhibitor, or on media containing 0, 1, 5, 10, or 20 mM norflurazon, which causes photooxidative damage. Neither compound increased the hypocotyl lengths of the seedlings, even though cotyledons of the norflurazon-treated seedlings exhibited photobleaching (data not shown).
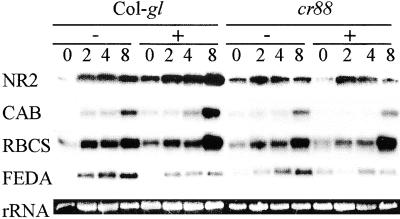
We also asked whether inhibition of photosynthesis would affect light-induced gene expression. Wild-type and cr88 seedlings were grown for 4 days in the dark in media containing 0 or 10 μM DCMU and were then exposed to red light for 2, 4, or 8 hr. Similar to our previous observations on plants grown in white light (Lin and Cheng, 1997), the amounts of red light–induced NR2, CAB, and RBCS mRNAs were less in cr88 than in the wild-type seedlings (Figure 1). DCMU did not cause a decrease in the steady state amounts of mRNAs of these genes; to the contrary, DCMU may have increased their expression. The increase in gene expression caused by DCMU is consistent with a previous report in which DCMU was shown to increase CAB transcription rates (Escoubas et al., 1995). In contrast, the expression of the gene encoding ferredoxin (FEDA) was decreased in response to DCMU, as also reported previously (Petracek et al., 1997).
Figure 1.
Effects of DCMU on Red Light Induction of NR2, CAB, and RBCS mRNA.
Each lane contains 5 μg of total RNA isolated from seedlings grown on medium in the dark for 4 days and then exposed to red light. The number above each lane indicates the hours of red light induction in the absence (−) or presence (+) of 10 μM DCMU. At left, NR2, CAB, RBCS, and FEDA indicate the RNAs hybridizing with their respective probes. Ethidium bromide–stained 28S rRNA served as a loading control.
These results demonstrate that inhibition of photosynthesis did not produce a phenocopy of the Hy phenotype of cr88 in red light, nor did it cause a decrease in light-induced expression of NR2, CAB, and RBCS genes. Thus, these defects of cr88 are unlikely to be secondary effects caused by decreased photosynthetic activity.
CR88 Is Located on Chromosome 2
CR88 was mapped to chromosome 2 by using the set of Arabidopsis restriction fragment length polymorphism markers described by Fabri and Schaffner (1994). Use of additional markers on chromosome 2 located cr88 between m246 and CDs3. Complementation tests between cr88 and other known hy mutants suggested that CR88 defines a new HY locus (Lin and Cheng, 1997). The map position of CR88 eliminates the possibility that it encodes one of the minor phytochromes, PHYC (Schmidt et al., 1997), PHYD (Schmidt et al., 1996), or PHYE (Schmidt et al., 1996), all of which map to other chromosomes. It also precludes CR88 being an allele of the recently identified long hypocotyl mutants elongated (elg) (Halliday et al., 1996), pef1 (Ahmad and Cashmore, 1996), and red1 (Wagner et al., 1997) or of the CAB underexpressed (cue) mutants (Li et al., 1995; Lopez-Juez et al., 1998) because none of these maps to chromosome 2.
Light Induction of NR2, CAB, and RBCS Is Defective in Red, Far-Red, and Blue Light Conditions
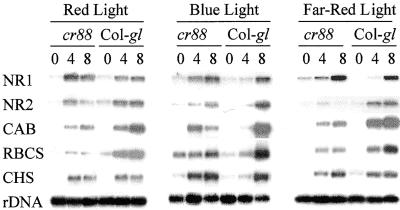
We showed previously that light-induced expression of NR2, CAB, and RBCS, but not NR1 and CHS, is impaired in cr88 after 8 hr of exposure to white light (Lin and Cheng, 1997). To further understand this defect, we examined the induction of light-regulated gene expression in different light spectra. Wild-type and cr88 seedlings were grown for 4 days in the dark and then exposed to red, blue, or far-red light for 4 or 8 hr. The steady state amounts of mRNAs for NR1, NR2, CAB, RBCS, and CHS in cr88 were compared with those in the wild type (Figure 2). In cr88, the mRNAs for NR2, CAB, and RBCS were less than those in the wild type in all light conditions, whereas the amounts of the mRNAs for NR1 and CHS at steady state were similar to or greater than those in the wild type. This result further confirms our earlier conclusion that the light regulation of NR2, CAB, and RBCS is in a pathway different from that of NR1 and CHS (Lin and Cheng, 1997) and that CR88 controls the expression of a subset of light-regulated genes that includes NR2, CAB, and RBCS. Neuhaus et al. (1997) also reported that the light-signaling pathway regulating CHS gene expression differs from the one that regulates CAB and RBCS. In addition, the reduction of light-regulated gene expression in broad spectra in cr88 is consistent with CR88 acting downstream of the convergence of the pathways that transduce light signals from different photoreceptors.
Figure 2.
RNA Gel Blot Analysis of Induction of NR1, NR2, CAB, RBCS, and CHS mRNA in cr88 and the Wild-Type Col-gl Seedlings after Exposure to Red, Blue, and Far-Red Light.
Each lane contains 5 μg of total RNA isolated from seedlings that were grown in the dark for 6 days before exposure to red, blue, or far-red light. The number above each lane indicates the hours of light exposure. At left, NR1, NR2, CAB, RBCS, CHS, and rDNA indicate the RNAs hybridizing with their respective probes.
Epistatic Relationships between cr88 and cop1 Mutations in Hypocotyl Elongation Are Allele Specific and Light Condition Dependent
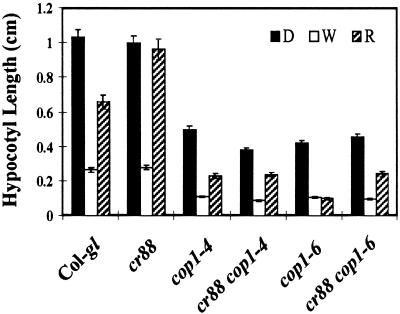
Previous physiological and genetic characterization of cr88 suggests that CR88 regulates a subset of COP1-repressed photomorphogenesis responses (Lin and Cheng, 1997). We hypothesized that CR88 plays a minor role in inhibiting hypocotyl elongation; the Hy phenotype of cr88 in red light is the result of the inability of COP1 to suppress HY5 completely under this condition. HY5 is known to play a major role in inhibiting hypocotyl elongation in the light and was shown recently to interact directly with the COP1 protein (Koornneef et al., 1980; Oyama et al., 1997). We examined the epistatic relationships between cr88 and two cop1 alleles, cop1-4 and cop1-6, to gain insight into the relationship between CR88 and COP1 in the light signal transduction pathway. Mutations in the cop1 and cr88 loci cause opposite phenotypes with respect to hypocotyl length in red light. Being constitutively photomorphogenic, cop1 exhibits short hypocotyls in all light conditions (Deng et al., 1991), whereas cr88 exhibits long hypocotyls only in red light (Lin and Cheng, 1997). The cr88 cop1-4 double mutant resembles the cop1-4 single mutant in all conditions, indicating that cop1-4 is epistatic to cr88 in deetiolation. In contrast, the epistatic relationship between cr88 and cop1-6 is dependent on the light conditions. In dark and white light, the hypocotyl lengths of the cr88 cop1-6 double mutant are equal to that of the cop1-6 single mutant. In red light, however, the double mutant exhibits intermediate hypocotyl length, indicating that cr88 and cop1-6 mutations partially suppress each other in red light (Figure 3). The allele-specific interaction of cr88 and cop1 mutations suggests that these two gene products may function in close proximity. Allele-specific and light condition–dependent epistatic relationships have also been observed between the hy5 and cop1 mutations (Ang and Deng, 1994).
Figure 3.
Hypocotyl Lengths of Col-gl, cr88, cop1-4, cr88 cop1-4, cop1-6, and cr88 cop1-6 Grown in the Dark and in White Light and Red Light.
Seedlings were grown on medium in the dark (D) or in white (W) or red (R) light for 4 days. The hypocotyl lengths of 25 seedlings from each treatment were measured and used to calculate the mean. The error bars represent the standard deviations.
cr88 and cop1 Mutations Show Contrasting Epistatic Relationships in Expression of NR2, RBCS, and CAB
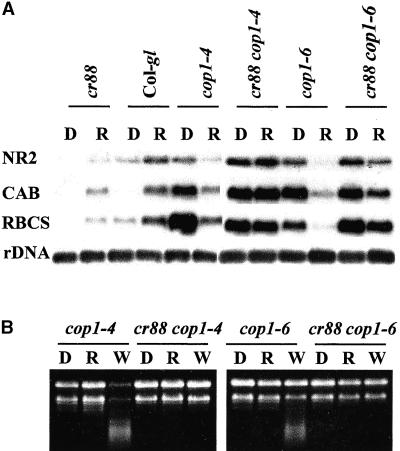
We examined the epistatic relationship of cr88 and cop1 with respect to expression of NR2, CAB, and RBCS genes. Seedlings were grown in the dark for 4 days and were then exposed to red light for 8 hr. The amounts of mRNA for the light-regulated genes NR2, CAB, and RBCS were compared (Figure 4A). All of these genes were expressed in dark-grown cop1-4 and cop1-6 and in the double mutants cr88 cop1-4 and cr88 cop1-6. Eight hours of red light exposure led to a considerable decrease in the mRNAs for these genes in the cop1-4 and cop1-6 mutants. In contrast, light-induced expression of these genes was restored in the double mutants cr88 cop1-4 and cr88 cop1-6, the amounts being as high as those in dark-grown cop1 mutants and much higher than that in cr88. The constitutive expression of these genes in the double mutants revealed contrasting epistatic relationships between cr88 and cop1 mutations. On the one hand, cop1 mutations suppressed the decreased expression of NR2, RBCS, and CAB in cr88. On the other hand, cr88 suppressed the light-triggered decreased expression of these genes in cop1 mutants.
Figure 4.
Total RNA Analysis and RNA Gel Blot Analysis of NR2, CAB, and RBCS mRNA in Dark-Grown Col-gl, cr88, cop1-4, cop1-6, cr88 cop1-4, and cr88 cop1-6 Seedlings after Exposure to Light.
(A) Each lane contains 5 μg of total RNA isolated from seedlings grown on medium in the dark for 4 days and harvested before (D) or 8 hr after (R) exposure to red light. At left, NR2, CAB, RBCS, and rDNA indicate the RNAs hybridizing with their respective probes.
(B) Each lane contains 5 μg of total RNA isolated from seedlings grown on medium in the dark for 4 days and harvested before (D) or 8 hr after exposure to red (R) or white (W) light. RNA was stained with ethidium bromide for visualization.
cr88 Mutation Restores the Greening Ability of the Dark-Grown cop1 Seedlings
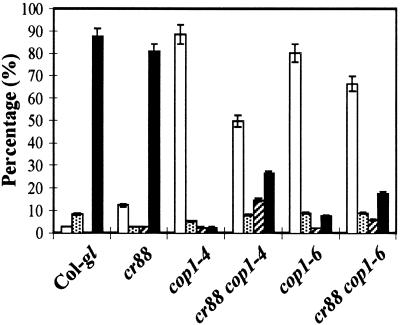
After growing in the dark for 4 days, most of the cop1 seedlings, unlike wild-type seedlings, are incapable of greening when transferred to light. The hy5 mutant can rescue this cop1-triggered defect (Ang and Deng, 1994). To examine whether cr88 could also rescue this defect, we compared the greening ability after transfer to light of 4- and 6-day-old dark-grown mutant seedlings. Consistent with the report of Ang and Deng (1994), only 2.5% of cop1-4 seedlings and 56% of cop1-6 seedlings retained the greening ability after 4 days in the dark (Table 1). Six days of growth in the dark further decreased the percentage of cop1-4 and cop1-6 individuals that retained the greening ability to 1.5 and 38.6%, respectively. The double mutants turned green in light after 4 or 6 days of growth in the dark. After 4 days in the dark, 97% of the double mutant seedlings turned green in the light. After 6 days in the dark, 90.7% of cr88 cop1-4 and 80.5% of cr88 cop1-6 seedlings still retained greening ability. Nevertheless, although the double mutants restored the greening ability, their cotyledons exhibited the yellow-green phenotype of cr88 (data not shown).
Table 1.
Greening Ability of Dark-Grown Col-gl, cr88, cop1-4, cop1-6, cr88 cop1-4, and cr88 cop1-6 Seedlings after Exposure to Light
| 4 Days in the Dark
|
6 Days in the Dark
|
|||
|---|---|---|---|---|
| Lines | Total Plants |
Greened (%)a | Total Plants |
Greened (%)a |
| Col-glb | 43 | 43 (100) | 81 | 81 (100) |
| cr88 | 45 | 45 (100) | 73 | 73 (100) |
| cop1-4 | 58 | 2 (3.5) | 64 | 1 (1.5) |
| cr88 cop1-4 | 87 | 85 (97.0) | 65 | 59 (90.7) |
| cop1-6 | 54 | 30 (56.0) | 67 | 26 (38.5) |
| cr88 cop1-6 | 71 | 69 (97.0) | 36 | 29 (80.5) |
The percentage of green seedlings in total plants scored is shown in parentheses.
Col-gl serves as the wild-type control.
Light Exposure of Dark-Grown cop1 Causes Plastid Destruction and RNA Degradation That Can Be Rescued by cr88
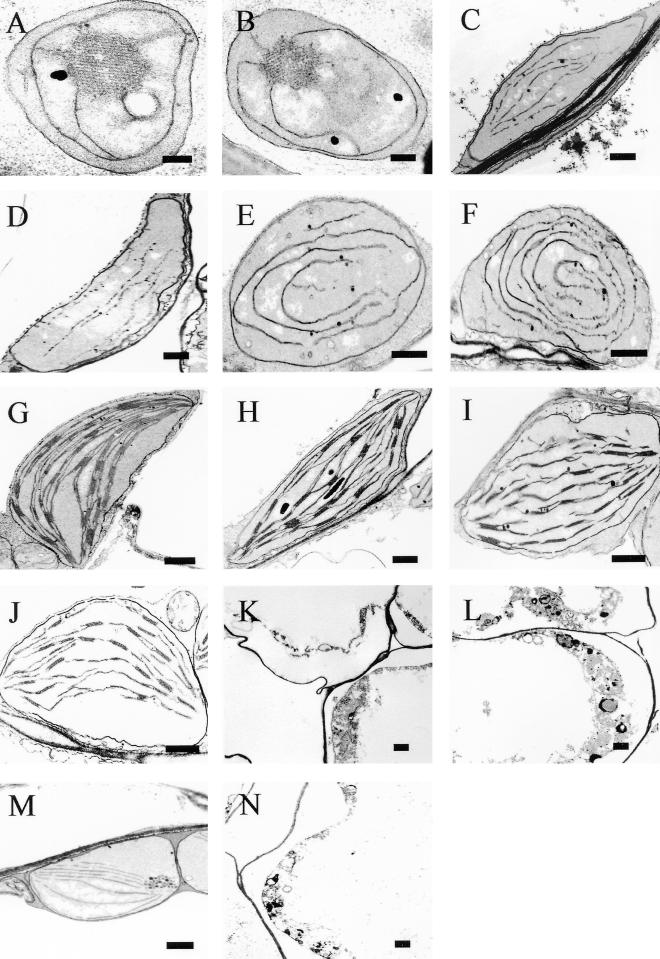
The loss of greening ability in dark-grown cop1-6 and cop1-4 on transfer to light suggested a defect in chloroplast development, and the restoration of greening ability to these mutants by cr88 suggested suppression of this defect. The defect and the restoration of plastid development may be revealed by examining the ultrastructure of the developing plastids. No plastid development was observed in dark-grown wild-type (Figure 5A) and cr88 seedlings (Figure 5B). Apparent development of the thylakoid membrane was found in the dark-grown seedlings of cop1-4 (Figure 5E) and cop1-6 (Figure 5F), as reported previously (Deng et al., 1991). The thylakoid membrane of the double mutants cr88 cop1-4 (Figure 5C) and cr88 cop1-6 (Figure 5D), although exhibiting less stacking than that of the cop1 single mutants, was partially developed, suggesting that the constitutive plastid development of cop1 was not suppressed by cr88. Chloroplasts were fully developed 6 days after transfer of the 6-day-old dark-grown seedlings of the wild type (Figure 5G) and of cr88 (Figure 5H) into light. Consistent with the previous report (Lin and Cheng, 1997), cr88 showed less thylakoid stacking. Transfer of the dark-grown seedlings of cop1-4 (Figure 5E) and cop1-6 (Figure 5F) to white light led to destruction of the chloroplasts and other cellular structures (Figures 5K and 5L, respectively). The destruction was not restricted to chloroplasts; indeed, almost no organelle within the cell could be recognized. In contrast, chloroplasts were well developed in the double mutants (Figures 5I and 5J), albeit with less stacking of the thylakoid membrane than in the wild-type (Figure 5G) chloroplasts. The destruction occurred as early as 8 hr after transfer to white light (Figure 5N) and was rescued by cr88 (Figure 5M).
Figure 5.
Plastid Development of Dark-Grown Col-gl, cr88, cop1-4, cr88 cop1-4, cop1-6, and cr88 cop1-6 Seedlings after Exposure to Light.
Representative plastids from seedlings grown in the dark for 4 days ([A] to [F]) and then transferred to white light for 4 days ([G] to [L]) or 8 hr ([M] and [N]).  .
.
(A) and (G) Col-gl.
(B) and (H) cr88.
(C), (I), and (M) cr88 cop1-4.
(D) and (J) cr88 cop1-6.
(E), (K), and (N) cop1-4.
(F) and (L) cop1-6.
During the investigation, we discovered that exposure to light of dark-grown cop1 mutants also causes general RNA degradation in addition to chloroplast destruction. We treated the 4-day-old dark-grown seedlings for 8 hr with white light or red light and then compared the integrity of the total RNA afterward (Figure 4B). The RNA extracted from 4-day-old dark-grown cop1-4 and cop1-6 seedlings was intact. In contrast, 8 hr of white light exposure led to degradation of the RNA. At this time, the plastids in cop1-4 seedlings were already showing severe degradation (Figure 5M), but no degradation was observed in the cr88 cop1-4 and cr88 cop1-6 double mutants. Although a general degradation of the RNA extracted from red light–treated cop1 seedlings was not visible by ethidium bromide staining (Figure 4B), a decrease in the amount of mRNA from specific genes was evident (Figure 4A).
Taken together, analyses at the phenotypic, ultrastructural, and RNA levels suggest that the cr88 mutation suppresses the loss of the greening ability caused by cop1 mutations by preventing the destruction of chloroplasts and other cellular structures in response to light.
cr88 Partially Suppresses the Loss of Negative Gravitropic Response of the Dark-Grown Seedlings of cop1
Hypocotyls of the dark-grown wild-type Arabidopsis seedlings grow upward. This phenomenon, called the negative gravitropic response, is attenuated in light (Hangarter, 1997). Although cop1 seedlings exhibit similar degrees of negative gravitropism as the wild type in light (Hou et al., 1993; D. Cao and C.-L. Cheng, unpublished results), these seedlings lost the negative gravitropic response in the dark (Figure 6), whereas cr88 seedlings retain the negative gravitropic response (Figure 6). To determine whether the cr88 mutation suppresses the loss of the negative gravitropic response caused by cop1 mutations, we grew single and double mutant seedlings in the dark for 4 days and then measured the growth angles of the seedlings relative to the horizontal level (Figure 6). Performing this experiment with both horizontal and vertical plates yielded consistent results. Figure 6 shows the results from seedlings grown on horizontal plates. Eighty-two percent of cr88 and 90% of the wild-type seedlings grew at angles between 75 and 90°. In contrast, 90% of cop1-4 and 80% of cop1-6 lay flat on the plates (growth angle at 0°). For the double mutant cr88 cop1-4, approximately 30% of the seedlings grew at angles ranging from 75 to 90°, whereas only 2% of cop1-4 grew at angles in this range. Similar but less pronounced reversal was observed between the double mutant cr88 cop1-6 and cop1-6. These results indicate that cr88 can partially suppress the loss of negative gravitropic response of the dark-grown cop1 seedlings.
Figure 6.
Hypocotyl Orientations of Dark-Grown Col-gl, cr88, cop1-4, cop1-6, cr88 cop1-4, and cr88 cop1-6 Seedlings.
Angles relative to the agar surface are 0° (white bars), 1 to 29° (stippled bars), 30 to 74° (cross-hatched bars), and 75 to 90° (black bars). Error bars represent standard deviations of the mean in percentage of seedlings that exhibited the indicated range of angles.
CR88 Controls Hypocotyl Length in Red Light Independent of HY5
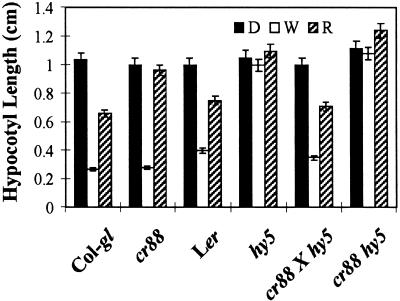
To test the hypothesis that CR88 and HY5 act in separate pathways to regulate hypocotyl length, we constructed a double mutant of cr88 and hy5. The seedlings were grown in the dark or in red or white light for 4 days, after which the hypocotyl lengths of the double and single mutants were compared (Figure 7). As expected, cr88 seedlings exhibited hypocotyls as long as those of the wild-type in white light and longer hypocotyls in red light; hy5 seedlings exhibited hypocotyls longer than wild-type hypocotyls in both white and red light. In white light, the hypocotyl length of the cr88 hy5 double mutant was similar to that of the hy5 single mutant. In red light, however, the hypocotyl length of cr88 hy5 was significantly greater than that of either hy5 (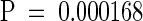 ) or cr88 (
) or cr88 (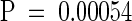 ). These results support our hypothesis that for inhibition of hypocotyl length, the function of CR88 is redundant to HY5 in all but red light spectra.
). These results support our hypothesis that for inhibition of hypocotyl length, the function of CR88 is redundant to HY5 in all but red light spectra.
Figure 7.
Hypocotyl Lengths of Col-gl, cr88, Landsberg erecta, hy5, and cr88 hy5 Grown in the Dark and in White and Red Light.
Col-gl and Landsberg erecta (Ler) are wild types for cr88 and hy5, respectively. Seedlings were grown on medium in the dark (D) or in white (W) or red (R) light for 4 days. The hypocotyl lengths of 25 seedlings from each line were measured for calculating the mean. The error bars represent standard deviations.
New Phenotypes Arise in cr88 hy5
The overall morphology of the mature cr88 hy5 double mutant is different from that of either parent (Figure 8). The mature cr88 hy5 plants were thinner and weaker and had fewer leaves than either cr88 or hy5 plants. The double mutants were spindly and could not stand upright when grown in soil. Seeds in 25 siliques sampled randomly from 10 plants for each mutant line were counted, and the standard deviations (±sd) were calculated. The cr88 hy5 double mutant yielded fewer seeds (26.4 ± 2.7) per silique, whereas the parents hy5 and cr88 yielded 35.8 ± 3.4 and 33.4 ± 3.4 seeds per silique, respectively. Although, like cr88, the cr88 hy5 double mutant exhibited the yellow-green phenotype, greening was delayed more severely than in cr88 (data not shown). Taken together, the new, additive morphology of the double mutant at mature stages suggests that CR88 and HY5 act in separate yet interacting pathways.
Figure 8.

Morphologies of Mature hy5, cr88, and cr88 hy5 Plants.
hy5 (left), cr88 (middle), and cr88 hy5 (right) plants were grown in soil for 1 month and then photographed.
cr88 hy5 Flowers Early
The time to flowering was measured as the number of rosette leaves present when the inflorescence was 2 cm long. Table 2 shows that cr88 flowered at a later developmental stage (10 leaves) than did the wild-type ecotype Columbia glabrous (Col-gl; 8.3 leaves), whereas hy5 flowered at a similar stage (7.8 leaves), and so did the wild-type ecotype Landsberg erecta (7.6 leaves). Although cr88 flowered later than hy5, the double mutant cr88 hy5 flowered when the plants only had four rosette leaves, a much earlier developmental stage than that of either parents or the F1 plants. Again, these results demonstrate that a new phenotype arises in the double mutant, suggesting that CR88 and HY5 act in separate yet interacting pathways.
Table 2.
Comparison of Rosette Leaf Number for Double Mutant cr88 hy5 and the Parents
| Linesa | Leaf Numberb |
|---|---|
| Col-glc | 8.3 ± 0.3 |
| cr88 | 10.0 ± 0.6 |
| Lerd | 7.6 ± 0.2 |
| hy5 | 7.8 ± 0.3 |
| cr88 × hy5; F1 | 11.2 ± 0.4 |
| cr88 hy5 | 4.0 ± 0.5 |
Plants were grown in soil under 16-hr-light/8-hr-dark cycles at 21°C.
Rosette leaf numbers were recorded at bolting. Numbers are the means of 20 to 25 plants ±sd.
The wild type for cr88.
Ler, Landsberg erecta, the wild type for hy5.
DISCUSSION
Inhibiting Photosynthesis Does Not Produce a Phenocopy of the Photomorphogenic Defects of cr88
cr88 plants exhibit a typical etiolated morphology in red light: long hypocotyls and closed and yellow cotyledons. They also exhibit a slow-greening phenotype in all light conditions. In addition, the light-induced expression of certain genes is impaired in red, blue, far-red, and white light (Lin and Cheng, 1997; Figure 2). We showed previously that 1% sucrose cannot eliminate differences in the extents of expression of NR2, RBCS, and CAB mRNA between cr88 and the wild-type seedlings (Lin and Cheng, 1997). However, the photomorphogenic defects and the impaired gene expression could be secondary effects of a possible decrease of photosynthetic activity in cr88. Neither blocking photosynthesis with DCMU nor photobleaching with norflurazon could abolish the inhibition of hypocotyl elongation in the wild-type seedlings grown in red light. Thus, simply inhibiting photosynthesis did not produce a phenocopy of the photomorphogenic defect of cr88. These results also suggest that no plastidic signal is required for the inhibition of hypocotyl elongation mediated by PhyB—although one apparently is required for phytochrome-mediated nuclear gene expression (Oelmueller and Mohr, 1986; Oelmueller et al., 1986; Oelmueller and Briggs, 1990; Susek et al., 1993; Lopez-Juez et al., 1998). Thus, the proposed plastidic signal probably acts downstream of CR88.
DCMU also inhibits the expression of FEDA mRNA (Figure 1), as has been reported (Petracek et al., 1997). In contrast, the red light–induced expression of NR2, CAB, and RBCS is not inhibited. Therefore, photosynthesis is required only for the light-induced expression of a subset of nuclear-encoded genes, the gene products of which function in chloroplasts. CAB and RBCS are not in this group. The impaired induction of NR2, CAB, and RBCS in cr88 by light is therefore unlikely to be caused by a presumed decrease in photosynthesis.
Epistatic Relationships between cr88 and cop1 and between cr88 and hy5
The only known null allele used in these analyses is hy5 (Koornneef et al., 1980). The cop1-4 and 1-6 alleles are weak mutants (Ang and Deng, 1994), and the nature of the cr88 mutation is unknown. Meaningful interpretation of the results for determining epistatic relationships was made possible by taking advantage of the pleiotropic phenotypes of cop1 and cr88 and the information of COP1 (Deng et al., 1992; Ang et al., 1998) and HY5 (Oyama et al., 1997; Chattopadhyay et al., 1998) at the structural and functional levels.
The epistatic relationships between cr88 and cop1 vary with respect to cop1 alleles chosen, physiological responses analyzed, and light conditions used. Similar variations have been observed in the analysis of epistasis between hy5 and cop1 mutations (Ang and Deng, 1994). Such similarities suggest that CR88 may be located at a similar hierarchical position as HY5, acting in close proximity with COP1, in the photomorphogenesis pathway.
Because CR88 and HY5 each regulates a subset of COP1-controlled processes, they are likely to function downstream of COP1. The additive relationship between cr88 and hy5 with respect to hypocotyl length in red light, together with the new phenotypes such as early flowering, suggest that CR88 and HY5 act in separate yet interacting pathways. An alternative explanation for the arising of the new phenotypes is that the two genes may interact differently in different developmental stages. The phenotypic differences between hy5 and cr88 indicate that HY5 plays a major role in controlling hypocotyl elongation, whereas CR88 plays a major role in promoting the greening process. The requirement for both genes in the red light inhibition of hypocotyl elongation and the ability of both cr88 and hy5 mutations to restore the greening ability of dark-grown cop1-4 and cop1-6 seedlings indicate that CR88 and HY5 are also required in certain common processes. The function of CR88 in inhibition of hypocotyl elongation is required in red light, possibly because of incomplete derepression of HY5 by COP1. Red light is less efficient than blue and far-red light in inhibiting hypocotyl elongation (McNellis and Deng, 1995). In the blue and far-red light spectra, the derepression of HY5 is complete, and the function of CR88 in inhibition of hypocotyl elongation becomes dispensable.
The above interpretation is consistent with the allele-specific interactions between cr88 and the two cop1 alleles with respect to hypocotyl length in red light. cop1-4 has a nonsense mutation that produces a truncated protein lacking the C-terminal domain of WD-40 repeats, a domain that is required for the interaction between COP1 and HY5 (McNellis et al., 1994; Ang et al., 1998). When HY5 is fully derepressed, the ability of CR88 to inhibit hypocotyl elongation is masked. Whereas the cop1-6 allele encodes a full-length protein with a five–amino acid insertion 5′ of the WD-40 domain (McNellis et al., 1994), this protein may still partially repress HY5 in red light. In this situation, a complete inhibition of hypocotyl elongation requires a functional CR88. The nonlinear relationship of COP1, CR88, and HY5 that we proposed in the pathway for transduction of light signals may partly explain the complex epistatic interactions between cr88 and cop1 and between hy5 and cop1 with respect to hypocotyl length.
The light-induced expression of NR2, CAB, and RBCS genes is defective in cr88 (Lin and Cheng, 1997; Figure 2). However, the constitutive expression of these genes caused by cop1 mutations is not suppressed by cr88 (Figure 4A). In view of the broad pleiotropism of cop1 mutants (Deng et al., 1992) and the role of COP1 as a repressor of photomorphogenesis (Torii et al., 1998; Stacey et al., 1999), COP1 most likely interacts with additional factors other than HY5 and CR88 to control downstream pathways. Some of these factors may override the control of CR88 over the expression of NR2, CAB, and RBCS genes in cop1 mutants in the dark.
Loss of Greening Ability of Dark-Grown cop1 and Its Reversal by cr88
Our results show that the loss of greening ability of cop1 seedlings after prolonged growth in the dark is accompanied by chloroplast destruction and degradation of RNA. Eight hours of white light treatment degraded all of the RNA (Figure 4B) and destroyed chloroplasts (Figure 5N). Although a causal relationship between RNA degradation and chloroplast destruction has not been established, chloroplast destruction probably causes a general degradation of the RNA. The cause of light-induced destruction seen in cop1 is unclear, but studies of protochlorophyllide reductase (POR) suggest that this light-activated enzyme may play a role. Barnes et al. (1996) showed that far-red light blocks the greening of cotyledons and inhibits POR synthesis. The plastids of dark-grown cop1 mutants lack an organized prolamellar body, a defect that can be restored by overexpressing POR (Sperling et al., 1998). We do not see an organized prolamellar body in the dark-grown plastids in double mutants of cr88 and cop1 (Figures 5C and 5D). It will be interesting to determine whether transgenic cop1plants that overexpress POR are protected from photodestruction.
The ability of cr88 to rescue light-induced chloroplast destruction is consistent with CR88 acting downstream of COP1. In addition to NR2, CAB, and RBCS, CR88 is likely to control the expression of other genes in the greening process. Some of the genes controlled by CR88 may play a role in causing the light-induced destruction in dark-grown cop1. Mutant alleles of HY5 also can restore the greening ability of dark-grown cop1 (Ang and Deng, 1994), probably also by preventing light-induced destruction of chloroplasts. HY5 is known to interact with COP1 and is likely to activate light-regulated gene expression of a subset of genes, including CHS (Ang et al., 1998). Interestingly, genes expressed in subnormal amounts in cr88, such as CAB and RBCS, are not affected in hy5 (Ang and Deng, 1994). Conversely, CHS expression is not affected in cr88 (Lin and Cheng, 1997) but is lower in hy5 (Ang et al., 1998). Mutation in either CR88 or HY5 can rescue the loss of greening ability of cop1, suggesting that both gene products contribute to the loss of greening ability in cop1.
In conclusion, we have further characterized cr88 with respect to long hypocotyls in red light and defective gene expression. Using morphological, cellular, and gene expression criteria for epistasis analyses, we determined that CR88 acts downstream of COP1 and in a branch separate from HY5 in the genetic hierarchy of the photomorphogenesis pathway. Most strikingly, cr88 rescues the light-induced destruction of chloroplast in dark-grown cop1 seedlings. The map position of CR88 demonstrates that it defines a new photomorphogenesis locus.
METHODS
Plant Material and Growth Conditions
The chlorate-resistant mutant line cr88 was isolated previously (Lin and Cheng, 1997). Seeds of Arabidopsis thaliana Columbia glabrous (Col-gl) were obtained from Lehle Seeds (Round Rock, TX). The two constitutive photomorphogenic cop1 mutant alleles, cop1-4 and cop1-6 (McNellis et al., 1994), were a gift from X.-W. Deng (Yale University, New Haven, CT). The hy5 mutant (Koornneef et al., 1980) used in all experiments has been renamed long hypocotyl hy5-1 and was obtained from the Arabidopsis Biological Research Center (Ohio State University, Columbus).
For growth on media, seeds were surface-sterilized by treating with 95% ethanol for 2 min and 1.5% sodium hypochlorite for 5 min. Seeds were then rinsed six times with sterile water. Seeds were sown on Petri dishes containing half-strength Murashige and Skoog salts (Sigma), 0.7% agar (Difco, Detroit, MI), and 1% sucrose. To achieve uniform germination, we incubated the plates in the dark at room temperature for 1 day, moved them to 4°C for 2 days, and grew them at 21°C. For treatments with 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) and norflurazon (Sandoz 9789; Novartis Crop Protection, Greensboro, NC), the stock solutions were filter-sterilized and added to the media.
For growth in soil, seeds were imbibed in 10 mM KNO3 in the dark at 4°C for 2 days before being sown on Jiffy Mix Plus (Jiffy Products of America, Inc., Batavia, IL). Plants were grown at 21°C under a 16-hr-light/8-hr-dark cycle in 800 μmol m−2 sec−1 white light for the indicated time.
Light sources used in light induction experiments were as described previously (Lin and Cheng, 1997). The fluence rates used were as follows: red, 19 μmol m−2 sec−1; blue, 11 μmol m−2 sec−1; far red, 10 μmol m−2 sec−1; and white, 100 μmol m−2 sec−1.
Construction of Double Mutants
To construct cr88 hy5-1, cr88 cop1-4, and cr88 cop1-6 double mutants, we crossed individuals homozygous for each mutant allele and allowed the F1 progeny to self-fertilize. Double mutants of cr88 and cop1 were identified easily from the F2 population because a new phenotype of short stature and yellow-green color arose in one-sixteenth of the population. These plants were considered to be double mutants, and their seeds were used directly in the analysis. No segregation of the new phenotype in the F3 population was observed from the 25 selected F2 individuals. To select the double mutant cr88 hy5-1, we identified F2 individuals homozygous for the phenotype of the cr88 mutant (yellow-green) and allowed them to self-fertilize. The F3 seedlings that exhibited long hypocotyls in white light (the phenotype of hy5 but not cr88) were considered homozygous for cr88 and hy5; no segregation of the yellow-green phenotype was observed in the F3 generation.
RNA Gel Blot Analysis
The procedure for RNA gel blot analysis and the sources for Arabidopsis nitrate reductase NR1 and NR2 and the small subunit of ribulose bisphosphate carboxylase (RBCS) were described by Cheng et al. (1991). The sources for cDNAs of Arabidopsis chlorophyll a/b binding protein (CAB), chalcone synthase (CHS), and the soybean 28S rRNA were as described previously (Lin and Cheng, 1997). The ferredoxin gene (FEDA) was described by Somers et al. (1990).
Linkage Analysis
The genetic location of cr88 was established by determining linkage between cr88 and restriction fragment length polymorphisms (Fabri and Schaffner, 1994). A homozygous cr88 plant (ecotype Col) was crossed to a wild-type plant (ecotype Landsberg erecta). The resulting F1 progeny were allowed to self-pollinate, generating F2 plants. For rough mapping, seeds were collected from 20 cr88 F2 plants to establish F3 families. Genomic DNA was isolated as described by Dellaporta et al. (1983). DNA was digested with diagnostic restriction enzymes, transferred to membranes, and hybridized with probes made from a set of markers described by Fabri and Schaffner (1994). DNA gel blot analysis was performed according to standard procedure (Ausubel et al., 1987). Additional DNA samples were isolated from 150 F2 individuals exhibiting the phenotype of the cr88 mutant and used in linkage analysis. Markers used were Mi320, Mi421 (Liu et al., 1996), and CDs3. All markers were obtained from the Arabidopsis Biological Research Center except for CDs3, which was kindly provided by G. Picard (University of Blasé Pascal, Aubière cedex, France).
Transmission Electron Microscopy
Fixation and embedding of Arabidopsis seedlings were performed as described previously (Lin and Cheng, 1997). Briefly, cotyledons were fixed in 2.5% glutaraldehyde in 100 mM sodium cacodylate buffer, pH 7.2, kept at room temperature for 1 hr, and then transferred to 4°C overnight. After postfixation in 1% osmium tetroxide for 1 hr, samples were stained with 2.5% uranyl acetate for 20 min. Samples were then dehydrated in acetone and embedded in Spurr's resin (Polysciences Incorporated, Warrington, PA). After microtomy, 95-nm-thick sections were poststained with 5% uranyl acetate for 8 min, followed with lead citrate for 7 min. Specimens were observed with a transmission electron microscope (model H7000; Hitachi Scientific Instruments, Mountain View, CA).
Acknowledgments
We thank Xing-Wang Deng for cop1 seeds; George Picard and Peter H. Quail for DNA probes; Mark A. Conkling, Wayne Carlson, and Rodney N. Nagoshi for critical reading of the manuscript; and Jean Ross for assisting with the electron microscopy work. This work was supported by grants from the Carver Foundation and by National Science Foundation Grant No. IBN 98-08823.
References
- Ahmad, M., and Cashmore, A.R. (1993). HY4 gene of A. thaliana encodes a protein with characteristics of a blue light photoreceptor. Nature 360 162–166. [DOI] [PubMed] [Google Scholar]
- Ahmad, M., and Cashmore, A.R. (1996). The pef mutants of Arabidopsis thaliana define lesions early in the phytochrome signaling pathway. Plant J. 10 1103–1110. [DOI] [PubMed] [Google Scholar]
- Ang, L.-H., and Deng, X.-W. (1994). Regulatory hierarchy of photomorphogenic loci: Allele-specific and light-dependent interaction between the HY5 and COP1 loci. Plant Cell 6 613–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang, L.-H., Chattopadhyay, S., Wei, N., Oyama, T., Okada, K., Batschauer, A., and Deng, X.-W. (1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1 213–222. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kinston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K., eds (1987). Current Protocols in Molecular Biology. (New York: Green Publishing/Wiley Interscience).
- Barnes, S.A., Nishizawa, N.K., Quaggio, R.B., Whitelam, G.C., and Chua, N.-H. (1996). Far-red light blocks greening of Arabidopsis seedlings via a phytochrome A–mediated change in plastid development. Plant Cell 8 601–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay, S., Ang, L.-H., Puente, P., Deng, X.-W., and Wei, N. (1998). Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, C.L., Acedo, G.N., Dewdney, J., Goodman, H.M., and Conkling, M.A. (1991). Differential expression of the two Arabidopsis nitrate reductase genes. Plant Physiol. 96 275–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory, J. (1993). Out of darkness: Mutants reveal pathways controlling light-regulated development on plants. Trends Genet. 9 167–172. [DOI] [PubMed] [Google Scholar]
- Chory, J., Peto, C., Feinbaum, R., Pratt, L., and Ausubel, F. (1989). Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light. Cell 58 991–999. [DOI] [PubMed] [Google Scholar]
- Dehesh, K., Francl, C., Parks, B.M., Seeley, K.A., Short, T.W., Tepperman, J.M., and Quail, P.H. (1993). Arabidopsis HY8 encodes phytochrome A. Plant Cell 5 1081–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta, S.L., Wood, J., and Hicks, J.B. (1983). A plant DNA minipreparation: Version II. Plant Mol. Biol. Rep. 1 19–21. [Google Scholar]
- Deng, X.-W. (1994). Fresh view of light signal transduction in plants. Cell 76 423–426. [DOI] [PubMed] [Google Scholar]
- Deng, X.-W., Caspar, T., and Quail, P.H. (1991). COP1: A regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes Dev. 5 1172–1182. [DOI] [PubMed] [Google Scholar]
- Deng, X.-W., Matsui, M., Wei, N., Wagner, D., Chu, A.M., Feldmann, K.A., and Quail, P.H. (1992). COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a Gβ homologous domain. Cell 71 791–801. [DOI] [PubMed] [Google Scholar]
- Escoubas, J.M., Lomas, M., Laroche, J., and Falkowski, P.G. (1995). Light intensity regulation of cab gene transcription is signaled by the redox state of the plastoquinone pool. Proc. Natl. Acad. Sci. USA 92 10237–10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabri, C.O., and Schaffner, A.R. (1994). An Arabidopsis thaliana RFLP mapping set to localize mutations to chromosomal regions. Plant J. 5 149–156. [Google Scholar]
- Genoud, T., Millar, A.J., Nishizawa, N., Kay, S.A., Schafer, E., Nagatani, A., and Chua, N.-H. (1998). An Arabidopsis mutant hypersensititive to red and far-red light signals. Plant Cell 10 889–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday, K., Devlin, P.F., Whitelam, G.C., Hanhart, C., and Koornneef, M. (1996). The ELONGATED gene of Arabidopsis acts independently of light and gibberellins in the control of elongation growth. Plant J. 9 305–312. [DOI] [PubMed] [Google Scholar]
- Hangarter, R.P. (1997). Gravity, light and plant form. Plant Cell Environ. 20 796–800. [DOI] [PubMed] [Google Scholar]
- Hoecker, U., Xu, Y., and Quail, P.H. (1998). SPA1: A new genetic locus involved in phytochrome A–specific signal transduction. Plant Cell 10 19–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker, U., Tepperman, J.M., and Quail, P.H. (1999). SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science 284 496–499. [DOI] [PubMed] [Google Scholar]
- Hou, Y., von Arnim, A.G., and Deng, X.-W. (1993). A new class of Arabidopsis constitutive photomorphogenic genes involved in regulating cotyledon development. Plant Cell 5 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, E., Bradley, M., Harberd, N.P., and Whitelam, G.C. (1994). Photoresponses of light-grown phyA mutants of Arabidopsis: Phytochrome A is required for the perception of daylength extensions. Plant Physiol. 105 141–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrick, R.E., and Kronenberg, G.H.M., eds (1994). Photomorphogenesis in Plants, 2nd ed. (Dordrecht, The Netherlands: Kluwer Academic Publishers).
- Koornneef, M., Rolff, E., and Spruit, C.J.P. (1980). Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana. Z. Pflanzenphysiol. 100 147–160. [Google Scholar]
- Kwok, S.F., Piekos, B., Misera, S., and Deng, X.-W. (1996). A complement of ten essential and pleiotropic Arabidopsis COP/DET/FUS genes is necessary for repression of photomorphogenesis in darkness. Plant Physiol. 110 731–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H.M., Culligan, K., Dixon, R.A., and Chory, J. (1995). CUE1: A mesophyll cell–specific positive regulator of light-controlled gene expression in Arabidopsis. Plant Cell 7 1599–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y., and Cheng, C.L. (1997). A chlorate-resistant mutant defective in the regulation of nitrate reductase gene expression in Arabidopsis defines a new HY locus. Plant Cell 9 21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.G., Mitsukawa, N., Lister, C., Dean, C., and Whittier, R.F. (1996). Isolation and mapping of a new set of 129 RFLP markers in Arabidopsis thaliana using recombinant inbred lines. Plant J. 10 733–736. [DOI] [PubMed] [Google Scholar]
- Lopez-Juez, E., Jarvis, R.P., Takeuchi, A., Page, A.M., and Chory, J. (1998). New Arabidopsis cue mutants suggest a close connection between plastid and phytochrome regulation of nuclear gene expression. Plant Physiol. 118 803–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis, T.W., and Deng, X.-W. (1995). Light control of seedling morphogenic pattern. Plant Cell 7 1749–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis, T.W., von Arnim, A.G., Araki, T., Komeda, Y., Misera, S., and Deng, X.-W. (1994). Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6 487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misera, S., Muller, A.J., Weiland-Heidecker, U., and Jurgens, G. (1994). The FUSCA genes of Arabidopsis: Negative regulators of light responses. Mol. Gen. Genet. 244 242–252. [DOI] [PubMed] [Google Scholar]
- Mullet, J.E. (1988). Chloroplast development and gene-expression. Annu. Rev. Plant Physiol. Plant Mol. Biol. 39 475–502. [Google Scholar]
- Neuhaus, G., Bowler, C., Hiratsuka, K., Yamagata, H., and Chua, N.-H. (1997). Phytochrome-regulated repression of gene expression requires calcium and cGMP. EMBO J. 16 2554–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni, M., Tepperman, J.M., and Quail, P.H. (1998). PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95 657–667. [DOI] [PubMed] [Google Scholar]
- Oelmueller, R., and Mohr, H. (1986). Photooxidative destruction of chloroplasts and its consequences for expression of nuclear genes. Planta 167 106–113. [DOI] [PubMed] [Google Scholar]
- Oelmueller, R., and Briggs, W.R. (1990). Intact plastids are required for nitrate-induced and light-induced accumulation of nitrate reductase activity and messenger RNA in squash cotyledons. Plant Physiol. 92 434–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelmueller, R., Levitan, I., Bergfeld, R., Rajasekhar, V.K., and Mohr, H. (1986). Expression of nuclear genes as affected by treatments acting on the plastids. Planta 168 482–492. [DOI] [PubMed] [Google Scholar]
- Osterlund, M.T., Ang, L.-H., and Deng, X.-W. (1999). The role of COP1 in repression of Arabidopsis photomorphogenic development. Trends Cell Biol. 9 113–117. [DOI] [PubMed] [Google Scholar]
- Oyama, T., Shimura, Y., and Okada, K. (1997). The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 11 2983–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks, B.M., and Quail, P.H. (1993). hy8, a new class of Arabidopsis mutant deficient in functional phytochrome A. Plant Cell 5 39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petracek, M.E., Dickey, L.F., Huber, S.C., and Thompson, W.F. (1997). Light-regulated changes in abundance and polyribosome association of ferredoxin mRNA are dependent on photosynthesis. Plant Cell 9 2291–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, J.W., Nagpal, P., Poole, D.S., Furuya, M., and Chory, J. (1993). Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis. Plant Cell 5 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter, R.S., Coomber, S.A., Bourett, T.M., Bartley, G.E., and Scolnik, P.A. (1994). Control of leaf and chloroplast development by the Arabidopsis gene pale cress. Plant Cell 6 1253–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, R., West, J., Gerda, C., Love, K., Balestrazzi, A., and Dean, C. (1996). Detailed description of four YAC contigs representing 17 Mb of chromosome 4 of Arabidopsis thaliana ecotype Columbia. Plant J. 9 755–765. [DOI] [PubMed] [Google Scholar]
- Schmidt, R., Love, K., West, J., Lenehan, Z., and Dean, C. (1997). Description of 31 YAC contigs spanning the majority of Arabidopsis thaliana chromosome 5. Plant J. 11 563–572. [DOI] [PubMed] [Google Scholar]
- Somers, D.E., Caspar, T., and Quail, P.H. (1990). Isolation and characterization of ferredoxin gene from Arabidopsis thaliana. Plant Physiol. 93 572–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling, U., Franck, F., van Cleve, B., Frick, G., Apel, K., and Armstrong, G.A. (1998). Etioplast differentiation in Arabidopsis: Both PORA and PORB restore the prolamellar body and photoactive protochlorophyllide-F655 to the cop1 photomorphogenic mutant. Plant Cell 10 283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey, M.G., Hicks, S.N., and von Arnim, A.G. (1999). Discrete domains mediate the light-responsive nuclear and cytoplasmic localization of Arabidopsis COP1. Plant Cell 11 349–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susek, R., Ausubel, F., and Chory, J. (1993). Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74 787–799. [DOI] [PubMed] [Google Scholar]
- Torii, K.U., McNellis, T.W., and Deng X.-W. (1998). Functional dissection of Arabidopsis COP1 reveals specific roles of its three structural modules in light control of seedling development. EMBO J. 17 5577–5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, D., Hoecker, U., and Quail, P.H. (1997). RED1 is necessary for phytochrome B–mediated red light–specific signal transduction in Arabidopsis. Plant Cell 9 731–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, N., and Deng, X.-W. (1992). COP9: A new genetic locus involved in light-regulated development and gene expression in Arabidopsis. Plant Cell 4 1507–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, N., Kwok, S.F., von Arnim, A.G., Lee, A., McNellis, T.W., Piekos, B., and Deng, X.-W. (1994). Arabidopsis COP8, COP10, and COP11 genes are involved in repression of photomorphogenic development in darkness. Plant Cell 6 629–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam, G.C., Johnson, E., Peng, J., Carol, P., Anderson, M.L., Cowl, J.S., and Harberd, N.P. (1993). Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5 757–768. [DOI] [PMC free article] [PubMed] [Google Scholar]