Abstract
Previous studies have presented indirect evidence that the transposase of the maize transposable element Activator (TPase) is active as an oligomer and forms inactive macromolecular complexes expressed in large amounts. Here, we have identified and characterized a dimerization domain at the C terminus of the protein. This domain is the most highly conserved region in the transposases of elements belonging to the Activator superfamily (hAT element superfamily) and contains a characteristic signature motif. The isolated dimerization domain forms extremely stable dimers in vitro. Interestingly, mutations in five of the six conserved residues of the signature motif do not affect in vitro dimerization, whereas mutations in other, less strictly conserved residues of the signature motif do. Loss of dimerization in vitro correlates with loss of TPase activity in vivo. As revealed by in situ immunofluorescence staining of mutant TPase proteins, the dimerization domain also is involved in forming inactive macromolecular aggregates when overexpressed, and the TPase contains one or more additional interaction functions.
INTRODUCTION
The maize transposon Activator (Ac), a small, autonomous element that transposes by a cut-and-paste mechanism (McClintock, 1951), encodes an 807–amino acid transposase protein (TPase; Kunze et al., 1987), which binds specifically to the terminal inverted repeats and repetitive subterminal sequence motifs of Ac (Kunze and Starlinger, 1989; Becker and Kunze, 1997). The transactive TPase is the only element-encoded protein required for transposition (Coupland et al., 1988); in numerous plant species, it is capable of mobilizing a nonautonomous Dissociation (Ds) element (reviewed in Kunze, 1996).
The transposition reactions of only a few eukaryotic transposable elements have been biochemically characterized. The best understood elements are the Drosophila P element and the Caenorhabditis elegans Tc1 transposon, for which in vitro transposition systems have been developed (Kaufman and Rio, 1992; Vos et al., 1996). However, little is known about the structures of the synaptic complexes, the transpososomes, of these and other eukaryotic transposons.
For some prokaryotic mobile elements, models for the molecular architecture of the transpososome have been suggested. Common to these models is the formation of a higher order nucleoprotein complex (the synaptic complex) as an intermediate structure during the transposition reaction; in the synaptic complex, the two ends of the transposon are brought into proximity by protein–DNA and protein–protein interactions. In addition to the transposase, the transpososome is said to facultatively encompass the reinsertion target site, other transposon-encoded proteins, and accessory host factors.
Because the transposition mechanism and in some cases the transposases of certain prokaryotic and eukaryotic elements are similar, presumably the structures of their transpososomes also are related, with protein–protein interactions playing a key role in their assembly. Indirect evidence for Ac TPase interactions has been obtained from DNA binding studies. TPase binds in a cooperative mode to its target sites in the Ac ends, a finding that can be interpreted to indicate stabilization of the protein–DNA complexes by the interaction between TPase molecules (Becker and Kunze, 1997). A second, more compelling argument for Ac TPase being active as an oligomeric protein comes from analyses of mutants. DNA binding–deficient, transpositionally inactive TPase mutants coexpressed with the wild-type TPase in plant cells containing a Ds reporter construct act as dominant inhibitors of transposition and thus supposedly interfere with TPase function by forming nonfunctional heterooligomers with the wild-type protein (Kunze et al., 1993).
Additional evidence for TPase interactions is the appearance of large nuclear TPase complexes resembling filaments. These structures have been observed in maize endosperm cells that are homozygous for an active Ac element (i.e., are carrying three copies of the element), in the nuclei of petunia protoplasts (Heinlein et al., 1994), transgenic tobacco plants (I. Kornacker and R. Kunze, unpublished data), and even in the nuclei of TPase-expressing insect cells (Hauser et al., 1988). Investigation of the Ac transposition reaction and its regulation by mainly genetic but also some biochemical studies has revealed that Ac is predominantly regulated post-transcriptionally. The TPase apparently catalyzes transposition only within a specific window of concentration, whereas at concentrations above a threshold level, TPase becomes autoinhibitory (Scofield et al., 1993). Heinlein et al. (1994) have proposed that TPase aggregation into the filament-like complexes is responsible for inactivation.
Functional dissection of the Ac TPase has revealed that the N-terminal 200 amino acids contain the DNA binding domain and nuclear transport functions but are dispensable for aggregate formation. Coimport of a nuclear transport–deficient TPase fragment (amino acids 189 to 807) into the nucleus when coexpressed with the wild-type TPase indicates that the ∼600 TPase residues at the C terminus contain a self-interaction function (Boehm et al., 1995).
Taken together, these results suggest that the Ac TPase has complex oligomerization and aggregation properties. In this study, we report the identification and characterization of an Ac TPase dimerization domain by using the yeast two-hybrid system, biochemical in vitro assays, in vivo functional tests, and in situ immunofluorescence staining. This dimerization domain is located close to the C terminus between residues 674 and 754 of the TPase. It is the most highly conserved of three closely related regions in the transposases of elements belonging to the Ac transposon superfamily (also called hAT elements, for hobo, Ac, and Tam3). We demonstrate that, in addition to facilitating dimerization, this domain harbors still another function that is essential for transposase activity.
RESULTS
TPase Specifically Self-Interacts in Yeast Cells
To identify and locate Ac TPase interaction domains, we used the yeast two-hybrid system. We fused the GAL4 DNA binding (BD) and transactivation (AD) domains to the N terminus of the full-length 807–amino acid Ac TPase (the fusion proteins are termed BD-TPase1–807 and AD-TPase1–807, respectively) and the TPase derivative TPase103–807, which is truncated at its N terminus. This derivative is fully functional in plant cells (Li and Starlinger, 1990; Kunze et al., 1995b). The AD and BD fusion protein expression plasmids were transformed in yeast cells, and LacZ activity was determined. We initially determined LacZ expression by using a standard colony lift assay (Breedon and Nasmyth, 1985) or a colorimetric assay of soluble extracts (Miller, 1972). Both assays easily detected the interaction of GAL4 and other positive control proteins (see Methods).
In cells coexpressing any combination of AD and BD TPase fusion proteins, LacZ activity was detectable but was ∼40-fold less than in transformants expressing wild-type GAL4 and fivefold less than in those expressing the simian virus SV40 T-antigen and p53 controls. Therefore, we used a more sensitive, semiquantitative dot blot assay for measuring LacZ activity (Essers and Kunze, 1996). In this assay, the coexpression of AD-TPase1–807 or AD-TPase103–807 with BD-TPase1–807 or BD-TPase103–807, but not the expression of any single protein, activated the lacZ gene (Figure 1). This result suggests that the wild-type Ac TPase protein and the functional derivative TPase103–807 specifically interact in yeast cells.
Figure 1.
The Ac TPase Protein Interacts in Yeast Cells.
Equal numbers of yeast cells expressing the indicated proteins were transferred to nylon membranes, and LacZ activity was determined. AD, GAL4 activation domain; AD-TPase1–807, AD fused to the N terminus of the wild-type Ac TPase; AD-TPase103–807, AD fused to the truncated TPase derivative TPase103–807; BD, GAL4 DNA binding domain; BD-TPase1–807, BD fused to the wild-type Ac TPase; BD-TPase103–807, BD fused to TPase103–807.
The TPase Interaction Domain Is Located near the C Terminus
Because the interactive properties of TPase103–807 do not differ from those of the full-length protein, we used the truncated form as one interaction partner in most of the subsequent experiments. Having generated a series of N-terminal TPase deletions fused with the GAL4-BD, we tested their ability to interact with the AD-TPase103–807 fusion. As controls, the BD-TPase hybrids were expressed individually and in combination with the unfused AD domain (Table 1). BD fusions with TPase protein fragments starting at residues 1, 103, 136, 268, 560, and 646 did not induce LacZ expression when expressed separately but did when coexpressed with AD-TPase103–807. These results suggest that the C-terminal 162 TPase residues contain a domain that mediates specific interaction with the TPase103–807 protein.
Table 1.
Analysis of Ac TPase Interactions in Yeast by Using the Two-Hybrid System
| Protein 2b
|
|||
|---|---|---|---|
| Protein 1a | No protein | AD | AD-TPase103–807 |
| BD | − | − | − |
| BD-TPase1–807 | − | − | + |
| BD-TPase103–807 | − | − | + |
| BD-TPase136–807 | − | − | + |
| BD-TPase268–807 | − | − | + |
| BD-TPase395–807 | + | + | + |
| BD-TPase560–807 | − | − | + |
| BD-TPase646–807 | − | − | + |
| BD-TPase662–807 | + | + | + |
| BD-TPase709–807 | + | + | + |
| BD-TPase743–807 | + | + | + |
| BD-TPase785–807 | NDc | + | + |
Proteins are designated as in the text.
LacZ expression was induced (+) or not induced (−) by coexpression of protein 1 and protein 2.
ND, not determined.
Surprisingly, BD-TPase395–807 and all BD fusions with TPase fragments in which 661 or more N-terminal amino acids were deleted induced LacZ expression in the absence of a fusion partner. These fusions did not support any conclusions about TPase interactions. Possibly, a TPase peptide that usually is hidden in these constructs and carries a transactivation function in yeast cells was exposed. Consistent with this idea, the BD-TPase395–663 fusion, which lacks the 144 C-terminal TPase residues, did not promote lacZ activation when expressed alone or with AD-TPase103–807 (data not shown).
Subsequently, we fused the TPase fragments deleted from the C terminus to the GAL4 activation and DNA binding domains. Pairwise coexpression of these constructs revealed that the 53 Ac TPase residues at the C terminus are not required for interaction, whereas when 98 amino acids were deleted, the fragment was no longer capable of inducing detectable LacZ expression (Table 2). We also investigated the possibility that more extensive C-terminal deletions might expose another otherwise hidden interaction function. However, none of the fusion proteins tested (AD-TPase103–249, AD-TPase103–337, AD-TPase103–458, or AD-TPase103–585) induced detectable LacZ activity. These experiments indicate that the Ac TPase contains a domain that mediates self-interaction in yeast cells close to its C terminus between residues 646 and 754.
Table 2.
Localization of the Interaction Domain of the Ac TPase
| Protein 2b
|
||||
|---|---|---|---|---|
| Protein 1a | AD | AD-TPase103–709 | AD-TPase103–754 | AD-TPase103–807 |
| BD | − | − | − | − |
| BD-TPase103–709 | − | − | − | − |
| BD-TPase103–754 | − | − | + | + |
| BD-TPase103–807 | − | − | + | + |
Proteins are designated as in the text.
LacZ expression was induced (+) or not induced (−) by coexpression of protein 1 and protein 2.
Earlier in vivo experiments had investigated the effects on transposition of inserting two amino acids at various positions throughout the TPase protein (Kunze et al., 1993; Behrens-Jung et al., 1994). We fused three mutant TPase-coding sequences to both the GAL4 activation and DNA binding domains and determined their potential for pairwise interaction in yeast. The insertions of a threonine-arginine dipeptide C-terminal to TPase residue 754 (TPase103–807/754TR) and an arginine-valine dipeptide C-terminal to TPase residue 771 (TPase103–807/771RV) did not interfere with TPase activity in vivo (Kunze et al., 1993). These two mutations also did not affect the self-interaction function of TPase (Table 3). In contrast, inserting an arginine-valine dipeptide C-terminal to TPase residue 709 (TPase103–807/709RV) abolished TPase function in vivo (Kunze et al., 1993) and also disrupted the interaction function in yeast (Table 3). This result corroborates the conclusion that the TPase protein contains a self-specific interaction domain between residues 646 and 754.
Table 3.
Interaction of Mutant TPase Proteins in Yeast
| Protein 2b
|
||||
|---|---|---|---|---|
| Protein 1a | AD-TPase103–709 | AD-TPase103–807/709RV | AD-TPase103–807/754TR | AD-TPase103–807/771RV |
| BD-TPase103–807 | + | − | + | + |
| BD-TPase103–807/709RV | − | − | − | − |
| BD-TPase103–807/754TR | + | − | + | + |
| BD-TPase103–807/771RV | + | − | + | + |
Proteins are designated as in the text.
LacZ expression was induced (+) or not induced (−) by coexpression of protein 1 and protein 2.
The TPase674–777 Interaction Domain Forms Exceptionally Stable Dimers
The two-hybrid system permits identification of protein interaction domains but yields no clue as to whether the interacting protein moieties form dimers or, for example, tetramers. Therefore, we used a chemical cross-linking approach to characterize the identified C-terminal TPase interaction domain in more detail. We expressed an Ac TPase674–777 fragment preceded by a six-histidine (His6) tag (His6-TPase674–777) in Escherichia coli and purified the protein under denaturating conditions by using metal affinity chromatography. After refolding of the protein, aliquots of the protein were cross-linked with ethylene glycol-bis(succinimidylsuccinate) (EGS), size fractionated by SDS-PAGE, and detected by protein gel blotting. The protein was efficiently cross-linked to the dimeric state with 0.5 mM EGS. A 12-fold increase in EGS concentration cross-linked most of the His6-TPase674–777 protein to the dimeric form; only a minor fraction appeared as higher multimers (Figure 2A). For a positive control, we used aldolase, which under the same reaction conditions is efficiently cross-linked into the expected tetramers; the negative control was lysozyme, which remains monomeric (data not shown). The results demonstrate that the Ac TPase674–777 fragment contains a dimerization domain. By taking into account the results from the interaction studies in yeast, the dimerization domain can be narrowed down to 81 TPase residues: 674 to 754.
Figure 2.
Dimerization of the His6-TPase674–777 Protein.
(A) The renatured His6-TPase674–777 protein was chemically cross-linked by incubation for 5 min at room temperature with various EGS concentrations (indicated above each lane), fractionated on an SDS-containing polyacrylamide gel, blotted, and detected by immunofluorescence staining. 1× to 4× indicates monomeric, dimeric, trimeric, and tetrameric His6-TPase674–777, respectively.
(B) Stability of His6-TPase674–777 dimers under denaturing conditions. Lane 1, His6-TPase674–777 column eluate in 8 M urea was loaded directly onto an SDS-containing polyacrylamide gel; lane 2, the eluate was heated to 95°C for 10 min before loading; and lane 3, the eluate was mixed with SDS sample buffer and heated to 95°C for 10 min before loading. The gels were blotted, and the proteins were visualized by immunofluorescence staining. Migration of size markers (in kilodaltons) is indicated at left.
To investigate the stability of the His6-TPase674–777 dimers, we analyzed them under various conditions. Protein solubilized and affinity-purified in 6 M guanidine hydrochloride, and eluted from the purification column in 8 M urea, was loaded directly onto an SDS–polyacrylamide gel and size fractionated. Even in 8 M urea, almost half of the protein persisted in a dimeric state (Figure 2B). After the eluate was boiled for 10 min, a small fraction of the dimers remained intact; complete dissociation into monomers required boiling the protein in Laemmli gel loading buffer for 10 min (Figure 2B). This exceptional stability of the His6-TPase674–777 dimers in vitro is remarkable; however, this finding does not necessarily mirror the strength of interaction in the context of the full-length TPase in vivo.
Effects of Dimerization Domain Mutations on in Vitro Dimerization
The dimerization domain is the most highly conserved domain among the transposases belonging to the class of hAT elements. Using the JPred program at the European Bioinformatics Institute (http://circinus.ebi.ac.uk:8081/; Cuff et al., 1998; Cuff and Barton, 1999) to predict the secondary structure of the aligned domains shown in Figure 3 revealed that some of the most stringently conserved residues are located in regions with a high probability for helical conformation. To investigate the functional significance of predicted helical regions and conserved amino acids, we conducted a mutational analysis. Using site-directed mutagenesis, we replaced individual amino acids with either alanine or proline. Alanine residues are likely to interfere with hydrophilic and hydrophobic interactions but do not change the secondary structure, whereas proline supposedly disrupts α helices. The mutant dimerization domain derivatives were expressed in the His6-TPase674–777 context and purified by affinity chromatography; the stability of the dimers was determined as described above.
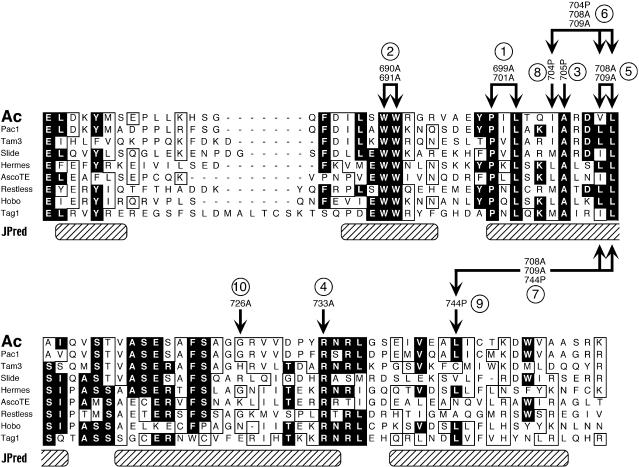
Figure 3.
Mutational Analysis of the Dimerization Domain of the Ac TPase.
Alignment of the conserved C-terminal domains of the transposases of the hAT elements is shown. The black boxes indicate >50% identical residues; the open boxes indicate >50% similar residues. The hatched boxes show regions with a high probability for helical structure predicted by JPred analysis. Arrows above the Ac TPase sequence indicate amino acid substitutions introduced in the various mutations used in this study. Circled numbers refer to the lanes of the gels shown in Figures 4 and 5. Ac, Ac TPase from maize (GenBank accession number X05425; residues 669 to 756); Pac1, putative Pac1 transposase from Pennisetum glaucum (GenBank accession number U02300; residues 649 to 736); Tam3, Tam3 transposase from Antirrhinum (GenBank accession number X55078; residues 630 to 717); Slide, Slide transposase from tobacco (GenBank accession number X97569; residues 584 to 672); Hermes, putative Hermes transposase from Musca domestica (GenBank accession number L34807; residues 523 to 609); AscoTE, putative transposase from an Ascobulus immersus transposon (GenBank accession number Y07695; residues 515 to 599); Restless, putative Restless transposase from Tolypocladium inflatum (GenBank accession number Z69893; residues 710 to 798); Hobo, putative hobo transposase from Drosophila melanogaster (GenBank accession number X04705; residues 552 to 637); Tag1, Tag1 transposase from Arabidopsis (GenBank accession number AF051562; residues 555 to 649).
Surprisingly, replacement of the highly conserved residues W690 and W691 [1] (bracketed numbers correspond to designations in Figure 3 and lane numbers in Figures 4 and 5) or P699 and L701 [2] by alanine and A705 [3] by proline, respectively, did not interfere with dimerization in vitro (Figure 4). In contrast, the mutations I704P [8], V708A/L709A [5], R733A [4], and L744P [9] caused a substantial reduction of dimer stability on SDS gels. Dimerization was not abolished completely, however; some residual protein dimers persisted, and the stability of these dimers apparently was not further decreased in the triple mutants I704P/V708A/L709A [6] and V708A/L709A/L744P [7].
Figure 4.
Stability of Mutant His6-TPase674–777 Derivative Dimers.
His6-TPase674–777 (lane WT) and derivatives thereof carrying the amino acid substitution mutations shown in Figure 3 (lanes 1 to 9) were purified by column chromatograph under denaturing conditions, eluted in 8 M urea, and loaded directly onto an SDS-containing 17.5% polyacrylamide gel. The gel was blotted, and the proteins were visualized by immunofluorescence staining. Migration of the size markers (in kilodaltons) is indicated at left. D, dimeric protein; M, monomeric protein.
Figure 5.
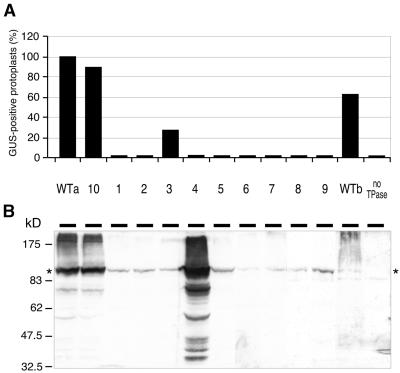
In Vivo Transpositional Activities of TPase Derivatives with Mutations in the Dimerization Domain.
(A) Bars show the average frequency of GUS-positive protoplasts, relative to wild-type TPase103–807. Each mutant was scored in two to six independent experiments. WTa, TPase103–807 expressed from plasmid pcATG10; WTb, TPase103–807 expressed from plasmid pcATG3-10. The designations of mutant proteins (lanes 1 to 10) refer to Figure 3. no TPase, untransfected petunia protoplasts.
(B) Protein gel blot showing the TPase expression levels in the transfected petunia protoplasts. Migration of size markers (in kilodaltons) is indicated at left. The asterisks indicate wild-type and mutant TPase103–807 proteins.
These results demonstrate that only a subset of the absolutely conserved residues in the TPase dimerization domain as well as the nonconserved I704 and L744 residues in putative helical regions are involved directly in the formation of Ac TPase dimers in vitro. Moreover, the more N-terminal conserved amino acids do not appear to be involved in dimerization but may have a different function.
Effects of Dimerization Domain Mutations on TPase Function in Vivo
To determine whether the ability to dimerize in vitro correlates with transpositional activity of the TPase in vivo, we introduced the above-mentioned dimerization domain mutations by site-directed mutagenesis into the TPase103–807 protein encoded in plasmid pcATG10 (Kunze et al., 1993; henceforth, mutant TPase103–807 proteins are designated with abbreviations, e.g., G726A for TPase103–807/G726A). Petunia protoplasts were then cotransfected with these plasmids and with the Ds excision reporter pTY/DIR. Because TPase-induced excision of the Ds results in reversion of the gene encoding β-glucuronidase (GUS), the frequency of GUS-positive protoplasts reflects the activity of the TPase derivative expressed in the cells. The results of the in vivo excision assays are shown in Figure 5A. The mutation A705P [3], which did not impair dimerization of the TPase674–777/A705P protein in vitro, also did not abolish activity of the TPase103–807/A705P protein in vivo, but Ds excision frequency decreased to ∼30% of that of the wild-type protein. All other mutations, including the two that exhibit normal dimer stability, W690A/W691A [2] and P699A/L701A [1], resulted in a total loss of Ds excision. To test whether the effects of the mutations on in vivo activity are specific, we generated an additional mutant, G726A [10] (Figure 3). This neutral substitution at a nonconserved position behaved like the wild-type protein (Figure 5A).
To exclude the possibility that the mutants lack in vivo activity because the proteins are unstable or are not expressed at all, we determined the amounts of (mutant) TPase in the transfected protoplasts by protein gel blotting at the time the protoplasts were harvested for GUS staining (Figure 5B). For expression of wild-type TPase103–807, we used two different plasmids: pcATG10 expresses large amounts of the protein (Figure 5B, lane WTa), whereas pcATG3-10 expresses much less (Figure 5B, lane WTb; Kunze et al., 1993). Although all mutants are based on pcATG10, the amounts of TPase derivatives that accumulated differed. The active mutant G726A [10] was expressed approximately as much as the wild-type protein. In contrast, the mutant R733A [4] accumulated to higher concentrations, whereas all other mutant TPase proteins were found at lower concentrations, similar to that of the wild-type TPase expressed from pcATG3-10. The observation that this lower wild-type TPase concentration is sufficient to induce approximately the same Ds excision frequencies as pcATG10 (Figure 5A) excludes the possibility that the low concentration of mutant proteins is responsible for their complete lack of activity. This conclusion is corroborated by the fact that the weakly expressed mutant A705P [3] is also active, whereas the strongly expressed mutant R733A [4] is completely inactive.
These results are consistent with those of earlier experiments with two–amino acid insertion mutants. Mutants 623TR, 663DR, 754TR, and 771RV in the N- and C-terminal vicinities of the dimerization domain, respectively, are as transpositionally active as the wild-type protein, whereas insertion of two amino acids within the dimerization domain (709RV) inactivates the TPase (Kunze et al., 1993; Behrens-Jung et al., 1994).
In summary, all mutations that affect dimerization are transpositionally inactive. Interestingly, the two mutations W690A/W691A [2] and P699A/L701A [1] are also inactive, although they do not impair dimerization in vitro. This suggests that the conserved TPase residues W690, W691, P699, and L701 are critically involved in the transposition reaction but are not required for dimerization.
Functioning of the Dimerization Domain Is Not Required in All TPase Molecules in the Transpososome
To investigate the role of the dimerization domain in more detail, we coexpressed inactive mutant TPases with wild-type TPase103–807 in petunia cells. Coexpression of W690A/W691A [2], I704P [8], V708A/L709A [5], or V708A/L709A/L744P [7] with the wild-type TPase did not impair the activity of the wild-type protein; accordingly, these mutants are recessive (Table 4). In previous experiments, we had shown that another transpositionally inactive mutant in the dimerization domain, 709RV, is also recessive (Kunze et al., 1993; Behrens-Jung et al., 1994). Recessiveness could be explained if these mutant proteins did not participate in the assembly of the transpososome. However, this possibility can be excluded: coexpressing the mutants W690A/W691A [2], I704P [8], V708A/L709A [5], or V708A/L709A/L744P [7] with 585TR, another recessive mutant that carries a two–amino acid insertion more than 100 residues N-terminal of the dimerization domain (Kunze et al., 1993; Behrens-Jung et al., 1994) restored the activity to mobilize a nonautonomous Ds element (Table 4). Hence, the dimerization function and also the uncharacterized function associated with residues W690/ W691 most likely are not required in each TPase molecule subunit of the transpososome.
Table 4.
Complementation Analysis of Mutant TPase Proteins in Petunia Protoplasts
| Protein 2
|
||||
|---|---|---|---|---|
| Protein 1a | No protein | TPase103–807 | TPase103–807/585TRb | TPase103–807/W690A/W691Ac |
| No protein | NAd | +e,f | −g | − |
| TPase103–807/W690A/W691A | − | +h | + | NA |
| TPase103–807/P699A/L701A | − | NDi | ND | − |
| TPase103–807/I704P | − | +h | + | ND |
| TPase103–807/V708A/L709A | − | +h | ±j | − |
| TPase103–807/V708A/L709A/L744P | − | +h | + | − |
| TPase103–807/R733A | − | −k | − | ND |
Proteins are designated as in the text.
Protoplasts were cotransfected with 10 μg of pcATG3-10/585TR and 10 μg of pcATG10/Protein 1.
Protoplasts were cotransfected with 5 μg of pcATG10/W690A/691A and 5 μg of pcATG10/Protein 1.
NA, not applicable.
(+), full induction of Ds excision (frequency of stained cells >80%; wild-type TPase is 100%).
Protoplasts were cotransfected with 18 μg of pcATG3-10 (weakly expressed wild-type TPase103–807).
(−), no induction of Ds excision (frequency of stained cells <2% is background).
Protoplasts were cotransfected with 3 μg of pcATG3-10 and 15 μg of pcATG10/Protein 1.
ND, not determined.
(±), low induction of Ds excision (frequency of stained cells slightly above background).
Protoplasts were cotransfected with 2 μg of pcATG10 (strongly expressed wild-type TPase103–807) and 10 μg of pcATG10/R733A.
Complementation was not achieved by coexpressing the two mutants W690A/W691A [2] and P699A/L701A [1] or by coexpressing W690A/W691A [2] with V708A/L709A [5]. In contrast, Ds excision was abolished almost completely when mutant R733A was coexpressed with the wild-type TPase (Table 4). The exceptionally high accumulation of this protein in the cells in comparison with the other mutants (Figure 5B), the reduced dimer stability (Figure 4), and the aberrant shape of the aggregates (Figure 6) may indicate that the mutation in R733A [4] alters the protein's structure in a way that affects its interaction properties. Because R733A [4] presumably is able to bind to the TPase target sites in the ends of Ds, the negative dominance could result from an overall distortion of the transpososome.
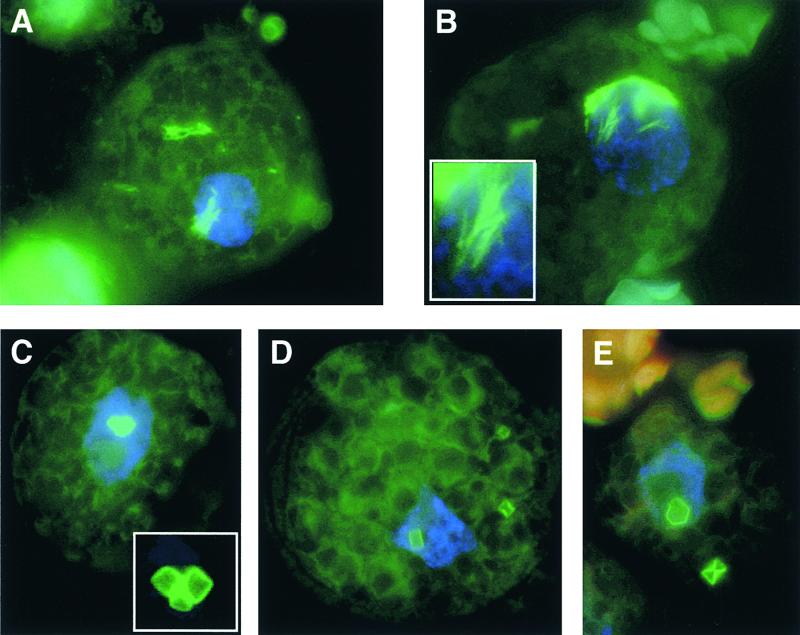
Figure 6.
Aggregate Formation of Wild-Type and Mutant TPase in Petunia Protoplasts.
The Ac TPase was detected by immunofluorescence staining. Nuclei were visualized with 4′,6-diamidino-2-phenylindole.
(A) Wild-type TPase103–807 forms filament-like aggregates in the cytoplasm and nucleus.
(B) Localization and appearance of TPase103–807/G726A aggregates are virtually indistinguishable from those of the wild-type TPase103–807.
(C) to (E) TPase103–807/R733A assembles in the cytoplasm and nucleus into compact aggregates reminiscent of hexagonal crystals.
The insets in (B) and (C) show the respective aggregates in more detail.
Taken together, these experiments provide additional evidence that Ac TPase is active as an oligomer; they also demonstrate the existence of three functionally distinct domains in the C terminus of the protein. An unknown function is encoded in the region at approximately amino acid 585. A second unknown function is encoded by the conserved residues W690, W691, P699, and L701. C-terminal to this function (or overlapping with it) is the dimerization domain, which is highly conserved among the transposases of the hAT superfamily.
The Dimerization Domain Is Involved in Formation of TPase Aggregates in Vivo
When expressed in large amounts in vivo, the Ac TPase forms characteristic filament-like aggregates that can be displayed by immunofluorescence staining (Heinlein et al., 1994). To investigate whether TPase interactions through the dimerization domain are responsible for or are involved in the aggregation process, we analyzed by immunofluorescence staining petunia protoplasts expressing wild-type TPase103–807, the active mutant G726A [10], or the dimerization-impaired mutant R733A [4]. As in previous experiments, the wild-type TPase103–807 assembled into rodlike or filament-like complexes. Because of the lack of a nuclear localization signal in the N-terminal 103 amino acids, the major fraction of the protein was not imported into the nucleus but remained in the cytoplasm (Figure 6A). The G726A [10] aggregates are virtually indistinguishable from those of the wild type (Figure 6B). In contrast, the substitution of alanine for arginine in the R733A [4] protein produced a marked change in the shape of the TPase aggregates: the protein assembled into very large, cubelike complexes reminiscent of crystalline structures (Figures 6C to 6E). These “crystals” were remarkably uniform in size and shape in hundreds of inspected cells. As with the wild-type protein, the mutant complexes accumulated in the cytoplasm or were associated with the nucleus.
Although the R733A [4] mutation is accompanied by reduced dimer stability in vitro and the mutant TPase is completely inactive in vivo, in vivo aggregation is not abolished, but the complexes do assume an altered shape. Thus, the dimerization domain plays a role in aggregate formation, although this process requires an additional, uncharacterized TPase domain.
DISCUSSION
Using the yeast two-hybrid system, we identified and mapped a dimerization domain near the C terminus of the Ac TPase. The observation that activation of the LacZ reporter gene induced by TPase self-interaction is very weak in comparison with the controls apparently conflicts with the results of in vitro interaction studies, which have shown that the dimers are exceptionally stable. The explanation for this discrepancy is presumably that at least one of the two fusion proteins contains the TPase103–807 protein, which can aggregate into macromolecular complexes (Heinlein et al., 1994). These complexes probably are unable to trigger transcriptional activation because they supposedly block the promoter or they get arrested in the cytoplasm (or both). The residual fraction of TPases still able to enter the nucleus would be depleted further because 50% of the TPase fusions would assemble into activation-deficient homodimers; of these, the BD-TPase/BD-TPase homodimers would block the GAL4 promoter.
The Dimerization Domain Contains a Signature Sequence for the hAT Element Transposases
The dimerization domain is highly conserved and specific for the transposases of members of the Ac, or hAT, superfamily of transposable elements. After two rounds of iteration runs in PSI-BLAST GenBank searches (Altschul et al., 1997; Zhang et al., 1998) with the perfectly conserved WWxxxxxxxPxLxxxAxxxL motif or TPase residues 666 to 737 (Figure 3), the targets found were almost exclusively transposases and putative transposases. Accordingly, this motif can be considered a signature sequence of the hAT element transposases.
A sequence evaluation of the aligned sequences shown in Figure 3A with the JPred prediction machine (Cuff et al., 1998; Cuff and Barton, 1999) indicated that several segments of the Ac dimerization domain have a high probability of exhibiting α-helical structures (see Figure 3). In several highly conserved positions, hydrophobic residues are found, but no apparent leucine zipper or heptad repeat of hydrophobic residues is present that would suggest a coiled-coil interaction structure. Also arguing against this type of interaction is the finding that mutating the conserved leucine 701 residue did not negatively affect dimerization in vitro.
Ac TPase Contains a Second Interaction Function
Using immunofluorescence staining, we found that a TPase derivative with a defective dimerization domain, R733A [4], still forms macromolecular aggregates in vivo and acts as a dominant inhibitor of transposition. Thus, the TPase must harbor at least one other interaction function in addition to the dimerization domain. Another negatively dominant mutant, TPase189–807, forms cytoplasmic complexes in petunia and tobacco cells that are microscopically indistinguishable from TPase103–807 aggregates (Boehm et al., 1995; R. Kunze and I. Kornacker, unpublished data). Accordingly, this second interaction function is located between TPase residues 189 and 674 and is the major determinant for formation of macromolecular aggregates.
The Role of Transposase Interactions in Transposition Reactions
The transposition complexes (transpososomes) of several prokaryotic transposons have been studied with biochemical analyses. These synaptic complexes involve both transposon ends, the transposase, and facultatively accessory element- or host-encoded proteins. Probably the most thoroughly studied case is bacteriophage Mu. In the Mu transposition intermediates, the two Mu ends are connected (synapsed) by a transposase (MuA) tetramer (Lavoie et al., 1991; Mizuuchi et al., 1995). The transposase of Tn7 consists of subunits of at least two different proteins, TnsA and TnsB, and the active transposase is a heterooligomeric protein composed of at least two protomers of each subunit. In addition, the Tn7 transpososome includes the Tn7-encoded TnsC protein, a key player in target site selection that supposedly interacts directly with TnsAB and either the TnsD or the TnsE protein (Sarnovsky et al., 1996; Stellwagen and Craig, 1997, 1998). For Tn10, transposase-mediated end pairing has been shown, and some have suggested that all chemical reactions of Tn10 transposition might be performed by only two transposase monomers, one per end (Bolland and Kleckner, 1996; Kennedy et al., 1998).
All of these transpososomes have a common feature: their assembly is facilitated through multiple protein–DNA and protein–protein interactions. The Tn5 transposase (Tnp), for example, contains two dimerization domains, one in the N-terminal half of the protein and the other at the C terminus. These two domains have been implicated in synapsis of the transposon ends, dimerization, and nonproductive multimerization (Weinreich et al., 1994; Mahnke Braam and Reznikoff, 1998; Mahnke Braam et al., 1999). Although their sequences are not similar, the Ac TPase and the Tn5-encoded Tnp share remarkably similar autoregulatory properties that are mediated by protein interactions. The multimerization domains of the IS911-encoded transposase (OrfAB) also have been studied in detail (Haren et al., 1998). This protein contains three multimerization domains involved in assembly of multimeric complexes.
In contrast, little is known about the architecture of eukaryotic transposable element transpososomes. Protein–protein contacts generally are assumed in these cases also to promote synapsis of the transposon ends. The crystal structure of the DNA binding domain of the Tc3 transposase in complex with the transposon DNA revealed that this domain forms a dimer with each monomer bound to a separate transposon end, implying that the dimer plays a role in synapsis (van Pouderoyen et al., 1997). The Drosophila mariner (Lohe et al., 1996) and P element transposases (Beall and Rio, 1998) supposedly are active in an oligomeric form. In the maize Enhancer/Suppressor-mutator transposon, the element-encoded TNPA protein binds to the transposon ends, dimerizes, and associates with transposase (TNPD), thus possibly constituting the core of a macromolecular transposition complex (Trentmann et al., 1993; Raina et al., 1998).
The Ac TPase is active as an oligomer too (Kunze et al., 1993; Behrens-Jung et al., 1994; this report), but with increasing expression levels, it associates into large, filament-like aggregates that are transpositionally inactive (Heinlein et al., 1994). Whether the formation of the filament-like TPase aggregates involves interaction interfaces that do not participate in the assembly of the active transpososome is largely a matter of speculation, as is the involvement of additional (host) proteins in this process. However, because aggregate formation also is observed in insect cells (Hauser et al., 1988), the requirement for specific plant proteins can be dismissed. Given that the substitution of a single amino acid in the dimerization domain (R733A) disrupts the formation of the characteristic filament-shaped aggregates and inactivates the protein, we assume that aggregate assembly is an ordered process, not a chaotic misfolding reaction, and involves the same protein interfaces as the assembly of the active oligomers.
Activation Function of the TPase C Terminus
Unexpectedly, deleting N-terminal TPase sequences resulted in unmasking a transcription activation activity of the 24 TPase residues at the C terminus. Unmasking of hidden functions by deletion analysis in the two-hybrid system has been reported in several studies (e.g., Sato et al., 1994; Wu and Chaconas, 1995). The TPase C terminus shares an acidic character (the terminal 24 amino acids carry a net charge of −7) with many transcription factors. Because TPase binds to multiple sites immediately upstream of the Ac transcription start sites (Kunze et al., 1987; Kunze and Starlinger, 1989), it is tempting to speculate about whether TPase acts in vivo as a transcriptional modulator of its own gene. TPase binding to the Ac promoter and a weak activation function could shield TPase expression from positional influences at various insertion sites and promote constitutive and low expression.
METHODS
Plasmids
For use in the yeast two-hybrid system, Activator (Ac) transposase (TPase) cDNA fragments were fused in-frame to the C termini of the GAL4 activation and DNA binding domain in plasmids pGAD424 and pGBT9, respectively (Bartel et al., 1993a). To express the TPase dimerization domains in Escherichia coli, a polymerase chain reaction–amplified Ac cDNA fragment 3176 to 3968 was cloned as a NdeI-BamHI fragment into pET15b (Novagen, Madison, WI; Studier et al., 1990). The resulting plasmid pORFa(H674-777) expresses the TPase residues 674 to 777 with a His6 tag attached at the N terminus.
Site-directed mutagenesis of the TPase dimerization domain was performed with plasmids pORFa(H674-777) and pcATG10 (Kunze et al., 1993) by using the Quick Change mutagenesis kit (Stratagene, La Jolla, CA). Some of the mutants were generated by a second round of mutagenesis on pORFa(H674-777/V708A/L709A) and pcATG10/V708A/L709A. In each construct, the entire sequence of the mutated gene encoding the TPase was verified by DNA sequencing. Details of plasmid constructions and primer sequences can be obtained from us on request.
Two-Hybrid System and LacZ Assay in Saccharomyces cerevisiae
The pGAD424 and pGBT9 derivatives that encoded proteins consisting of GAL4-BD and GAL4-AD domains fused with Ac TPase fragments were transformed in yeast strain Y526 (Bartel et al., 1993b). LacZ activity in the transformants was determined as described previously (Essers and Kunze, 1996). Positive controls were the yeast GAL4 protein, the Ustilago maydis bE21–136 and bW11–186 proteins fused to the GAL4-BD and GAL4-AD (encoded by plasmids pGB-E2136 and pW1186-GA; Kämper et al., 1995), and a simian virus SV40 T-antigen fragment (residues 84 to 708) and a p53 fragment (residues 72 to 390) fused to the GAL4-BD and GAL4-AD (encoded by plasmids pVA3 and pTD1; Chien et al., 1991).
Expression and Purification of Protein in E. coli
TPase fragments for biochemical interaction experiments were expressed in E. coli BL21(DE3), essentially as described by Feldmar and Kunze (1991) and Kunze et al. (1995a). Recombinant protein was extracted and purified under denaturing conditions by metal affinity chromatography on TALON (Clontech, Palo Alto, CA) or NTA (Qiagen, Hilden, Germany) resin, according to the manufacturers' recommendations, except that the TALON elution buffers 1 and 2 contained 20 mM Tris-HCl, pH 6.3, and 20 mM Tris-HCl, pH 5.2, respectively. Refolding reactions were performed as described (Feldmar and Kunze, 1991).
TPase Interaction Assays and Chemical Cross-Linking
For interaction analysis, renatured proteins were subjected to different treatments. Protein eluted from the metal-affinity column in buffers 1 and 2 was immediately loaded onto a 17.5% acrylamide and 0.1% SDS gel and size fractionated. The protein eluate was incubated for 10 min at 95°C before electrophoresis or was mixed with 1 volume of SDS sample buffer (2% SDS and 0.5% DTT, 70 mM Tris-HCl, pH 6.8, 10% glycerol, and 0.005% bromophenol blue) and incubated for 10 min at 95°C before electrophoresis.
Chemical cross-linking of renatured proteins was performed by incubating 2 μg of protein for 5 to 10 min at room temperature with 0 to 6 mM ethylene glycol-bis(succinimidylsuccinate) (EGS). The reaction was stopped by adjusting the sample to contain 50 mM l-lysine and 1% SDS and incubating for 20 min at room temperature. Before SDS gel analysis, all proteins were precipitated by adding 5 volumes of ice-cold acetone and incubating for 20 min at −70°C and then were collected by centrifugation (40 min at 15,000g at 4°C). The sediments were dried and dissolved in SDS sample buffer by incubating for 10 min at 95°C; this was followed by electrophoresis on a 17.5% acrylamide and 0.1% SDS gel. Proteins were detected by protein gel blotting as described previously (Kunze et al., 1993).
Dissociation Excision Assay in Petunia Cells and Protein Gel Blotting
Transpositional activity of wild-type and mutant TPase proteins in petunia cells was assessed as described by Houba-Hérin et al. (1990) and Heinlein et al. (1994). Briefly, petunia protoplasts were cotransfected with a TPase expression plasmid and the Dissociation (Ds) excision reporter plasmid pTY/DIR (F. Ros and R. Kunze, unpublished data). Genes encoding mutant TPase103–807 were generated by site-directed mutagenesis on pcATG10 (Kunze et al., 1993). After 3 days of incubation at 26°C, two aliquots of the transfected protoplasts were plated on nitrocellulose filters for β-glucuronidase (GUS) staining; another aliquot was fixed on glass slides for immunofluorescence staining. The remaining cells were sedimented by centrifugation and resuspended in SDS sample buffer for protein gel blot analysis.
In Situ Immunofluorescence Staining of TPase
Immunofluorescence detection of TPase protein in Petunia hybrida cells was performed by a modification of the procedure described by Heinlein et al. (1994). All steps were performed at room temperature. The secondary antibody (anti-rabbit IgG fluorescein isothiocyanate conjugate; Sigma) was diluted 1:80 in 10% fetal calf serum in PBS-T (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4, pH 7.3, 0.5% Tween 20, and 0.2% Triton X-100). After two washes with PBS-T, the nuclei were stained with 0.1 μg/mL 4′,6-diamidino-2-phenylindole in PBS-T and then washed again with PBS-T. Embedding and microscopic examination of the specimen were performed as described (Heinlein et al., 1994), except that photographs were taken with a digital camera.
Acknowledgments
We thank Francesca Ros (University of Munich) for providing plasmid pTY/DIR and for help with and advice regarding the Ds excision assay, and Iris Heidmann (Max-Planck-Institut für Züchtungsforschung, Cologne) for the generous supply of petunia seeds. This work was supported by Deutsche Forschungsgemeinschaft through SFB 190, TP B8.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, P., Chien, C.-T., Sternglanz, R., and Fields, S. (1993a). Using the two-hybrid system to detect protein–protein interactions. In Cellular Interactions in Development: A Practical Approach, D.A. Hartley, ed (Oxford: Oxford University Press), pp. 153–179.
- Bartel, P., Chien, C.-T., Sternglanz, R., and Fields, S. (1993. b). Elimination of false positives that arise in using the two-hybrid system. BioTechniques 14 920–924. [PubMed] [Google Scholar]
- Beall, E.L., and Rio, D.C. (1998). Transposase makes critical contacts with, and is stimulated by, single-stranded DNA at the P element termini in vitro. EMBO J. 17 2122–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, H.-A., and Kunze, R. (1997). Maize Activator transposase has a bipartite DNA binding domain that recognizes subterminal motifs and the terminal inverted repeats. Mol. Gen. Genet. 254 219–230. [DOI] [PubMed] [Google Scholar]
- Behrens-Jung, U., Kühn, S., and Kunze, R. (1994). The Ac transposase consists of several, functionally distinct domains. Maize Genet. Coop. Newsl. 68 20–21. [Google Scholar]
- Boehm, U., Heinlein, M., Behrens, U., and Kunze, R. (1995). One of three nuclear localization signals of maize Activator (Ac) transposase overlaps the DNA-binding domain. Plant J. 7 441–451. [DOI] [PubMed] [Google Scholar]
- Bolland, S., and Kleckner, N. (1996). The three chemical steps of Tn10/IS10 transposition involve repeated utilization of a single active site. Cell 84 223–233. [DOI] [PubMed] [Google Scholar]
- Breedon, L., and Nasmyth, K. (1985). Regulation of the yeast HO gene. Cold Spring Harbor Symp. Quant. Biol. 50 643–650. [DOI] [PubMed] [Google Scholar]
- Chien, C.-T., Bartel, P.L., Sternglanz, R., and Fields, S. (1991). The two-hybrid system: A method to identify and clone genes for proteins that interact with a protein of interest. Proc. Natl. Acad. Sci. USA 88 9578–9582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coupland, G., Baker, B., Schell, J., and Starlinger, P. (1988). Characterization of the maize transposable element Ac by internal deletions. EMBO J. 7 3653–3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuff, J.A., and Barton, G.J. (1999). Evaluation and improvement of multiple sequence methods for protein secondary structure prediction. Proteins 34 508–519. [DOI] [PubMed] [Google Scholar]
- Cuff, J.A., Clamp, M.E., Siddiqui, A.S., Finlay, M., and Barton, G.J. (1998). JPred: A consensus secondary structure prediction server. Bioinformatics 14 892–893. [DOI] [PubMed] [Google Scholar]
- Essers, L., and Kunze, R. (1996). A sensitive, quick and semi-quantitative LacZ assay for the two-hybrid system. Trends Genet. 12 449–450. [DOI] [PubMed] [Google Scholar]
- Feldmar, S., and Kunze, R. (1991). The ORFa protein, the putative transposase of maize transposable element Ac, has a basic DNA binding domain. EMBO J. 10 4003–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haren, L., Polard, P., Ton Hoang, B., and Chandler, M. (1998). Multiple oligomerization domains in the IS911 transposase: A leucine zipper motif is essential for activity. J. Mol. Biol. 283 29–41. [DOI] [PubMed] [Google Scholar]
- Hauser, C., Fusswinkel, H., Li, J., Oellig, C., Kunze, R., Müller-Neumann, M., Heinlein, M., Starlinger, P., and Doerfler, W. (1988). Overproduction of the protein encoded by the maize transposable element Ac in insect cells by a baculovirus vector. Mol. Gen. Genet. 214 373–378. [DOI] [PubMed] [Google Scholar]
- Heinlein, M., Brattig, T., and Kunze, R. (1994). In vivo aggregation of maize Activator (Ac) transposase in nuclei of maize endosperm and petunia protoplasts. Plant J. 5 705–714. [DOI] [PubMed] [Google Scholar]
- Houba-Hérin, N., Becker, D., Post, A., Larondelle, Y., and Starlinger, P. (1990). Excision of a Ds-like maize transposable element (AcΔ) in a transient assay in petunia is enhanced by a truncated coding region of the transposable element Ac. Mol. Gen. Genet. 224 17–23. [DOI] [PubMed] [Google Scholar]
- Kämper, J., Reichmann, M., Romeis, T., Bölker, M., and Kahmann, R. (1995). Multiallelic recognition: Nonself-dependent dimerization of the bE and bW homeodomain proteins in Ustilago maydis. Cell 81 73–83. [DOI] [PubMed] [Google Scholar]
- Kaufman, P.D., and Rio, D.C. (1992). P element transposition in vitro proceeds by a cut-and-paste mechanism and uses GTP as a cofactor. Cell 69 27–39. [DOI] [PubMed] [Google Scholar]
- Kennedy, A.K., Guhathakurta, A., Kleckner, N., and Haniford, D.B. (1998). Tn10 transposition via a DNA hairpin intermediate. Cell 95 125–134. [DOI] [PubMed] [Google Scholar]
- Kunze, R. (1996). The Activator (Ac) element of Zea mays L. In Transposable Elements, H. Saedler and A. Gierl, eds (Heidelberg, Germany: Springer-Verlag), pp. 161–194.
- Kunze, R., and Starlinger, P. (1989). The putative transposase of transposable element Ac from Zea mays L. interacts with subterminal sequences of Ac. EMBO J. 8 3177–3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze, R., Stochaj, U., Laufs, J., and Starlinger, P. (1987). Transcription of transposable element Activator (Ac) of Zea mays L. EMBO J. 6 1555–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze, R., Behrens, U., Courage-Franzkowiak, U., Feldmar, S., Kühn, S., and Lütticke, R. (1993). Dominant transposition-deficient mutants of maize Activator (Ac) transposase. Proc. Natl. Acad. Sci. USA 90 7094–7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze, R., Fusswinkel, H., and Feldmar, S. (1995a). Expression of plant proteins in baculoviral and bacterial systems. In Methods in Cell Biology—Plant Cell Biology (Part B), D.W. Galbraith, D.P. Bourque, and H.J. Bohnert, eds (Orlando, FL: Academic Press), pp. 461–479. [DOI] [PubMed]
- Kunze, R., Kühn, S., Jones, J.D.G., and Scofield, S.R. (1995. b). Somatic and germinal activities of maize Activator (Ac) transposase mutants in transgenic tobacco. Plant J. 8 45–54. [Google Scholar]
- Lavoie, B.D., Chan, B.S., Allison, R.G., and Chaconas, G. (1991). Structural aspects of a higher order nucleoprotein complex: Induction of an altered DNA structure at the Mu-host junction of the Mu type 1 transpososome. EMBO J. 10 3051–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M.-G., and Starlinger, P. (1990). Mutational analysis of the N terminus of the protein of maize transposable element Ac. Proc. Natl. Acad. Sci. USA 87 6044–6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohe, A.R., Sullivan, D.T., and Hartl, D.L. (1996). Subunit interactions in the mariner transposase. Genetics 144 1087–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahnke Braam, L.A., and Reznikoff, W.S. (1998). Functional characterization of the Tn5 transposase by limited proteolysis. J. Biol. Chem. 273 10908–10913. [DOI] [PubMed] [Google Scholar]
- Mahnke Braam, L.A., Goryshin, I.Y., and Reznikoff, W.S. (1999). A mechanism for Tn5 inhibition: Carboxyl-terminal dimerization. J. Biol. Chem. 274 86–92. [DOI] [PubMed] [Google Scholar]
- McClintock, B. (1951). Chromosome organization and genic expression. Cold Spring Harbor Symp. Quant. Biol. 16 13–47. [DOI] [PubMed] [Google Scholar]
- Miller, J.H. (1972). Experiments in Molecular Genetics. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Mizuuchi, M., Baker, T.A., and Mizuuchi, K. (1995). Assembly of phage Mu transpososomes: Cooperative transitions assisted by protein and DNA scaffolds. Cell 83 375–385. [DOI] [PubMed] [Google Scholar]
- Raina, R., Schlappi, M., Karunanandaa, B., Elhofy, A., and Fedoroff, N. (1998). Concerted formation of macromolecular Suppressor–mutator transposition complexes. Proc. Natl. Acad. Sci. USA 95 8526–8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnovsky, R.J., May, E.W., and Craig, N.L. (1996). The Tn7 transposase is a heteromeric complex in which DNA breakage and joining activities are distributed between different gene products. EMBO J. 15 6348–6361. [PMC free article] [PubMed] [Google Scholar]
- Sato, T., Hanada, M., Bodrug, S., Irie, S.J., Iwama, N., Boise, L.H., Thompson, C.B., Golemis, E., Fong, L., Wang, H.G., and Reed, J.C. (1994). Interactions among members of the Bcl-2 protein family analyzed with a yeast two-hybrid system. Proc. Natl. Acad. Sci. USA 91 9238–9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield, S.R., English, J.J., and Jones, J.D.G. (1993). High level expression of the Activator (Ac) transposase gene inhibits the ex-cision of Dissociation (Ds) in tobacco cotyledons. Cell 75 507–517. [DOI] [PubMed] [Google Scholar]
- Stellwagen, A.E., and Craig, N.L. (1997). Avoiding self—Two Tn7-encoded proteins mediate target immunity in Tn7 transposition. EMBO J. 16 6823–6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen, A.E., and Craig, N.L. (1998). Mobile DNA elements: Controlling transposition with ATP-dependent molecular switches. Trends Biochem. Sci. 23 486–490. [DOI] [PubMed] [Google Scholar]
- Studier, F.W., Rosenberg, A.H., Dunn, J.J., and Dubendorff, J.W. (1990). Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185 60–89. [DOI] [PubMed] [Google Scholar]
- Trentmann, S.M., Saedler, H., and Gierl, A. (1993). The transposable element En/Spm-encoded TNPA protein contains a DNA binding and a dimerization domain. Mol. Gen. Genet. 238 201–208. [DOI] [PubMed] [Google Scholar]
- van Pouderoyen, G., Ketting, R.F., Perrakis, A., Plasterk, R.H., and Sixma, T.K. (1997). Crystal structure of the specific DNA-binding domain of Tc3 transposase of C. elegans in complex with transposon DNA. EMBO J. 16 6044–6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos, J.C., De Baere, I., and Plasterk, R.H.A. (1996). Transposase is the only nematode protein required for in vitro transposition of Tc1. Genes Dev. 10 755–761. [DOI] [PubMed] [Google Scholar]
- Weinreich, M.D., Gasch, A., and Reznikoff, W.S. (1994). Evidence that the cis preference of the Tn5 transposase is caused by nonproductive multimerization. Genes Dev. 8 2363–2374. [DOI] [PubMed] [Google Scholar]
- Wu, Z.G., and Chaconas, G. (1995). A novel DNA binding and nu-clease activity in domain III of Mu transposase: Evidence for a catalytic region involved in donor cleavage. EMBO J. 14 3835–3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Schaffer, A.A., Miller, W., Madden, T.L., Lipman, D.J., Koonin, E.V., and Altschul, S.F. (1998). Protein sequence similarity searches using patterns as seeds. Nucleic Acids Res. 26 3986–3990. [DOI] [PMC free article] [PubMed] [Google Scholar]