Abstract
The production of cell wall–degrading enzymes (wall depolymerases) by plant pathogenic fungi is under catabolite (glucose) repression. In Saccharomyces cerevisiae, the SNF1 gene is required for expression of catabolite-repressed genes when glucose is limiting. An ortholog of SNF1, ccSNF1, was isolated from the maize pathogen Cochliobolus carbonum, and ccsnf1 mutants of HC toxin–producing (Tox2+) and HC toxin–nonproducing (Tox2−) strains were created by targeted gene replacement. Growth in vitro of the ccsnf1 mutants was reduced by 50 to 95% on complex carbon sources such as xylan, pectin, or purified maize cell walls. Growth on simple sugars was affected, depending on the sugar. Whereas growth on glucose, fructose, or sucrose was normal, growth on galactose, galacturonic acid, maltose, or xylose was somewhat reduced, and growth on arabinose was strongly reduced. Production of HC toxin was normal in the Tox2+ ccsnf1 mutant, as were conidiation, conidial morphology, conidial germination, and in vitro appressorium formation. Activities of secreted β-1,3-glucanase, pectinase, and xylanase in culture filtrates of the Tox2+ ccsnf1 mutant were reduced by 53, 24, and 65%, respectively. mRNA expression was downregulated under conditions that induced the following genes encoding secreted wall-degrading enzymes: XYL1, XYL2, XYL3, XYL4, XYP1, ARF1, MLG1, EXG1, PGN1, and PGX1. The Tox2+ ccsnf1 mutant was much less virulent on susceptible maize, forming fewer spreading lesions; however, the morphology of the lesions was unchanged. The Tox2− ccsnf1 mutant also formed fewer nonspreading lesions, which also retained their normal morphology. The results indicate that ccSNF1 is required for biochemical processes important in pathogenesis by C. carbonum and suggest that penetration is the single most important step at which ccSNF1 is required. The specific biochemical processes controlled by ccSNF1 probably include, but are not necessarily restricted to, the ability to degrade polymers of the plant cell wall and to take up and metabolize the sugars produced.
INTRODUCTION
A major barrier to the penetration and spread of potential pathogenic organisms is the plant cell wall, and all of the major groups of cellular plant pathogens are known to make extracellular enzymes that can degrade cell wall polymers. Although the involvement of wall-degrading enzymes and their genes in penetration, pathogen ramification, plant defense induction, and symptom expression has been studied extensively, conclusive evidence for or against a role for any particular enzyme activity in any aspect of pathogenesis has been difficult to obtain (Walton, 1994).
The major obstacle to addressing the function of wall-degrading enzymes has been redundancy: all of the pathogens that have been studied in detail have multiple genes for any particular enzyme activity. Thus, most fungal strains mutated in wall-degrading enzyme genes—by either conventional (e.g., Cooper, 1987) or molecular (e.g., Scott-Craig et al., 1990) methods—retain at least some residual enzyme activity. For example, the pea pathogen Nectria haematococca (Fusarium solani f sp pisi) has four functional pectate lyase genes, the maize pathogen Cochliobolus carbonum and the rice pathogen Magnaporthe grisea each have at least four xylanase genes, and the cosmopolitan pathogen Botrytis cinerea has as many as five endopolygalacturonase genes (Apel-Birkhold and Walton, 1996; Guo et al., 1996; Wu et al., 1997; ten Have et al., 1998). Even strains of fungi that carry multiple mutations retain residual enzyme activity and are still pathogenic (Apel-Birkhold and Walton, 1996; Scott-Craig et al., 1998; J.S. Scott-Craig and J.D. Walton, unpublished results). Despite this redundancy, single genes of a particular class have been shown, in two cases, to contribute to the virulence of pathogenic fungi—particular constitutive pectinases are virulence factors for Aspergillus flavus on cotton bolls and for B. cinerea on tomato (Shieh et al., 1997; ten Have et al., 1998).
The gamut of extracellular wall-degrading enzymes produced by the ascomycete C. carbonum includes pectinases, xylanases, cellulases, mixed-linked (β-1,3–β-1,4) glucanases, β-1,3-glucanases, proteases, xylosidases, arabinosidase, and undoubtedly others. None of the strains generated to date with single mutations in any of the genes encoding these enzymes has reduced virulence. Furthermore, with only a few exceptions, the mutants still grow as well as wild type on the appropriate substrate in vitro (Scott-Craig et al., 1990; Apel et al., 1993; Schaeffer et al., 1994; Sposato et al., 1995; Apel-Birkhold and Walton, 1996; Murphy and Walton, 1996; Görlach et al., 1998; Nikolskaya et al., 1998; Scott-Craig et al., 1998; Wegener et al., 1999).
An alternative approach to the isolation and disruption of individual genes encoding wall-degrading enzymes would be to identify the genetic regulatory elements for which mutation results in the simultaneous loss or downregulation of multiple enzymes. If a mutant that had been globally impaired in its ability to make wall-degrading enzymes were still pathogenic, this would bring into serious doubt a significant role for such enzymes in pathogenesis (Walton, 1994).
In culture, the expression of most wall-degrading enzymes by most fungi, including plant pathogens, is inhibited by glucose or other simple sugars in a well-studied metabolic process known as catabolite or glucose repression (Ruijter and Visser, 1997). Most (perhaps all) of the examined extracellular enzyme activities of C. carbonum are subject to catabolite repression (Walton and Cervone, 1990; Van Hoof et al., 1991; Holden and Walton, 1992; Ransom and Walton, 1997). In yeast, release from catabolite repression requires a protein kinase called Snf1p. Snf1p is necessary for the expression of glucose-repressed genes such as that encoding invertase (SUC2) when glucose is limiting. That is, glucose-repressed genes remain repressed in a snf1 mutant even in the absence of glucose (Celenza and Carlson, 1984; Hardie et al., 1998; Östling and Ronne, 1998; Treitel et al., 1998). A major function of Snf1p is to phosphorylate Mig1p, a DNA binding transcriptional repressor. The ortholog of MIG1 in filamentous fungi is called creA (Ronne, 1995). Phosphorylation of Mig1p inhibits its binding to the promoters of the genes it represses and also promotes its movement out of the nucleus into the cytoplasm (DeVit et al., 1997). Snf1p controls the response to glucose through additional mechanisms, because it also activates by phosphorylation the transcriptional activators Sip4p and Cat8p (Lesage et al., 1996; Vincent and Carlson, 1998).
Orthologs of SNF1 are present in many other organisms, including mammals and plants. Its counterpart in mammals is AMP-dependent protein kinase (Hardie et al., 1998). A SNF1 ortholog is involved in the response of Arabidopsis to glucose (Bhalerao et al., 1999). To our knowledge, the biology of SNF1 genes has not been studied previously in filamentous fungi.
Because SNF1 is required for derepression of catabolite-repressed genes in yeast, mutation of the orthologous gene in C. carbonum might cause irreversible downregulation of catabolite-repressed wall-degrading enzymes. Accordingly, snf1 mutants might be useful for testing whether wall-degrading enzymes are virulence factors in pathogenic fungi.
RESULTS
Isolation of C. carbonum SNF1 (ccSNF1)
Two degenerate oligonucleotide primers were synthesized on the basis of conserved regions of Snf1p-related sequences of Saccharomyces cerevisiae, Candida glabrata, and tobacco NPK5 (Celenza and Carlson, 1984; Muranaka et al., 1994; Petter and Kwon-Chung, 1996). Amplification by polymerase chain reaction (PCR) with C. carbonum genomic DNA as template yielded a product of the predicted size (400 bp). The PCR product was used to isolate the SNF1 gene and cDNA copies from C. carbonum genomic and cDNA libraries, respectively. The C. carbonum ccsnf1 cDNA clone was cloned into pGEM7 to create plasmid pSnf1c-3. The C. carbonum gene (designated ccSNF1) has introns of 60 and 51 bp. The 3′ untranslated region is 159 bp long, and the 5′ untranslated region is 120 bp (data not shown).
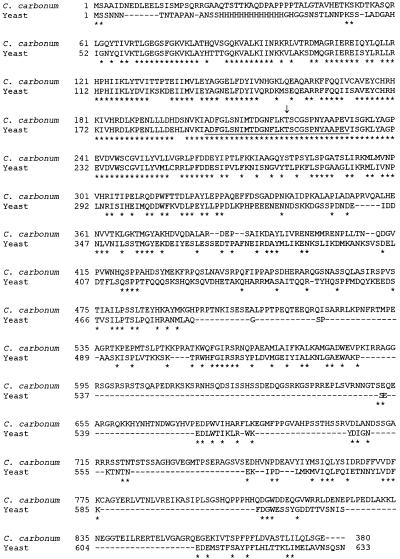
The open reading frame of the product of ccSNF1, ccSnf1p, is 880 amino acids and has a molecular mass of 98 kD. ccSnf1p has ∼40% overall identity with yeast Snf1p. As shown in Figure 1, the similarity is very strong at the N terminus, a region of the protein that includes the “activation segment,” which is conserved in all known related protein kinases. This block of amino acids is 100% conserved between Snf1p and ccSnf1p (Johnson et al., 1996; Hardie et al., 1998). At the C terminus, however, the similarity between Snf1p and ccSnf1p is weak to nonexistent, with the possible exception of a few blocks of amino acids that can only be aligned with the introduction of many large gaps (Figure 1).
Figure 1.
Comparison of Deduced Amino Acid Sequences of C. carbonum ccSNF1 and Yeast SNF1.
Residues that are identical in both sequences are indicated with asterisks. The protein kinase activation segment situated between Asp-Phe-Gly and Ala-Pro-Glu is underlined (Johnson et al., 1996; Hardie et al., 1998). The arrow indicates the phosphorylated threonine in Snf1p. The sequence of ccSNF1 has GenBank accession number AF159253. Dashes indicate introduced gaps in the amino acid sequences.
ccSnf1p is the largest (98 kD) of the known Snf1p-related proteins. Those of various yeasts are ∼70 kD, those of plants are ∼56 kD, and those of Caenorhabditis elegans, Drosophila, and mammals are ∼63 kD. ccSnf1p also differs from other proteins related to Snf1p in that the PSORT program (Nakai and Kanehisa, 1992) strongly predicts localization in the nucleus. The computed probability of ccSnf1p having a nuclear localization is 78%, whereas that of yeast Snf1p is only 48%. The difference is attributable to the presence of three regions of basic amino acids in ccSnf1p that are absent in yeast Snf1p: PTKKPRA starting at amino acid 547, PKIRRAG at amino acid 587, and PEDRKSK at amino acid 607 (Figure 1). Of 14 additional Snf1p-like proteins analyzed by PSORT, most were predicted to be in the cytoplasm, with nuclear localization probabilities ranging from 9% (Drosophila and rice) to 56% (C. glabrata).
Hybridization of a ccSNF1 probe to a blot of C. carbonum genomic DNA digested with various restriction enzymes (ClaI, EcoRV, NcoI, or SacI) resulted in a single band in each case (data not shown), indicating the existence of a single copy of the gene in C. carbonum.
Complementation of a Yeast snf1 Mutant by ccSNF1
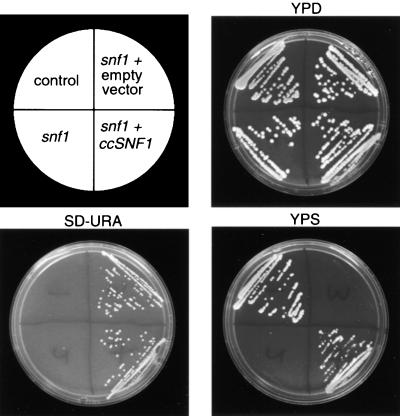
Yeast snf1 mutants cannot grow on a medium containing sucrose as the sole source of carbon, because the Snf1 protein kinase is essential for derepression of invertase (Celenza and Carlson, 1984). To determine whether the ccSNF1 gene can complement the snf1 mutation, we subcloned the 2.94-kb ccsnf1 cDNA of pSnf1c-3 into the yeast vector pVT100-U under the control of the constitutive alcohol dehydrogenase ADH1 promoter. Both transformed and untransformed cells, including the wild-type reference strain MG106, grew on glucose (YPD medium; see Methods), whereas only yeast transformed with either pSnf1c-4 or pVT100-U grew on synthetic dextrose minus uracil (SD-URA) medium, which contains glucose but lacks uracil. Only the transformant expressing ccSNF1 also grew on YPS medium (see Methods), which contains sucrose as a carbon source (Figure 2). Thus, despite their differences in size and in C terminus sequence, ccSnf1p can functionally replace yeast Snf1p.
Figure 2.
Complementation of Yeast snf1 by ccSNF1.
The control reference strain was MG106. snf1 denotes the yeast snf1 mutant MCY1846. The snf1 + empty vector is MCY1846 transformed with vector pVT100-U. snf1 + ccSNF1 is MCY1846 transformed with vector pSnf1c-4 (pVT100-U containing ccSNF1). SD-URA, glucose medium lacking uracil (see Methods); YPD, YP medium plus glucose (dextrose) as carbon source; YPS, YP medium plus sucrose as carbon source.
Targeted Disruption of ccSNF1
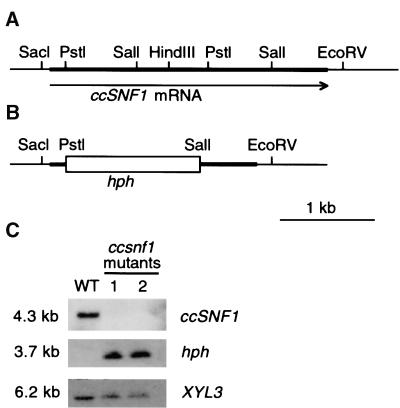
A ccsnf1 mutant in an HC toxin–producing (Tox2+) background was generated by DNA-mediated transformation. Two hygromycin-resistant transformants (T688 and T669) from two separate transformation experiments were isolated and purified, and both were determined by DNA gel blot analysis to have undergone gene replacement by homologous integration at the ccSNF1 locus (Figures 3A and 3B). Genomic DNA from both transformants and the wild type was digested with SacI, blotted, and probed with the 2.1-kb PstI-SalI internal ccSNF1 fragment. Neither transformant showed hybridization, whereas the wild type exhibited the expected hybridization to a band of 4.3 kb (Figure 3C). A similar blot was probed with the Escherichia coli hph gene encoding hygromycin phosphotransferase. As predicted, the hph gene hybridized, yielding a band of 3.7 kb with DNA from the two mutant strains, but it did not hybridize with the DNA from the wild-type strain. A control probe (a genomic fragment of XYL3) hybridized with the wild type and both transformants (Figure 3C).
Figure 3.
Construction and Analysis of ccsnf1 Disruption Transformants.
(A) Map of the ccSNF1 gene.
(B) An internal 2.1-kb PstI-SalI fragment of ccSNF1 was replaced with hph.
(C) DNA gel blotting of wild type (WT) and two ccsnf1 disruptant transformants (lane 1, T688; lane 2, T669). DNA was digested with SacI. Similar blots were probed with ccSNF1, hph, or XYL3 as a control.
Expression of Wall-Degrading Enzyme Activities and mRNAs in the ccsnf1 Mutant
In preliminary experiments, both ccsnf1 mutants (T688 and T689) displayed similar phenotypes with regard to growth and enzymatic activities; therefore, only ccsnf1 mutant T688 was used in further experiments. Strains 367-2A (wild type) and T688 (ccsnf1) were grown in liquid still culture with purified maize cell walls as the sole carbon source. After 7 days of growth, β-1,3-glucanase, α-1,4-polygalacturonase, and β-1,4-xylanase activities in the culture filtrates were measured. All three of these activities are due to multiple enzymes encoded by different genes (Schaeffer et al., 1994; Apel-Birkhold and Walton, 1996; Scott-Craig et al., 1998). Compared with those in the wild type, the respective enzyme activities in the mutant were reduced by 53% ± 4%, 24% ± 6%, and 65% ± 7% (means of four independent replicates ±sd).
The expression of particular structural genes encoding wall-degrading enzymes was analyzed further by RNA gel blotting. Expression of XYL3 (encoding endo-β-1,4-xylanase 3) and ARF1 (encoding α-arabinosidase; Y. Cheng, S. Wegener, and J.D. Walton, unpublished results) was undetectable in the ccsnf1 mutant under conditions that ordinarily would induce these genes (Figure 4). Expression of XYL1, XYL2, XYL4, XYP1, PGX1, PGN1, MLG1, and EXG1 (encoding endo-β-1,4-xylanases 1, 2, and 4; β-xylosidase; exo-α-1,4-polygalacturonase; endo-α-1,4-polygalacturonase; mixed-linked [β-1,3–β-1,4] glucanase; and exo-β-1,3-glu-canase, respectively) was moderately to strongly reduced in the ccsnf1 mutant (Figure 4). Expression of ccRPD3 (encoding a histone deacetylase related to yeast Rpd3p; S. Wegener and J.D. Walton, unpublished results) and GPD1 (encoding glyceraldehyde-3-phosphate dehydrogenase) was not affected by the ccsnf1 mutation (Figure 4).
Figure 4.
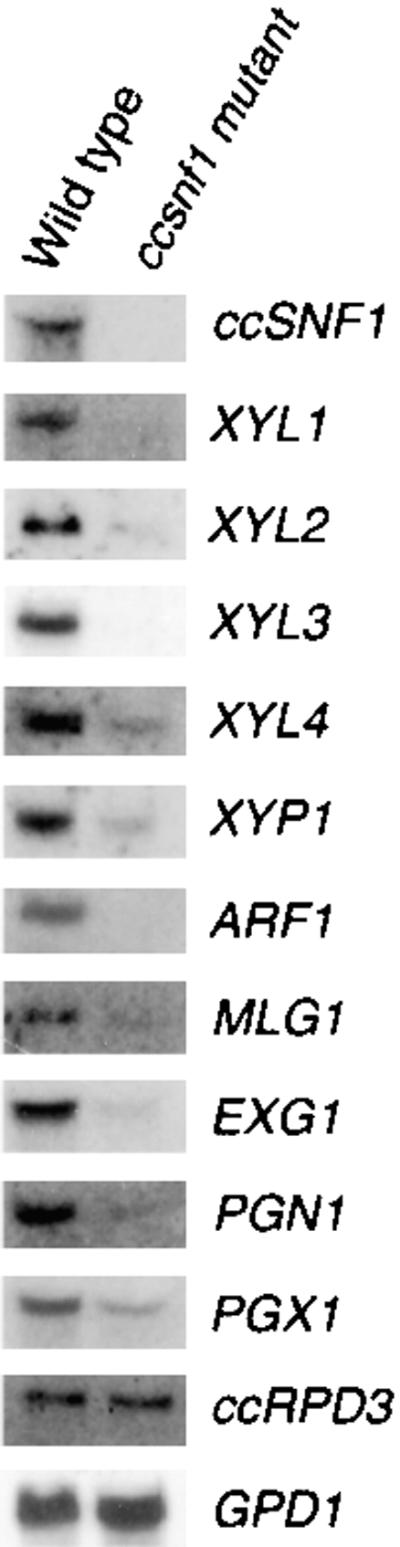
RNA Expression of Wall-Degrading Enzyme Genes in the Wild Type and in a ccsnf1 Mutant of C. carbonum.
Fungi (367-2A and T688) were grown for 7 days in liquid still culture containing, as the sole carbon source, 2% xylan (for expression of XYL1, XYL2, XYL3, XYL4, XYP1, and ARF1), 2% pectin (PGN1 and PGX1), or 2% maize cell walls (ccSNF1, MLG1, EXG1, ccRPD3, and GPD1). Total RNA was fractionated on an agarose gel and transferred to a nylon membrane, and the membrane was hybridized with probes of the indicated genes.
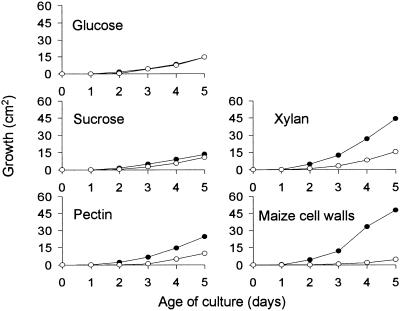
Growth of the ccsnf1 Mutant on Complex and Simple Carbon Sources
T688 and the wild-type strain 367-2A grew at the same rate on glucose and almost at the same rate on sucrose (Figure 5)—in contrast to yeast snf1 mutants, which grow on glucose but not sucrose (Celenza and Carlson, 1984; Figure 2). This apparent contradiction can be explained by considering that expression of secreted invertase is subject to glucose repression in yeast but not in C. carbonum. When tested, invertase activity in the wild type was the same whether grown on glucose or sucrose, and invertase activity in the ccsnf1 mutant was not markedly different from that of the wild type (data not shown). When grown on glucose, conidia of the ccsnf1 mutant were normal in numbers and morphology. Conidia of the ccsnf1 mutant germinated and formed appressoria on glass slides at the same rate as the wild type did (data not shown; Horwitz et al., 1999).
Figure 5.
Growth of C. carbonum Wild Type and a ccsnf1 Mutant on Glucose, Sucrose, or Complex Carbon Sources.
Fungi (367-2A and T688) were grown on agar in 15-cm-diameter Petri plates. Basal salts medium was supplemented with glucose, sucrose, xylan, pectin, or maize cell walls at 2% (w/v) as the sole carbon source. Filled circles, wild type; open circles, ccsnf1 mutant.
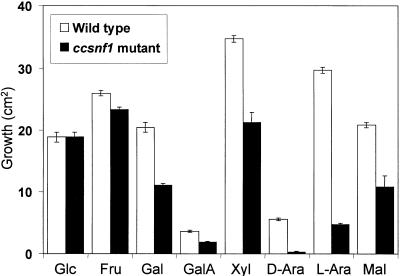
Compared with the wild type, the ccsnf1 mutant was moderately to severely impaired in its ability to grow on pectin, xylan, or maize cell walls (Figure 5). Its growth on maize cell walls, in which the carbon source was derived directly from the host of C. carbonum, was particularly impaired. Reduced growth on complex substrates could be due either to a decreased ability to degrade the substrates or to an inability to take up and metabolize the released sugars. To test the latter possibility, we compared growth of the ccsnf1 mutant with that of the wild type on various simple sugars as the sole carbon source. The wild type grew best on xylose and reasonably well on, in descending order, l-arabinose, fructose, maltose, galactose, and glucose; growth on galacturonic acid or d-arabinose was relatively poor (Figure 6). The ccsnf1 mutant was able to grow almost as well as the wild type on fructose but showed less growth on galactose, galacturonic acid, xylose, or maltose. Growth of the mutant was most strikingly reduced on l- or d-arabinose (Figure 6). Among the substrates on which the wild type grew well, the growth of the ccsnf1 mutant was most reduced on l-arabinose, one of the most abundant sugars in maize cell walls (Kato and Nevins, 1984). These results indicate that ccSNF1 is required not just for the expression of extracellular depolymerases but also for uptake and/or metabolism of the products of those enzymes.
Figure 6.
Growth of C. carbonum Wild Type and ccsnf1 Mutant on Simple Sugars.
Fungi (367-2A and T688) were grown on agar in 15-cm-diameter Petri plates. Basal salts medium was supplemented with glucose (Glc), fructose (Fru), galactose (Gal), galacturonic acid (GalA), xylose (Xyl), d-arabinose (D-Ara), l-arabinose (L-Ara), or maltose (Mal) at 2% (w/v) as the sole carbon source. Growth was measured after 6 days.
Effect of the ccsnf1 Mutation on Pathogenicity
The Tox2+/ccsnf1 mutant was tested for pathogenicity by spray inoculation of maize plants of genotype hm1/hm1 (sensitive to HC toxin). The Tox2+/ccsnf1 mutant caused fewer lesions (<15%) than did the wild type. Most of the lesions caused by the mutant were similar to those caused by the wild type in appearance and rate of expansion but did not develop as uniformly (Figure 7). Furthermore, whereas the wild type eventually killed the entire seedling, the Tox2+/ccsnf1 mutant never did so.
Figure 7.
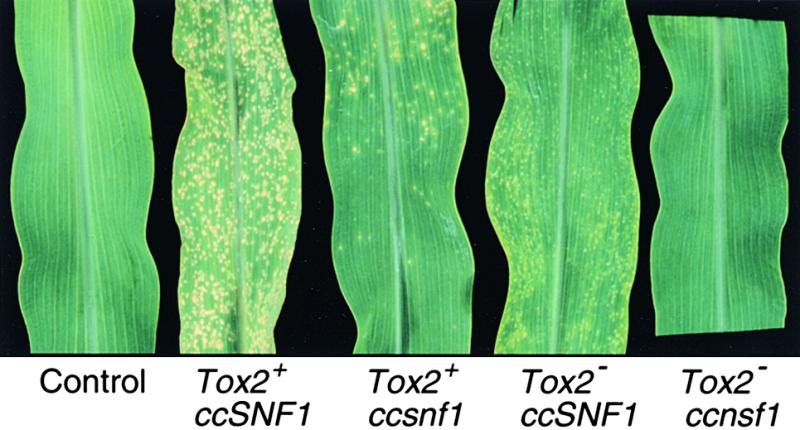
Pathogenicity Assays of ccsnf1 Mutants.
Plants were genotype hm1/hm1 (inbred Pr). Tox2+ indicates a strain that produces HC toxin; Tox2− indicates a strain that does not produce toxin.
To determine whether the reduced virulence of the ccsnf1 mutant was the result of reduced HC toxin production, we analyzed culture filtrates of the wild type and the ccsnf1 mutant. Toxin production was normal in the ccsnf1 mutant (Figure 8), indicating that synthesis of HC toxin does not require ccSNF1 and that the reduced virulence of the mutant is not due to a defect in toxin biosynthesis or secretion.
Figure 8.
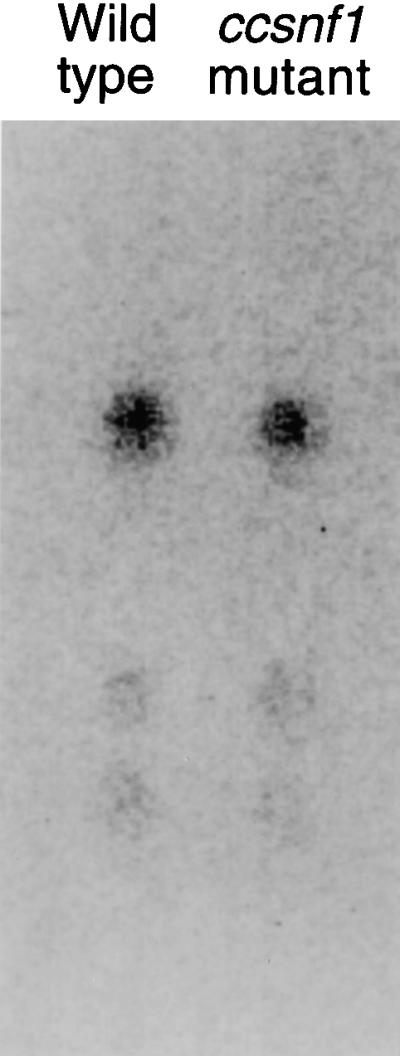
HC Toxin Production by Tox2+ Wild Type and Tox2+/ccsnf1 Mutant.
The results of thin-layer chromatography of HC toxin isolated from culture filtrates are shown. Fungi (3672A and mutant T688) were grown for 14 days in liquid culture containing glucose as a carbon source. The culture filtrates were extracted with chloroform, fractionated by silica gel thin-layer chromatography, and detected with an epoxide-specific reagent. The uppermost (and darkest) spot in both lanes is HC toxin I, the major form of HC toxin.
HC toxin–nonproducing (Tox2−) isolates of C. carbonum successfully penetrate resistant maize leaves (genotype Hm1/−) but cause only small necrotic lesions that do not expand (Panaccione et al., 1992; Ahn and Walton, 1997). A ccsnf1 mutant of a wild-type Tox2− isolate was made in the same way as the Tox2+/ccsnf1 mutant, and disruption was confirmed by DNA gel blot analysis. The Tox2−/ccsnf1 mutant that was analyzed in detail (T707) showed the same in vitro growth phenotypes as the Tox2+/ccsnf1 mutant (data not shown). Like the Tox2+/ccsnf1 mutant, the Tox2−/ccsnf1 mutant caused many fewer lesions than did the wild-type Tox2−/ccSNF1 strain (Figure 7).
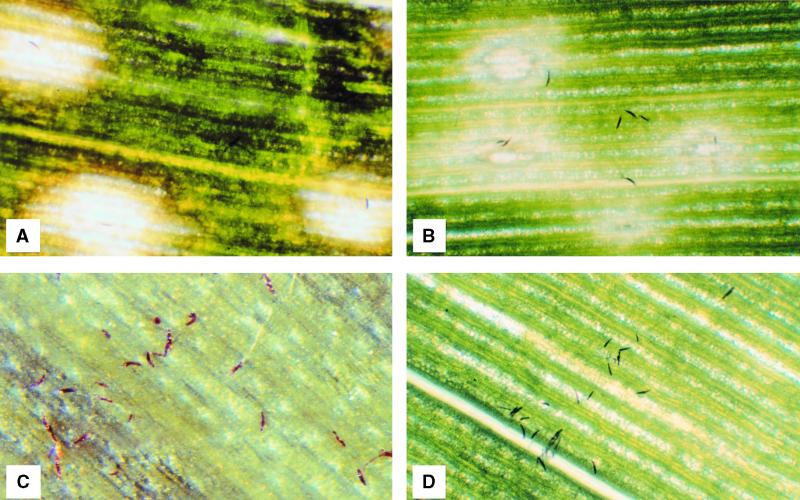
Microscopical examination revealed that spores of the mutant adhered, germinated, and formed appressoria at the same rate as did those of the wild type. Whereas the efficiency of penetration (as manifested by the appearance of either compatible or incompatible macroscopic lesions) by adhered spores of the wild type was quite high (ranging from 50 to 80% in different experiments), infection efficiency by both the Tox2+/ccsnf1 and Tox2−/ccsnf1 mutants was much lower (Figures 9A to 9D).
Figure 9.
Infection by Wild Type and ccsnf1 Mutants.
Plants were of genotype hm1/hm1. Photographs were taken at ×30 magnification 3 days after spray inoculation.
(A) Wild type (Tox2+/ccSNF1). Three lesions, each with a spore in its center, are shown. Although not clearly visible, the fungus already has spread well outside the necrotic zones, and the entire leaf will be dead in another 48 hr.
(B) Wild type (Tox2−/ccSNF1). The lesions are typical of an incompatible reaction. Penetration and necrotic lesions have formed, but the fungus fails to spread further. The leaf will remain alive.
(C) and (D) Tox2+/ccsnf1 and Tox2−/ccsnf1 mutants, respectively. Whereas penetration efficiency by ccSNF1 wild-type isolates is high (cf. [A] and [B]), penetration by ccsnf1 mutants is much reduced. Most of the spores shown in (C) and (D) have germinated, but none has penetrated the leaf to give either a compatible (cf. [A]) or an incompatible (cf. [B]) reaction.
Yeast snf1 mutants have reduced thermotolerance (Thompson-Jaeger et al., 1991). Because the temperature in the greenhouse where the pathogenicity tests were performed sometimes exceeded 27°C, the tests were repeated, keeping the temperature consistently below 24°C, and an identical reduction in virulence was obtained. Also compared was growth on agar plates of the wild type and the ccsnf1 mutant. Both isolates grew equally well at 26°C, both were equally reduced in growth at 30°C, and neither grew at 37°C. Therefore, the reduced growth and virulence of the ccsnf1 mutant was not attributable to decreased thermotolerance.
DISCUSSION
The ccSNF1 gene of C. carbonum is structurally and functionally related to SNF1 of yeast. Its overall amino acid similarity is strong within the region conserved among all known Snf1 proteins, and SNF1 and its counterpart in C. carbonum, ccSNF1, are required for the expression of catabolite (glucose)-repressed genes in both organisms. ccSNF1 can complement growth on sucrose of a yeast snf1 mutant. The genes actually regulated by SNF1 and ccSNF1 differ in yeast and C. carbonum because yeast does not make xylanase or many other wall-degrading enzymes, and invertase is not subject to SNF1-mediated catabolite repression in C. carbonum. HC toxin synthesis is not regulated by ccSNF1, indicating that HC toxin, although necessary, is not sufficient for full virulence of C. carbonum.
The ccsnf1 mutant of C. carbonum grew at the same rate as the wild type on glucose and was also normal in other respects, such as colony morphology, conidiation, and appressorium formation. This argues that ccSNF1 is not required for essential housekeeping functions in C. carbonum. The growth of the ccsnf1 mutant was impaired on complex polysaccharide substrates as well as on certain simple sugars and, most importantly, the virulence of the mutant on maize was decreased. Analysis of Tox2+/ccsnf1 and Tox2−/csnf1 mutants indicated a specific role for ccSNF1 in penetration.
As with any regulatory gene, the reduced virulence of the ccsnf1 mutant may reflect a defect in some process unrelated to expression of extracellular depolymerases or the ability to utilize their breakdown products. SNF1 in yeast and related genes in other organisms also regulate other cellular processes, such as glycogen, sterol, and fatty acid biosynthesis, and fatty acid β-oxidation (Hardie et al., 1998). However, unlike wall-degrading enzymes, these processes have not previously been implicated by experimental data in the process of fungal pathogenesis. More likely, the ccsnf1 mutant has reduced virulence because of its reduced ability to express wall-degrading enzymes or to utilize the products of those enzymes.
These results complement earlier studies in which structural enzyme genes were directly mutated. A striking difference is the much more drastic change in growth and pathogenicity phenotype of the ccsnf1 mutant in comparison with any combination of mutated structural genes. That is, the reduction in growth of the ccsnf1 mutant was stronger than would have been predicted by the degree to which the measurable enzyme activities were decreased, as compared with the results obtained by mutations in the structural genes for the enzymes themselves. For example, although Xyl1p accounts for ∼80% of the total extracellular endo-β-1,4-xylanase activity of C. carbonum grown in vitro, mutation of XYL1 does not reduce growth on xylan or maize cell walls (Apel et al., 1993; Apel-Birkhold and Walton, 1996). In contrast, XYL1, XYL2, and XYL4 are only partially downregulated, and total xylanase activity is decreased by only 65% in the ccsnf1 mutant, yet growth on xylan is decreased by >60%. Similarly, the simultaneous disruption of PGN1 and PGX1 causes a >95% loss of extracellular polygalacturonase activity in C. carbonum yet results in only a 40 to 60% decrease in the growth of the mutant on pectin (Scott-Craig et al., 1998; J.S. Scott-Craig and J.D. Walton, unpublished results). In contrast, disruption of ccSNF1 only partially downregulates PGN1 and PGX1 expression and decreases total pectinase activity by only 24%, yet growth of the mutant is decreased by >50%.
These comparative results are consistent with the hypothesis that ccSNF1 is also necessary for expression of the enzymes needed for utilization of xylan or pectin, such as the enzymes needed for the transport and intracellular catabolism of xylose or galacturonic acid. This hypothesis is supported by the data shown here indicating that ccsnf1 mutants have a reduced ability to grow on simple sugars such as xylose or galacturonic acid. Studies in other systems also support this hypothesis. For example, catabolism of the products of pectinase in Erwinia chrysanthemi requires at least two transport proteins and seven cytoplasmic enzymes, which are coregulated with the extracellular pectinases (Hugouvieux-Cotte-Pattat et al., 1996). Moreover, glucose, acting at least partially through SNF1, regulates the transcription of hexose transporters in yeast (Carlson, 1998).
ccSNF1 is required for the expression of wall-degrading enzymes and for growth on simple sugars, but our results do not indicate which is more important for the virulence of C. carbonum. Wall-degrading enzymes might be important for the actual process of penetration. Species of Cochliobolus do not require either melanization or the formation of appressoria to cause disease and therefore have been presumed, by default, to penetrate enzymatically and not by mechanical force (Horwitz et al., 1999). A decreased ability to produce wall-degrading enzymes, therefore, would be predicted to result in decreased penetration, which is what was observed for the ccsnf1 mutant. Alternatively, or in addition, C. carbonum might require free sugars as a source of nutrition to sustain the metabolic activity necessary for penetration. These sugars could conceivably come from the plant leaf surface or epidermis, although the concentrations of free sugars (other than glucose or sucrose, which are utilized equally well by the ccsnf1 mutant and the wild type) are probably quite low in these environments. A more likely source of sugars, such as arabinose or xylose, comes from the action of extracellular depolymerases on the maize cell wall. Therefore, even if the proximal cause of the decreased virulence of the ccsnf1 mutant is a defect in the uptake and metabolism of simple sugars, the ultimate cause still would be its reduced expression of wall-degrading enzymes.
Insofar as wall-degrading enzymes are important for virulence of C. carbonum, determining which enzymes in particular are important should be valuable. Reduced virulence of the ccsnf1 mutant could result from downregulation of one enzyme, of all enzymes of a particular class, or of many enzymes partially. Our results do not distinguish between these alternatives, because all of the enzymes studied were downregulated. Previous studies have excluded many individual structural genes from making a major contribution to virulence. Of those that have not been directly tested, ARF1 is intriguing as a possible candidate for a solo virulence gene because (1) its expression is completely dependent on ccSNF1; (2) growth of the ccsnf1 mutant is most strongly impaired on arabinose; (3) arabinose is a major component of maize cell walls; and (4) α-arabinosidase has been implicated as a virulence factor in two other diseases (Howell, 1975; Rehnstrom et al., 1994).
METHODS
Fungal Cultures, Media, and Growth Conditions
The wild-type HC toxin–producing (Tox2+) isolate of Cochliobolus carbonum, designated 367-2A, was derived from isolate SB111 (ATCC 90305) and maintained on V8 juice–agar plates (Apel et al., 1993). The wild-type HC toxin–nonproducing (Tox2−) isolate was 164R1, a progeny of SB111 (Walton, 1987). The fungus was grown in liquid media or agar plates containing mineral salts, 0.2% yeast extract, and trace elements (Van Hoof et al., 1991). Carbon sources were 2% (w/v) glucose, sucrose, oat spelt xylan (Fluka, Buchs, Switzerland), citrus pectin (catalog no. P-9135; Sigma), or maize cell walls (Sposato et al., 1995). For quantifying growth on agar plates, 5 μL of a conidial suspension (104 conidia per mL) in 0.1% Tween 20 was pipetted onto the center of the plate. Plates were incubated under fluorescent lights at 21°C.
Nucleic Acid Manipulations
The C. carbonum genomic and cDNA libraries have been described previously (Scott-Craig et al., 1990). DNA and total RNA were extracted from lyophilized mats (Apel et al., 1993; Pitkin et al., 1996). The methods used for DNA and RNA electrophoresis, gel blotting, probe labeling, and hybridization also have been described elsewhere (Apel-Birkhold and Walton, 1996). For RNA gel blots, the Cochliobolus heterostrophus GPD1 gene, encoding glyceraldehyde-3-phosphate dehydrogenase, was used as a loading control (Van Wert and Yoder, 1992).
Polymerase chain reaction (PCR) was performed in a thermocycler (MJ Research, Callahan, CA) by using Taq DNA polymerase (Gibco BRL) and two degenerate oligonucleotide primers based on the conserved regions of SNF1 genes (sense, 5′-CAYCCNCAYATHATHAA-3′; antisense, 5′-TCNGGNGCNGCRTARTT-3′; where Y is T or C; R is G or A; H is T, C, or A; and N is A, T, G, or C). Touchdown PCR (Don et al., 1991) with these primers and C. carbonum genomic DNA as template was performed under the following conditions: initial denaturation at 94°C for 3 min, followed by 45 cycles of denaturation at 94°C for 1 min, annealing for 1 min, and polymerization at 72°C for 2 min. The annealing temperature ranged from 60 to 45°C with a decrease of 1°C every three cycles. This was followed by 10 cycles of denaturation at 94°C for 1 min, annealing at 45°C for 1 min, and polymerization at 72°C for 2 min. The PCR product was cloned into the EcoRV site of pBluescriptII KS+ (Stratagene, La Jolla, CA).
The transcriptional start site of ccSNF1 was determined by using the 5′ rapid amplification of cDNA ends system, version 2.0, following the manufacturer's (Gibco BRL) instructions (Frohman et al., 1988). An oligonucleotide of sequence 5′-GCCCTCGCCCAGGGTGC-3′ was used to prime the first-strand cDNA synthesis, which then was amplified with PCR by using a nested primer of the sequence 5′-GCCGAGACGCTGGCTCG-3′.
Automated fluorescence DNA sequencing was conducted at the Department of Energy Plant Research Laboratory Sequencing Facility (Michigan State University). Sequence data were analyzed with Lasergene software (DNASTAR, Inc., Madison, WI).
Functional Complementation of Yeast snf1
Saccharomyces cerevisiae was grown at 30°C on plates containing YPD (1% yeast extract, 2% [w/v] peptone, 2% [w/v] glucose, and 2% [w/v] agar). The ura reference strain was MG106 (MATa ade2-1 can1-100 his3-1115 leu2-3112 trp1-1 ura3-1). The ccSNF1 cDNA was excised from plasmid pSnf1c-3 with SacI and XbaI and then cloned into pVT100-U, a yeast expression vector with the alcohol dehydrogenase ADH1 promoter and terminator and the URA3 selectable marker. The resulting construct, pSnf1c-4, or the empty vector pVT100-U as a control was used to transform yeast MCY1846 (MATa snf1Δ10 lys2-801 ura3-52) cells as described by Gietz and Woods (1994). Transformants were first selected on synthetic dextrose minus uracil (SD-URA) medium, which contains 0.67% yeast nitrogen base without amino acids (Difco Laboratories, Detroit, MI), 0.062% −Leu/−Trp/−Ura dropout supplement (Clonetech Laboratories, Palo Alto, CA), 0.01% Leu, 0.002% tryptophan, 2% (w/v) glucose, and 2% (w/v) agar at 30°C. The transformed cells were grown in liquid YPD medium overnight. The cultures then were streaked on YPS agar plates, which contain 1% yeast extract, 2% peptone, 2% (w/v) sucrose, and 2% (w/v) agar. The plates were photographed after 2 days at 30°C.
Disruption of ccSNF1
The gene replacement vector was derived from a 3.3-kb SacI-EcoRV genomic fragment containing ccSNF1 (Figure 3A) cloned into pBluescriptII KS+. An internal 2.1-kb PstI-SalI fragment was deleted and replaced with a 1.4-kb PstI-SalI fragment containing the hph gene conferring hygromycin resistance. To create convenient restriction sites, the HpaI fragment from plasmid CB1003 (Carroll et al., 1994) was cloned into the EcoRV site of pBluescriptII KS+.
The replacement vector was linearized at the BssHII-SacI sites before transformation of C. carbonum 367-2A or 164R1 protoplasts (Scott-Craig et al., 1990; Pitkin et al., 1996). Mycelium for protoplasts for transformation was obtained from germinating conidia (Apel et al., 1993). Transformants were purified by two rounds of single-spore isolation.
Enzyme Assays
Total xylanase, pectinase, and β-1,3-glucanase activities were assayed by measuring the release of reducing sugars from the indicated substrate (Lever, 1972). Thirty microliters of culture supernatant was assayed in a 300-μL reaction volume containing 1.0% oat spelt xylan, polygalacturonic acid, or β-1,3-glucan (laminarin) and 50 mM sodium acetate, pH 5.0, at 37°C for 30 min. A 25- or 100-μL aliquot of the reaction mixture was mixed with 1.5 mL of a working solution of p-hydroxybenzoic acid hydrazide, the mixture was heated at 100°C for 10 min, and its absorbance was measured at 410 nm (Lever, 1972).
HC Toxin Analysis
HC toxin was extracted with chloroform from culture filtrates of C. carbonum grown for 14 days, fractionated by thin-layer chromatography on silica by using a solvent system of dichloromethane:acetone (1:1 [v/v]), and detected with an epoxide-specific colorimetric reagent (Meeley and Walton, 1991).
Pathogenicity Assay
Pathogenicity was tested by spray-inoculating 18-day-old plants of the susceptible inbred maize line Pr (genotype hm1/hm1) with a suspension of conidia (104 per mL) in 0.1% Tween 20. After inoculation in the afternoon, the plants were covered with plastic bags overnight. The plants were grown in a greenhouse and monitored daily for 8 days or until they died.
Acknowledgments
We thank Joe Leykam (Michigan State University Macromolecular Facility) for synthesis of oligonucleotides and Marian Carlson (Columbia University, New York, NY) for the yeast snf1 mutant MCY1486. This research was supported by the U.S. Department of Agriculture National Research Initiative Competitive Grants Program, the U.S. Department of Energy Division of Energy Biosciences, and the Michigan State University Research Excellence Fund.
References
- Ahn, J.-H., and Walton, J.D. (1997). A fatty acid synthase gene required for production of the cyclic tetrapeptide HC-toxin, cyclo(d-pro-lyl-l-alanyl-d-alanyl-l-2-amino-9,10-epoxi-8-oxodecanoyl). Mol. Plant-Microbe Interact. 10 207–214. [DOI] [PubMed] [Google Scholar]
- Apel, P.C., Panaccione, D.G., Holden, F.R., and Walton, J.D. (1993). Cloning and targeted gene disruption of XYL1, a β-1,4-xylanase gene from the maize pathogen Cochliobolus carbonum. Mol. Plant-Microbe Interact. 6 467–473. [DOI] [PubMed] [Google Scholar]
- Apel-Birkhold, P.C., and Walton, J.D. (1996). Cloning, disruption, and expression of two endo-β-1,4-xylanase genes, XYL2 and XYL3, from Cochliobolus carbonum. Appl. Environ. Microbiol. 62 4129–4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalerao, R.P., Salchert, K., Bakó, L., Okrész, L., Szabados, L., Muranaka, T., Machida, Y., Schell, J., and Koncz, C. (1999). Regulatory interaction of PRL1 WD protein with Arabidopsis SNF1-like protein kinases. Proc. Natl. Acad. Sci. USA 96 5322–5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, M. (1998). Regulation of glucose utilization in yeast. Curr. Opin. Genet. Dev. 8 560–564. [DOI] [PubMed] [Google Scholar]
- Carroll, A.M., Sweigard, J.A., and Valent, B. (1994). Improved vectors for selecting resistance to hygromycin. Fungal Genet. Newsl. 41 22. [Google Scholar]
- Celenza, J.L., and Carlson, M. (1984). Cloning and genetic mapping of SNF1, a gene required for expression of glucose-repressible genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 4 49–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, R.M. (1987). The use of mutants in exploring depolymerases as determinants of pathogenicity. In Genetics and Plant Pathogenesis, P.R. Day and G.J. Jellis, eds (Oxford, UK: Blackwell), pp. 261–281.
- DeVit, M.J., Waddle, J.A., and Johnston, M. (1997). Regulated nuclear translocation of the Mig1 glucose repressor. Mol. Biol. Cell 8 1603–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Don, R.H., Cox, P.T., Wainwright, B.J., Baker, K., and Mattick, J.S. (1991). ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 19 4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohman, M.A., Dush, M.K., and Martin, G.R. (1988). Rapid production of full-length cDNAs from rare transcripts: Amplification using a single-specific oligonucleotide primer. Proc. Natl. Acad. Sci. USA 85 8998–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, R.D., and Woods, R.A. (1994). High efficiency transformation in yeast. In Molecular Genetics of Yeast: Practical Approaches, J.A. Johnston, ed (Oxford, UK: Oxford University Press), pp. 121–134.
- Görlach, J.M., Van Der Knaap, E., and Walton, J.D. (1998). Cloning and targeted disruption of MLG1, a gene encoding two of three extracellular mixed-linked glucanases of Cochliobolus carbonum. Appl. Environ. Microbiol. 64 385–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, W.J., Gonzalez-Candelas, L., and Kolattukudy, P.E. (1996). Identification of a novel pelD gene expressed uniquely in planta by Fusarium solani f sp pisi (Nectria haematococca, mating type VI) and characterization of its protein product as an endo-pectate lyase. Arch. Biochem. Biophys. 332 305–312. [DOI] [PubMed] [Google Scholar]
- Hardie, D.G., Carling, D., and Carlson, M. (1998). The AMP-activated/Snf1 protein kinase subfamily: Metabolic sensors of the eukaryotic cell? Annu. Rev. Biochem. 67 821–855. [DOI] [PubMed] [Google Scholar]
- Holden, F.R., and Walton, J.D. (1992). Xylanases from the fungal maize pathogen Cochliobolus carbonum. Physiol. Mol. Plant Pathol. 40 39–47. [Google Scholar]
- Horwitz, B.A., Sharon, A., Lu, S.-W., Ritter, V., Sandrock, T.M., Yoder, O.C., and Turgeon, B.G. (1999). A G protein alpha subunit from Cochliobolus heterostrophus involved in mating and appressorium formation. Fungal Genet. Biol. 26 19–32. [DOI] [PubMed] [Google Scholar]
- Howell, H.E. (1975). Correlation of virulence with secretion in vitro of three wall-degrading enzymes in isolates of Sclerotinia fructigena obtained after mutagen treatment. J. Gen. Microbiol. 90 32–40. [DOI] [PubMed] [Google Scholar]
- Hugouvieux-Cotte-Pattat, N., Reverchon, S., Nasser, W., Condemine, G.T., and Robert-Baudouy, J. (1996). Regulation of pectinase biosynthesis in Erwinia chrysanthemi. In Pectins and Pectinases, J. Visser and A.G.J. Voragen, eds (Amsterdam: Elsevier), pp. 311–330.
- Johnson, L.N., Noble, M.E., and Owen, D.J. (1996). Active and inactive protein kinases: Structural basis for regulation. Cell 85 149–158. [DOI] [PubMed] [Google Scholar]
- Kato, Y., and Nevins, D.J. (1984). Enzymic dissociation of Zea shoot cell wall polysaccharides. Plant Physiol. 75 740–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage, P., Yang, X., and Carlson, M. (1996). Yeast SNF1 protein kinase interacts with SIP4, a C6 zinc cluster transcriptional activator: A new role for SNF1 in the glucose response. Mol. Cell. Biol. 16 1921–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever, M. (1972). A new reaction for colorimetric determination of carbohydrates. Anal. Biochem. 47 273–279. [DOI] [PubMed] [Google Scholar]
- Meeley, R.B., and Walton, J.D. (1991). Enzymatic detoxification by maize of the host-selective toxin HC-toxin. Plant Physiol. 97 1080–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muranaka, T., Banno, H., and Machida, Y. (1994). Characterization of tobacco protein kinase NPK5, a homolog of Saccharomyces cerevisiae SNF1 that constitutively activates expression of the glucose-repressible SUC2 gene for a secreted invertase of S. cerevisiae. Mol. Cell. Biol. 14 2958–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, J.M., and Walton, J.D. (1996). Three extracellular proteases from Cochliobolus carbonum: Cloning and targeted disruption of ALP1. Mol. Plant-Microbe Interact. 9 290–297. [DOI] [PubMed] [Google Scholar]
- Nakai, K., and Kanehisa, M. (1992). A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics 14 897–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolskaya, A.N., Pitkin, J.W., Schaeffer, H.J., Ahn, J.H., and Walton, J.D. (1998). EXG1p, a novel exo-β-1,3-glucanase from the fungus Cochliobolus carbonum, contains a repeated motif present in other proteins that interact with polysaccharides. Biochim. Biophys. Acta 1425 632–636. [DOI] [PubMed] [Google Scholar]
- Östling, J., and Ronne, H. (1998). Negative control of the Mig1p repressor by Snf1p-dependent phosphorylation in the absence of glucose. Eur. J. Biochem. 252 162–168. [DOI] [PubMed] [Google Scholar]
- Panaccione, D.G., Scott-Craig, J.S., Pocard, J.A., and Walton, J.D. (1992). A cyclic peptide synthetase gene required for pathogenicity of the fungus Cochliobolus carbonum on maize. Proc. Natl. Acad. Sci. USA 89 6590–6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petter, R., and Kwon-Chung, K.J. (1996). Disruption of the SNF1 gene abolishes trehalose utilization in the pathogenic yeast Candida glabrata. Infect. Immun. 64 5269–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitkin, J.W., Panaccione, D.G., and Walton, J.D. (1996). A putative cycle peptide efflux pump encoded by the TOXA gene of the plant pathogenic fungus Cochliobolus carbonum. Microbiology 142 1557–1565. [DOI] [PubMed] [Google Scholar]
- Ransom, R.F., and Walton, J.D. (1997). Purification and characterization of extracellular β-xylosidase and α-arabinosidase from the plant pathogenic fungus Cochliobolus carbonum. Carbohydr. Res. 297 357–364. [Google Scholar]
- Rehnstrom, A.L., Free, S.J., and Pratt, R.G. (1994). Isolation, characterization and pathogenicity of Sclerotinia trifoliorum arabinofuranosidase-deficient mutants. Physiol. Mol. Plant Pathol. 44 199–206. [Google Scholar]
- Ronne, H. (1995). Glucose repression in fungi. Trends Genet. 11 12–17. [DOI] [PubMed] [Google Scholar]
- Ruijter, G.J.G., and Visser, J. (1997). Carbon repression in Aspergilli. FEMS Microbiol. Lett. 151 103–114. [DOI] [PubMed] [Google Scholar]
- Schaeffer, H.J., Leykam, J., and Walton, J.D. (1994). Cloning and targeted gene disruption of EXG1, encoding exo-β-1,3-glucanase, in the phytopathogenic fungus Cochliobolus carbonum. Appl. Environ. Microbiol. 60 594–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott-Craig, J.S., Panaccione, D.G., Cervone, F., and Walton, J.D. (1990). Endopolygalacturonase is not required for pathogenicity of Cochliobolus carbonum on maize. Plant Cell 2 1191–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott-Craig, J.S., Cheng, Y.Q., Cervone, F., De Lorenzo, G., Pitkin, J.W., and Walton, J.D. (1998). Targeted mutants of Cochliobolus carbonum lacking the two major extracellular polygalacturonases. Appl. Environ. Microbiol. 64 1497–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh, M.T., Brown, R.L., Whitehead, M.P., Cary, J.W., Cotty, P.J., Cleveland, T.E., and Dean, R.A. (1997). Molecular genetic evidence for the involvement of a specific polygalacturonase, P2c, in the invasion and spread of Aspergillus flavus in cotton bolls. Appl. Environ. Microbiol. 63 3548–3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sposato, J.P., Ahn, J.-H., and Walton, J.D. (1995). Characterization and disruption of a gene in the maize pathogen Cochliobolus carbonum encoding a cellulase lacking a cellulose binding domain and hinge region. Mol. Plant-Microbe Interact. 8 602–609. [DOI] [PubMed] [Google Scholar]
- ten Have, A., Mulder, W., Visser, J., and van Kan, J.A.L. (1998). The endopolygalacturonase gene Bcpg1 is required for full virulence of Botrytis cinerea. Mol. Plant-Microbe Interact. 11 1009–1016. [DOI] [PubMed] [Google Scholar]
- Thompson-Jaeger, S., Francois, J., Gaughran, J.P., and Tatchell, K. (1991). Deletion of SNF1 affects the nutrient response of yeast and resembles mutations which activate the adenylate cyclase pathway. Genetics 129 697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treitel, M.A., Kuchin, S., and Carlson, M. (1998). Snf1 protein kinase regulates phosphorylation of the Mig1 repressor in Saccharomyces cerevisiae. Mol. Cell. Biol. 18 6273–6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoof, A., Leykam, J., Schaeffer, H.J., and Walton, J.D. (1991). A single β-1,3-glucanase secreted by the maize pathogen Cochliobolus carbonum acts by an exolytic mechanism. Physiol. Mol. Plant Pathol. 39 259–267. [Google Scholar]
- Van Wert, S.L., and Yoder, O.C. (1992). Structure of the Cochliobolus heterostrophus glyceraldehyde-3-phosphate dehydrogenase gene. Curr. Genet. 22 29–35. [DOI] [PubMed] [Google Scholar]
- Vincent, O., and Carlson, M. (1998). Sip4, a Snf1 kinase-dependent transcriptional activator, binds to the carbon source-responsive element of gluconeogenic genes. EMBO J. 17 7002–7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton, J.D. (1987). Two enzymes involved in biosynthesis of the host-selective phytotoxin HC-toxin. Proc. Natl. Acad. Sci. USA 84 8444–8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton, J.D. (1994). Deconstructing the cell wall. Plant Physiol. 104 1113–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton, J.D., and Cervone, F. (1990). Endopolygalacturonase from the maize pathogen Cochliobolus carbonum. Physiol. Mol. Plant Pathol. 36 351–359. [Google Scholar]
- Wegener, S., Ransom, R.F., and Walton, J.D. (1999). A unique eukaryotic β-xylosidase from the phytopathogenic fungus Cochliobolus carbonum. Microbiology 145 1089–1095. [DOI] [PubMed] [Google Scholar]
- Wu, C., Ham, K.-S., Darvill, A.G., and Albersheim, P. (1997). De-letion of two endo-β-1,4-xylanase genes reveals additional isozymes secreted by the rice blast fungus. Mol. Plant-Microbe Interact. 10 700–708. [Google Scholar]