Abstract
Using reporter assays in tobacco protoplasts and yeast, we investigated the function of the acidic C-terminal activation domains of tomato heat stress transcription factors HsfA1 and HsfA2. Both transcription factors contain short, essential peptide motifs with a characteristic pattern of aromatic and large hydrophobic amino acid residues embedded in an acidic context (AHA motifs). The prototype is the AHA1 motif of HsfA2, which has the sequence DDIWEELL. Our mutational analysis supports the important role of the aromatic and large hydrophobic amino acid residues in the core positions of the AHA motifs. The pattern suggests the formation of an amphipathic, negatively charged helix as the putative contact region with components of the basal transcription complex. In support of this concept, proline or positively charged residues in or adjacent to the AHA motifs markedly reduce or abolish their activity. Both AHA motifs of HsfA1 and HsfA2 contribute to activator potential, and they can substitute for each other; however, there is evidence for sequence and positional specificity.
INTRODUCTION
Eukaryotic heat stress–inducible genes share conserved palindromic promoter elements with the consensus motif AGAAn–nTTCT (Pelham, 1982; Pelham and Bienz, 1982; Nover, 1987). They represent the recognition sites for the corresponding heat stress transcription factors (HSFs), which are encoded by small multigene families. Similar to other transcription factors, HSFs have a modular structure with an N-terminal DNA binding domain characterized by a central helix-turn-helix motif, an adjacent domain with heptad hydrophobic repeats (HR-A/B) involved in oligomerization, a cluster of basic amino acid residues essential for nuclear import (nuclear localization signal), and a C-terminal activation domain (CTAD; reviewed in Wu, 1995; Nover et al., 1996; Morimoto, 1998; Scharf et al., 1998a).
In contrast to the modules for DNA binding, oligomerization, and nuclear import, which are more or less conserved in yeast, animal, and plant HSFs, our understanding of the functional elements within the CTAD is limited. Two important functional aspects can be considered: (1) the qualitative aspect, that is, the search for elements involved in heat stress regulation, and (2) the quantitative aspect, that is, the identification of activator motifs assumed to interact with components of the general transcription complex. The second aspect is central to our work.
Despite some general similarities, many details of heat stress regulation are organism specific (Nover and Scharf, 1997; Morimoto, 1998; Scharf et al., 1998a). Phosphorylation of HSFs in yeast and vertebrates, the role of the C-terminal HR-C region as an intramolecular repressor domain, and the interaction with chaperones of the heat stress protein Hsp70 and Hsp90 families may contribute to the maintenance and/or generation of the inactive state. Recently, a small repressor protein (HSBP1) that binds to the HR-A/B region of animal HSFs was identified (Satyal et al., 1998).
The plant system of HSFs has a number of characteristics not found in other organisms (Scharf et al., 1990, 1998a). During the heat stress response, new HSFs are expressed as heat stress–inducible proteins, and there is increasing evidence for functional interaction between HSFs. One prominent example is HsfA2 in tomato, which, depending on the heat stress conditions, exists in three different forms: (1) a soluble, cytoplasmic form that is present under control conditions; (2) a nuclear form that is found at the onset of heat stress and that requires interaction with HsfA1 for efficient nuclear accumulation; and (3) a high molecular weight storage form that is present in the cytoplasmic chaperone complexes (heat stress granules) found in long-term heat-stressed cells (Scharf et al., 1998b). The dynamic changes of the cellular levels and localization of HsfA2 exemplify the intriguing complexity of the plant system of HSFs.
While searching for potential activator motifs in the CTAD of tomato HSFs, we identified two peptide motifs with a central tryptophan residue (Treuter et al., 1993). By comparing this motif with the functionally essential portions of the activator regions of several other mammalian and yeast transcription factors (i.e., p53, Fos, Jun, RelA, VP16, NRF1, RXRa, Gcn4p, and Gal4p), we identified similar peptide motifs with a characteristic composition of aromatic, large hydrophobic, and acidic amino acid residues (AHA motifs; reviewed in Nover and Scharf, 1997).
We present here a detailed analysis of the AHA1 and AHA2 motifs of the tomato HsfA1 and HsfA2 proteins. Although both HSFs show significant differences in the arrangement of functional modules in their CTADs, there is evidence that both AHA motifs of HsfA1 and HsfA2 contribute to the activator potential and that they can substitute for each other. With few exceptions, the results are very similar irrespective of the test system (tobacco protoplasts versus yeast) or the molecular context of the activator, that is, whether the CTAD is fused to its own DNA binding domain or to that of the yeast activator GAL4p.
RESULTS
The C-Terminal Activation Domain of HsfA2 Contains Two Independent Activator Motifs
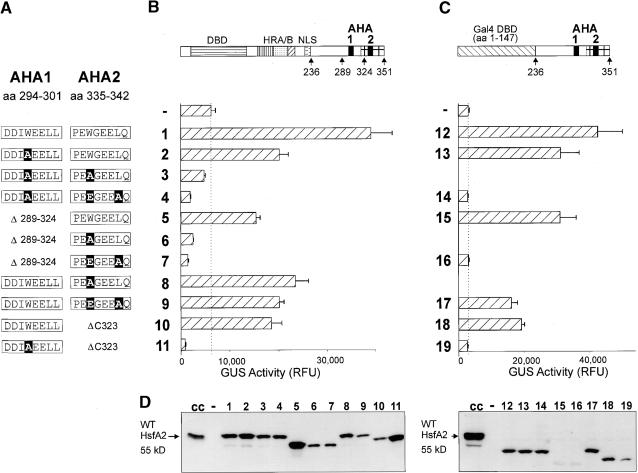
A series of deletions and amino acid substitutions in the CTAD of HsfA2 was used to investigate the role of the two AHA motifs in the activator function of the protein (see Figure 1A for details of the constructs). Two different forms were tested in tobacco protoplasts cotransformed with the appropriate β-glucuronidase (GUS) reporter constructs. On the one hand, we tested mutant forms of the CTAD in the native context of HsfA2 (Figure 1B). On the other hand, hybrid proteins containing the HsfA2 CTAD (amino acid residues 236 to 351) fused to the yeast Gal4p DNA binding domain (amino acid residues 1 to 147) were investigated (Figure 1C). Due to the endogenous HSFs of the protoplasts, the basal GUS activity is higher in Figure 1B and markedly reduced by cotransformation with constructs encoding inactive forms of HSFs (e.g., constructs 4, 6, 7, and 11). As expected, this repressor effect, which is due to the competition with the endogenous HSFs, is lacking in the results shown in Figure 1C, which were obtained with the hybrid Gal4p constructs.
Figure 1.
Functional Equivalence of the Two AHA Motifs in the C-Terminal Activation Domain of HsfA2.
(A) Details of the amino acid (aa) sequences in the AHA1 and AHA2 regions are shown. Point mutations are printed in white letters on a black background.
(B) and (C) Tobacco protoplasts were transformed with the indicated activator constructs (see block diagrams at top) and the appropriate reporter constructs, that is, phsp17GUS (B) and pGal4DBSGUS (C). The activator constructs (C) encode fusion proteins of the yeast Gal4p DNA binding domain (amino acid residues 1 to 147) with the HsfA2 CTAD (amino acid residues 236 to 351). The dotted lines in (B) and (C) mark the level of GUS activity in samples transformed with the reporter only, that is, the activity of the endogenous HSFs. Error bars indicate the absolute deviation from the mean values. DBD, DNA binding domain; HRA/B, hydrophobic heptad repeat region for oligomerization; NLS, nuclear localization signal; RFU, relative fluorescence units.
(D) The expression levels were monitored by protein gel blot analysis. Lanes 1 to 19 correspond to constructs 1 to 19 in (B) and (C). For controls, we included samples from heat stress–induced tomato cell cultures (cc) and from tobacco protoplasts transformed with the empty vector only (−, background control). WT, wild type.
The results with both types of constructs were very similar, demonstrating that the observed properties of the CTAD elements were independent of the N-terminal DNA binding domain. Only one of the two AHA motifs was required and sufficient to give a functional activator. Deletion or functional knock-out by mutation of either one resulted in a 20 to 50% reduction in activation potential. Severe defects in the activity of full-length HsfA2 were observed only when the tryptophan residues of both AHA motifs were substituted (cf. results with construct 1 versus 3 and 12 versus 14). This effect of the tryptophan substitution was also seen with the HsfA2ΔC323 deletion forms, which possessed only one AHA motif (cf. activity of construct 10 versus 11 or 18 versus 19).
Generally, the expression levels of all forms of HsfA2 were comparable (see signals from protein gel blots shown in Figure 1D). There were two exceptions; however, these did not affect our conclusions. Both concerned the deletion forms with the AHA2 motif only (constructs 5, 6, 7, 15, and 16). The strong, negative effect of mutant forms 6 and 7 on the endogenous activity level and the comparison of activities between constructs 15 and 16 clearly indicate that, similar to AHA1, substitution of the tryptophan residue in AHA2 abolishes its function as activator motif.
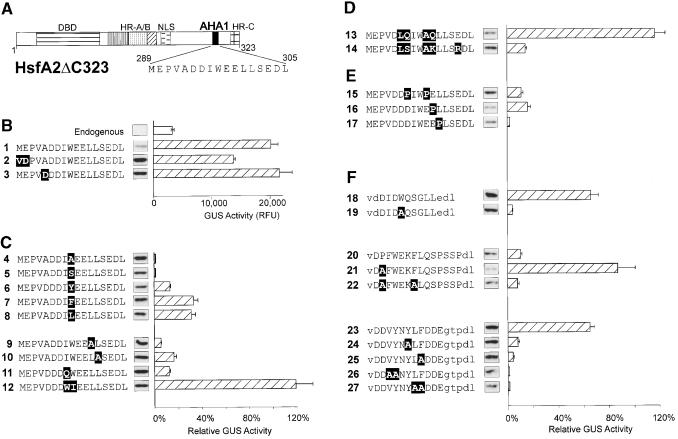
Analysis of AHA Mutant Forms in HsfA2ΔC323
For a more detailed characterization of the AHA1 motif of HsfA2ΔC323, a series of mutant proteins was tested in the same reporter assay with phsp17GUS. Figure 2 presents the results in groups according to the types of mutants. In all cases, the construct number together with details of the amino acid sequences are indicated at left. Similar to those shown in Figure 1, the expression levels shown in Figure 2 were controlled by protein gel blot analysis (see insets at the bottom of each column).
Figure 2.
Mutational Analysis of AHA Motifs in the Context of HsfA2ΔC323.
The activator potential of the indicated HsfA2ΔC323 mutants was tested in tobacco protoplasts cotransformed with the phsp17GUS reporter plasmid. The basic structure (A) and the partial amino acid sequences of the AHA1 motif in groups (B) to (F) identify the individual constructs. Point mutations are indicated with white letters on black background. For group (F) with heterologous AHA motifs replacing the AHA1 motif of HsfA2, the capital letters represent the amino acid sequences of the indicated parts of HsfA1 (constructs 18 to 22; see details given in Figure 3B) and yeast Gal4p (amino acid residues 862 to 872), respectively. Lowercase letters in the sequence data indicate flanking sequences, including the VD (SalI site) and DL (BglII site) motifs used for linker insertion. The relative GUS activities are indicated by the hatched bars. They refer to the activity of the basal constructs (B) set as 100%. Results with constructs 4, 5, and 7 to 10 refer to the activity of construct 1, results with constructs 18 to 27 refer to construct 2, whereas constructs 6 and 11 to 17 refer to construct 3. The expression levels are indicated by the signals from protein gel blot analysis shown between the AHA sequence and the corresponding GUS activity bar. Error bars indicate the absolute deviation from the mean values. DBD, DNA binding domain; NLS, nuclear localization signal; RFU, relative fluorescence units.
To facilitate construction, we created two types of mutants in which a unique SalI site encoding a VD motif was placed in front of the codons for the AHA1 motif, that is, HsfA2.2 with ME289/290→VD and HsfA2.3 with VA292/293→VD (Figures 2A and 2B, constructs 1 to 3). Compared with HsfA2.1 with the wild-type AHA1 motif (construct 1), HsfA2.2 (construct 2) had 30% reduced activity, whereas HsfA2.3 (construct 3) had slightly higher activity (Figure 2B). For direct comparison of the mutant forms compiled in groups C to F, the individual activity values are always given as the percentage of GUS activity measured, with the corresponding basal construct set as 100% (for details, see legend to Figure 2).
The results can be summarized as follows. The central tryptophan and the adjacent large hydrophobic residues (isoleucine and leucine) in the DDIWEELL core motif are essential (group C). Substitution of W297 by Y, F, or L (constructs 6 to 8) led to a reduction of the activity to 15 to 25%, whereas the mutant forms with W→A or W→S (constructs 4 and 5) were inactive. Low or no activity was also observed with the two L→A mutants (constructs 9 and 10) and mutant 11 with I→Q. Interestingly, inversion of the central IW motif (IW→WI, construct 12) had no negative effect on the activator function.
The two central clusters of acidic residues in the AHA1 element (group D) are evidently dispensable. The HsfA2 form with the mutations DD→LQ and EE→AQ (construct 13) was even more active than the wild-type form, but introduction of positive charges in this region strongly reduced activity (construct 14).
With the proline substitution mutants presented in group E, we investigated the significance of the DDIWEELL motif as part of a putative amphipathic helix. In support of this concept, introduction of a proline residue in any position markedly reduced the activator potential (constructs 15 to 17). The particularly pronounced negative effect of the L→P substitution (construct 17) was probably due to both the introduction of proline and the elimination of the important leucine residue (see also construct 9).
In group F, we summarized information about HsfA2 versions containing heterologous activator motifs in the position originally occupied by AHA1. These are the two putative AHA motifs of HsfA1 (see details in Figure 3) and the corresponding motif from the C terminus of yeast Gal4p, respectively (Leuther et al., 1993; Melcher and Johnston, 1995). For orientation, the amino acid positions in the original proteins (Gal4p and HsfA1) are indicated in the legend to Figure 2 and in Figure 3B, respectively.
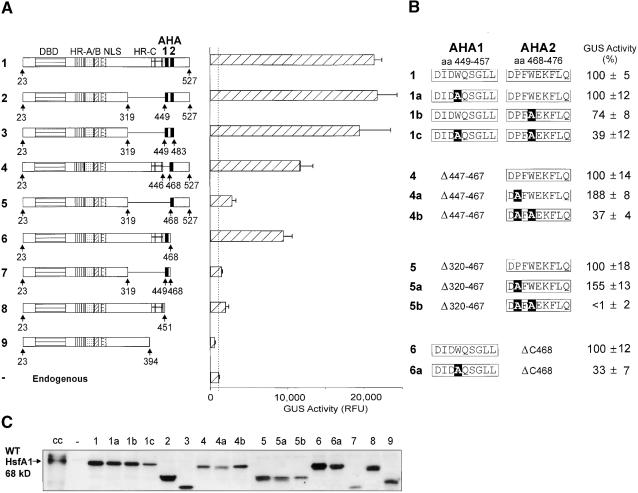
Figure 3.
Functional Analysis of the CTAD of HsfA1.
(A) A series of internal and C-terminal deletion constructs was tested in tobacco protoplasts using the phsp17GUS reporter. The basic structure of the deletion forms is indicated by the block diagrams on the left, whereas the GUS activities are given in the hatched bars at right. The dotted line marks the level of GUS activity in samples transformed with the reporter only, that is, the activity of the endogenous HSFs. Error bars indicate the absolute deviation from the mean values. DBD, DNA binding domain; NLS, nuclear localization signal; RFU, relative fluorescence units.
(B) Details of the functional analysis of the two AHA motifs are presented using constructs 1, 4, 5, and 6. The activity of the basal construct is set as 100%, and the GUS activity of the substitution forms is referred to this value. For calculation, the endogenous level of GUS activity (1000 RFU; see [A]) was subtracted. Point mutations are printed in white letters on a black background. aa, amino acid residues.
(C) Similar to Figure 1, the expression levels of the HsfA1 forms are indicated by the signals from the corresponding protein gel blot analysis. cc, cell cultures; WT, wild type.
The AHA1 motif of HsfA1 was active in the hybrid protein (construct 18), whereas the AHA2 motif was not (construct 20). In view of the results with the proline mutants (group E), we hypothesized that the proline residue immediately adjacent to the FWEKFL motif of AHA2 might be responsible for the low activity. Indeed, mutating P→A created a much more active form (construct 21) that was used to investigate the role of the aromatic residues. As expected, both HsfA2 hybrids with the W→A exchange in the AHA1 motif and the F→A mutant of the gain-of-function version of the AHA2 motif of HsfA1 were inactive (constructs 19 and 22, respectively).
With respect to the pattern of aromatic and hydrophobic residues, the well-known activator peptide motif of yeast Gal4p is very similar to the AHA motifs of HsfA2. In support of a functional similarity, a construct with the indicated Gal4p motif replacing AHA1 in HsfA2ΔC323 had high activity (construct 23), whereas all four mutant forms of the Gal4p AHA motif with alanine substitutions of the aromatic or hydrophobic residues were inactive (constructs 24 to 27).
Two AHA Motifs Contribute to the Activator Potential of HsfA1
As already mentioned, the original identification of peptide motifs with a central tryptophan residue in both HsfA1 and HsfA2 was based on C-terminal deletions (Treuter et al., 1993). Despite some important differences in structure and properties, the close similarities of the core regions of the HSFs prompted us to investigate the role of the two putative AHA motifs identified in HsfA1 by using a similar series of internal and C-terminal deletions (Figure 3). The block diagrams (Figure 3A) help to identify the functional parts of HsfA1 deleted or preserved in the individual constructs.
The results from the GUS reporter assay in tobacco protoplasts clearly confirmed that the two tryptophan-containing AHA motifs in HsfA1 also play an essential role in activation. The results were basically similar to those obtained with the HsfA2 constructs (Figure 1). Deletion of the entire central part of HsfA1, including the HR-C region, did not affect the activity of the corresponding HsfA1 forms (cf. construct 1 encoding the wild-type HsfA1 with constructs 2 and 3). Even the additional deletion of the entire C terminus, generating a minimum form of HsfA1 with the two adjacent AHA motifs linked directly to the N-terminal half of the molecule, had almost wild-type activity (construct 3). Only deletion of either one or both of the AHA motifs led to a reduction or total loss of activity (constructs 4 to 9). However, there was a remarkable difference of activity between the two forms of HsfA1 containing either one of the AHA motifs plus the HR-C region (constructs 4 and 6) and the corresponding forms lacking the HR-C region (constructs 5 and 7). The former retained ∼50% of the activator potential, whereas the latter possessed ⩽10%. A C-terminal deletion form possessing the HR-C region only (construct 8) had similarly low residual activity. The strong synergistic effect of either one of the AHA motifs with the HR-C region was probably due to the acidic surrounding provided by HR-C, which is lacking in constructs 5 and 7.
In Figure 3B, we present results with selected amino acid substitution forms in the background of constructs 1, 4, 5, and 6. Alanine substitution of either one of the central tryptophan residues in full-length HsfA1 had no effect (construct 1a) or led to a modest reduction in GUS activity (construct 1b). Only substitution of both tryptophan residues (construct 1c) had a more pronounced effect (39% residual activity). With the following two groups of mutant forms, we tested the function of the AHA2 motif alone in deletion constructs 4 and 5, respectively. After our previous observation about the negative influence of the proline residue in front of the core FWEKFL motif in HsfA2 (see constructs 20 to 22 in Figure 2F), we investigated the activator potential of mutant forms with P→A substitution (constructs 4a and 5a) as well as that of double substitution forms with P,W→A,A (constructs 4b and 5b) in HfsA1. In both cases, replacement of the proline residue resulted in a significant increase in GUS activity compared with the form with the wild-type AHA2 motif. On the other hand, simultaneous replacement of the tryptophan residue caused a severe reduction or total loss of the activator function. Finally, W→A substitution in the AHA1 motif of construct 6 led to the expected reduction of activity by 67% (constructs 6 versus 6a). The overall results with the two AHA motifs of HsfA1 are basically similar to those obtained with HsfA2 (Figure 1). However, the residual activity of the W→A substitution forms was much higher, indicating additional contributions to the activator potential (see note added in proof).
Function of HsfA2 and Its Mutant Forms in Yeast
In support of the central role of AHA motifs for the function of transcription-activating proteins in general, we wanted to test selected mutant forms of HsfA2 in yeast. Fortunately, two very different test situations for HSFs can be used in this organism. On the one hand, yeast strains with a disruption of their own HSF1 gene allow tests for survival in the presence of HsfA2 and its mutant forms, respectively (Figure 4). On the other hand, the generation of fusion proteins of the HsfA2 CTAD and the DNA binding domain of yeast Gal4p provided the basis for a more detailed investigation of the activator potential with a Gal4p-dependent lacZ reporter construct (Figure 5). Although the fusion point between the Gal4 DNA binding domain (amino acid residues 1 to 147) and the CTAD of HsfA2 (amino acid residues 98 to 351) is different, this set of constructs is similar to that used in the protoplast assays (Figure 1C).
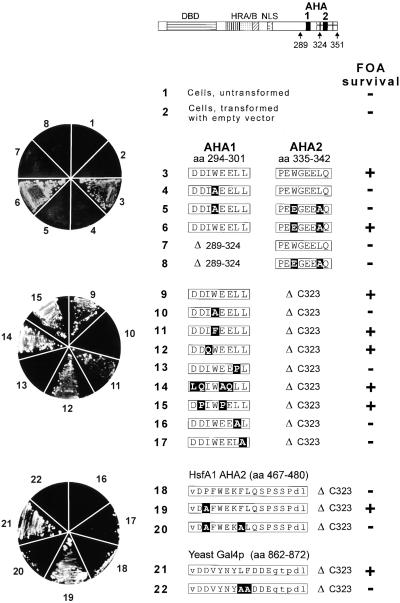
Figure 4.
HsfA2 Forms with an Active CTAD Support Growth of Yeast Strains in Which the Endogenous HSF1 Gene Is Disrupted.
Cells of the yeast strain RSY4 containing a chromosomal disruption of the HSF1 gene (Boscheinen et al., 1997) and a copy of the yeast HSF1 gene on a URA3 plasmid were transformed with a TRP1 plasmid coding for the indicated form of HsfA2. The URA3 plasmid was eliminated by plating on media containing FOA. FOA survival is shown by the pictures of agar plates at left. Details of the HsfA2 cassettes in the yeast vector pMBI are given by the block diagram on top and the sequence information for constructs 3 to 22 (for further explanation, see Figures 1 and 2). Point mutations are printed in white letters on a black background. aa, amino acid residues; DBD, DNA binding domain; HRA/B, hydrophobic heptad repeat region for oligomerization; NLS, nuclear localization signal; (+), survival on FOA plates; (−), no survival.
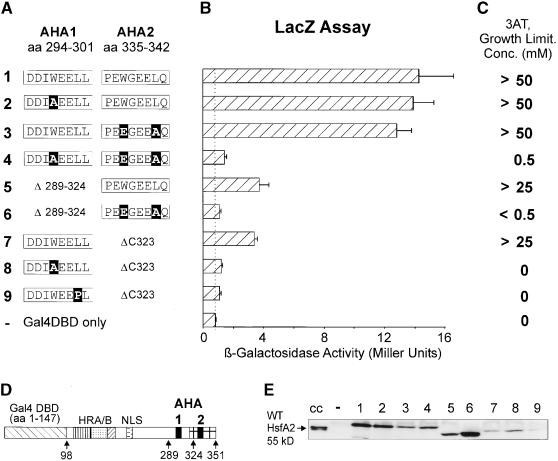
Figure 5.
Reporter Assays in the Yeast Strain YRG2.
The basic structure of the yeast Gal4DBD (amino acid residues 1 to 147) × HsfA2 CTAD (amino acid residues 98 to 351) together with the relevant sequence details of the AHA1 and AHA2 motifs ([D] and [A], respectively) identify the structure of the hybrid activator protein. Point mutations are printed in white letters on a black background. aa, amino acid residues; DBD, DNA binding domain; HRA/B, hydrophobic heptad repeat region for oligomerization; NLS, nuclear localization signal.
(B) β-Galactosidase levels based on the expression of a chromosomal lacZ reporter construct. The dotted line marks the level of LacZ activity in yeast strains transformed with a vector coding for the Gal4p DNA binding domain only. Error bars indicate the absolute deviation from the mean values.
(C) Growth on histidine-free medium due to the Gal4p-dependent expression of the HIS3 gene. The expression level can be estimated from the concentration of 3-AT required to stop growth of a given yeast strain on histidine-free medium.
(E) Protein gel blot analysis for monitoring expression of the indicated Gal4DBD × HsfA2 fusion proteins. Lanes 1 to 9 correspond to constructs 1 to 9. For control, samples from heat stress–induced tomato cell cultures (cc) and from yeast cells transformed with the Gal4DBD vector (−) were included. WT, wild type.
The yeast Hsf1 protein is essential for survival and growth irrespective of control or heat stress conditions (Sorger and Pelham, 1988; Wiederrecht et al., 1988). We demonstrated previously that tomato HsfA1 and HsfA2 are able to replace yeast Hsf1 in this survival function and that an intact DNA binding domain and a functional activation domain are required for this substitution (Boscheinen et al., 1997). A selected set of AHA mutant forms of HsfA2 was tested with respect to the capability to replace yeast Hsf1 in its survival function (see experimental details given in the legend to Figure 4). At left in Figure 4 is shown the growth behavior of yeast strains expressing the indicated HsfA2 forms in the presence of 5-fluoroorotic acid (FOA) used to eliminate the URA3 plasmid encoding yeast Hsf1. In all cases, the inability to survive FOA treatment indicated nonfunctional forms of HsfA2 and not a deficiency of expression, as was shown by protein gel blot analysis before FOA treatment (data not shown).
The relevant details of the amino acid sequences of the HsfA2 forms are indicated at right in Figure 4. Constructs 3 to 8 encode full-length HsfA2 proteins that possess both AHA motifs (constructs 3 to 6) or that contain an internal deletion of AHA1 (constructs 7 and 8). Constructs 9 to 22 are based on the C-terminal deletion form, HsfA2ΔC323 containing the AHA1 motif only. With one noticeable exception, the results were basically similar to those obtained with the tobacco protoplast assays (Figures 1 and 2). Constructs 4 and 7 containing only the functional AHA2 motif could not grow in the presence of FOA; that is, in contrast to the tobacco protoplast assay, the AHA1 and AHA2 motifs are not equivalent in this test situation. However, from results with the ensuing lacZ reporter assays (Figure 5), it was evident that this is a characteristic of the survival assay and not a general defect of the AHA2 function in yeast.
Interestingly, the results with the inactivity of the AHA2 motif of HsfA1 in the HsfA2 background (construct 18) and the possibility to generate a gain-of-function mutant by substitution of P469 by A (construct 19) were also reproduced in the yeast survival assay. Plasmids encoding HsfA2 forms 18 and 20 were unable to support growth on FOA-containing media, whereas the yeast strain carrying construct 19 with the P→A exchange was able to grow.
For the Gal4 DNA binding domain—dependent reporter assay, we used the YRG2 yeast strain with two chromosomal Gal4p-dependent reporter genes (lacZ and HIS3; see Methods). Details of constructs and results are presented in Figure 5A. In the lacZ reporter assay (Figure 5B), functional disruption by point mutations of either one of the two AHA motifs in the full-length background of the HsfA2 CTAD did not affect the activity (constructs 1 to 3). However, deletion of either the AHA1 or AHA2 motifs caused a 70% reduction in activator potential (constructs 5 and 7). As was expected from the results obtained with tobacco protoplasts, mutation of both AHA motifs in the core positions and/or deletion of them abolished activity in yeast (constructs 4, 6, 8, and 9) as well. An alternative to the β-galactosidase assay is the evaluation of growth on histidine-free media in the presence of 3-amino-1,2,4-triazol (3-AT) as a competitive inhibitor of histidine biosynthesis (Figure 5C). The results were similar to those in Figure 5B, demonstrating that the levels of 3-AT, required to limit growth of the corresponding yeast strains, are valuable indicators for the activator potential.
DISCUSSION
HSF Activation Domains
According to the amino acid composition, activation domains are usually classified as acidic, glutamine rich, or proline rich (reviewed in Tjian and Maniatis, 1994; Triezenberg, 1995; Goodrich et al., 1996; Ptashne and Gann, 1997; Sauer and Tjian, 1997; Kadonaga, 1998). Although not all activation domains can be easily assigned to one of these classes, the classification is useful and reflects basic differences between classes of transcription factors (Seipel et al., 1992; Künzler et al., 1994; Ponticelli et al., 1995). However, acidic activation domains may also be rich in proline residues, and in some cases, several types of activation domains coexist in the same transcription activator (e.g., c-Myc or CREB).
Following this basic concept, most HSFs from vertebrates, Drosophila, and plants have acidic CTADs, which are usually also enriched in proline residues. However, in plants, there are two classes of HSFs that differ in the structure of their oligomerization domains as well as their CTADs. Class A HSFs, on which we have focused, are characterized by an insertion of 21 amino acid residues in the oligomerization domain and an acidic CTAD, whereas class B HSFs have no insertion and possess a CTAD that is neutral or positively charged but not glutamine or proline rich. Interestingly, only class A but not class B HSFs can functionally replace the yeast Hsf1 protein in the corresponding gene disruption strain (Boscheinen et al., 1997).
AHA Motifs
Irrespective of the molecular context used for the classification, many if not all activation domains contain short peptide motifs (AHA motifs) with characteristic patterns of aromatic and large hydrophobic amino acid residues (Hahn, 1993; Regier et al., 1993; Tjian and Maniatis, 1994; Triezenberg, 1995; Nover and Scharf, 1997). These activator modules are assumed to represent the putative contact sites for the interaction with components of the basal transcription complex. Tjian and Maniatis (1994) proposed a model of cohesive interfaces, that is, interacting surfaces with a mutually corresponding pattern of aromatic/hydrophobic amino acid residues between activator protein and its target protein(s). In support of this concept, mutant forms with exchanges of the aromatic and/or hydrophobic residues do not interact with components of the basal transcription complex in vitro and are deficient in reporter assays in vivo. This was shown for several mammalian activator proteins such as VP16 (Barlev et al., 1995; Hengartner et al., 1995; Shen et al., 1996; Uesugi et al., 1997), Sp1 (Gill et al., 1994), p53 (Lin et al., 1994; Kussie et al., 1996), RelA (Blair et al., 1994; Schmitz et al., 1994), and E1A (Geisberg et al., 1994; Molloy et al., 1999) as well as for the yeast activator proteins Gal4p (Melcher and Johnston, 1995) and Gcn4p (Jackson et al., 1996; Drysdale et al., 1998).
The activator domains of tomato HsfA1 and HsfA2 contain two AHA motifs. Although the functional significance remains to be investigated, inspection of the CTADs of other class A plant HSFs, for example, from Arabidopsis and tobacco, available from the databases demonstrates the remarkable conservation of AHA motifs in all cases (Table 1). This may be true even for other HSFs, for example, from yeast, Drosophila, and mammals, that have similar clusters of aromatic and hydrophobic residues in their CTADs (Chen et al., 1993; Newton et al., 1996; Wisniewski et al., 1996).
Table 1.
AHA Motifs in the Acidic C-Terminal Activation Domains of Class A Plant HSFs
| HSFsa | AHA Motifsb | References (GenBank Accession Numbers) |
|---|---|---|
| Lp-HsfA1 (527 aa) | AHA1 (448) –ADIDWQSGLLDEIQ– | Treuter et al., 1993 (CAA47869) |
| AHA2 (466) –VGDPFWEKFLQSPS– | ||
| Lp-HsfA (351 aa) | AHA1 (292) –VADDIWEELLSEDL– | Treuter et al., 1993 (CAA47870) |
| AHA2 (333) –KTPEWGEELQDLVD– | ||
| At-HsfA1a (Hsf1, 495 aa) | AHA (432) –SNFEFLEEYMPESP– | Hübel and Schöffl, 1994 (CAB10555.1) |
| At-HsfA1b (Hsf3, 520 aa) | AHA (417) –IQDPFWEQFFSVEL– | Prandl et al., 1998 (CAA74397) |
| At-HsfA4a (Hsf21, 401 aa) | AHA1 (255) –SSIAIWENLVSDSC– | (CAA16745.1) |
| AHA2 (338) –ANDGFWQQFFSENP– | ||
| At-HsfA3 (272 aa) | AHA (254) –LDDGFWEELLSDES– | (CAB41311.1) |
| At-HsfA4b (466 aa) | AHA (413) –VNDVFWEQFLTERP– | (CAB10177.1) |
| At-HsfA1c (406 aa) | AHA (365) –YGEGFWEDLLNEGQ– | (AB022223) |
| Nt-HsfA4 (408 aa) | AHA1 (256) –SSLTFWENVLQDVD– | (BAA83711.1) |
| AHA2 (341) –VNDIFWEQFLTENP– |
Abbreviations of plant names are as follows: Lp, Lycopersicon peruvianum; At, Arabidopsis thaliana; NT, Nicotiana tabacum. For orientation, the size of each HSF is given in parentheses; aa, amino acid residues.
The position of the first amino acid residue of the AHA motif is given in parentheses. Boldface letters mark the relevant aromatic/hydrophobic amino acid residues. They are underlined if their functional significance can be derived from results in this study. For similar AHA motifs in other transcription factors of mammals and yeast, see the summary by Nover and Scharf (1997).
Both AHA motifs of tomato HsfA1 and HsfA2 contribute to the overall activity. In all cases, aromatic residues (i.e., tryptophan and phenylalanine) and adjacent leucine residues are essential. Although substitution of the acidic residues in the core of the motifs has no effect on the activator potential, this does not argue against the functional relevance of the acidic context. Even a mutant form of the HsfA2 AHA1 motif in which the two acidic dipeptide motifs in the core (underlined) were replaced, that is, VDDDIWEELLSEDL→VDLQIWAQLLSEDL (Figure 2D, construct 13), is still embedded in its acidic surrounding. Only introduction of positively charged residues (K and R), for example, in the AHA1 mutant 14 with the VDLSIWAKLLSRDL motif, abolished activity. The important role of the acidic context is probably also the reason for the low activity of some HsfA1 deletion forms lacking the acidic HR-C region adjacent to either the AHA1 or AHA2 motif (Figure 3; cf. the high activity of constructs 4 and 6 with the low activity of constructs 5 and 7).
Following the concept of cohesive interfaces (Tjian and Maniatis, 1994), the detailed pattern and quality of aromatic and/or hydrophobic residues must be decisive for the recognition of the correct target protein(s) and the strength of interaction. In support of this concept, the W→F, W→Y, and W→L mutant forms of HsfA2ΔC323 (see Figure 2, constructs 6 to 8) have reduced activity (10 to 35% of the wild-type form). Similarly, a V147→L exchange in the activator module of the E1A CR3 region destroys the activator potential and the interaction with the TATA box binding protein (TBP; Geisberg et al., 1994; Molloy et al., 1999). It is tempting to speculate that the putative interaction domains of the target proteins in the basal transcription complex are positively charged and contain corresponding patterns of hydrophobic binding sites.
Helical Conformation of AHA Motifs
The intriguing pattern of aromatic/hydrophobic and hydrophilic amino acid residues in the AHA motifs suggests the potential for formation of an amphipathic helix, as originally predicted by Ptashne (1988). Evidently, such a helical conformation may result from a type of induced fit in a hydrophobic solvent (Schmitz et al., 1994; Massari et al., 1996) or by interaction with components of the transcription complex. This was reported for VP16 interacting with TBP (Shen et al., 1996) or with hTAF31 (Uesugi et al., 1997) and for CREB interacting with the CREB binding protein (CBP) (Radhakrishnan et al., 1997; Parker et al., 1998) as well as for c-Myc after binding to TBP (McEwan et al., 1996).
Although structural data are lacking, predictions for the AHA1 region of HsfA2 indicate the formation of such an amphipathic helix. In support of this prediction, all proline substitution forms (Figure 2E, constructs 15 to 17) have very low or no activity; conversely, the low activity of the wild-type AHA2 motif of HsfA1 can be markedly stimulated by replacing the proline residue in the PFWEKFL motif with alanine (see Figure 2F, constructs 20 to 22, and data presented in Figure 3B). Similar results regarding the detrimental effects of helix-destabilizing proline substitutions in the activator motifs were reported for the AHA motif of RelA (Blair et al., 1994), the human glucocorticoid receptor τ1 region (Dahlman-Wright et al., 1995), and the AD1 motif of E2A (Massari et al., 1996). An interesting peculiarity in this respect was reported for yeast Gal4p (Ansari et al., 1998). Introduction of proline residues in the AHA motif (e.g., V864→P or L868→P; see sequence information in Figure 2, construct 23) did not affect the activator function but markedly diminished the interaction with the repressor protein Gal80p. Evidently, in this case, a helical conformation of the Gal4p AHA motif is essential for interaction with Gal80p but not for the contacts to the basal transcription complex.
Another example of an AHA motif representing the interaction site with a corepressor is p53 binding to its repressor protein MDM2 (Lin et al., 1994; Thut et al., 1995). Crystal structure analysis of a 15–amino acid peptide motif of p53 (amino acid residues 15 to 29) bound to the 109-residue N-terminal domain of MDM2 reveals an amphipathic helix in which F19, W23, and L26 are exposed to MDM2 (Kussie et al., 1996). Substitutions of the F, W, or L residues in p53 abolish its function as a transactivator and eliminate the interaction with the TAF31–TAF80 complex (Lin et al., 1994; Thut et al., 1995).
Generally, activation can be envisaged as presentation of the hydrophobic surface of the AHA motifs by a conformational change and/or release of a corepressor. Xiao et al. (1994) discussed a regulatory model involving competition of AHA-like motifs of human Hsf1 with similar motifs adjacent to the C-terminal domain of the RNAPII large subunit. In brief, a preformed transcription complex is blocked in elongation because AHA motifs in the RNAPII interact with the recognition sites in the basal transcription complex. By competition for these recognition sites, Hsf1 triggers the release from the elongation block connected with C-terminal domain phosphorylation as an important step in the transition to productive elongation (Mason and Lis, 1997). A similar competition model was discussed for the VP16-induced opening of the TFIIB conformation, which is necessary for recruitment of other components of the basal transcription complex (Roberts and Green, 1994).
Complex Promoters
The modular structure of transcription factors in general and the similarity of AHA motifs in the core of acidic, glutamine-rich, or proline-rich activation domains provide the basis for functional tests in different heterologous expression systems. For the sake of simplicity and to avoid background problems (cf. Figures 1B and 1C), hybrid constructs are frequently used with the activator domain fused to a heterologous DNA binding domain. To support the general validity of our conclusions, we investigated the activator potential of tomato HsfA1 and HsfA2 in the normal molecular context, that is, with the HsfA1 or HsfA2 DNA binding domains as well as with fusion proteins containing the yeast Gal4p DNA binding domain. As expected, the results were basically similar, irrespective of the expression system used, that is, tobacco protoplasts or yeast.
However, these simplified test systems can provide only partial insight into activator function. Evidence from in vitro and in vivo experiments supports the concept that in the reality of complex promoters, usually several DNA binding proteins contribute synergistically to the assembly of the transcription complex, the composition of which varies with the particular activator/repressor cocktail available in a given cell or situation (Seipel et al., 1992; Chen et al., 1994; Künzler et al., 1994; Thanos and Maniatis, 1995; Blau et al., 1996; Tansey and Herr, 1997; Holstege et al., 1998; Ellwood et al., 1999). There is experimental evidence for the essential role of a synergistic assembly of activator and helper proteins in an “enhanceosome,” which represents a starting point for building a transcription complex with high processivity (Thanos and Maniatis, 1995; Blau et al., 1996; Carey, 1998).
In many transcription-activating proteins, two or more AHA motifs contribute to the overall activity (Blair et al., 1996). In the artificial reporter assays used to investigate corresponding mutant forms, the AHA motifs frequently appear to be functionally equivalent. Basically, this is also true for the AHA motifs of HsfA1 and HsfA2, but there are two noticeable exceptions. First, the AHA2 motif of HsfA2 is defective in one of the test situations in yeast (survival function). It is tempting to speculate that AHA1 and AHA2 make contacts to different components of the basal transcription complex. For the unknown genes, whose expression is required for growth and survival under nonstress conditions, contacts of the AHA1 motif of HsfA2 are particularly important for assembly of the transcription complex. In this function, it cannot be replaced by AHA2. Pull-down experiments with different HsfA2 forms may help us to identify components of the basal transcription complex responsible for this specificity and to find out whether these differences are also relevant for plants.
In line with these arguments, Drysdale et al. (1998) investigated the role of the seven clusters of aromatic/hydrophobic amino acid residues in the N-terminal activation domain of yeast Gcn4p. However, they found for all of them very similar capacity for interaction with different TBP-associated factors (TAFs), Srb proteins, or components of the Adap/Gcn5p complex in vitro. By contrast, the cAMP-regulated transcription factor CREB has two totally different activator motifs: (1) a Q-rich motif with critical hydrophobic residues interacting with hTAF135/dTAF110, and (2) the kinase inducible domain, which after phosphorylation is able to form an amphipathic helix with a hydrophobic surface binding to CBP (Nakajima et al., 1997; Parker et al., 1998; Felinski and Quinn, 1999).
The second characteristic is connected with the function of the AHA2 motif in HsfA1. It is evidently active in its native context but not if it is placed in the AHA1 position of HsfA2 (Figure 2, constructs 20 to 22). The reason is the proline residue immediately adjacent to the aromatic/hydrophobic amino acid clusters. A P→A substitution restores activity, and this gain-of-function effect is also observed if the same constructs are tested in the yeast survival assay (Figure 4, constructs 18 to 20). Although the AHA2 motif is always active in the different forms of HsfA1 with various internal and C-terminal deletions, their activity can be increased by P→A exchange (see Figure 3B). In combination with our observations about the negative effects of proline residues in the HsfA2ΔC323 background (Figure 2E, constructs 15 to 17), we hypothesize that the potential for a helical conformation may be essential for the protein contacts between AHA motifs and components of the basal transcription complex but that, depending on the molecular context, proline residues in activation domains also contribute to an open, flexible conformation that can easily adapt to the appropriate surface of the target protein in the basal transcription complex.
In view of the expected complexity of protein interactions required for a regulated assembly of a highly productive heat stress transcription complex, the identification and mutational analysis of the central activator modules in the CTADs of tomato HsfA1 and HsfA2 are only the starting point for identification of the putative target proteins of the transcription complex and for a detailed characterization of the intriguing interactions between different HSFs in the course of the plant heat stress response.
METHODS
General Materials and Methods
Yeast strains with disruption of the heat stress transcription factor HSF1 gene and the procedure for functional replacement of the yeast Hsf1 by tomato heat stress transcription factors were described (Boscheinen et al., 1997). Antisera against HsfA1 and HsfA2, the use of tobacco (Nicotiana plumbaginifolia) mesophyll protoplasts for transient expression, and functional characterization of HSFs as well as the corresponding plant expression vectors have been described previously (Treuter et al., 1993; Lyck et al., 1997; Scharf et al., 1998b). Measurement of the β-glucuronidase (GUS) reporter activity is based on the method described by Jefferson (1987), with several modifications. Protoplasts were harvested by centrifugation, then resuspended in 50 μL of GUS extraction buffer (Jefferson, 1987), and immediately frozen in liquid nitrogen. For measurement of GUS activity, samples were thawed, and 25 μL of the lysate plus 25 μL of 1 mM methylumbelliferyl glucuronide (MUG) solution were incubated for 1 to 3 hr at 37°C in a microtiter plate. Fluorescence was measured in a FLUOstar microtiter plate reader (BMG LabTechnologies GmbH, Offenburg, Germany ). All GUS activity values result from the mean of at least three independent transformations. Error bars in Figures 1 to 3 and 5 indicate the absolute deviation from the mean values. The relative GUS activity of 10,000 relative fluorescence units (RFU) represents the cleavage of 5 pmol of MUG in 1 min by an aliquot of cell extract corresponding to 8000 protoplasts.
Plant Expression Vectors
Plant expression vectors were derivatives of pRT101 (Töpfer et al., 1988). For convenient deletion or combination of functional parts of HsfA1 and HsfA2, we introduced unique SalI (-GTC GAC-) sites in different regions of the cDNA (Figures 1 and 3) by using site-directed mutagenesis (Kunkel, 1985). Introduction of the SalI sites created a series of VD mutants, which in transactivation assays were shown to have wild-type activity (data not shown). The C-terminal deletion forms resulting from exonuclease III digestion were described previously (Treuter et al., 1993).
The functional dissection of the AHA motif was based on the C-terminally truncated version of HsfA2ΔC323, which lacks the AHA2 motif. Two additional SalI sites were introduced immediately downstream of the codons for the central DDIWEELL motif. The three HsfA2ΔC323 variants (HsfA2.1, HsfA2.2, and HsfA2.3) were used to generate the whole set of AHA1 mutants by inserting the appropriate synthetic oligonucleotides or by polymerase chain reaction–based mutagenesis.
For the generation of pBDGal4 (amino acid residues 1 to 147)–HsfA2 C-terminal activation domain (CTAD; amino acid residues 236 to 351) fusion constructs, polymerase chain reaction fragments with 5′ XhoI and 3′ NotI sites were generated from the corresponding pRTHsfA2 derivatives and inserted between SalI and NotI sites of pBDGal4/pBI221ΔGUS (kindly provided by E. Czarnecka-Verner, Department of Microbiology and Cell Sciences, University of Florida, Gainesville).
Plant Reporter Constructs
For the transient reporter assay in tobacco mesophyll protoplasts, we used the Hsf-dependent reporter phsp17GUS (Schöffl et al., 1989), which contains the promoter region (base pairs −321 to −12) from the soybean hsp17.3B gene fused to the minimal 35S cauliflower mosaic virus promoter upstream of the GUS gene (Treuter et al., 1993). For tests with the pBDGal4–HsfA2 CTAD fusion proteins, the reporter was pGal4DBS–GUS (kindly provided by E. Czarnecka-Verner), which contains 10 copies of the GAL4 promoter 17-mer binding site (Ma et al., 1988) inserted upstream of the minimal 35S cauliflower mosaic virus promoter.
Yeast Expression Vectors
For generation of the yeast Hsf1 expression plasmids (Figure 4), the cassette with the alcohol dehydrogenase (ADH1) promoter/terminator from the 2μ pAD5Δ vector (Ballester et al., 1989; Boscheinen et al., 1997) was introduced into the Cen/Ars vector pRS414 (Stratagene). The new vector (pMBI) was cut with SalI and AvrII to allow insertion of the XhoI-XbaI fragments from the corresponding pRTHsfA2 constructs.
The series of fusion constructs of the HsfA2 CTAD (amino acid residues 98 to 351) and the Gal4p DNA binding domain (amino acid residues 1 to 147) shown in Figure 5 were created by polymerase chain reaction amplification of the corresponding HsfA2 fragment with primers generating a 5′ XhoI and a 3′ PstI site, respectively. The XhoI/PstI fragments were inserted between SalI and PstI of the pBDGal4 vector (Stratagene).
Yeast Reporter Assays
The Saccharomyces cerevisiae YRG2 strain (Stratagene) carries two chromosomal reporters (HIS3 and lacZ genes) under control of Gal4p-inducible promoters. After transformation with the yeast expression vectors coding for the pBDGal4–HsfA2 CTAD fusion proteins, the transactivation potential was determined either by measuring the β-galactosidase activity or evaluating growth on histidine-free media in the presence of the indicated concentrations of 3-amino-1,2,4-triazol (3-AT).
NOTE ADDED IN PROOF
As expected, the residual activity of the AHA motifs of HsfA1 forms with W→A substitution (Figure 3, constructs 1c, 4b, and 6a) could be abolished by also replacing the adjacent hydrophobic/aromatic amino acid residues, that is, for AHA1: W452, L456, L457→AAA and for AHA2: F470, W471 F474, L475→AAAA.
Acknowledgments
We thank Gisela Englich for excellent technical assistance and Kapil Bharti, Karsten Melcher, Joachim Klein, and Klaus-Dieter Scharf for many helpful discussions and comments during preparation of the manuscript. An essential part of the experimental work was performed by E.T. while a co-worker at the Institute of Plant Biochemistry (Halle, Germany). This work was supported by grants from the Deutsche Forschungsgemeinschaft to L.N. (Grant No. SFB 474) and from the Fonds der Chemischen Industrie.
References
- Ansari, A.Z., Reece, R.J., and Ptashne, M. (1998). A transcriptional activating region with two contrasting modes of protein interaction. Proc. Natl. Acad. Sci. USA 95 13543–13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballester, R., Michaeli, T., Ferguson, K., Xu, H.P., McCormick, F., and Wigler, M. (1989). Genetic analysis of mammalian GAP expressed in yeast. Cell 59 681–686. [DOI] [PubMed] [Google Scholar]
- Barlev, N.A., Candau, R., Wang, L., Darpino, P., Silverman, N., and Berger, S.L. (1995). Characterization of physical interaction of the putative transcriptional adapter, ADA2, with acidic activation domains and TATA-binding protein. J. Biol. Chem. 270 19337–19344. [DOI] [PubMed] [Google Scholar]
- Blair, W.S., Bogert, H.P., Madore, S.J., and Cullen, B.R. (1994). Mutational analysis of the transcription activation domain of RelA: Identification of a highly synergistic minimal acidic activation module. Mol. Cell. Biol. 14 7226–7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair, W.S., Fridell, R.A., and Cullen, B.R. (1996). Synergistic enhancement of both initiation and elongation by acidic transcription activation domains. EMBO J. 15 1658–1665. [PMC free article] [PubMed] [Google Scholar]
- Blau, J., Xiao, H., McCracken, S., O'Hare, P., Greenblatt, J., and Bentley, D. (1996). Three functional classes of transcriptional activation domains. Mol. Cell. Biol. 16 2044–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boscheinen, O., Lyck, R., Queitsch, C., Treuter, E., Zimarino, V., and Scharf, K.D. (1997). Heat stress transcription factors from tomato can functionally replace HSF1 in the yeast Saccharomyces cerevisiae. Mol. Gen. Genet. 255 322–331. [DOI] [PubMed] [Google Scholar]
- Carey, M. (1998). The enhanceosome and transcriptional synergy. Cell 92 5–8. [DOI] [PubMed] [Google Scholar]
- Chen, J.-L., Attardi, L.D., Verrijeer, C.P., Yokomori, K., and Tjian, R. (1994). Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell 79 93–105. [DOI] [PubMed] [Google Scholar]
- Chen, Y.Q., Barlev, N.A., Westergaard, O., and Jakobsen, B.K. (1993). Identification of the C-terminal activator domain in yeast heat shock factor—Independent control of transient and sustained transcriptional activity. EMBO J. 12 5007–5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlman-Wright, K., Baumann, H., McEwan, I., Almlöf, T., Wright, A., Gustafsson, J.A., and Hard, T. (1995). Structural characterization of a minimal functional transactivation domain from the human glucocorticoid receptor. Proc. Natl. Acad. Sci. USA 92 1699–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drysdale, C.M., Jackson, B.M., McVeigh, R., Klebanow, E.R., Bai, Y., Kokubo, T., Swanson, M., Nakatani, Y., Weil, P.A., and Hinnebusch, A.G. (1998). The Gcn4p activation domain interacts specifically in vitro with RNA polymerase II holoenzyme, TFIID and the Adap, Gcn5p coactivator complex. Mol. Cell. Biol. 18 1711–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellwood, K., Huang, W., Johnson, R., and Carey, M. (1999). Multiple layers of cooperativity regulate enhanceosome-responsive RNA polymerase II transcription complex assembly. Mol. Cell. Biol. 19 2613–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felinski, E.A., and Quinn, P.G. (1999). The CREB constitutive activation domain interacts with TATA-binding protein-associated factor 110 (TAF110) through specific hydrophobic residues in one of the three subdomains required for both activation and TAF110 binding. J. Biol. Chem. 274 11672–11678. [DOI] [PubMed] [Google Scholar]
- Geisberg, J.V., Lee, W.S., Berk, A.J., and Ricciardi, R.P. (1994). The zinc finger region of the adenovirus E1A transactivating domain complex with the TATA box binding protein. Proc. Natl. Acad. Sci. USA 91 2488–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill, G., Pascal, E., Tseng, Z.H., and Tjian, R. (1994). A glutamine-rich hydrophobic patch in transcription factor SP1 contacts the dTAF(II)110 component of the Drosophila TFIID complex and mediates transcriptional activation. Proc. Natl. Acad. Sci. USA 91 192–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich, J.A., Cutler, G., and Tjian, R. (1996). Contacts in context: Promoter specificity and macromolecular interactions in transcription. Cell 84 825–830. [DOI] [PubMed] [Google Scholar]
- Hahn, S. (1993). Structure and function of acidic transcription activators. Cell 72 481–483. [DOI] [PubMed] [Google Scholar]
- Hengartner, C.J., Thompson, C.M., Zhang, J., Chao, D.M., Lian, S., Koleske, A.J., Okamura, S., and Young, R.A. (1995). Association of an activator with an RNA polymerase II holoenzyme. Genes Dev. 9 897–910. [DOI] [PubMed] [Google Scholar]
- Holstege, F.C.P., Jenning, E.G., Wyrick, J.J., Lee, T.I., Hengartner, C.J., Green, M.R., Golub, T.R., Lander, E.S., and Young, R.A. (1998). Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95 717–728. [DOI] [PubMed] [Google Scholar]
- Hübel, A., and Schöffl, F. (1994). Arabidopsis heat shock factor: Isolation and characterization of the gene and the recombinant protein. Plant Mol. Biol. 26 353–362. [DOI] [PubMed] [Google Scholar]
- Jackson, B.M., Drysdale, C.M., Natarajan, K., and Hinnebusch, A.G. (1996). Identification of seven hydrophobic clusters in Gcn4 making redundant contributions to transcriptional activation. Mol. Cell. Biol. 16 5557–5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson, R.A. (1987). Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol. Biol. Rep. 5 387–405. [Google Scholar]
- Kadonaga, J.T. (1998). Eukaryotic transcription: An interlaced network of transcription factors and chromatin-modifying machines. Cell 92 307–313. [DOI] [PubMed] [Google Scholar]
- Kunkel, T.A. (1985). Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. USA 82 488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Künzler, M., Braus, G.H., Georgiev, O., Seipel, R., and Schaffner, W. (1994). Functional differences between mammalian transcription activation domains at the yeast GAL1 promoter. EMBO J. 13 641–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussie, P.H., Gorina, S., Marechal, V., Elenbaas, B., Moreau, J., Levine, A.J., and Pavletich, N.P. (1996). Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science 274 948–953. [DOI] [PubMed] [Google Scholar]
- Leuther, K.K., Salmeron, J.M., and Johnston, S.A. (1993). Genetic evidence that an activation domain of GAL4 does not require acidity and may form a β-sheet. Cell 72 575–585. [DOI] [PubMed] [Google Scholar]
- Lin, J., Chen, J., Elenbaas, B., and Levine, A.J. (1994). Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to MDM2 and the adenovirus 5 E1B 55-kD protein. Genes Dev. 8 1235–1246. [DOI] [PubMed] [Google Scholar]
- Lyck, R., Harmening, U., Höhfeld, I., Scharf, K.D., and Nover, L. (1997). Intracellular distribution and identification of the nuclear localization signals of two tomato heat stress transcription factors. Planta 202 117–125. [DOI] [PubMed] [Google Scholar]
- Ma, J., Przibilla, E., Hu, J., Bogorad, L., and Ptashne, M. (1988). Yeast activators stimulate plant expression. Nature 334 631–633. [DOI] [PubMed] [Google Scholar]
- Mason, P.B., and Lis, J.T. (1997). Cooperative and competitive protein interactions at the hsp70 promoter. J. Biol. Chem. 272 33227–33233. [DOI] [PubMed] [Google Scholar]
- Massari, M.E., Jennings, P.A., and Murre, C. (1996). The AD1 transactivation domain of E2A contains a highly conserved helix which is required for its activity in both Saccharomyces cerevisiae and mammalian cells. Mol. Cell. Biol. 16 121–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan, I.J., Dahlman-Wright, K., Ford, J., and Wright, A.P.H. (1996). Functional interaction of the c-Myc activation domain with the TATA binding protein: Evidence for an induced fit model of transactivation domain. Biochemistry 35 9584–9593. [DOI] [PubMed] [Google Scholar]
- Melcher, K., and Johnston, S.A. (1995). Gal4 interacts with TATA-binding protein and coactivators. Mol. Cell. Biol. 15 2839–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molloy, D.P., Smith, K.J., Milner, A.E., and Gallimore, P.H. (1999). The structure of the site on adenovirus early region 1A responsible for binding to TATA-binding protein determined by NMR spectroscopy. J. Biol. Chem. 274 3503–3512. [DOI] [PubMed] [Google Scholar]
- Morimoto, R.I. (1998). Regulation of the heat shock transcriptional response: Cross talk between family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 12 3788–3796. [DOI] [PubMed] [Google Scholar]
- Nakajima, T., Uchida, C., Anderson, S.F., Parvi, J.D., and Montminy, M. (1997). Analysis of a cAMP-responsive activator reveals a two-component mechanism for transcriptional induction via signal-dependent factors. Genes Dev. 11 738–747. [DOI] [PubMed] [Google Scholar]
- Newton, E.M., Knauf, U., Green, M., and Kingston, R.E. (1996). The regulatory domain of human heat shock factor 1 is sufficient to sense heat stress. Mol. Cell. Biol. 16 839–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover, L. (1987). Expression of heat shock genes in homologous and heterologous systems. Enzyme Microb. Technol. 9 130–144. [Google Scholar]
- Nover, L., and Scharf, K.-D. (1997). Heat stress proteins and transcription factors. Cell. Mol. Life Sci. 53 80–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover, L., Scharf, K.D., Gagliardi, D., Vergne, P., Czarnecka-Verner, E., and Gurley, W.B. (1996). The Hsf world: Classification and properties of plant heat stress transcription factors. Cell Stress Chap. 1 215–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, D., Jhala, U.S., Radhakrishnan, I., Yaffe, M.B., Reyes, C., Shulman, A.I., Cantley, L.C., Wright, P.E., and Montminy, M. (1998). Analysis of an activator:coactivator complex reveals an essential role for secondary structure in transcriptional activation. Mol. Cell 2 353–359. [DOI] [PubMed] [Google Scholar]
- Pelham, H.R.B. (1982). A regulatory upstream promoter element in the Drosophila hsp70 heat-shock gene. Cell 30 517–528. [DOI] [PubMed] [Google Scholar]
- Pelham, H.R.B., and Bienz, M. (1982). A synthetic heat-shock promoter element confers heat-inducibility on the Herpes simplex virus thymidine kinase gene. EMBO J. 1 1473–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponticelli, A.S., Pardee, T.Z.S., and Struhl, K. (1995). The glutamine-rich activation domains of human Sp1 do not stimulate transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 15 983–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prandl, R., Hinderhofer, K., Eggers-Schumacher, G., and Schöffl, F. (1998). Hsf3, a new heat shock factor from Arabidopsis thaliana, derepresses the heat shock response and confers thermotolerance when overexpressed in transgenic plants. Mol. Gen. Genet. 258 269–278. [DOI] [PubMed] [Google Scholar]
- Ptashne, M. (1988). How eukaryotic transcriptional activators work. Nature 335 683–689. [DOI] [PubMed] [Google Scholar]
- Ptashne, M., and Gann, A. (1997). Transcriptional activation by recruitment. Nature 386 569–577. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan, I., Perez-Alvarado, G.C., Parker, D., Dyson, H.J., Montminy, M.R., and Wright, P.E. (1997). Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: A model for activator:coactivator interactions. Cell 91 741–752. [DOI] [PubMed] [Google Scholar]
- Regier, J.L., Shen, F., and Triezenberg, S.J. (1993). Pattern of aromatic and hydrophobic amino acids critical for one of two subdomains of the VP16 transcriptional activator. Proc. Natl. Acad. Sci. USA 90 883–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, S.G.E., and Green, M.R. (1994). Activator-induced conformational change in general transcription factor TFIIB. Nature 371 717–720. [DOI] [PubMed] [Google Scholar]
- Satyal, S.H., Chen, D., Fox, S.G., Kramer, J.M., and Morimoto, R.I. (1998). Negative regulation of the heat shock transcriptional response by HSBP1. Genes Dev. 12 1962–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer, F., and Tjian, R. (1997). Mechanisms of transcriptional activation: Differences and similarities between yeast, Drosophila and man. Curr. Opin. Genet. Dev. 7 176–181. [DOI] [PubMed] [Google Scholar]
- Scharf, K.-D., Rose, S., Zott, W., Schöffl, F., and Nover, L. (1990). Three tomato genes code for heat stress transcription factors with a region of remarkable homology to the DNA-binding domain of the yeast HSF. EMBO J. 9 4495–4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf, K.-D., Höhfeld, I., and Nover, L. (1998. a). Heat stress response and heat stress transcription factors. J. Biosci. 23 313–329. [Google Scholar]
- Scharf, K.-D., Heider, H., Höhfeld, I., Lyck, R., Schmidt, E., and Nover, L. (1998. b). The tomato Hsf system: HsfA2 needs interaction with HsfA1 for efficient nuclear import and may be localized in cytoplasmic heat stress granules. Mol. Cell. Biol. 18 2240–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz, M.L., dos Santo Silva, M.A., Altman, H., Czisch, M., Holak, T.A., and Baeuerle, P.A. (1994). Structural and functional analysis of the NFκB p65 C terminus. An acidic and modular transactivation domain with the potential to adapt an alpha helical conformation. J. Biol. Chem. 269 25613–25620. [PubMed] [Google Scholar]
- Schöffl, F., Rieping, M., Baumann, G., Bevan, M.W., and Angermüller, S. (1989). The function of heat shock promoter elements in the regulated expression of chimaeric genes in transgenic tobacco. Mol. Gen. Genet. 217 246–253. [DOI] [PubMed] [Google Scholar]
- Seipel, K., Georgiev, O., and Schaffner, W. (1992). Different activation domains stimulate transcription from remote (“enhancer”) and proximal (“promoter”) positions. EMBO J. 11 4961–4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, F., Triezenberg, S.J., Hensley, P., Porter, D., and Knutson, J.R. (1996). Transcriptional activation domain of the herpes virus protein VP16 becomes conformationally constrained upon interaction with basal transcription factors. J. Biol. Chem. 271 4827–4837. [DOI] [PubMed] [Google Scholar]
- Sorger, P.K., and Pelham, H.R.B. (1988). Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell 54 855–864. [DOI] [PubMed] [Google Scholar]
- Tansey, W.P., and Herr, W. (1997). Selective use of TBP and TFIIB revealed by a TATA-TBP-TFIIB array with altered specificity. Science 275 829–831. [DOI] [PubMed] [Google Scholar]
- Thanos, D., and Maniatis, T. (1995). Virus induction of human IFNβ gene expression requires the assembly of an enhanceosome. Cell 83 1091–1100. [DOI] [PubMed] [Google Scholar]
- Thut, C.J., Chen, J.L., Klemm, R., and Tjian, R. (1995). p53 transcriptional activation mediated by coactivators TAFII 40 and TAFII 60. Science 267 100–104. [DOI] [PubMed] [Google Scholar]
- Tjian, R., and Maniatis, T. (1994). Transcriptional activation—A complex puzzle with few easy pieces. Cell 77 5–8. [DOI] [PubMed] [Google Scholar]
- Töpfer, R., Schell, J., and Steinbiss, H.H. (1988). Versatile cloning vectors for transient gene expression and direct gene transfer in plant cells. Nucleic Acids Res. 16 8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treuter, E., Nover, L., Ohme, K., and Scharf, K.-D. (1993). Promoter specificity and deletion analysis of three heat stress transcription factors of tomato. Mol. Gen. Genet. 240 113–125. [DOI] [PubMed] [Google Scholar]
- Triezenberg, S.J. (1995). Structure and function of transcriptional activation domains. Curr. Opin. Genet. Dev. 5 190–196. [DOI] [PubMed] [Google Scholar]
- Uesugi, M., Nyanguile, O., Lu, H., Levine, A.J., and Verdine, G.L. (1997). Induced α-helix in the VP16 activation domain upon binding to a human TAF. Science 277 1310–1313. [DOI] [PubMed] [Google Scholar]
- Wiederrecht, G., Seto, D., and Parker, C.S. (1988). Isolation of the gene encoding the S. cerevisiae heat shock transcription factor. Cell 54 841–853. [DOI] [PubMed] [Google Scholar]
- Wisniewski, J., Orosz, A., Allada, R., and Wu, C. (1996). The C-terminal region of Drosophila heat shock factor (Hsf) contains a constitutively functional transactivation domain. Nucleic Acids Res. 24 367–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C. (1995). Heat stress transcription factors. Annu. Rev. Cell Biol. 11 441–469. [DOI] [PubMed] [Google Scholar]
- Xiao, H., Friesen, J.D., and Lis, J.T. (1994). A highly conserved domain of RNA polymerase II shares a functional element with acidic activation domains of upstream transcription factors. Mol. Cell. Biol. 14 7507–7516. [DOI] [PMC free article] [PubMed] [Google Scholar]