Abstract
In many organisms, including plants, nucleic acid bases and derivatives such as caffeine are transported across the plasma membrane. Cytokinins, important hormones structurally related to adenine, are produced mainly in root apices, from where they are translocated to shoots to control a multitude of physiological processes. Complementation of a yeast mutant deficient in adenine uptake (fcy2) with an Arabidopsis cDNA expression library enabled the identification of a gene, AtPUP1 (for Arabidopsis thaliana purine permease1), belonging to a large gene family (AtPUP1 to AtPUP15) encoding a new class of small, integral membrane proteins. AtPUP1 transports adenine and cytosine with high affinity. Uptake is energy dependent, occurs against a concentration gradient, and is sensitive to protonophores, potentially indicating secondary active transport. Competition studies show that purine derivatives (e.g., hypoxanthine), phytohormones (e.g., zeatin and kinetin), and alkaloids (e.g., caffeine) are potent inhibitors of adenine and cytosine uptake. Inhibition by cytokinins is competitive (competitive inhibition constant 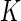 i =
i = 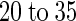 μ
μ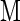 ), indicating that cytokinins are transported by this system. AtPUP1 is expressed in all organs except roots, indicating that the gene encodes an uptake system for root-derived nucleic acid base derivatives in shoots or that it exports nucleic acid base analogs from shoots by way of the phloem. The other family members may have different affinities for nucleic acid bases, perhaps functioning as transporters for nucleosides, nucleotides, and their derivatives.
), indicating that cytokinins are transported by this system. AtPUP1 is expressed in all organs except roots, indicating that the gene encodes an uptake system for root-derived nucleic acid base derivatives in shoots or that it exports nucleic acid base analogs from shoots by way of the phloem. The other family members may have different affinities for nucleic acid bases, perhaps functioning as transporters for nucleosides, nucleotides, and their derivatives.
INTRODUCTION
Nucleic acid bases are essential for a wide spectrum of metabolic processes, not the least of which is nucleic acid synthesis. Derivatives of nucleic acid bases and nucleotides play potentially important roles in energization, cell division, senescence, and defense reactions. Alkaloids, such as theobromine, caffeine, and nicotine, are structurally closely related to nucleic acid bases. Other important purine derivatives are cytokinins, which serve as hormones that control many processes in the plant (Chen et al., 1985; Chen, 1997).
Many examples of nucleic acid base and nucleoside uptake in plants are known, but the respective transporter genes have not been identified. Specific transport systems for uracil and guanine have been described in Chlorella fusca (Knutsen, 1972; Pettersen and Knutsen, 1974). Uptake of adenine by cell cultures of Acer pseudoplantanus and of uridine by Lemna gibba have also been demonstrated (Doreé, 1973; Nakashima and Tsuzuki, 1976).
Adenosine, guanosine, cytidine, and uridine are taken up against a concentration gradient into petunia pollen (Kamboj and Jackson, 1984, 1985, 1987). In contrast, uptake of thymidine in the same system occurs by facilitated diffusion at much lower rates. These data are consistent with a role for nucleosides in germinating pollen—mainly in RNA synthesis and DNA repair—which is consistent with the supply of nucleoside precursors from the carpel after pollen germination (van der Donk, 1974; Jackson and Linskens, 1978, 1980).
During germination, storage reserves from the endosperm are metabolized and translocated into the developing seedling. Besides secretion of sucrose and amino acids, adenine, adenosine, and guanosine are also exported from isolated endosperm tissue into the medium. Ricinus communis cotyledons separated from the endosperm take up endosperm-derived secretion products, including purine and pyrimidine bases, nucleosides, and AMP, with high efficiency, but not ATP (Kombrink and Beevers, 1983). Uptake of adenine may also play an important role for ATP synthesis. In seeds, ATP accumulates soon after hydration, a result of the conversion of adenine to AMP by adenine phosphoribosyltransferase (Moreland et al., 1974; Lee and Moffat, 1994). Exogenous adenine is readily taken up and converted into AMP and ATP (Lee and Moffat, 1994). Thus, efficient adenine uptake from the endosperm may be essential for supplying the germinating seedling with sufficient ATP during early stages of development.
Purine-related alkaloids, such as caffeine, are translocated in the plant and found as constituents of xylem sap (Mazzafera and Gonçalves, 1999). Also, as shown by grafting experiments, nicotine is produced in tobacco roots and then is transported to leaves (Dawson, 1942). Therefore, transport systems for alkaloids must be present in plants.
External application of cytokinins leads to turnover inside plant cells, indicating the presence of import mechanisms (Fusseder et al., 1989). Roots are considered to serve as the principal sites of cytokinin production, whereas the shoot depends on importing these hormones by way of the transpiration stream (Letham and Palni, 1983; Horgan, 1992). In addition, reflux of cytokinins from shoot to root through the phloem has been observed (Weiler and Ziegler, 1981).
Compared with the biosynthesis of nucleic acid bases and their derivatives, little is known about the molecular basis of transport mechanisms in eukaryotes. Only in bacteria and fungi have carrier genes for nucleic acid bases been identified. The Escherichia coli PurP is responsible for energized high-affinity adenine uptake (Burton, 1994). Bacterial transporters are related to the Emericella nidulans UapA purine permease and the UapC uric acid–xanthine permease (Gorfinkiel et al., 1993; Diallinas et al., 1995, 1998). Homologs of this family also have been identified in mammals, but their function has not been demonstrated (Faaland et al., 1998). Also related to this family is a plant membrane protein, leaf permease1 (LPE1), which seems to be involved in chloroplast function (Schultes et al., 1996). Again, a function in nucleic acid base transport has not been demonstrated. Thus far, the best-studied systems for nucleic acid base transport are the yeast ScFCY2 purine–cytosine permease, which mediates proton-coupled uptake of adenine, hypoxanthine, guanine, and cytosine (Weber et al., 1990; Bloch et al., 1992; Brethes et al., 1992; Pinson et al., 1996), and the yeast uracil permease FUR4 (Jund et al., 1988; Galan et al., 1996; Marchal et al., 1998).
As a first step toward identifying transporters for nucleic acid bases and their derivatives, such as cytokinins or secondary metabolites, the purine–cytosine transport—deficient yeast mutant fcy2 was used to clone plant transporter genes by functional complementation. This approach led to identification and characterization of a new class of polytopic membrane proteins that mediate transport of nucleic acid bases and their derivatives.
RESULTS
Cloning of Putative Adenine/Cytokinin Transporters
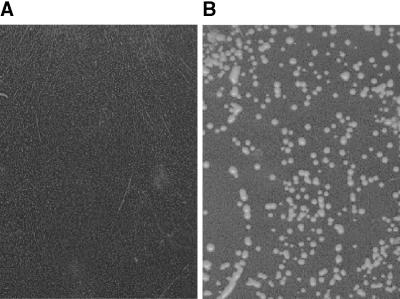
The yeast fcy2 mutant cannot grow on media containing adenine or cytosine as the sole nitrogen source because of the lack of a common uptake system (Polak and Grenson, 1973). To create uracil auxotrophy, we introduced a deletion into the URA3 gene in the fcy2 mutant MG887 by gene replacement. Heterologous complementation of the resulting strain MG887-1 with a cDNA library from Arabidopsis and subsequent selection on adenine-containing medium led to the identification of two independent cDNA clones mediating growth on medium containing adenine as the sole nitrogen source (Figure 1). Retransformation of the mutant with the isolated plasmids demonstrated that no mutation or reversion at a second site was responsible for suppression. DNA sequence analysis showed that both cDNAs encode the same gene product.
Figure 1.
Functional Complementation of MG887-1 by AtPUP1 (in pDR195).
Growth was on minimal medium containing 7.4 mM adenine as the sole nitrogen source.
(A) MG887-1pDR195 is the fcy2 mutant transformed with pDR195, which served as the control.
(B) MG887-1pAtPUP1 is the fcy2 mutant transformed with AtPUP1 in pDR195.
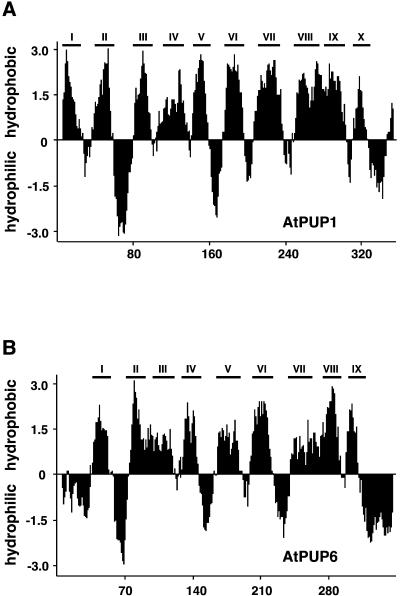
The longer clone (1251 bp), designated AtPUP1 (for Arabidopsis thaliana purine permease1), was sequenced completely. AtPUP1 contains an open reading frame (ORF) of 1068 bp encoding a protein of 356 amino acids with a calculated molecular mass of 39 kD. Hydrophobicity analyses (Figure 2A), performed with THMM1.0 (Sonnhammer et al., 1998), predicted 10 putative membrane-spanning domains, demonstrating that AtPUP1 is a member of a new class of small, highly hydrophobic membrane proteins. In database searches, no similarities were found to other known transporters, including nucleic acid base transporters from other organisms. However, 14 putative ORFs derived from the Arabidopsis genome project, which have 16.9 to 64.0% amino acid identity to AtPUP1, were identified (GenBank accession numbers U78721, AL021713, AC004135, AB010072, AC000132, AC005967, AC006434, AC007635, and AC007519). Even for the most distant members of the family, alignments show clearly related regions (data not shown).
Figure 2.
Prediction of Putative Membrane-Spanning Domains of AtPUP1 and AtPUP6.
Hydropathy plots were performed with a window of 11 amino acids (Kyte and Doolittle, 1982). Predicted membrane-spanning domains were confirmed by using THMM1.0 (Sonnhammer et al., 1998) and are marked as bars designated I to X (A) and I to IX (B).
(A) AtPUP1.
(B) AtPUP6.
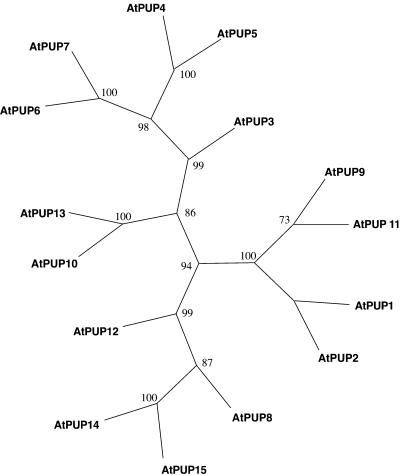
The AtPUP genes are distributed over at least four different chromosomes (Table 1). The bacterial artificial chromosome clone AL021713 on chromosome 4 contains a repeat of five closely related PUP genes, indicating recent gene amplifications. In addition, a pseudogene sharing 90% identity to AtPUP5 is located between the ORFs encoding AtPUP5 and AtPUP6. The predicted PUP proteins have hydrophobicity patterns highly similar to that of AtPUP1; however, AtPUP6 and AtPUP7 lack the first putative N-terminal membrane-spanning domain, potentially indicating that they are targeted to different subcellular compartments (Figure 2B). Only AtPUP genes 2, 6, 7, 14, and 15 contain a single intron within the coding sequence, whereas the paralogs lack introns (Table 1). A phylogenetic tree based on a comparison of the 15 PUP-like sequences is depicted in Figure 3. Related sequences were found in rhododendron (GenBank accession number AF022896), tomato (GenBank accession numbers AI488700 and AI780992), cotton (GenBank accession number AI729914), and rice (GenBank accession numbers C99477, AU30775, and D46617).
Table 1.
Structural Features of PUP Transporter Genes and Proteins
| Gene | Chromosome | Intron (Contained) | Lengtha/No. of Hydrophobic Domains | % Identity to AtPUP1 | Molecular Massb (kD) |
|---|---|---|---|---|---|
| AtPUP1 | NDc | ND | 357/10 | 100 | 39.2 |
| AtPUP2 | 2 | yes | 358/10 | 64.0 | 38.1 |
| AtPUP3 | 4 | no | 373/10 | 29.5 | 41.2 |
| AtPUP4 | 4 | no | 419/10 | 30.6 | 46.3 |
| AtPUP5 | 4 | no | 387/10 | 27.5 | 42.2 |
| AtPUP6 | 4 | yes | 349/9 | 30.4 | 38.6 |
| AtPUP7 | 4 | yes | 345/9 | 33.0 | 37.8 |
| AtPUP8 | 1 | no | 383/10 | 16.9 | 43.2 |
| AtPUP9 | 1 | no | 382/10 | 32.3 | 42.3 |
| AtPUP10 | 5 | no | 358/10 | 26.4 | 39.6 |
| AtPUP11 | 2 | no | 361/10 | 30.9 | 39.1 |
| AtPUP12 | 1 | no | 384/10 | 16.6 | 43.7 |
| AtPUP13 | 4 | no | 373/10 | 28.2 | 41.5 |
| AtPUP14 | 1 | yes | 393/10 | 19.4 | 43.3 |
| AtPUP15 | 1 | yes | 389/10 | 19.9 | 43.3 |
Lengths are given in amino acids.
Calculated molecular mass.
ND, not determined.
Figure 3.
Tree Based on the Maximum Parsimony Analysis of the PUP Protein Sequences (Swofford, 1998).
The complete alignment was based on 465 sites, with 351 being phylogenetically informative. Percentage bootstrap values of 1000 replicates are given at each branch point.
Biochemical Properties of AtPUP1
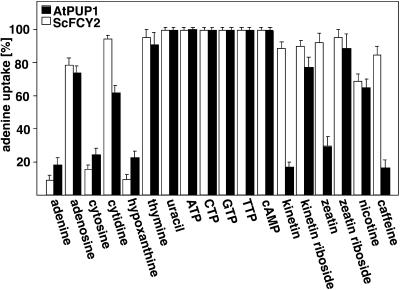
Radiotracer uptake studies performed in MG887-1 expressing AtPUP1 showed that AtPUP1 has a Km of 30 ± 5 μM for adenine. Adenine uptake is energy dependent (addition of 1% glucose to the medium led to a sevenfold increase of uptake rate; data not shown), sensitive to protonophores and to H+-ATPase inhibitors (Table 2), and increases with decreasing pH (Table 3), potentially indicating proton-coupled transport. In agreement with this hypothesis, uptake occurs against a 15-fold concentration gradient (data not shown). To study the substrate specificity of AtPUP1 in comparison with ScFCY2, we determined adenine uptake at a 10-fold molar excess of various nucleic acid bases and derivatives (Figure 4). Cytosine and hypoxanthine, but not thymine and uracil, are efficient competitors for adenine uptake. Guanine was not tested because of its low solubility.
Table 2.
Influence of Inhibitors and Protonophores on the Adenine/Cytosine Uptake Rate in MG887-1pAtPUP1a
| Inhibitors | Adenine Uptake Rate (nmol mg DW−1 min−1)b |
Cytosine Uptake Rate (nmol mg DW−1 min−1) |
|---|---|---|
| Without inhibitor | 1.33 | 0.68 |
| Diethylstilbestrol (100 μM) | 0.046 | 0.027 |
| N, N′-Dicyclohexylcarbodiimide (100 μM) | 0.75 | 0.36 |
| Carbonyl cyanide m-chlorphenyl-hydrazone (100 μM) |
0.19 | 0.12 |
| 2,4-Dinitrophenol (100 μM) | 0.55 | 0.37 |
Adenine and cytosine concentrations were 100 μM.
DW, dry weight.
Table 3.
pH Dependence of Adenine (100 μM) and Cytosine (25 μM) Uptake Rate in MG887-1pAtPUP1
| pH | Adenine Uptake Rate (nmol mg DW−1 min−1)a |
Cytosine Uptake Rate (nmol mg DW−1 min−1) |
|---|---|---|
| 4 | 1.14 | 0.35 |
| 5 | 0.76 | 0.34 |
| 6 | 0.48 | 0.14 |
| 7 | 0.24 | 0.07 |
| 8 | 0 | 0 |
DW, dry weight.
Figure 4.
Substrate Specificity of AtPUP1 (in MG887-1) and ScFCY2 (Strain 1278b).
Specificity was determined by competition for uptake of 14C-adenine (25 μM) with a 10-fold molar excess of nucleic acid bases, nucleosides, nucleotides, and derivatives. Data represent the mean of three independent experiments ±sd. Comparable results were obtained when 14C-cytosine was used as the substrate.
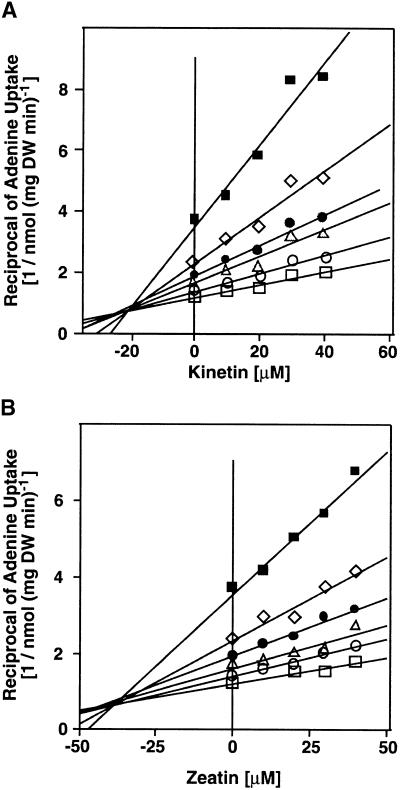
Plants produce a wide spectrum of secondary metabolites, for example, alkaloids, some of which (e.g., nicotine, theobromine, and caffeine) share structural similarities with purines. Cytokinins also share structural similarities with purines. Both nicotine and caffeine are mobile within the vascular tissue of the plant (Dawson, 1942; Mazzafera and Gonçalves, 1999). In competition assays, the order of inhibition was adenine/kinetin/caffeine ⩾ cytosine/zeatin/hypoxanthine > cytidine/nicotine > kinetin riboside/adenosine/zeatin riboside > thymine. In comparison, the nucleotides ATP, CTP, GTP, and TTP did not compete significantly for adenine uptake. Inhibition of adenine uptake by kinetin and zeatin is competitive (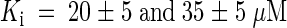 , respectively; Figures 5A and 5B), indicating that these compounds may serve as transported substrates. Cytosine also acts as a competitive inhibitor (
, respectively; Figures 5A and 5B), indicating that these compounds may serve as transported substrates. Cytosine also acts as a competitive inhibitor (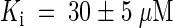 ). AtPUP1 suppresses the cytosine uptake deficiency in the yeast mutant, proving that cytosine is an actual substrate (
). AtPUP1 suppresses the cytosine uptake deficiency in the yeast mutant, proving that cytosine is an actual substrate (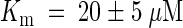 ). Nicotine, the least-related analog tested, inhibits adenine transport by ScFCY2 and AtPUP1. However, it cannot be excluded that inhibition is due to toxicity. In comparison, yeast ScFCY2 shares many similarities regarding broad specificity toward purines and pyrimidines and selectivity toward nucleosides, but plant-specific compounds, such as caffeine or cytokinins, are not recognized by the yeast protein (Figure 4).
). Nicotine, the least-related analog tested, inhibits adenine transport by ScFCY2 and AtPUP1. However, it cannot be excluded that inhibition is due to toxicity. In comparison, yeast ScFCY2 shares many similarities regarding broad specificity toward purines and pyrimidines and selectivity toward nucleosides, but plant-specific compounds, such as caffeine or cytokinins, are not recognized by the yeast protein (Figure 4).
Figure 5.
Competitive Inhibition of Adenine Transport by Kinetin and Zeatin.
Uptake of 14C-adenine (nmol adenine per mg dry weight [DW] per minute) was determined in the presence of the competitors kinetin (A) and zeatin (B) at different concentrations of adenine (5 μM, filled squares; 10 μM, diamonds; 15 μM, filled circles; 20 μM, triangles; 30 μM, circles; and 40 μM, squares). Results are shown as Dixon plots (reciprocal of uptake rate versus inhibitor concentration). Competitive inhibition mode was derived from Lineweaver–Burke plots (data not shown).
RNA and DNA Gel Blot Analyses of the Expression of Nucleic Acid Base Transporters
DNA gel blot analysis was conducted to demonstrate that AtPUP1 is actually a plant gene. At high stringency, only a single locus was detected in Arabidopsis (Figure 6A), whereas under reduced stringency, additional loci were detected, which is consistent with the presence of a quite divergent gene family (Figure 6B). The expression of AtPUP1 was analyzed under high-stringency conditions in different organs (Figure 6C). Expression of AtPUP1 was greatest in leaves, stems, and flowers. Lower expression was found in developing siliques, whereas no expression was detectable in roots.
Figure 6.
DNA and RNA Gel Blot Analyses of AtPUP1.
(A) and (B) High-stringency DNA gel blot (A) and low-stringency DNA gel blot (B), obtained by using Arabidopsis genomic DNA digested with different restriction enzymes and hybridized with the radiolabeled AtPUP1 cDNA. The positions of length markers are given at right in kilobases.
(C) RNA gel blot. RNA (30 μg) from different organs was isolated and hybridized with the radiolabeled AtPUP1 cDNA. Control hybridization with a 25S rRNA probe is shown below.
DISCUSSION
Suppression Cloning of Transporters
The major approach by which plant transporters have been cloned uses the suppression of uptake deficiencies in yeast mutants by functional expression of cDNA libraries (Frommer and Ninnemann, 1995). Detailed knowledge of the transport system itself is not a prerequisite. In fact, these systems are so sensitive they can even identify secondary activities of transporters that are irrelevant under physiological conditions. The extraordinary sensitivity of this suppression system is best shown in the case of NTR1. Originally identified as a histidine transporter because it complemented a histidine uptake–deficient yeast strain (Frommer et al., 1994), NTR1 was shown by a more detailed analysis to serve as a high-affinity oligopeptide transporter, with histidine transport representing only a physiologically irrelevant side activity (Rentsch et al., 1995). Because adenine is the basic structure for many compounds in the plant, attempts were made to complement a yeast adenine transport–deficient mutant. Such an approach should thus be ideal to clone transporters not only for adenine but also for the wide variety of purine analogs produced and transported in plants. This rationale is further supported by the finding that nucleic acid base transporters, such as the yeast protein ScFCY2, have low selectivity and accept purines, pyrimidines, and even nucleotides as substrates (Grenson, 1969; Pickering and Woods, 1972). Because carrier-mediated transport of cytokinins or alkaloids must be a new capacity developed during plant evolution, one might suspect that cytokinin transport systems have evolved from nucleic acid base transporters. Thus, suppression of an adenine uptake deficiency was first used to identify adenine transporters with the hope of also finding transporters for adenine analogs.
Identification of a Functional Plant Nucleic Acid Base Transporter
By using the yeast fcy2 mutant as a sensitive complementation system, a new family (PUP) of relatively small and highly hydrophobic membrane proteins with nine to 10 putative membrane-spanning domains able to transport adenine was identified. This family consists of at least 15 quite divergent members in Arabidopsis, with related genes being present in various other plant species (e.g., rice, tomato, and rhododendron). No PUP-related sequences were found in any other kingdom of organisms until now. Given their complementation of an uptake deficiency, one may postulate that PUP proteins are located at the plant plasma membrane. The PUP family does not share significant sequence homologies with the nucleoside permease/YSPL (animal nucleic acid base transporter)/LPE nucleic acid base transporter family found in all kingdoms (Faaland et al., 1998). The putative maize nucleic acid base transporter LPE1 might serve as an organellar permease because the effects of LPE deficiency are consistent with a plastidic localization (Schultes et al., 1996). For most transporters found in plants, homologs have been identified in animals; thus, further progress in the human genome project may lead to the identification of candidates for mammalian nucleic acid base transporters related to the PUP proteins that have been postulated based on transport studies.
The adenine transporter AtPUP1 might function as a plasma membrane proton cotransporter because uptake is energy dependent, occurs against a concentration gradient, and is stimulated by acidification and inhibited by protonophores. High-affinity uptake (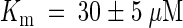 ) of adenine was shown directly for yeast expressing AtPUP1. Moreover, adenine analogs such as hypoxanthine and the pyrimidine cytosine are efficient competitors.
) of adenine was shown directly for yeast expressing AtPUP1. Moreover, adenine analogs such as hypoxanthine and the pyrimidine cytosine are efficient competitors.
The expression pattern and transport properties of AtPUP1 suggest what its possible roles within the plant might be. AtPUP1 is expressed in all tissues of the plant, except for roots, and functions in yeast as an uptake rather than an efflux system. Because cytokinin, caffeine, and nicotine are produced in roots and translocated to shoots, AtPUP1 may play a role in the uptake of these compounds from xylem sap into shoot tissues. Further studies are necessary to determine the transport properties and physiological functions of the other members of the gene family. Because adenine and zeatin can act cooperatively in flower initiation, AtPUP1 may also play a role here (Nitsch, 1968). Furthermore, nodules of tropical legumes generally export symbiotically fixed nitrogen in the form of ureides, which are produced by oxidation of de novo–synthesized purines. The greater concentration of xanthine dehydrogenase in the uninfected cells suggests that xanthine or a precursor to xanthine, rather than uric acid, is the intermediate that moves from infected to uninfected cells during ureide biogenesis (Datta et al., 1991). Thus, PUP proteins may also play a role in the intercellular transport of xanthine in nodules. A detailed analysis of all members of the PUP family regarding substrate specificity and expression is required to determine the potential in vivo function of the different paralogs.
Transport of Cytokinins and Caffeine by PUP Proteins
Besides nucleic acid bases and nucleotides, derivatives such as cytokinin and alkaloids such as caffeine or nicotine are translocated in higher plants. Nucleic acid base transport plays an important role in pollen tube growth and seed germination. Cytokinin transport affects cell division, flower induction, seed germination, de novo bud formation, and senescence.
Cytokinins inhibit adenine uptake by AtPUP1 competitively with a competitive inhibition constant Ki for kinetin of 20 ± 5 μM and for zeatin of 35 ± 5 μM. Comparable values for competitive inhibition were obtained when radiolabeled cytosine was used as a substrate. Cytosine was transported with a Km similar to the Ki. These data support the hypothesis that AtPUP1 transports cytokinins. However, they do not exclude the possibility of competitive inhibition from nontransported competitive inhibitors. Further experiments using radiolabeled cytokinins or electrophysiological measurements in Xenopus oocytes are required to prove unambiguously that AtPUP1 transports cytokinins.
However, the affinity of AtPUP1 for cytokinins as compared with concentrations in the nanomolar range for free bases, and five to 10 times greater concentrations of ribosides found in the xylem sap, may be taken as an argument against a physiological role of AtPUP1 in cytokinin transport in vivo. In yeast, a closely related member of the uracil permease family is responsible for uridine transport (Wagner and Beck, 1993). Conceivably, therefore, members of the PUP family are involved not only in nucleic acid base transport but also in nucleoside transport.
An argument for an actual function of AtPUP1 in cytokinin transport comes from the finding that the enzymes thought to be responsible for zeatin biosynthesis also have comparatively low affinities for their substrates. Adenine phosphoribosyltransferase, which converts adenine into AMP, also accepts zeatin as a substrate with an affinity of ∼3 μM (AtTAP2) (Schnorr et al., 1996). Adenine phosphoribosyltransferase mutants are male sterile and are impaired in their metabolism of cytokinins (Gaillard et al., 1998). Similarly, adenosine kinase phosphorylates adenosine as well as isopentenyladenosine. Feeding Physcomitrella spp with tritiated isopentenyladenosine also resulted in the conversion of cytokinins toward their nucleotides (von Schwartzenberg et al., 1998). Together, these results demonstrate that at least two different enzymes that accept adenine or adenosine are also involved in cytokinin metabolism in vivo, despite their seemingly low affinities relative to the actual concentrations found in the plant. Thus, the results described here do not exclude the possibility that PUP proteins play a role in cytokinin transport in vivo. The actual physiological function of AtPUP1 in cytokinin transport remains to be shown, for example, by antisense repression in transgenic Arabidopsis plants.
The purine analog caffeine, which is translocated in the xylem, has also been implicated in hormonal functions. Analysis of the substrate specificity of AtPUP1 also demonstrated that caffeine competes for adenine uptake. Interestingly, transport of caffeine across the blood–brain barrier is also mediated by a side activity of the yet to be identified mammalian adenine transport protein (McCall et al., 1982).
In summary, complementation of a yeast adenine transport mutant allowed the identification of a new superfamily of transport proteins that play potential roles in the physiologically well-characterized transport of adenine. Such transport activities are potentially important in supplying pollen, germinating seeds, or the sieve elements with adenine for maintaining high ATP concentrations. A function in the transport of nucleic acid bases and hormones is also possible. Alternatively, members of the PUP protein family may be involved in transporting other secondary metabolites, such as caffeine and nicotine, the latter being synthesized in roots and translocated to leaves.
METHODS
Yeast Strains
To introduce uracil auxotrophy into the fcy2 mutant MG887 (Mat a fcy2) (Grenson, 1969), we excised the truncated URA3 gene (Δura3) with EcoRI and SmaI from pΔura3 (see below), and we used the Δura3 fragment for transformation of MG887. A Δura3 mutant was selected on 5-fluoro-orotic acid and named MG887-1 (Mat a fcy2 ura3). The yeast strain MG887-1 was transformed with an expression library derived from Arabidopsis thaliana seedlings (Dohmen et al., 1991; library in the episomal plasmid pFL61 containing the URA3 gene [Minet et al., 1992]). Transformants were selected on yeast nitrogen base without ammonium and amino acids, supplemented with 1 mg/mL adenine (7.4 mM) or cytosine (9 mM) as sole nitrogen sources. Colonies able to grow under selective conditions were reselected in liquid medium, plasmid DNA was isolated, and plasmids were reintroduced into MG887-1. The cDNAs described were able to restore growth of the mutant on adenine-containing media. Markedly better growth after complementation was obtained when AtPUP1 was expressed under control of the PMA1 promoter in pDR195 (Rentsch et al., 1995).
Plant Material and DNA Work
Arabidopsis ecotype C24 was grown in soil in the greenhouse. Yep24 (New England Biolabs, Beverly, MA) was digested with NcoI and ApaI to delete an internal 173-bp fragment of the URA3 gene, treated with T4 polymerase, and blunt-ligated to yield pΔura3 (W.N. Fischer and W.B. Frommer, unpublished results). Both strands of AtPUP1 were sequenced with T7 polymerase (Pharmacia, Freiburg, Germany). The sequence was deposited in GenBank under accession number AF078531. DNA and RNA gel blot analyses were performed as described previously (Rentsch et al., 1996). The 25S rRNA gene served as a control (Lauter et al., 1996).
Transport Measurements
For standard uptake studies, yeast cells were harvested at 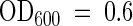 , washed, and resuspended in 100 mM potassium phosphate, pH 4.5, to a final
, washed, and resuspended in 100 mM potassium phosphate, pH 4.5, to a final 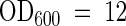 . To start the reaction, we added 100 μL of cell suspension to 100 μL of buffer containing 18.5 kBq 14C-labeled adenine or cytosine (Amersham) and unlabeled analogs as indicated. Samples of 50 μL were removed after 20, 60, 120, and 180 sec, transferred to 4 mL of ice-cold water, filtered on glass fiber filters, and washed with 8 mL of water. Uptake was linear for the time period chosen (initial uptake rates were derived from these measurements). Glucose dependence was determined in the presence or absence of 200 μM glucose. The pH dependence was determined in the presence of 1% glucose in phosphate-buffered solutions. Inhibitor sensitivity was determined under comparable conditions at substrate concentrations of 100 μM. To determine the capacity of AtPUP1 to accumulate adenine or cytosine against a concentration gradient, we measured uptake over a period of 80 min. Aliquots were removed every 10 min, and cells were centrifuged, resuspended in 200 μL of buffer, filtered on glass fiber filters, and washed. Radioactivity was determined in sediment and supernatant by liquid scintillation spectrometry (Beckman). Transport measurements were repeated independently; the reported results represent the mean of at least three experiments. The almost isogenic parental strain 1278b (Mat α) served as a control (Grenson, 1969).
. To start the reaction, we added 100 μL of cell suspension to 100 μL of buffer containing 18.5 kBq 14C-labeled adenine or cytosine (Amersham) and unlabeled analogs as indicated. Samples of 50 μL were removed after 20, 60, 120, and 180 sec, transferred to 4 mL of ice-cold water, filtered on glass fiber filters, and washed with 8 mL of water. Uptake was linear for the time period chosen (initial uptake rates were derived from these measurements). Glucose dependence was determined in the presence or absence of 200 μM glucose. The pH dependence was determined in the presence of 1% glucose in phosphate-buffered solutions. Inhibitor sensitivity was determined under comparable conditions at substrate concentrations of 100 μM. To determine the capacity of AtPUP1 to accumulate adenine or cytosine against a concentration gradient, we measured uptake over a period of 80 min. Aliquots were removed every 10 min, and cells were centrifuged, resuspended in 200 μL of buffer, filtered on glass fiber filters, and washed. Radioactivity was determined in sediment and supernatant by liquid scintillation spectrometry (Beckman). Transport measurements were repeated independently; the reported results represent the mean of at least three experiments. The almost isogenic parental strain 1278b (Mat α) served as a control (Grenson, 1969).
Acknowledgments
We are grateful to John Ward for critical reading of the manuscript. We thank Wolf-Nicolas Fischer for providing pΔura3. We gratefully acknowledge support by Deutsche Forschungsgemeinschaft (Schwerpunktprogramm “CO2 and transport” and Sonderforschungsbereich 446 B2).
References
- Bloch, J.C., Sychrova, H., Souciet, J.L., Jund, R., and Chevallier, M.R. (1992). Determination of a specific region of the purine–cytosine permease involved in the recognition of its substrates. Mol. Microbiol. 6 2989–2997. [DOI] [PubMed] [Google Scholar]
- Brethes, D., Napias, C., Torchut, E., and Chevallier, J. (1992). Purine–cytosine permease of Saccharomyces cerevisiae: Effect on external pH on nucleobase uptake and binding. Eur. J. Biochem. 210 785–791. [DOI] [PubMed] [Google Scholar]
- Burton, K. (1994). Adenine transport in Escherichia coli. Proc. R. Soc. Lond. B 225 153–157. [DOI] [PubMed] [Google Scholar]
- Chen, C.M. (1997). Cytokinin biosynthesis and interconversion. Physiol. Plant. 101 665–673. [Google Scholar]
- Chen, C.M., Ertl, J.R., Leisner, S.M., and Chang, C.C. (1985). Localization of cytokinin biosynthetic sites in pea plants and carrot roots. Plant Physiol. 78 510–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta, D.B., Triplett, E.W., and Newcomb, E.H. (1991). Localization of xanthine dehydrogenase in cowpea root nodules: Implications for the interaction between cellular compartments during ureide biogenesis. Proc. Natl. Acad. Sci. USA 88 4700–4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson, R.F. (1942). Accumulation of nicotine in reciprocal grafts of tomato and tobacco. Am. J. Bot. 29 66–71. [Google Scholar]
- Diallinas, G., Gorfinkiel, L., Arst, H.N.J., Checchetto, G., and Scazzocchio, C. (1995). Genetic and molecular characterization of a gene encoding a wide specificity purine permease of Aspergillus nidulans reveals a novel family of transporters conserved in prokaryotes and eukaryotes. J. Biol. Chem. 270 8610–8622. [DOI] [PubMed] [Google Scholar]
- Diallinas, G.J.V., Sophianopoulou, V., Rosa, A., and Scazzocchio, C. (1998). Chimeric purine transporters of Aspergillus nidulans define a domain critical for function and specificity conserved in bacterial, plant and metazoan homologues. EMBO J. 17 3827–3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen, R.J., Straffer, A.W.M., Honer, C.B., and Hollenberg, C.P. (1991). An efficient transformation procedure enabling long-term storage of competent cells of various yeast genera. Yeast 7 691–692. [DOI] [PubMed] [Google Scholar]
- Doreé, M. (1973). Étude de l'absorption de l'adénine et des adénines-N6-substituées par les cellules d'Acer pseudoplantanus. Physiol. Vég. 11 267–290. [Google Scholar]
- Faaland, C.A., Race, J.E., Ricken, G., Warner, F.J., Williams, W.J., and Holtzmann, E.J. (1998). Molecular characterization of two novel transporters from human and mouse kidney and from LLC-PK1 cells reveals a novel conserved family that is homologous to bacterial and Aspergillus nucleobase transporters. Biochim. Biophys. Acta 1442 353–360. [DOI] [PubMed] [Google Scholar]
- Frommer, W.B., and Ninnemann, O. (1995). Heterologous expression of genes in bacterial, fungal, animal, and plant cells. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46 419–444. [Google Scholar]
- Frommer, W.B., Hummel, S., and Rentsch, D. (1994). Cloning of an Arabidopsis histidine transporting protein related to nitrate and peptide transporters. FEBS Lett. 347 185–189. [DOI] [PubMed] [Google Scholar]
- Fusseder, A., Ziegler, P., Peters, W., and Beck, E. (1989). Turnover of O-glucosides of dihydrozeatin and dihydrozeatin-9-riboside during the cell growth cycle of photoautotrophic cell suspension cultures of Chenopodium rubrum. Bot. Acta 102 335–340. [Google Scholar]
- Gaillard, C., Moffatt, B.A., Blacker, M., and Laloue, M. (1998). Male sterility associated with APRT deficiency in Arabidopsis thaliana results from a mutation in the gene APT1. Mol. Gen. Genet. 257 348–353. [DOI] [PubMed] [Google Scholar]
- Galan, J.M., Moreau, V., André, B., Volland, C., and Haguenauer-Tsapis, R. (1996). Ubiquitination mediated by the Npi1p/Rsp5p ubiquitin–protein ligase is required for endocytosis of the yeast uracil permease. J. Biol. Chem. 271 10946–10952. [DOI] [PubMed] [Google Scholar]
- Gorfinkiel, L., Diallinas, G., and Scazzocchio, C. (1993). Sequence and regulation of the uapA gene encoding a uric acid–xanthine permease in the fungus Aspergillus nidulans. J. Biol. Chem. 268 23376–23381. [PubMed] [Google Scholar]
- Grenson, M. (1969). The utilization of exogenous pyrimidines and the recycling of the uridine-5′-phosphate derivatives in Saccharomyces cerevisiae, as studied by means of mutants affected in pyrimidine uptake and metabolism. Eur. J. Biochem. 11 249–260. [DOI] [PubMed] [Google Scholar]
- Horgan, R. (1992). Present and future prospects of cytokinin research. In Physiology and Biochemistry of Cytokinins in Plants, M. Kamínek, D.W.S. Mok, and E. Zazímalová, eds (The Hague: SPB Academic Publishers), pp. 3–13.
- Jackson, J.H., and Linskens, H.F. (1978). Evidence for DNA repair after ultraviolet irradiation of Petunia hybrida pollen. Mol. Gen. Genet. 161 117–120. [Google Scholar]
- Jackson, J.H., and Linskens, H.F. (1980). DNA repair in pollen: Range of mutagens inducing repair, effect of replication inhibitors and changes in thymidine nucleotide metabolism during repair. Mol. Gen. Genet. 180 517–522. [Google Scholar]
- Jund, R., Weber, E., and Chevallier, M.R. (1988). Primary structure of the uracil transport protein of Saccharomyces cerevisiae. Eur. J. Biochem. 171 417–424. [DOI] [PubMed] [Google Scholar]
- Kamboj, R.K., and Jackson, J.F. (1984). Divergent transport mechanisms for pyrimidine nucleosides in Petunia pollen. Plant Physiol. 75 499–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamboj, R.K., and Jackson, J.F. (1985). Pyrimidine nucleoside uptake by Petunia pollen. Plant Physiol. 79 801–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamboj, R.K., and Jackson, J.F. (1987). Purine nucleoside transport in Petunia pollen is an active, carrier-mediated system not sensitive to nitrobenzylthioinosine and not renewed during pollen tube growth. Plant Physiol. 84 688–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutsen, G. (1972). Uptake of uracil by synchronous Chlorella fusca. Physiol. Plant. 27 300–309. [Google Scholar]
- Kombrink, E., and Beevers, H. (1983). Transport of purine and pyrimidine bases and nucleosides from endosperm to cotyledons in germinating castor bean seedlings. Plant Physiol. 73 370–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte, J., and Doolittle, F.R. (1982). A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157 693–698. [DOI] [PubMed] [Google Scholar]
- Lauter, F., Ninnemann, O., Bucher, M., Riesmeier, J., and Frommer, W.B. (1996). Preferential expression of an ammonium transporter and of two putative nitrate transporters in root hairs of tomato. Proc. Natl. Acad. Sci. USA 93 8139–8144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, D., and Moffat, B.A. (1994). Adenine salvage activity during callus induction and plant growth. Physiol. Plant. 90 739–747. [Google Scholar]
- Letham, D.S., and Palni, L.M.S. (1983). The biosynthesis and metabolism of cytokinins. Annu. Rev. Plant Physiol. 34 163–197. [Google Scholar]
- Marchal, C., Haguenauer-Tsapis, R., and Urban-Grimal, D. (1998). A PEST-like sequence mediates phosphorylation and efficient ubiquitination of yeast uracil permease. Mol. Cell. Biol. 18 314–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzafera, P., and Gonçalves, K.V. (1999). Nitrogen compounds in the xylem sap of coffee. Phytochemistry 50 383–386. [Google Scholar]
- McCall, A.L., Millington, W.R., and Wurtman, R.J. (1982). Blood–brain barrier transport of caffeine: Dose-related restriction of adenine transport. Life Sci. 31 270–271. [DOI] [PubMed] [Google Scholar]
- Minet, M., Dufour, M.-E., and Lacroute, F. (1992). Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNA. Plant J. 2 417–422. [DOI] [PubMed] [Google Scholar]
- Moreland, D.E., Hussey, G.G., Shriner, C.R., and Farmer, F.S. (1974). Adenosine phosphates in germinating radish (Raphanus sativus L.) seeds. Plant Physiol. 54 560–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima, H., and Tsuzuki, T. (1976). Uptake of uridine by a long-day duckweed Lemna gibba G3. Plant Cell Physiol. 17 701–711. [Google Scholar]
- Nitsch, C. (1968). Effects on growth substances on the induction of flowering of a short day plant in vitro. In Biochemistry and Physiology of Plant Growth Substances, F. Wightman and G. Setterfield, eds (Ottawa, Canada: Runge), pp. 1385–1398.
- Pettersen, R., and Knutsen, G. (1974). Uptake of guanine by synchronized Chorella fusca: Characterization of the transport system in autospores. Arch. Microbiol. 96 233–246. [DOI] [PubMed] [Google Scholar]
- Pickering, W.R., and Woods, R.A. (1972). The uptake and incorporation of purines by wild-type Saccharomyces cerevisiae and a mutant resistant to 4-aminopyrazolo-(3,4-d)-pyrimidine. Biochim. Biophys. Acta 264 45–58. [DOI] [PubMed] [Google Scholar]
- Pinson, B., Pillois, X., Brethes, D., Chevallier, J., and Napias, C. (1996). In vivo phosphorylation of the purine/cytosine permease from the plasma membrane of the yeast Saccharomyces cerevisiae. Eur. J. Biochem. 239 439–444. [DOI] [PubMed] [Google Scholar]
- Polak, A., and Grenson, M. (1973). Evidence for a common transport system for cytosine, adenine and hypoxanthine in Saccharomyces cerevisiae and Candida albicans. Eur. J. Biochem. 32 276–282. [DOI] [PubMed] [Google Scholar]
- Rentsch, D., Laloi, M., Rouhara, I., Schmelzer, E., Delrot, S., and Frommer, W.B. (1995). NTR1 encodes a high affinity oligopeptide transporter in Arabidopsis. FEBS Lett. 370 264–268. [DOI] [PubMed] [Google Scholar]
- Rentsch, D., Hirner, B., Schmelzer, E., and Frommer, W.B. (1996). Salt stress–induced proline transporters and salt stress–repressed broad specificity amino acid permeases identified by suppression of a yeast amino acid permease–targeting mutant. Plant Cell 8 1437–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnorr, K.M., Gaillard, C., Biget, E., Nygaard, P., and Laloue, M. (1996). A second form of adenine phosphoribosyltransferase in Arabidopsis thaliana with relative specificity toward cytokinins. Plant J. 9 891–898. [DOI] [PubMed] [Google Scholar]
- Schultes, N.P., Brutnell, T.P., Allen, A., Dellaporta, S.L., Nelson, T., and Chen, J. (1996). Leaf permease1 gene of maize is required for chloroplast development. Plant Cell 8 463–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer, E.L.L., von Heijne, G., and Krogh, A. (1998). A hidden Markov model for predicting transmembrane helices in protein sequences. In Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology, J. Glasgow, T. Littlejohn, F. Major, R. Lathrop, D. Sankoff, and C. Sensen, eds (Menlo Park, CA: AAAI Press), pp. 175–182. [PubMed]
- Swofford, D.L. (1998). PAUP: Phylogenetic Analysis Using Parsimony (and Other Methods). (Sunderland, MA: Sinauer Associates).
- van der Donk, J.A.V.M. (1974). Synthesis of RNA and protein as a function of time and type of pollen tube–style interactions in Petunia hybrida L. Mol. Gen. Genet. 134 93–98. [Google Scholar]
- von Schwartzenberg, K., Kruse, S., Reski, R., Moffat, B., and Laloue, M. (1998). Cloning and characterization of an adenosine kinase from Physcomitrella involved in cytokinin metabolism. Plant J. 13 249–257. [DOI] [PubMed] [Google Scholar]
- Wagner, B.M., and Beck, E. (1993). Cytokinins in the perennial herb Urtica dioica L. as influenced by its nitrogen status. Planta 190 511–518. [Google Scholar]
- Weber, E., Rodriguez, C., Chevallier, M.R., and Jund, R. (1990). The purine–cytosine permease gene of Saccharomyces cerevisiae: Primary structure and deduced protein sequence of the FCY2 gene product. Mol. Microbiol. 4 585–596. [DOI] [PubMed] [Google Scholar]
- Weiler, E., and Ziegler, H. (1981). Determination of phytohormones in the phloem exudate from tree species by radioimmunoassay. Planta 152 168–170. [DOI] [PubMed] [Google Scholar]