Abstract
The SIN3 corepressor and RPD3 histone deacetylase are components of the evolutionarily conserved SIN3/RPD3 transcriptional repression complex. Here we show that the SIN3/RPD3 complex and the corepressor SMRTER are required for Drosophila G2 phase cell cycle progression. Loss of the SIN3, but not the p55, SAP18, or SAP30, component of the SIN3/RPD3 complex by RNA interference (RNAi) causes a cell cycle delay prior to initiation of mitosis. Loss of RPD3 reduces the growth rate of cells but does not cause a distinct cell cycle defect, suggesting that cells are delayed in multiple phases of the cell cycle, including G2. Thus, the role of the SIN3/RPD3 complex in G2 phase progression appears to be independent of p55, SAP18, and SAP30. SMRTER protein levels are reduced in SIN3 and RPD3 RNAi cells, and loss of SMRTER by RNAi is sufficient to cause a G2 phase delay, demonstrating that regulation of SMRTER protein levels by the SIN3/RPD3 complex is a vital component of the transcriptional repression mechanism. Loss of SIN3 does not affect global acetylation of histones H3 and H4, suggesting that the G2 phase delay is due not to global changes in genome integrity but rather to derepression of SIN3 target genes.
Posttranslational acetylation of evolutionarily conserved lysine residues within the N-terminal tails of histones has been implicated in the regulation of transcription (33). In general, histone acetylation levels are correlated with transcription levels; nucleosomes located near active genes contain hyperacetylated histones, while those located near inactive genes contain hypoacetylated histones (5, 20). Histone acetylation levels are determined by the relative activities of various histone acetyltransferases (HATs) and histone deacetylases (HDACs) that display specificity for particular lysine residues (33). Thus, targeting of an HDAC to a given promoter provides a mechanism for transcriptional repression (29, 55). Histone deacetylation may repress transcription by strengthening histone tail-DNA interactions and thereby blocking access of transcriptional regulators to the DNA template or by removing acetyl moieties on histone tails that are important for the interaction of transcriptional regulators with chromatin (17, 25, 37, 63, 67).
SIN3 and the RPD3 deacetylase are components of a multiprotein complex that represses the transcription of many eukaryotic genes (3). The SIN3/RPD3 complex does not directly bind DNA but is targeted to specific genes through protein-protein interactions between SIN3 and DNA-binding proteins or corepressors that interact with DNA-binding proteins. The mammalian SIN3/RPD3 complex (which we refer to as the SIN3/HDAC1 complex and which contains SIN3A and/or SIN3B and HDAC1 and/or HDAC2) is involved in the regulation of transcription by nuclear hormone receptors (NHRs), the Myc/Mad/Max family of transcription factors, and a variety of other transcription factors (12, 18, 21, 28, 35, 44). NHRs and Myc/Mad/Max proteins participate in both activation and repression of genes. In the absence of hormone, type II NHRs, including the thyroid hormone receptor and the retinoic acid receptor, bind their cognate DNA sequences and repress transcription (15, 47). Early studies indicated that repression is mediated by targeting of the SIN3/HDAC1 complex through association of SIN3 with the corepressors SMRT and N-CoR, which, in turn, bind unliganded NHRs (1, 21, 44, 72). The preponderance of evidence suggested a model in which conversion of NHRs from repressors to activators involved reversal of repression, by ligand-dependent dissociation of the SIN3/RPD3 complex, and recruitment of coactivator complexes that possess intrinsic HAT activity (15, 47). However, involvement of the SIN3/HDAC1 complex in transcriptional repression by unliganded NHRs has recently come into question (65). While a Xenopus N-CoR/SIN3/RPD3 complex has been purified, mammalian SIN3 and HDAC1 do not purify with endogenous SMRT-containing complexes (27). Other HDACs, including HDAC3, associate with SMRT and N-CoR complexes and have been implicated in repression by NHRs (24, 38).
Aspects of the corepressor-to-coactivator conversion model have been addressed by using a Drosophila system. Ecdysteroid hormones, such as ecdysone, control Drosophila metamorphosis by activating transcription through the Ecdysone receptor (EcR), a member of the type II NHR family (51). Drosophila SMRTER, the functional homologue of SMRT and N-CoR, binds EcR and SIN3 to mediate repression in the absence of a hormone (62). Heterozygous EcR and SIN3 mutant flies show synthetic lethality and developmental phenotypes, providing in vivo evidence for a functional link between EcR and SIN3. Furthermore, SIN3, RPD3, and SMRTER colocalize at numerous loci in Drosophila salivary gland polytene chromosomes and the level of binding of SIN3 and RPD3 to ecdysone-regulated loci decreases upon ecdysone-induced transcriptional activation and increases coincident with a reduction in transcription (48). Taken together, these findings suggest that repression of EcR-regulated genes is relieved by dissociation of the SIN3/RPD3 complex upon ecdysone binding.
Studies suggest that, in addition to affecting histone acetylation levels, dissociation of the SIN3/RPD3 complex upon gene activation affects the stability of SIN3-interacting proteins. For example, the interaction between SIN3 and the p53 tumor suppressor protein is not only important for the ability of p53 to repress transcription but is also important for protection of p53 from proteasome-mediated degradation (43, 83).
Histone acetylation has also been implicated in regulation of progression through the cell cycle (39). In yeast, proper acetylation of histones H3 and H4 is essential for progression through the G2/M phase of the cell cycle. Loss of certain HATs that preferentially acetylate histones H3 or H4 or mutation or deletion of conserved lysine residues in the N-terminal tail of histone H4 leads to arrest in the G2/M phase (22, 41, 42, 71, 79). Similarly, chemical inhibitors of HDACs have been reported to have antiproliferative effects on mammalian cells, including arrest of the cell cycle in the G1 and/or G2 phases (14, 30, 31, 36, 45, 49, 52, 58, 74, 75, 76). These observations highlight the importance of the balance of histone acetylation-deacetylation during the cell cycle. The SIN3/RPD3 deacetylase complex may participate in regulation of G2 cell cycle progression, as ecdysone treatment of Drosophila tissue culture cells causes arrest in the G2 phase of the cell cycle (4, 9, 11, 19).
Thus, in this study, we examined the cell cycle requirement for individual components of the SIN3/RPD3 complex and the corepressor SMRTER. In addition to SIN3 and RPD3, the SIN3/RPD3 complex contains p55 (also known as chromatin assembly factor 1 [CAF-1] and RbAp46/48) and SIN3-associated polypeptides 18 (SAP18) and 30 (SAP30) (3, 18, 34, 35, 80, 82). p55 is a component of numerous complexes involved in histone metabolism, including CAF-1, nucleosome-remodeling HDAC (Mi-2/NuRD), and nucleosome remodeling factor, and is thought to target these complexes to histone H4 (40, 61, 64, 66, 68, 73, 81). SAP30 directly interacts with SIN3 and RPD3, and in yeast, SAP30 mutants display many, but not all, of the phenotypes observed in SIN3 and RPD3 mutants (35, 82). Finally, in mammalian cells, SAP18 binds to SIN3 and enhances SIN3/RPD3-mediated transcriptional repression (80).
By using an RNA interference (RNAi) approach to eliminate specific proteins in Drosophila tissue culture cells, we show that progression through the G2 phase of the cell cycle requires SIN3 and SMRTER but not p55, SAP18, and SAP30, suggesting that SIN3/RPD3 complex components play distinct roles in vivo. RPD3 RNAi cells do not display a distinct cell cycle phenotype but are growth impaired, possibly reflecting roles for RPD3 at multiple points in the cell cycle, including G2 phase progression. Global histone acetylation levels are increased in RPD3 RNAi cells, but this is likely due to the activity of RPD3-containing complexes other than SIN3/RPD3, as global acetylation levels are not affected in SIN3 RNAi cells. Surprisingly, the G2 phase delay caused by loss of SIN3 or SMRTER is independent of EcR, as the delay occurs in SIN3/EcR and SMRTER/EcR double-RNAi cells. The SIN3/RPD3 complex appears to act through SMRTER to control cell cycle progression, as loss of SIN3 or RPD3 leads to a reduction in the level of SMRTER protein. This is consistent with a role for the SIN3/RPD3 complex in protecting corepressors from proteolysis.
MATERIALS AND METHODS
Cell culture.
Drosophila Schneider cell line 2 (S2) cells were cultured at 22°C in Schneider's Drosophila medium (Life Technologies) containing 10% fetal bovine serum (FBS), 100 U of penicillin per ml, and 100 μg of streptomycin per ml. For ecdysone treatment, 20-hydroxyecdysone (Sigma) was dissolved in dimethyl sulfoxide and added to cells in culture medium at a concentration of 10−6 M.
dsRNA production.
Individual DNA fragments, approximately 700 to 1,200 bp in length and containing sequences encoding the protein to be targeted by RNAi, were amplified by PCR from Drosophila melanogaster genomic DNA and cloned in both orientations into the pCRII-TOPO cloning vector by using the TOPO TA cloning kit (Invitrogen). The following primer sets (oriented 5′-3′) were used in a standard PCR: SIN3 (GAATTTGAAGACCACAACCTCG and GATGGCGATATGTCCGGCAC), RPD3 (GACCGGCACCAAAGTAAACC and CTTGGTCATCTCATCGGCAG), SMRTER (TGAACTACCTGCCACCACAC and AATGGCAACCATGGTCTGCC), SAP30 (ACATCGCGCTGTCGAAAGAA and CAGGTGGTGTCGTTGCCAAG), SAP18 (TTGATATAGTTATCGAAAAGAGC and AGTTCGTGTTACTTGTATTCCAC), p55 (TCACACCATCTGCTTGTGGG and AGATTGTACAATCTGCTGAC), STG (AACACCAGCAGTTCGAGTAG and GCATAGGCTTTGCTGAAGTC), PP1-87B (AGTACTTGGACTCGTATGG and GAGGACAGCAATCTGTCGAAG), and EcR (CTATGACCACAGCTCGGAC and TCGGTTGGGGGCGCCATTAC). Sense and antisense clones were used as templates to generate single-stranded RNA (ssRNA) with a Ribomax kit (Promega). ssRNA was resuspended in annealing buffer (5 mM KCl, 10 mM NaH2PO4), and equal quantities of sense and antisense ssRNAs were annealed by heating to 95°C for 5 min and slow cooling for 12 to 18 h to generate double-stranded RNA (dsRNA). dsRNA was stored at −80°C.
RNAi.
RNAi was carried out on the basis of the protocol of Clemens et al. (10). Briefly, 2 × 106 cells were plated into a 60-mm-diameter dish. After 1 h, FBS-containing medium was removed and replaced with 2 ml of serum-free medium. Approximately 40 μg of dsRNA (as little as 10 μg has been tested and found to be effective) was added per dish and mixed by swirling. After 30 min, 4 ml of medium containing 10% FBS, 100 U of penicillin per ml, and 100 μg of streptomycin per ml was added. Cells were assayed at a specified day following addition of dsRNA. To determine the growth curves of RNAi cells, cells were mixed and counted each day following the addition of dsRNA for a total of 4 days.
Western blotting or reverse transcription (RT)-PCR analysis was routinely carried out for both single- and double-RNAi-treated cells to evaluate the level of the targeted protein or mRNA, respectively. Representative examples are shown in Fig. 1. Western blot assays were performed as described below, and RT reactions were carried out in accordance with standard procedures by using total RNA extracted from cells with Trizol reagent (Life Technologies) (57).
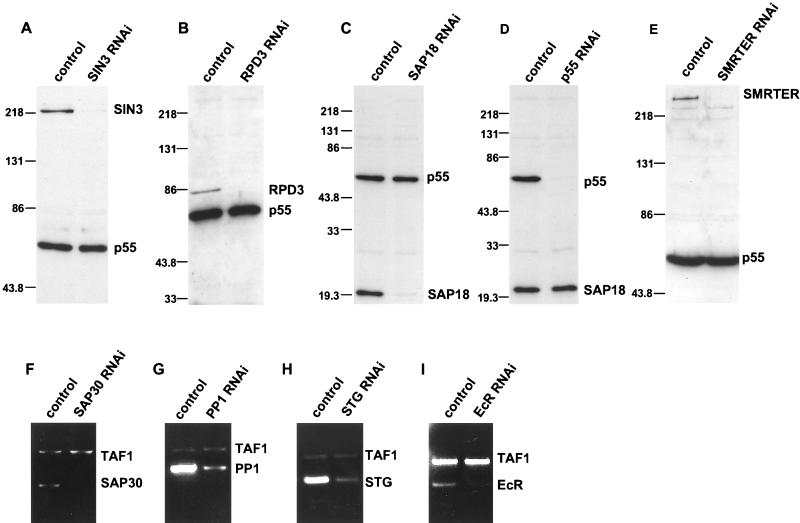
FIG. 1.
Specific genes can be targeted by RNAi. (A to E) Western blot assays of whole-cell extracts from control or RNAi cells. Each blot was probed with two antibodies, one specific for the protein targeted by RNAi, i.e., SIN3 (A), RPD3 (B), SAP18 (C), p55 (D), or SMRTER (E), and a second specific for a protein that was not targeted by RNAi, i.e., p55 (A to C and E) or SAP18 (D). Protein molecular weight markers are indicated in thousands to the left of each panel. (F to I) Ethidium bromide-stained agarose gels of RT-PCR products. Total RNA was isolated from control and RNAi cells and subjected to RT-PCR analysis. Two primer sets were used in each reaction mixture, one set for the mRNA targeted by RNAi, SAP30 (F), PP1 (G), STG (H), and EcR (I), and a second set specific for the TAF1 (formally designated TAFII250) mRNA that was not targeted by RNAi.
Western blot analysis.
Western blot analysis was performed in accordance with standard protocols (57). To prepare whole-cell extracts, cells were pelleted by centrifugation and then lysed in Laemmli sample buffer (Bio-Rad) at a concentration of 1.5 × 104 cells/μl of buffer. Protein concentration was determined with the Bio-Rad Dc protein assay reagent in accordance with the manufacturer's protocol. Extract (5 to 20 μg) was fractionated by sodium dodecyl sulfate (SDS)-8 or 10% polyacrylamide gel electrophoresis (PAGE), transferred to Immobilon P polyvinylidene difluoride membrane (Millipore), and probed with various rabbit primary antibodies, followed by donkey anti-rabbit horseradish peroxidase-conjugated immunoglobulin G (IgG; 1:3,000; Amersham Pharmacia Biotech), and detected with ECL reagents (Amersham Pharmacia Biotech). Primary antibodies included IgG-purified anti-SIN3 (1:500) and anti-RPD3 (1:500), anti-SMRTER (1:2,000; kindly provided by R. Evans), anti-SAP18 (1:2,000), and anti-p55 (1:6,000; kindly provided by R. Kamakaka) (48, 62, 64). The SAP18 polyclonal antibody was generated in rabbits by using a full-length recombinant Drosophila SAP18 protein as the antigen.
For anti-histone Western blot assays, crude whole-cell acid-soluble protein extracts were prepared as follows. Cells (2 × 107 to 5 × 107) were pelleted by centrifugation and resuspended in 1 ml of 1× phosphate-buffered saline (PBS)-10 mM sodium butyrate. Sulfuric acid was added to a final concentration of 0.4 N. Cells were incubated on ice for 30 min and then centrifuged at 12,000 × g for 10 min at 4°C. The supernatant was dialyzed against 0.1 N acetic acid for 1 to 3 h; this was followed by two changes of distilled water for 1 to 3 h and then overnight incubation at 4°C. Dialyzed supernatant was subjected to trichloroacetic acid (TCA) precipitation to isolate acid-soluble proteins. A 200-μl volume of 100% TCA was added to approximately 1 ml of dialyzed supernatant, and the solution was mixed and placed on ice for 30 min. Precipitated proteins were isolated by centrifugation at 12,000 × g for 15 min. All but 10 μl of the liquid was removed, 1 ml of ice-cold acetone was added, and the pellet in acetone was centrifuged at 12,000 × g for 2 min. The acetone was removed, and the pellet was allowed to briefly air dry. The TCA pellet was resuspended in 100 to 200 μl of Laemmli sample buffer (Bio-Rad). Protein concentration was determined by using the Bio-Rad Dc protein assay reagent. A 12-μg extract sample was separated by SDS-15% PAGE, transferred to Immobilon P polyvinylidene difluoride membrane (Millipore), probed with various rabbit primary antibodies, followed by donkey anti-rabbit horseradish peroxidase-conjugated IgG (1:3,000; Amersham Pharmacia Biotech), and detected with ECL reagents (Amersham Pharmacia Biotech). All histone antibodies were obtained from Upstate Biotechnology, including anti-phos H3 (1:2,000), anti-H3Ac9/14 (1:10,000), anti-H4Ac8 (1:600), anti-H4Ac12 (1:1,000), and anti-H4Ac5/8/12/16 (1:2,000).
FACS analysis.
To prepare cells for fluorescence-activated cell sorter (FACS) analysis, 106 cells were washed twice with 1× PBS and resuspended in 2 ml of 1× PBS-0.25% Triton X-100-4 μg of propidium iodide per ml and 10 μl of RNase A (10 mg/ml) was added. Stained cells were analyzed with a Becton Dickinson FACScan machine, and the data were analyzed with CELLQuest software.
Immunofluorescence assay.
Cells were seeded onto coverslips at a density of 5 × 105/ml and fixed in 2% formaldehyde in 1× PBS for 10 min. After a brief wash with 1× PBS, cells were blocked with 1% bovine serum albumin-0.1% Triton X-100 in 1× PBS for 30 min at 25°C, incubated with rhodamine-conjugated phalloidin antibody (1:100; Molecular Probes) in 1× PBS-0.1% Triton X-100-1% normal goat serum for 1 h at 25°C, washed three times for 10 min each time with 1× PBS, and mounted onto slides with Vectashield mounting medium (Vector Laboratories, Inc.).
RESULTS
SIN3 is required for progression through the G2 phase of the cell cycle.
To determine the physiological requirements for SIN3 in Drosophila cells, we have used RNAi methodology to reduce the level of SIN3 protein in S2 tissue culture cells (10). RNAi causes degradation of a specific mRNA, which, in turn, causes a reduction in the level of the encoded protein. In brief, RNAi was carried out by adding dsRNA, corresponding to an ∼930-nucleotide region of the SIN3 mRNA, to S2 cells and culturing the cells for 3 days prior to analysis. We refer to these cells as SIN3 RNAi cells. Western blot analysis of whole-cell extracts with an antibody directed against SIN3 showed that RNAi treatment resulted in near elimination of the SIN3 protein (Fig. 1A). In accord with an earlier report, the RNAi effect in Drosophila tissue culture cells appears to be all or none (16). Immunofluorescence staining of SIN3 RNAi cells with SIN3 antibody showed that ∼95% of the cells had nearly complete loss of expression, while ∼5% of the cells appeared to be unaffected and expressed SIN3 at or near wild-type levels. Furthermore, the RNAi effect was specific for SIN3, as Western blot analysis revealed that the level of other components of the SIN3/RPD3 complex in SIN3 RNAi cells was not reduced (Fig. 1A; see also Fig. 8; data not shown).
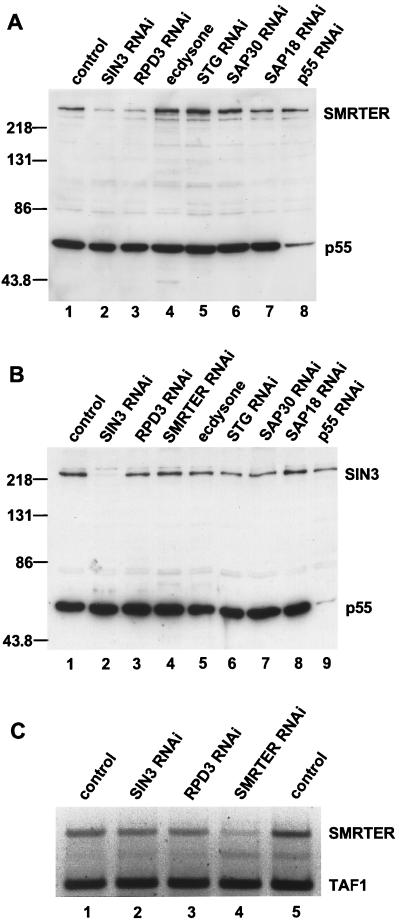
FIG. 8.
SMRTER protein, but not mRNA, levels are reduced in cells lacking SIN3 or RPD3. Treatments are indicated above the lanes. (A and B) Western blot analysis of whole-cell extracts from control and RNAi cells. (A) Western blot probed with antibodies to SMRTER and p55. (B) Western blot probed with antibodies to SIN3 and p55. Protein molecular weight markers are indicated in thousands to the left of each blot. (C) Ethidium bromide staining of agarose gels of RT-PCR products to examine SMRTER mRNA levels. As a loading control, TAF1 mRNA levels were determined for each sample. RT-PCR products are labeled on the right of the panel.
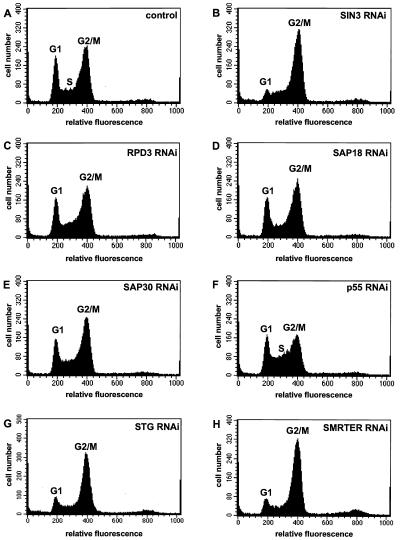
To monitor the physiological effect of the loss of SIN3, we analyzed SIN3 RNAi cells for morphology (Fig. 2), growth rate (Fig. 3), and cell cycle progression (Fig. 4). Loss of SIN3 caused morphological changes in Drosophila S2 cells. Control cells were round and had a fairly smooth surface, whereas many SIN3 RNAi cells were flattened and had long, thin projections (Fig. 2A and B). Counting of control and SIN3 RNAi cells for 4 days following addition of dsRNA revealed that loss of SIN3 increased the doubling time of S2 cells from ∼1 day to ∼2 days (Fig. 3). To examine this phenotype in more detail, the cell cycle distribution of SIN3 RNAi cells was determined by FACS analysis. By comparison with mock RNAi-treated cells, fewer SIN3 RNAi cells had a DNA content of 2N and more SIN3 RNAi cells had a DNA content of 4N, suggesting that SIN3 is required for progression through the G2/M phase of the cell cycle (Fig. 4A and B). SIN3 RNAi-treated Drosophila Kc167 tissue culture cells were also delayed in the G2/M phase, indicating that SIN3 plays a role in cell cycle progression in multiple cell types (data not shown). Note that we refer to the cell cycle phenotype of SIN3 RNAi cells as a G2 phase delay and not a G2 phase arrest because it is unclear whether SIN3 RNAi cells ever progress through G2 phase and initiate mitosis. The distinction between cell cycle delay and arrest is difficult to assess since the RNAi effect is transient and SIN3 protein levels begin to increase 5 to 6 days after addition of dsRNA (10; data not shown).
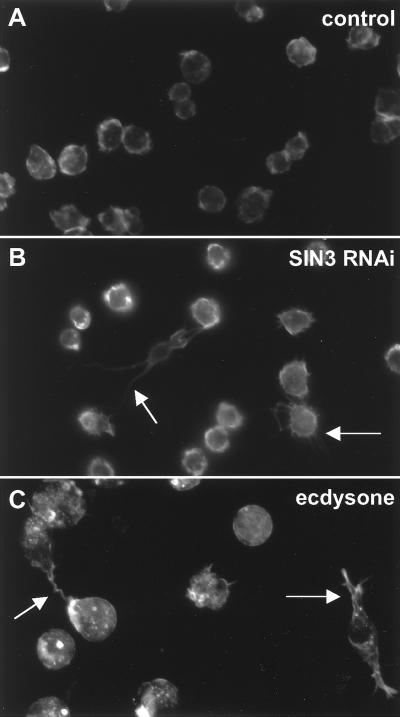
FIG. 2.
Loss of SIN3 results in altered cellular morphology. On the third day following the addition of dsRNA, RNAi cells were fixed and stained with a phalloidin antibody to visualize the cell surface. Panels: A, control; B, SIN3 RNAi; C, ecdysone treated. The arrows in panels B and C indicate abnormal cells that contain long, thin projections.
FIG. 3.
Loss of some SIN3/RPD3 complex subunits and SMRTER affects cell growth. Growth curves for control and RNAi cells are indicated by different symbols and labeled at the day 4 time point. Cell numbers were determined each day following addition of dsRNA for a total of 4 days. Results of a single experiment are shown, but identical relative growth rates for the seven samples were observed in multiple independent experiments.
FIG. 4.
Loss of some SIN3/RPD3 complex subunits, SMRTER, and STG affects cell cycle progression. Control and RNAi cells were analyzed by FACS on the third day following addition of dsRNA (A to H). G1 phase (2N DNA content, relative fluorescence of 200) and G2/M phase (4N DNA content, relative fluorescence of 400) peaks are indicated. S phase (2N to 4N DNA content, relative fluorescence of 200 to 400) is indicated in panels A and F. The gene targeted by RNAi is indicated in the upper right corner of each panel.
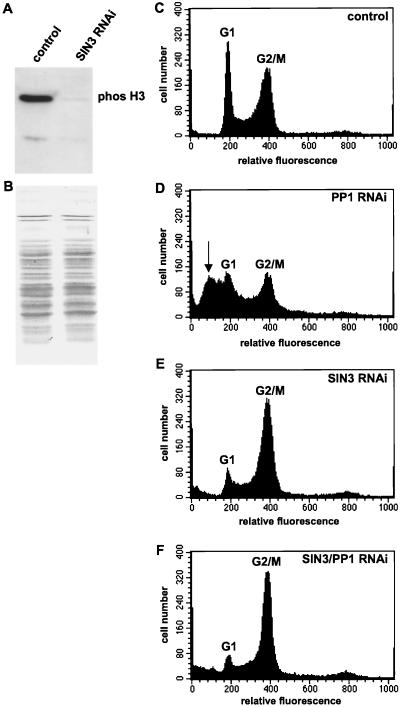
To determine if SIN3 RNAi cells were delayed in G2 or M phase, we examined the phosphorylation state of histone H3. One hallmark of initiation of mitosis is phosphorylation of serine 10 of histone H3 (23, 70). Western blot analysis of whole-cell acid-soluble protein extracts with an antibody directed against phosphorylated serine 10 of histone H3 (anti-phos H3) indicated that SIN3 RNAi cells have extremely low levels of phosphorylated histone H3 relative to asynchronously dividing control cells (Fig. 5A). We also fluorescently stained SIN3 RNAi cells with the phos H3 antibody and determined that 5.5% of the control cells (n = 868) and 1.5% of the SIN3 RNAi cells (n = 1,465) were stained with the phos H3 antibody. Thus, SIN3 RNAi cells do not initiate mitosis, indicating that SIN3 is required for G2 phase cell cycle progression.
FIG. 5.
SIN3 RNAi cells are delayed in the G2 phase of the cell cycle prior to initiation of mitosis. Whole-cell acid-soluble protein extracts from control and SIN3 RNAi cells were subjected to Western blot analysis with an antibody specific for histone H3 phosphorylated at serine 10 (A). India ink was used to visualize proteins bound to the membrane probed in panel A, demonstrating that equal quantities of protein were contained in each sample (B). Control and RNAi cells were analyzed by FACS on the third day following addition of dsRNA (C to F). G1 phase (2N DNA content, relative fluorescence of 200) and G2/M phase (4N DNA content, relative fluorescence of 400) peaks are indicated. The arrow in panel D indicates the sub-G1 peak representing cells with a DNA content of less than 2N. The gene(s) targeted by RNAi is indicated in the upper right corner of each panel.
Positioning of SIN3 upstream of protein phosphatase 1 (PP1) also supports a role for SIN3 prior to mitosis. In Caenorhabditis elegans, PP1/Glc7 was shown to be responsible for dephosphorylation of serine 10 of histone H3, allowing for chromatin decondensation at the end of mitosis (23). Null mutants of Drosophila PP1-87B die at the larval stage, and their cells fail to exit mitosis and exhibit overcondensed chromatin (2, 13). Loss of PP1 in S2 cells by PP1-87B RNAi strongly disrupted the cell cycle (Fig. 1G and 5D). FACS analysis revealed a reduction in both the G1 and G2/M phase peaks and a new sub-G1 phase peak, which may be due to progression through mitosis prior to completion of DNA replication. Interestingly, the FACS profile of SIN3/PP1-87B double-RNAi cells was similar to the SIN3 RNAi profile, indicating that SIN3 acts upstream of PP1 in cell cycle progression (Fig. 5E and F).
Not all components of the SIN3/RPD3 complex are required for progression through the G2 phase of the cell cycle.
To address the mechanism underlying the SIN3 requirement for G2 cell cycle progression, we asked whether other components of the SIN3/RPD3 complex are also required for this process. RNAi was carried out for each of the other subunits of the complex, RPD3, SAP18, SAP30, and p55. Western blot analysis, using antibodies directed against RPD3, SAP18, or p55 showed, in each case, that addition of dsRNA drastically reduced protein expression (Fig. 1B, C, and D). Similarly, RT-PCR analysis of SAP30 RNAi cells showed that the level of SAP30 mRNA was drastically reduced (Fig. 1F).
As with SIN3, we examined the morphology, growth rate, and cell cycle profile of RPD3, SAP18, SAP30, and p55 RNAi cells. Unlike loss of SIN3, loss of other SIN3/RPD3 complex subunits did not cause drastic changes in cell morphology (data not shown). Some RPD3 RNAi cells had small projections, but none were as prominent as those observed in SIN3 RNAi cells. Loss of some subunits of the SIN3/RPD3 complex slowed the rate of growth relative to that of control cells (Fig. 3). SAP18 and SAP30 RNAi cells exhibited growth rates similar to that of control cells. RPD3 RNAi cells had a moderate reduction in growth rate, doubling in ∼1.5 days, compared to ∼1 day for control cells. Finally, p55 RNAi cells showed a strong reduction in growth rate, equivalent to that seen in SIN3 RNAi cells.
Similar to and most likely a reflection of the growth rate, loss of individual components of the SIN3/RPD3 complex resulted in distinct cell cycle phenotypes (Fig. 4C, D, E, and F). p55 RNAi cells (which exhibited the slowest growth) showed the strongest deviation from the control cell cycle profile (Fig. 4F). The number of cells in both the G1 and G2/M phases was reduced, with a concomitant increase in the number of cells in S phase, possibly reflecting the role of p55 in chromatin assembly during DNA replication (64). More subtle effects were observed in RPD3, SAP18, and SAP30 RNAi cells, each having a slight reduction in the number of cells in the G1 phase (Fig. 4C, D, and E). While the cell cycle distribution of RPD3 RNAi cells was similar to that of control cells, the reduced growth rate of these cells suggests a delay at multiple points in the cell cycle, presumably including both the G1 and G2/M phases. Chemical inhibitors of deacetylases cause both G1 and G2 phase arrests, supporting a role for RPD3 at these points in the cell cycle (14, 30, 74, 75). Taken together, the growth rate and cell cycle distribution data indicate that SIN3, RPD3, and p55 play regulatory roles during the cell cycle but SAP18 and SAP30 do not.
Loss of RPD3, but not SIN3, affects global histone acetylation.
Since the SIN3/RPD3 complex possesses HDAC activity, we examined whether global effects on histone acetylation contribute to the cell cycle defect of SIN3 or RPD3 RNAi cells. Yeast RPD3-null mutants exhibit a global increase in acetylation at lysine 5 (K5) and K12 of histone H4 and K9/18 and K14 of histone H3 (54). Furthermore, treatment of mammalian tissue culture cells with HDAC inhibitors, including sodium butyrate, trichostatin A, and trapoxin, leads to a global increase in histone acetylation levels and arrest in the G1 and G2 phases of the cell cycle (14, 30, 31, 36, 45, 49, 52, 58, 74, 75, 76).
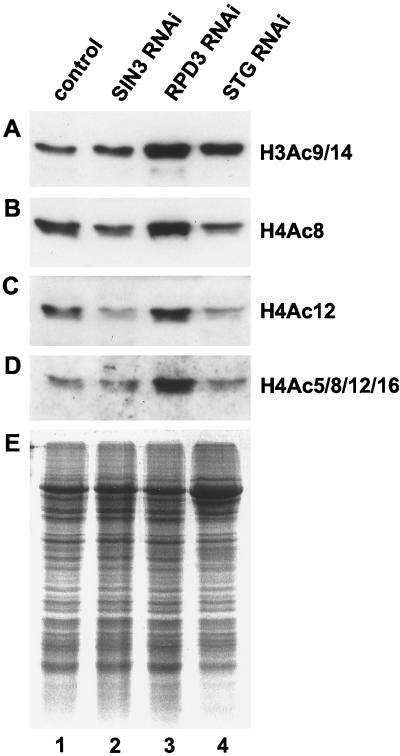
Histone acetylation levels in SIN3 and RPD3 RNAi cells were monitored by Western blot analysis of whole-cell acid-soluble protein extracts with antibodies against specific acetylated histone lysine residues. As predicted by studies with yeast, loss of RPD3 caused an increase in the acetylation of K8 and K12 of histone H4 and K9/14 of histone H3 (Fig. 6, compare lanes 1 and 3). Surprisingly, histone acetylation levels in SIN3 RNAi cells were equivalent or even slightly reduced relative to levels in control asynchronously dividing cells (Fig. 6, compare lanes 1 and 2).
FIG. 6.
Loss of RPD3, but not SIN3, results in a global increase in histone acetylation. (A to D) Western blot analysis of whole-cell acid-soluble protein extracts from control and RNAi cells with antibodies specific for acetylated histones, H3Ac9/14 (A), H4Ac8 (B), H4Ac12 (C), and H4Ac5/8/12/16 (D). (E) Samples identical to those probed in panels A to D were subjected to SDS-PAGE analysis, and proteins were visualized by staining with Gelcode Blue Stain Reagent (Pierce), demonstrating that the same quantity of protein was contained in each sample.
In plants, global histone acetylation levels are not constant throughout the cell cycle; rather, they vary depending on the cell cycle phase (26). Since SIN3 RNAi cells are delayed in the G2 phase, the overall level of acetylated histones may be decreased relative to the level in control asynchronous cells. Therefore, we compared the histone acetylation level of SIN3 RNAi cells with that of cells independently blocked in the G2 phase by String (STG) RNAi. STG is the Drosophila homologue of the yeast mitotic regulator Cdc25 phosphatase that is required for G2 phase cell cycle progression (50). STG mRNA levels were drastically reduced by RNAi, and FACS analysis revealed that STG RNAi cells were delayed in the G2 phase of the cell cycle (Fig. 1H and 4G). STG RNAi cells showed a small reduction in the acetylation of K8 and K12 of histone H4, indicating that G2 cells probably have lower overall histone H4 acetylation levels than do control cells and demonstrating that in Drosophila cells, acetylation levels vary through the cell cycle (Fig. 6, compare lanes 1 and 4). The histone acetylation levels of SIN3 and STG RNAi cells were very similar. Thus, unlike loss of RPD3, loss of SIN3 does not cause a global increase in histone acetylation. Therefore, morphological and cell cycle phenotypes associated with loss of SIN3 may result from gene-specific changes in histone acetylation levels.
The corepressor SMRTER is required for progression through the G2 phase of the cell cycle.
Genetic and biochemical studies functionally link the SMRTER corepressor and the SIN3/RPD3 complex (48, 62). Therefore, SMRTER RNAi was carried out to determine whether SMRTER is involved in cell cycle progression (Fig. 1E). Similar to that of SIN3 RNAi cells, the growth rate of SMRTER RNAi cells was strongly reduced (Fig. 3). In addition, FACS analysis of SMRTER RNAi cells revealed that SMRTER is required for G2/M phase progression of the cell cycle (Fig. 4H). Thus, SMRTER most likely recruits the SIN3/RPD3 complex to genes that need to be repressed for progression through the G2 phase of the cell cycle.
SIN3 and SMRTER function independently of EcR during the cell cycle.
In accord with published observations, we found that treatment of S2 cells with ecdysone caused a rapid and severe G2/M phase cell cycle block (Fig. 7C) (4, 8, 11, 19, 53). Thus, the SIN3/RPD3 complex, SMRTER, and EcR are required at similar points in the cell cycle. In addition, ecdysone treatment of S2 cells caused changes in cell morphology, including the generation of long, thin projections (Fig. 2C). Phenotypic similarities between SIN3 RNAi cells, SMRTER RNAi cells, and ecdysone-treated cells and biochemical and genetic links between the SIN3/RPD3 complex and EcR suggested that the SIN3/RPD3 complex represses EcR-regulated genes to allow progression through the G2 phase of the cell cycle. In other words, loss of the SIN3/RPD3 complex or SMRTER by RNAi allows activation of EcR-regulated genes, which mimics activation of EcR-regulated genes by ecdysone.
FIG. 7.
The G2 phase cell cycle delay resulting from loss of SIN3 or SMRTER is unaffected by loss of EcR. Control, RNAi, and ecdysone-treated cells were analyzed by FACS on the third day following addition of dsRNA (A, B, and D to H). Cells in panel C were analyzed by FACS 1 day following addition of ecdysone. For cells analyzed in panel D, ecdysone was added 2 days following the addition of EcR dsRNA, 1 day prior to FACS analysis. This provided sufficient time for the EcR RNAi effect to occur prior to ecdysone treatment. G1 phase (2N DNA content, relative fluorescence of 200) and G2/M phase (4N DNA content, relative fluorescence of 400) peaks are indicated. The gene(s) targeted by RNAi is indicated in the upper right corner of each panel. Ethidium bromide staining of agarose gels of RT-PCR products (I to L) was used to examine the expression of ecdysone-regulated genes, Eip71CD (I), Eip55E (J), E74A (K), and E75A (L), in control asynchronously dividing cells (lane1), SIN3 RNAi cells (lane 2), RPD3 RNAi cells (lane 3), SMRTER RNAi cells (lane 4), and ecdysone-treated cells (lane 5). As a loading control, TAF1 mRNA levels were determined for each sample. RT-PCR products are labeled on the right of each panel.
To address this proposition, we examined whether EcR is required for the ecdysone-induced G2 phase cell cycle block. EcR RNAi reduced the level of EcR mRNA but did not cause a cell cycle defect, suggesting that EcR is not required for cell cycle progression in the absence of a steroid (Fig. 1I and 7B). In contrast, the G2 phase block caused by ecdysone treatment was suppressed by EcR RNAi, indicating that EcR is required for the cell cycle phenotype of ecdysone-treated cells (Fig. 7D).
However, two lines of evidence suggest that the SIN3/RPD3 complex functions independently of EcR during progression through the G2 phase of the cell cycle. First, FACS analysis revealed that SIN3/EcR and SMRTER/EcR double-RNAi cells were delayed in the G2 phase of the cell cycle (Fig. 7F and H). Thus, while EcR is required for the ecdysone-induced G2 phase block, EcR is not required for the G2 phase delay caused by loss of SIN3 or SMRTER. Second, SIN3, RPD3, and SMRTER RNAi had different effects on gene expression than ecdysone treatment (Fig. 7I to L). The steady-state transcription levels of four genes (Eip71CD, Eip55E, E74A, and E75A) that are regulated by ecdysone during Drosophila development were examined by RT-PCR (8, 59). Relative to that in control asynchronously dividing cells, Eip71CD and Eip55E transcription was upregulated in SIN3 and RPD3 RNAi cells but not in ecdysone-treated cells while E74A and E75A transcription was upregulated in ecdysone-treated cells but not in SIN3 and RPD3 RNAi cells. Substantial changes in transcription were not observed in SMRTER RNAi cells. At least in the case of these four genes in S2 cells, these results suggest that loss of SIN3 or RPD3 is not sufficient to derepress transcription of genes that are responsive to ecdysone. In summary, it appears that the SIN3/RPD3 complex and EcR do not work in concert to repress transcription of ecdysone-inducible genes to allow progression through the G2 phase of the cell cycle.
SIN3- and RPD3-dependent regulation of SMRTER protein levels is required for progression through the G2 phase of the cell cycle.
SIN3 has been directly and indirectly implicated in regulation of the stability of transcription factors (46, 78, 83). Thus, we were interested in determining if SMRTER levels are affected by loss of the SIN3/RPD3 complex. To test this possibility, we examined SMRTER expression levels in SIN3 RNAi cells. Western blot analysis of extracts prepared from control and SIN3 RNAi cells revealed that SMRTER protein levels were reduced in SIN3 RNAi cells (Fig. 8A, lanes 1 and 2). This reduction occurred posttranscriptionally, as SMRTER mRNA levels were not reduced in SIN3 RNAi cells (Fig. 8C, lanes 1 and 2). SMRTER protein levels, but not mRNA levels, were reduced in RPD3 RNAi cells (Fig. 8A, lane 3, and C, lane 3), and were unchanged in p55, SAP18, SAP30, and STG RNAi cells (Fig. 8A, lanes 8, 7, 6, and 5, respectively). STG RNAi cells were included as a control for cells blocked in the G2 phase independently of SIN3/RPD3 complex activity.
In contrast to SMRTER protein levels, SIN3 protein levels were not affected by loss of any other SIN3/RPD3 complex subunit, by loss of SMRTER or STG, or by ecdysone treatment (Fig. 8B). Levels of RPD3, p55, SAP18, and SAP30 were also unchanged in SIN3 RNAi cells relative to those in control cells (Fig. 1; data not shown). In addition, transcriptional activation of EcR-regulated genes by ecdysone treatment did not lead to a reduction in SMRTER levels, indicating that SMRTER levels are not linked to ecdysone gene activation in this cell type (Fig. 8A, lane 4). Thus, SMRTER levels appear to be regulated by a posttranscriptional mechanism in response to loss of SIN3 or RPD3. The SIN3/RPD3 complex may regulate translation of the SMRTER mRNA, but given the documented role of SIN3 in the protection of transcription factors from proteosomal degradation, it is likely that loss of SIN3 or RPD3 leads to degradation of SMRTER and that stabilization of SMRTER by the SIN3/RPD3 complex is required for progression through the G2 phase of the cell cycle.
DISCUSSION
This study demonstrates that the SIN3/RPD3 complex is essential for G2 phase cell cycle progression. Elimination of SIN3 causes cells to be delayed in the cell cycle with a 4N DNA content prior to initiation of mitosis. Loss of SIN3 does not cause a global change in the acetylation level of histones H3 and H4, suggesting that the G2 phase delay is due to local, not global, changes in chromatin structure that presumably alter the expression of a limited number of genes. The SMRTER corepressor probably recruits the SIN3/RPD3 complex to these genes, as loss of SMRTER causes a G2 phase delay and SMRTER protein levels are reduced in response to loss of SIN3 or RPD3. Hormone-bound EcR blocks G2 phase progression but does so independently of the SIN3/RPD3 complex and SMRTER, leaving open the identity of the DNA-binding protein that recruits the SIN3/RPD3/SMRTER assemblage. In addition, this establishes at least two transcriptional repression mechanisms that are essential for progression through the G2 phase. SAP18 and SAP30 are not required for the cell cycle regulatory activity of the SIN3/RPD3 complex, indicating that they are not essential for stability of the complex, recruitment of the complex to some promoters, or the deacetylase activity of the complex. These findings provide in vivo evidence for (i) distinct roles for individual components of the SIN3/RPD3 complex, (ii) a functional link between the SIN3/RPD3 complex and the SMRTER corepressor, and (iii) a role for the SIN3/RPD3 complex, not just SIN3, in regulation of the level of corepressor proteins. Furthermore, these findings point to the SIN3/RPD3 complex as an essential regulator of progression through the G2 phase of the cell cycle and suggest that the SIN3/RPD3 complex is a target of deacetylase inhibitors that cause G2 phase arrest of cancer cells (31, 36, 45, 58).
RNAi reveals distinct roles for components of the SIN3/RPD3 complex.
We have found that loss of individual components of the SIN3/RPD3 complex leads to distinct cellular phenotypes. Elimination of SIN3 or RPD3 by RNAi reduces the rate of cell growth, causes cells to delay in the G2 phase of the cell cycle, and results in reduced levels of the corepressor SMRTER. However, G1 phase cell cycle progression is not affected in SIN3 RNAi cells but may be affected in RPD3 RNAi cells; cell morphology is affected in SIN3, but not RPD3, RNAi cells; and global histone acetylation levels are not affected in SIN3 RNAi cells but are affected in RPD3 RNAi cells. Phenotypic differences between SIN3 and RPD3 RNAi cells are presumably due to inactivation of other RPD3-containing complexes. RPD3 is a component of the Mi-2/NuRD complex and has been shown to interact with a number of repressors in the absence of SIN3 (7, 32, 61, 68, 73, 81). Thus, a strong G2 phase delay in RPD3 RNAi cells may be masked by cell cycle defects resulting from inactivation of RPD3 activities that are independent of SIN3. SIN3-independent activities of RPD3 are clearly demonstrated by the global change in histone acetylation observed in RPD3 RNAi cells but not in SIN3 RNAi cells (Fig. 6). On the other hand, it is possible that differences are due to loss of SIN3 activities that are independent of RPD3. Mutations in SIN3 that prohibit RPD3 binding diminish, but do not abolish, transcriptional repression in mammalian cells, and SIN3 binds sites on Drosophila polytene chromosomes that are not bound by RPD3, suggesting that SIN3 can function independently of RPD3 (18, 34, 48, 72). Thus, elimination of SIN3 and RPD3 by RNAi has uncovered multiple roles for these proteins. Many of these roles are independent of one another, with the exception of roles in the regulation of SMRTER protein levels and in G2 cell cycle progression, which appear to require both proteins, presumably in the context of the biochemically defined SIN3/RPD3 complex.
Similar to loss of SIN3 and RPD3, loss of p55 causes a growth rate reduction. However, p55 RNAi cells are delayed in the S phase of the cell cycle and SMRTER protein levels are not affected. Phenotypes observed in p55 RNAi cells may be due to the functioning of p55 as a component of the Mi-2/NuRD, CAF-1, or nucleosome remodeling factor complex (40, 61, 64, 68, 81). Finally, in contrast to loss of SIN3, RPD3, and p55, loss of SAP18 and SAP30 does not cause cell morphology changes, cell growth defects, cell cycle phenotypes, or loss of SMRTER protein. Thus, in these cellular processes, SAP18 and SAP30 are not essential for recruitment of the SIN3/RPD3 complex to promoters, deacetylation of substrates by the SIN3/RPD3 complex, or stability of the SIN3/RPD3 complex. The lack of a requirement for SAP30 for some SIN3/RPD3 activities is consistent with the observation that inactivation of SAP30 by antibody microinjection does not affect repression mediated by artificial tethering of SIN3 to a promoter (35). Our data suggest that if SAP18 and SAP30 are important for SIN3/RPD3 complex function, then they play gene-specific roles that do not involve regulation of progression through the G2 phase of the cell cycle.
Progression through the G2 phase of the cell cycle requires the SIN3/RPD3 complex.
Global changes in the histone acetylation level cause cell cycle phenotypes, including arrest in the G2 phase. Mutation of N-terminal lysine residues that are normally acetylated in histone H4 results in a G2/M phase delay (41, 42). HDAC inhibitors such as trichostatin A, trapoxin, and sodium butyrate have been reported to cause a cell cycle arrest in the G1 and/or G2 phase (14, 30, 31, 36, 45, 49, 52, 58, 74, 75, 76). Mutation of HATs such as GCN5, ELP1, and SAS3 results in arrest in the G2/M phase of the cell cycle (22, 71, 79). In each of these cases, the cell cycle phenotype has been attributed to global changes in histone H3 or H4 acetylation. In one case, the global change in genome integrity has been shown to activate the RAD9-dependent DNA damage checkpoint pathway that induces a delay in the G2 phase (41). Furthermore, the mammalian homologue of RPD3, HDAC1, exists in a complex with RAD9 (6). We found that loss of SIN3 causes a G2 phase delay but does not cause global changes in histone acetylation, raising the question of the mechanism underlying the cell cycle delay.
The RAD9-dependent DNA damage checkpoint pathway does not appear to be activated by loss of SIN3. The checkpoint signaling mechanism sequentially involves sensing of DNA damage by RAD proteins, activation of the CHK1 protein kinase (known as Grapes in Drosophila) by phosphorylation, inactivation of the Cdc25 phosphatase (known as STG in Drosophila) by CHK1-dependent phosphorylation, and inhibition of the Cdc2-cyclin B complex by phosphorylation of Cdc2 (50, 56, 69, 77). Disruption of RAD9 suppresses the G2/M phase delay caused by mutating the four conserved lysines that are acetylated in the N-terminal tail of histone H4 (41). However, we found that SIN3/RAD9 double-RNAi cells were delayed in the G2 phase, suggesting that the delay in SIN3 RNAi cells is independent of RAD9 (data not shown). In addition, we have examined the steady-state levels of mRNAs encoding components of the RAD9 pathway, as well as the p53 pathway, that cause growth arrest in the G2/M phase (60). Grapes, String, p53, and Gadd45 mRNA levels were not affected in SIN3 RNAi cells (data not shown). The cyclin B protein levels in SIN3 RNAi cells were reduced, but this is not sufficient to cause the G2 phase delay, as cyclin B RNAi cells were not delayed in the G2 phase (data not shown). Thus, other than a reduction in the level of SMRTER protein, events that occur downstream of loss of the SIN3/RPD3 complex that lead to the G2 phase delay are unclear, but they do not appear to be caused by global changes in genome integrity.
A role for the SIN3/RPD3 complex in regulating corepressor levels.
In addition to a role in transcriptional repression, SIN3 has recently been shown to play a role in regulating protein stability. SIN3 interacts with the p53 tumor suppressor protein, and SIN3 and p53 are localized, along with deacetylated histones, to a p53-regulated promoter (43). A recent report has shown that SIN3 stabilizes p53 and protects it from proteasome-mediated degradation in mammalian cells (83). Therefore, SIN3 appears to be required for deacetylase activity at a promoter, as well as stability of the repressor involved in recruitment of the SIN3/RPD3 complex to a promoter. In addition, the corepressor N-CoR, but not SMRT, appears to be subject to regulated proteolysis by mSiah2, which binds ubiquitin-conjugating enzymes (78). Interestingly, Drosophila SIN3 mutants were isolated in a genetic screen as dominant enhancers of a phenotype caused by ectopic expression of Sina, the Drosophila homologue of mSiah2, suggesting that SIN3 counteracts the activity of Sina (46). Thus, SIN3 has been directly and indirectly implicated in regulation of the protein stability of transcription factors that are targeted for degradation.
We found that SMRTER protein levels are regulated by SIN3 and RPD3. This regulation occurs posttranscriptionally, as SMRTER message levels are not affected in SIN3 or RPD3 RNAi cells. The SIN3/RPD3 complex may regulate SMRTER translation or stability. Given the previously described role for SIN3 in regulation of the proteosomal degradation of transcriptional regulators, it is most likely that the SIN3/RPD3 complex regulates SMRTER stability. This would provide the first evidence that the protein stabilization function of SIN3 is carried out as a component of the SIN3/RPD3 complex and that the deacetylase activity of the SIN3/RPD3 complex is important for this function. Sina does not appear to play a role in the regulation of SMRTER protein levels, as SMRTER levels are reduced in SIN3/Sina double-RNAi cells and the cells were delayed in the G2 phase of the cell cycle (data not shown).
This study demonstrates that RNAi is a powerful approach to investigate of the cellular requirements for individual proteins and to determine the epistatic relationship between proteins that function in a given cellular process. The RNAi approach can now be used to identify factors that function along with the SIN3/RPD3 complex to regulate progression through the G2 phase of the cell cycle, including DNA-binding factors that recruit the SIN3/RPD3 complex.
Acknowledgments
We thank R. Evans and R. Kamakaka for providing reagents; B. Stearman for assistance with FACS analysis; R. Kamakaka for assistance with histone purification; N. Dhillon, R. Kamakaka, and M. Lilly for providing thoughtful advice throughout the course of this work; and the anonymous reviewers for suggestions that greatly improved the manuscript.
This work was supported by the Intramural Program in the National Institute of Child Health and Human Development and by startup funds from the University of Wisconsin—Madison Medical School and Department of Pharmacology.
REFERENCES
- 1.Alland, L., R. Muhle, H. Hou, Jr., J. Potes, L. Chin, N. Schreiber-Agus, and R. A. DePino. 1997. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature 387:49-55. [DOI] [PubMed] [Google Scholar]
- 2.Axton, J. M., V. Dombradi, P. T. Cohen, and D. M. Glover. 1990. One of the protein phosphatase 1 isozymes in Drosophila is essential for mitosis. Cell 63:33-46. [DOI] [PubMed] [Google Scholar]
- 3.Ayer, D. E. 1999. Histone deacetylases: transcriptional repression with SINers and NuRDs. Trends Cell Biol. 9:193-198. [DOI] [PubMed] [Google Scholar]
- 4.Berger, E., R. Ringler, S. Alahiotis, and M. Frank. 1978. Ecdysone-induced changes in morphology and protein synthesis in Drosophila cell cultures. Dev. Biol. 62:498-511. [DOI] [PubMed] [Google Scholar]
- 5.Bone, J. R., J. Lavender, R. Richman, M. J. Palmer, B. M. Turner, and M. I. Kuroda. 1994. Acetylated histone H4 on the male X chromosome is associated with dosage compensation in Drosophila. Genes Dev. 8:96-104. [DOI] [PubMed] [Google Scholar]
- 6.Cai, R. L., Y. Yan-Neale, M. A. Cueto, H. Xu, and D. Cohen. 2000. HDAC1, a histone deacetylase, forms a complex with Hus1 and Rad9, two G2/M checkpoint Rad proteins. J. Biol. Chem. 275:27909-27916. [DOI] [PubMed] [Google Scholar]
- 7.Chen, G., J. Fernandez, S. Mische, and A. J. Courey. 1999. A functional interaction between the histone deacetylase RPD3 and the corepressor Groucho in Drosophila development. Genes Dev. 13:2218-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cherbas, L., and P. Cherbas. 1981. The effects of ecdysteroid hormones on Drosophila melanogaster cell lines. Adv. Cell Cult. 1:91-124. [Google Scholar]
- 9.Cherbas, L., M. M. D. Koehler, and P. Cherbas. 1989. Effects of juvenile hormone on the ecdysone response of Drosophila Kc cells. Dev. Genet. 10:177-188. [DOI] [PubMed] [Google Scholar]
- 10.Clemens, J. C., C. A. Worby, N. Simonson-Leff, M. Muda, T. Maehama, B. A. Hemmings, and J. E. Dixon. 2000. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl. Acad. Sci. USA 97:6499-6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Courgeon, A. M. 1972. Effects of α- and β-ecdysone on in vitro diploid cell multiplication in Drosophila melanogaster. Nat. New Biol. 238:250-251. [DOI] [PubMed] [Google Scholar]
- 12.Cress, W. D., and E. Seto. 2000. Histone deacetylases, transcriptional control and cancer. J. Cell. Physiol. 184:1-16. [DOI] [PubMed] [Google Scholar]
- 13.Dombradi, V., J. M., Axton, H. M. Barker, and P. T. Cohen. 1990. Protein phosphatase 1 activity in Drosophila mutants with abnormalities in mitosis and chromosome condensation. FEBS Lett. 275:39-43. [DOI] [PubMed] [Google Scholar]
- 14.Fallon, R. J., and R. P. Cox. 1979. Cell cycle analysis of sodium butyrate and hydroxyurea, inducers of ectopic hormone production in HeLa cells. J. Cell. Physiol. 100:251-262. [DOI] [PubMed] [Google Scholar]
- 15.Glass, C. K., and M. G. Rosenfeld. 2000. The coregulator exchange in transcriptional function of nuclear receptors. Genes Dev. 14:121-141. [PubMed] [Google Scholar]
- 16.Hammond, S. M., E. Bernstein, D. Beach, and G. J. Hannon. 2000. An RNA-directed nuclease mediates posttranscriptional gene silencing in Drosophila cells. Nature 404:293-296. [DOI] [PubMed] [Google Scholar]
- 17.Hassan, A. H., K. E. Neely, and J. L. Workman. 2001. Histone acetyltransferase complexes stabilize SWI/SNF binding to promoter nucleosomes. Cell 104:817-827. [DOI] [PubMed] [Google Scholar]
- 18.Hassig. C. A., T. C. Fleischer, A. N. Billin, S. L. Schreiber, and D. E. Ayer. 1997. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell 89:341-347. [DOI] [PubMed] [Google Scholar]
- 19.Hatt, P. J., M. Moriniere, H. Oberlander, and P. Porcheron. 1994. Roles for insulin and ecdysteroids in differentiation of an insect cell line of epidermal origin. In Vitro Cell Dev. Biol. 30A:717-720. [DOI] [PubMed] [Google Scholar]
- 20.Hebbes, T. R., A. W. Thorne, and C. Crane-Robinson. 1988. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 7:1395-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heinzel, T., R. M. Lavinsky, T. M. Mullen, M. Soderstrom, C. D. Laherty, J. Torchia, W. M. Yang, G. Brard, S. D. Ngo, J. R. Davie, E. Seto, R. N. Eisenman, D. W. Rose, C. K. Glass, M. G. Rosenfeld. 1997. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature 387:43-48. [DOI] [PubMed] [Google Scholar]
- 22.Howe, L., D. Auston, P. Grant, S. John, R. G. Cook, J. L. Workman, and L. Pillus. 2001. Histone H3 specific acetyltransferases are essential for cell cycle progression. Genes Dev. 15:3144-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsu, J. Y., Z. W. Sun, X. Li, M. Reuben, K. Tatchell, D. K. Bishop, J. M. Grushcow, C. J. Brame, J. A. Caldwell, D. F. Hunt, R. Lin, M. M. Smith, and C. D. Allis. 2000. Mitotic phosphorylation of histone H3 is governed by Ipl/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102:279-291. [DOI] [PubMed] [Google Scholar]
- 24.Huang, E. Y., J. Zhang, E. A. Miska, M. G. Guenther, T. Kouzarides, and M. A. Lazar. 2000. Nuclear receptor corepressors partner with class II histone deacetylases in a Sin3-independent repression pathway. Genes Dev. 14:45-54. [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobson, R. H., A. G. Ladurner, D. S. King, and R. Tjian. 2000. Structure and function of the human TAFII250 double bromodomain module. Science 288:1372-1373. [DOI] [PubMed] [Google Scholar]
- 26.Jasencakova, Z., A. Meister, J. Walter, B. M. Turner, and I. Schubert. 2000. Histone H4 acetylation of euchromatin and heterochromatin is cell cycle dependent and correlated with replication rather than with transcription. Plant Cell 12:2087-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones, P. L., L. M. Sachs, N. Rouse, P. A. Wade, and Y.-B. Shi. 2001. Multiple N-CoR complexes contain distinct deacetylases. J. Biol. Chem. 276:8807-8811. [DOI] [PubMed] [Google Scholar]
- 28.Kadosh, D., and K. Struhl. 1997. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell 89:365-371. [DOI] [PubMed] [Google Scholar]
- 29.Kadosh, D., and K. Struhl. 1998. Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol. Cell. Biol. 18:5121-5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kijima, M., M. Yoshida, K. Sugita, S. Horinouchi, and T. Beppu. 1993. Trapoxin, an antitumor cyclic tetrapeptide, is an irreversible inhibitor of mammalian histone deacetylase. J. Biol. Chem. 268:22429-22435. [PubMed] [Google Scholar]
- 31.Kim, Y. B., S. W. Ki, M. Yoshida, and S. Horinouchi. 2000. Mechanism of cell cycle arrest caused by histone deacetylase inhibitors in human carcinoma cells. J. Antibiot. 53:1191-1200. [DOI] [PubMed] [Google Scholar]
- 32.Knoepfler, P. S., and R. N. Eisenman. 1999. Sin meets NuRD and other tails of repression. Cell 99:447-450. [DOI] [PubMed] [Google Scholar]
- 33.Kuo, M.-H., and C. D. Allis. 1998. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays 20:615-626. [DOI] [PubMed] [Google Scholar]
- 34.Laherty, C., W. M. Yang, J. M. Sun, J. R. Davie, E. Seto, R. N. Eisenman. 1997. Histone deacetylases associated with the mSIN3 corepressor mediate Mad:Max transcriptional repression. Cell 89:349-356. [DOI] [PubMed] [Google Scholar]
- 35.Laherty, C. D., A. N. Billin, R. M. Lavinsky, G. S. Yochim, A. C. Bush, J. M. Sun, T. M. Mullen, J. R. Davie, D. W. Rose, C. K. Glass, M. G. Rosenfeld, D. E. Ayer, and R. N. Eisenman. 1998. SAP30, a component of the mSIN3 corepressor complex involved in N-CoR-mediated repression by specific transcription factors. Mol. Cell 2:33-42. [DOI] [PubMed] [Google Scholar]
- 36.Lallemand, F., D. Courilleau, C. Buquet-Fagot, A. Azeddine, M.-N. Montagne, and J. Mester. 1999. Sodium butyrate induces G2 arrest in the human breast cancer cells MDA-MB-231 and renders them competent for DNA replication. Exp. Cell Res. 247:432-440. [DOI] [PubMed] [Google Scholar]
- 37.Lee, D. Y., J. J. Hayes, D. Pruss, and A. Wolffe. 1993. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell 72:73-84. [DOI] [PubMed] [Google Scholar]
- 38.Li, J., J. Wang, J. Wang, Z. Nawaz, J. M. Liu, J. Qin, and J. Wong. 2000. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 19:4342-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Magnaghi-Jaulin, L., S. Ait-Si-Ali, and A. Harel-Bellan. 2000. Histone acetylation and control of the cell cycle. Prog. Cell Cycle Res. 4:41-47. [DOI] [PubMed] [Google Scholar]
- 40.Martinez-Balbas, M. A., T. Tsukiyama, D. Gdula, and C. Wu. 1998. Drosophila NURF-55, a WD repeat protein involved in histone metabolism. Proc. Natl. Acad. Sci. USA 95:132-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Megee, P. C., B. A. Morgan, and M. M. Smith. 1995. Histone H4 and the maintenance of genome integrity. Genes Dev. 9:1716-1727. [DOI] [PubMed] [Google Scholar]
- 42.Morgan, B. A., B. A. Mittman, and M. M. Smith. 1991. The highly conserved N-terminal domains of histones H3 and H4 are required for normal cell cycle progression. Mol. Cell. Biol. 11:4111-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murphy, M., J. Ahn, K. K. Walker, W. H. Hoffman, R. M. Evans, A. J. Levine, and D. L. George. 1999. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Dev. 13:2490-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagy, L., H. Y. Kao, D. Chakravarti, R. J. Lin, A. Hassig, D. E. Ayer, S. L. Schreiber, and R. M. Evans. 1997. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell 89:373-380. [DOI] [PubMed] [Google Scholar]
- 45.Nakajima, H., Y. B. Kim, H. Terano, M. Yoshida, and S. Horinouchi. 1998. FR901228, a potent antitumor antibiotic, is a novel histone deacetylase inhibitor. Exp. Cell Res. 241:126-133. [DOI] [PubMed] [Google Scholar]
- 46.Neufeld, T. P., A. H. Tang, and G. M. Rubin. 1998. A genetic screen to identify components of the sina signaling pathway in Drosophila eye development. Genetics 148:277-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perlmann, T., and R. M. Evans. 1997. Nuclear receptors in Sicily: all in the famiglia. Cell 90:391-397. [DOI] [PubMed] [Google Scholar]
- 48.Pile, L. A., and D. A. Wassarman. 2000. Chromosomal localization links the SIN3-RPD3 complex to the regulation of chromatin condensation, histone acetylation and gene expression. EMBO J. 19:6131-6140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qiu, L., A. Burgess, D. P. Fairlie, H. Leonard, P. G. Parsons, and B. G. Gabrielli. 2000. Histone deacetylase inhibitors trigger a G2 checkpoint in normal cells that is defective in tumor cells. Mol. Biol. Cell 11:2069-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reed, B. H. 1995. Drosophila development pulls the strings of the cell cycle. Bioessays 17:553-556. [DOI] [PubMed] [Google Scholar]
- 51.Riddiford, L. M., P. Cherbas, and J. W. Truman. 2001. Ecdysone receptors and their biological actions. Vitam. Horm. 60:1-73. [DOI] [PubMed] [Google Scholar]
- 52.Riggs, M. G., R. G. Whittaker, J. R. Neumann, V. M. Ingram. 1977. n-Butyrate causes histone modification in HeLa and Friend erythroleukaemia cells. Nature 268:462-464. [DOI] [PubMed] [Google Scholar]
- 53.Rosset, R. 1978. Effects of ecdysone on a Drosophila cell line. Exp. Cell Res. 111:31-36. [DOI] [PubMed] [Google Scholar]
- 54.Rundlett, S. E., A. A. Carmen, R. Kobayashi, S. Bavykin, B. M. Turner, and M. Grunstein. 1996. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl. Acad. Sci. USA 93:14503-14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rundlett, S. E., A. A. Carmen, N. Suka, B. M. Turner, and M. Grunstein. 1998. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature 392:831-835. [DOI] [PubMed] [Google Scholar]
- 56.Russell, P. 1998. Checkpoints on the road to mitosis. Trends Cell Biol. 23:399-402. [DOI] [PubMed] [Google Scholar]
- 57.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 58.Sambucetti, L. C., D. D. Fischer, S. Zabludoff, P. O. Kwon, H. Chamberlin, N. Trogani, H. Xu, and D. Cohen. 1999. Histone deacetylase inhibition selectively alters the activity and expression of cell cycle proteins leading to specific chromatin acetylation and antiproliferative effects. J. Biol. Chem. 274:34940-34947. [DOI] [PubMed] [Google Scholar]
- 59.Savakis, C., G. Demetri, and P. Cherbas. 1980. Ecdysteroid-inducible polypeptides in a Drosophila cell line. Cell 22:665-674. [DOI] [PubMed] [Google Scholar]
- 60.Taylor, W. R., and G. R. Stark. 2001. Regulation of the G2/M transition by p53. Oncogene 20:1803-1815. [DOI] [PubMed] [Google Scholar]
- 61.Tong, J. K., C. A. Hassig, G. R. Schnitzler, R. E. Kingston, and S. L. Schreiber. 1998. Chromatin deacetylation by an ATP-dependent nucleosome remodeling complex. Nature 395:917-921. [DOI] [PubMed] [Google Scholar]
- 62.Tsai, C.-C., H.-Y. Kao, T.-P. Yao, M. McKeown, and R. M. Evans. 1999. SMRTER, a Drosophila nuclear receptor coregulator, reveals that EcR-mediated repression is critical for development. Mol. Cell 4:175-186. [DOI] [PubMed] [Google Scholar]
- 63.Tse, C., T. Sera, A. P. Wolffe, and J. C. Hansen. 1998. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol. Cell. Biol. 18:4629-4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tyler, J. K., M. Bulger, R. T. Kamakaka, R. Kobayashi, and J. T. Kadonaga. 1996. The p55 subunit of Drosophila chromatin assembly factor 1 is homologous to a histone deacetylase-associated protein. Mol. Cell. Biol. 16:6149-6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Urnov, F. D., and A. P. Wolffe. 2001. A necessary good: nuclear hormone receptors and their chromatin templates. Mol. Endocrinol. 15:1-16. [DOI] [PubMed] [Google Scholar]
- 66.Verreault, A., P. D. Kaufman, R. Kobayashi, and B. Stillman. 1998. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr. Biol. 8:96-108. [DOI] [PubMed] [Google Scholar]
- 67.Vettese-Dadey, M., P. A. Grant, T. R. Hebbes, C. Crane-Robinson, C. D. Allis, and J. L. Workman. 1996. Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J. 15:2508-2518. [PMC free article] [PubMed] [Google Scholar]
- 68.Wade, P. A., P. L. Jones, D. Vermaak, and A. P. Wolffe. 1998. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr. Biol. 8:843-846. [DOI] [PubMed] [Google Scholar]
- 69.Walworth, N. C. 2000. Cell-cycle checkpoint kinases: checking in on the cell cycle. Curr. Opin. Cell Biol. 12:697-704. [DOI] [PubMed] [Google Scholar]
- 70.Wei, Y., L. Yu, J. Bowen, M. A. Gorovsky, and C. D. Allis. 1999. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell 97:99-109. [DOI] [PubMed] [Google Scholar]
- 71.Wittschieben, O. B., J. Fellows, W. Du, D. J. Stillman, and J. Q. Svejstrup. 2000. Overlapping roles for the histone acetyltransferase activities of SAGA and Elongator in vivo. EMBO J. 12:3060-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wong, C. W., and M. L. Privalsky. 1998. Transcriptional repression by the SMRT-mSin3 corepressor: multiple interactions, multiple mechanisms, and a potential role for TFIIB. Mol. Cell. Biol. 18:5500-5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xue, Y., J. Wong, G. T. Moreno, M. K. Young, J. Cote, and W. Wang. 1998. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell 2:851-861. [DOI] [PubMed] [Google Scholar]
- 74.Yamada, K., and G. Kimura. 1985. Formation of proliferative tetraploid cells after treatment of diploid cells with sodium butyrate in rat 3Y1 fibroblasts. J. Cell. Physiol. 122:59-63. [DOI] [PubMed] [Google Scholar]
- 75.Yoshida, M., and T. Beppu. 1988. Reversible arrest of proliferation of rat 3Y1 fibroblasts in both G1 and G2 phases by trichostatin A. Exp. Cell Res. 177:122-131. [DOI] [PubMed] [Google Scholar]
- 76.Yoshida, M., M. Kijima, M. Akita, and T. Beppu. 1990. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 265:17174-17179. [PubMed] [Google Scholar]
- 77.Yu, K. R., R. B. Saint, and W. Sullivan. 2000. The Grapes checkpoint coordinates nuclear envelope breakdown and chromatin condensation. Nat. Cell Biol. 2:609-615. [DOI] [PubMed] [Google Scholar]
- 78.Zhang, J., M. G. Guenther, R. W. Carthew, and M. A. Lazar. 1998. Proteasomal regulation of nuclear corepressor-mediated repression. Genes Dev. 12:1775-1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang, W., J. R. Bone, D. G. Edmondson, B. M. Turner, and S. Y. Roth. 1998. Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J. 17:3155-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang, Y., R. Iratni, H. Erdjument-Bromage, P. Tempst, and D. Reinberg. 1997. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell 89:357-364. [DOI] [PubMed] [Google Scholar]
- 81.Zhang, Y., G. LeRoy, H. P. Seelig, W. S. Lane, and D. Reinberg. 1998. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95:279-289. [DOI] [PubMed] [Google Scholar]
- 82.Zhang, Y., Z.-W. Sun, R. Iratni, H. Erdjument-Dromage, P. Tempst, M. Hampsey, and D. Reinberg. 1998. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol. Cell 1:1021-1031. [DOI] [PubMed] [Google Scholar]
- 83.Zilfou, J. T., W. H. Hoffman, M. Sank, D. L. George, and M. Murphy. 2001. The corepressor mSIN3a interacts with the proline-rich domain of p53 and protects p53 from proteasome-mediated degradation. Mol. Cell. Biol. 21:3974-3985. [DOI] [PMC free article] [PubMed] [Google Scholar]