Abstract
Ubiquitin-mediated degradation targets cell cycle regulators for proteolysis. Much of the ubiquitin pathway's substrate specificity is conferred by E3 ubiquitin ligases, and cullins are core components of some E3s. CUL-4A encodes one of six mammalian cullins and is amplified and/or overexpressed in breast cancer, which suggests a role in regulating cell cycle progression. To examine CUL-4A's physiologic function, we generated a CUL-4A deletion mutation in mice. No viable CUL-4A−/− pups and no homozygous mutant embryos as early as 7.5 days postcoitum (dpc) were recovered. However, CUL-4A−/− blastocysts are viable, hatch, form an inner cell mass and trophectoderm, and implant (roughly 4.5 dpc), indicating that CUL-4A−/− embryos die between 4.5 and 7.5 dpc. Despite 87% similarity between the Cul-4A and Cul-4B cullins, the CUL-4A−/− lethal phenotype indicates that CUL-4A has one or more distinct function(s). Surprisingly, 44% fewer heterozygous pups were recovered than expected by Mendelian genetics, indicating that many heterozygous embryos also die during gestation due to haploinsufficiency. Taken together, our findings indicate that appropriate CUL-4A expression is critical for early embryonic development.
Ubiquitin-mediated degradation plays a critical role in controlling the turnover of cell cycle regulators (reviewed in references 11 and 66). The ATP-dependent attachment of ubiquitin to a ubiquitin-activating enzyme (E1) activates ubiquitin for transfer to a ubiquitin-conjugating enzyme (E2) and then to a ubiquitin ligase (E3), which transfers ubiquitin to a substrate protein (reviewed in references 17 and 46). Repetition of this ubiquitin transferase reaction results in the attachment of a polyubiquitin chain to the substrate, which is then recognized by the 26S proteasome and degraded. Much of the ubiquitin pathway's substrate specificity derives from E3 ligases, so regulating E3 activity is likely to be important for controlling ubiquitin-mediated degradation. Cullins are a core component of a subset of E3 ligases (described below). In this report, we describe in vivo studies that examine the physiologic function of the CUL-4A cullin.
Multisubunit RING E3 ligases contain a RING finger protein, an E2, an adaptor protein, a substrate recognition subunit, and a cullin (reviewed in reference 46). An extensively studied type of multisubunit RING E3 is SCF (Skp1, Cdc53/cullin, F-box protein), which was initially characterized for Saccharomyces cerevisiae (reviewed in references 11 and 66). Skp1 (the adaptor protein [2]), Cdc53 (the cullin [26, 34, 42, 55, 68]), and Rbx1 (the RING finger protein [3, 23, 52, 56]) make up the SCF core. This core binds various F-box proteins (such as Cdc4, Grr1, and Met30), which are responsible for substrate recognition (reviewed in references 11 and 66). The resulting SCF complexes (SCFCdc4, SCFGrr1, and SCFMet30) target for degradation a variety of cell cycle regulators, including Sic1, Far1, Cdc6, Cln1, Cln2, Gic2, and Swe1.
Mammals encode six cullins, Cul-1, Cul-2, Cul-3, Cul-4A, Cul-4B, and Cul-5 (26), and these appear to be components of SCFs or related complexes. For example, Cul-1 is a component of SCFSkp2, which is related to the yeast SCF and is implicated in the ubiquitination of p21, p27, and E2F-1 (6, 30, 32, 33, 40, 60, 64, 72). SCFβ-TRCP shares the same core components as SCFSkp2 and includes the β-TRCP F-box protein, which targets for degradation IκBα and β-catenin (16, 27, 57, 61, 62, 69, 70). An SCF containing Cul-1 and the F-box protein, Cdc4, targets cyclin E for ubiquitin-mediated degradation (36, 59).
Cul-2 is a component of VCB, an E3 whose structure is similar to that of SCF. Along with Cul-2, VCB contains Rbx1, elongin B (a ubiquitin-like protein), and elongin C, which is similar to Skp1 (20, 23, 29, 31, 40, 44). Analogous to Skp1 and F-box proteins, elongin C interacts with SOCS-box proteins, such as elongin A and the von Hippel-Lindau tumor suppressor protein (VHL), and like F-box proteins, these are involved in substrate recognition (1, 12, 24, 25, 58, 73). In response to serum starvation, cells lacking VHL fail to exit the cell cycle and fail to accumulate the cyclin-dependent kinase inhibitor, p27, suggesting a link between p27 stability and an E3 containing VHL (43). This complex also targets a hypoxia-inducible transcription factor, HIF-1α, for degradation by the proteasome (35). In addition, an E3 containing Cul-2, elongin B, and elongin C interacts with another SOCS-box protein, SOCS1, to target the TEL-JAK2 oncoprotein for ubiquitin-mediated degradation (21, 28, 45).
Somewhat less is known about the other mammalian cullins and the complexes they form. Like Cul-1 and Cul-2, Cul-3, Cul-4A, and Cul-5 interact with Rbx1 (40). Cul-3 binds cyclin E and, when overexpressed, stimulates cyclin E ubiquitination (54). Cul-5 is the vasopressin-activated calcium-mobilizing (VACM-1) receptor and plays a role in regulating cellular signaling (4, 5). Also, Cul-5 and Rbx1 form complexes with elongins B and C and a SOCS-box protein (elongin A, SOCS1, WSB1, or MUF1), and each immunoprecipitated complex has E3 activity in vitro (22). Recently it was shown that a complex containing Cul-5, elongins B and C, Rbx1, and the adenovirus proteins E4orf6 and E1B55K functions as an E3 to promote p53 ubiquitination and turnover (47).
Thus, cullins and their E3s are involved in the ubiquitin-mediated degradation of cell cycle regulators, and recent findings suggest a similar function for CUL-4A. CUL-4A mRNA is cell cycle regulated and is high during the G1- to S-phase transition (9). This gene is amplified and/or overexpressed in 11 breast cancer cell lines and in 31 primary breast cancers, which also suggests a role in regulating cell cycle progression (7). When CUL-4A was overexpressed in normal mammary epithelial cells, and after exposure to ionizing radiation, these cells do not accumulate with G2/M DNA content and do not accumulate p53, indicating that CUL-4A does have a cell cycle-regulatory function (15). In addition, CUL-4A protein associates with UV-damaged DNA-binding protein (DDB), and CUL-4A overexpression was recently shown to promote the ubiquitination and destabilization of the p48 subunit (DDB2) of DDB (8, 38, 53).
In vivo studies demonstrate that cullins are required for normal development. Inactivation of a Caenorhabditis elegans cullin, CUL-1, causes hyperplasia in all tissues of the developing embryo (26). Mice that are deficient for CUL-1 fail to develop past 5.5 days postcoitum (dpc) (10, 67), and CUL-3-null mice fail to develop past 7.5 dpc (54). However, the role of the CUL-4A cullin during development has not been examined.
Here, we report that CUL-4A−/− animals are inviable, and no homozygous mutant embryos were recovered as early as 7.5 dpc. However, CUL-4A−/− blastocysts are viable and are capable of hatching and forming both an inner cell mass and trophectoderm. Approximately 20% of the 7.5-dpc implantation sites appear empty, suggesting that the mutant embryos are able to implant but die between 4.5 and 7.5 dpc. Despite 87% similarity between the Cul-4A and Cul-4B cullins, the CUL-4A−/− lethal phenotype indicates that CUL-4A has one or more distinct functions. In addition, fewer heterozygous pups were observed than expected by Mendelian genetics, indicating that many of the CUL-4A+/− embryos also die. Therefore, appropriate CUL-4A expression is critical for normal early embryonic development.
MATERIALS AND METHODS
Disruption of CUL-4A in embryonic stem cells.
To generate a deletion allele of CUL-4A, genomic clones were isolated by screening a lambda 129SVJ mouse genomic library (Stratagene, La Jolla, Calif.) with a mouse CUL-4A cDNA that includes its entire coding sequence (P. Moeljadi and K. T. Chun, unpublished data). One genomic clone contains approximately 5 kb of 5′ untranslated sequence, the first two coding exons, the intervening intron, and the downstream 11 kb. From this clone, the 3.5-kb KpnI-NotI fragment and the downstream 4.4-kb NotI-PstI fragment were each subcloned separately into pBS (Stratagene) and used to generate a targeting construct (Fig. 1A). The NotI-PstI fragment contains the first two coding exons and the intervening intron. The KpnI site of the upstream 3.5-kb KpnI-NotI fragment was converted with oligonucleotides to an EcoRV site, and the resulting EcoRV-NotI fragment was excised from pBS and inserted into the PmeI and NotI sites of pTNLOX1-3 (a generous gift from Loren Field, Indiana University School of Medicine). Into the resulting plasmid, a loxP site encoded by synthetic oligonucleotides was inserted into the NotI site to generate pCUL-4A5′. The inserted oligonucleotides were designed such that the NotI site is destroyed at the upstream end of the insert and retained at the downstream end. In the final targeting construct, this loxP site lies 0.9 kb upstream of the first coding exon. Next, a 1.7-kb NotI-AscI fragment containing Neor transcribed from the PGK promoter and flanked by loxP sites (“floxed”) was excised from pTNLOX1-3, its ends were made blunt with Klenow, and NheI linkers were attached. This modified fragment was inserted into the 4.4-kb NotI-PstI CUL-4A fragment in pBS at an NheI site in the intron between the first and second coding exons and oriented such that Neor was transcribed in the direction opposite to that of CUL-4A. The resulting NotI-PstI fragment was excised with NotI and SalI (pBS polylinker site) and inserted into the NotI-SalI sites of pCUL-4A5′ to generate the final targeting construct (Fig. 1A).
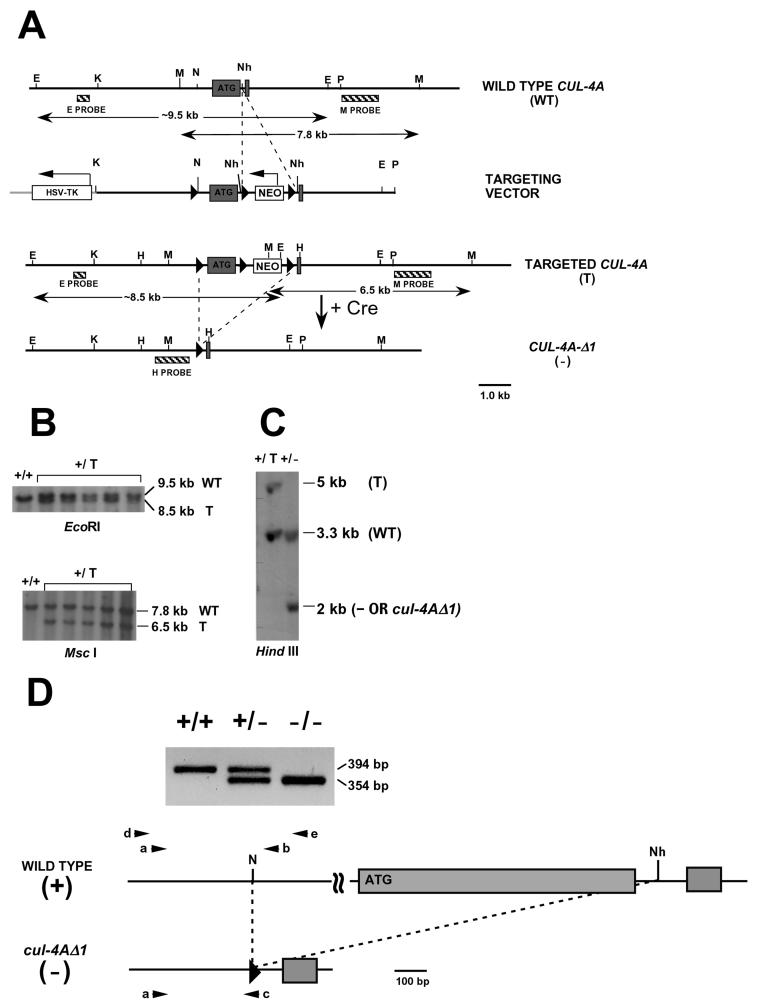
FIG. 1.
Targeted disruption of the mouse CUL-4A gene. (A) Partial restriction maps of the CUL-4 locus, targeting vector, targeted locus, and cul-4AΔ1 deletion. Exons are indicated by black boxes and “ATG” denotes the first coding exon. Restriction enzyme sites are denoted as follows: EcoRI, E; KpnI, K; MscI, M; NotI, N; NheI, Nh; PstI, P; HindIII, H. For the targeting vector, a single loxP site (black triangle) and a floxed PGK-neo cassette (black triangles flanking “NEO,” neomycin phosphotransferase) were inserted at NotI and NheI sites, respectively, as shown. An HSV-TK (Herpes simplex virus thymidine kinase) cassette lies next to the upstream arm of the CUL-4A fragment in the targeting vector. The single-headed arrows show the direction of transcription for the selection markers. In the maps for wild-type and targeted alleles, double-headed arrows underliethe distinguishing restriction enzyme fragments identified by Southern analysis. Probes are denoted by hatched boxes. Cre-mediated deletion removed the first coding exon and Neor cassette and generated the cul-4AΔ1 deletion allele. (B) Southern analyses of targeted ES cell clones digested with either EcoRI or MscI. The genotype of each clone is labeled at the top of each autoradiogram. (C) Southern analysis of ES cell clones after Cre-mediated recombination and digestion with HindIII. The genotype of each clone is labeled at the top of the autoradiogram. (D) Genotyping by PCR. Ethidium bromide-stained gel of PCR products from samples with the corresponding genotypes labeled at the top of the gel. Where the PCR primers, denoted by black triangles, anneal in the wild-type and/or cul-4AΔ1 alleles is depicted with maps of the corresponding alleles. Note that the scale in these two maps is 10-fold smaller than the maps in panel A.
Generation of cul-4AΔ1 mutant mice.
Mouse embryonic stem (ES) cell clones hemizygous for cul-4AΔ1 (Fig. 1A) were injected separately into C57BL/6 blastocysts and implanted into C57BL/6 pseudopregnant females (Jackson Laboratories, Bar Harbor, Maine). From one clone, two highly chimeric males resulted, which were backcrossed to C57BL/6 females, and offspring were genotyped by Southern analysis and PCR (details below) to detect germ line transmission from both males.
Antibody generation and immunoblot analysis.
Anti-Cul-4A antiserum was generated in a New Zealand White rabbit (Covance Research Products Inc., Denver, Pa.) using 6-His-tagged Cul-4A (6-His-Cul-4A) immunogen expressed from pET28a-CUL-4AΔN306 in Escherichia coli BL21(DE3) (Novagen, Madison, Wisc.). To construct this plasmid, the 2.7-kb StuI-NotI fragment containing the coding sequence for the 453 carboxy-terminal amino acids (deleting the first 306 codons, and leaving approximately 60% of the full-length Cul-4A) from the mouse CUL-4A cDNA was inserted into the NheI (made blunt with Klenow) and NotI sites of pET28a (Novagen). Protein expression was induced with 1.0 mM isopropyl-β-d-thiogalactopyranoside for 3 h at 37°C, and cells were harvested and then lysed with a French press in 50 mM NaH2PO4 (pH 8.0)-300 mM NaCl-10-μg/ml aprotinin-2-μg/ml pepstatin-5-μg/ml leupeptin-100-μg/ml Pefablock. Under these conditions, the 6-His-Cul-4A protein is insoluble, so this fraction was pelleted at 10,000 × g, solubilized in protein sample buffer with 2% sodium dodecyl sulfate (SDS), and subjected to SDS-polyacrylamide gel electrophoresis. The 6-His-Cul-4A protein was then electroeluted, concentrated, and injected into rabbits.
For immunoblots, cells were lysed for 30 min on ice in 50 mM Tris-HCl (pH 7.5)-10 mM EDTA-1% SDS-1 mM dithiothreitol-2 mM phenylmethylsulfonyl fluoride-15-μg/ml aprotinin-2-μg/ml pepstatin-5-μg/ml leupeptin followed by probe sonication for 10 s at 4°C. Protein lysates were cleared by centrifugation at 16,000 × g, 4°C, for 20 min, and protein concentration was measured by the Bradford assay (Bio-Rad, Hercules, Calif.), 10 μg of total cell lysate was fractionated by SDS-polyacrylamide gel electrophoresis, and proteins were transferred to a polyvinylidene difluoride membrane (Millipore; Bedford, Mass.) in 10 mM 3-[cyclohexylamino]-1-propanesulfonic acid (pH 11.0)-10% methanol. Membranes were blocked for 1 h in blocking buffer (10 mM Tris-HCl [pH 7.4], 50 mM NaCl, 0.2% Tween 20, 5% nonfat dry milk), washed three times for 10 min each in KPBS (pH 7.3) (137 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, 1.5 mM KH2PO4) supplemented with 0.2% Tween 20, probed for 1 h with 1/3,000-diluted anti-Cul-4A antiserum in blocking buffer, washed, probed with 1/10,000-diluted anti-rabbit immunoglobulin G antibody conjugated to horseradish peroxidase (A-6154; Sigma-Aldrich Co., St. Louis, Mo.) in blocking buffer, and then washed. Antibodies were visualized by enhanced chemiluminescence (NEN Life Science Products, Boston, Mass.). Actin was detected with antiactin monoclonal antibody (H5441; Sigma-Aldrich Co.) diluted 1/4,000, followed by anti-mouse immunoglobulin G antibody conjugated to horseradish peroxidase (NA931; Amersham Life Science; Buckinghamshire, United Kingdom) diluted 1/10,000.
A deletion mutation in the CUL-4A cDNA was generated by PCR and subcloned into pEF-PGKpac for expression in PLB-985 cells, a myelomonoblastic human cell line (65, 71). This mutation removes the first 266 codons and encodes a predicted 59-kDa truncated protein.
Timed pregnancies.
To generate timed pregnancies, CUL-4A+/− females were injected intraperitoneally with pregnant mare's serum gonadotropin (5 IU per animal; Sigma-Aldrich Co. or Professional Compounding Centers of America, Houston, Tex.), followed by injection with human chorionic gonadotropin (5 IU per animal; Sigma-Aldrich Co. or Serono Laboratories [Randolph, Mass.]) 48 h later and mating overnight with CUL-4A+/− males (18). The next morning, males were removed and females were examined for the presence of a vaginal plug. Females were sacrificed at either 7.5, 8.5, or 10.5 dpc to isolate embryos. These were genotyped by PCR (see below).
Isolation of 0.5-dpc embryos and in vitro culturing.
Four- to six-week-old heterozygous females were superovulated by treatment with gonadotropins as described above. At 0.5 dpc, females were sacrificed, and embryos were isolated from the oviducts and cultured for 4 days in a drop of M16 medium under mineral oil (18). For blastocyst outgrowths, individual blastocysts were placed on gelatin in 1 ml of ES cell medium (18) supplemented with leukemia inhibitory factor and cultured for 4 more days. Outgrowths were photographed, washed with phospate-buffered saline, and lysed for genotyping by PCR (described below).
Southern and PCR analyses for genotyping.
To genotype ES cell lines and identify targeted clones, 10 μg of genomic DNA was digested with EcoRI, separated on a 0.5% agarose gel in 1× Tris-borate-EDTA buffer, blotted onto a nylon membrane (Magnagraph; Osmonics, Westborough, Mass.), and probed with a 365-bp PCR fragment (E probe, Fig. 1A) that hybridizes upstream of the targeted region of CUL-4A. The 9.5-kb band corresponds to the wild-type allele, and the 8.5-kb band corresponds to the targeted allele (Fig. 1B). To demonstrate further that the targeted alleles are at the CUL-4A locus, genomic DNA was digested with MscI, separated by electrophoresis, blotted, and probed with a 1.2-kb PstI-EcoRI fragment (M probe; Fig. 1A) that hybridizes downstream of the targeted region of CUL-4A. The 7.8-kb fragment corresponds to the wild-type allele, and the 6.5-kb fragment corresponds to the targeted allele (Fig. 1B).
To genotype ES cell lines after Cre-mediated deletion, genomic DNA from each clone was digested with HindIII, separated by electrophoresis, blotted, and probed with a 1.2-kb PCR fragment that corresponds to the region 2.4 to 1.2 kb upstream of the first coding exon (H probe, Fig. 1A). The 5-kb fragment corresponds to the targeted allele (no deletion), the 3.3-kb fragment corresponds to the wild-type allele, and the 2-kb fragment corresponds to the deletion allele (cul-4AΔ1) (Fig. 1C).
To genotype weaned pups, genomic DNA was purified from approximately 0.5 cm of tail and analyzed by either Southern analysis or PCR. For Southern analysis, 10 or 20 μg of DNA were digested with HindIII and probed with the H probe as described above (Fig. 1A). For PCR analysis, genomic DNA was used as the template in a reaction where wild-type and mutant alleles were detected simultaneously. One oligonucleotide primer (primer A: TGCCAAGAGACACAAGGTGCTTCG) hybridizes to both wild-type and cul-4AΔ1 alleles (Fig. 1D). Primer B (CCAACGTGCAGAGGTTACCG) hybridizes to the wild-type allele only, and primer C (GGCCGCATAACTTCGTATAATGTATGCTATACGAAGTTATC) binds to the loxP site present only in the cul-4AΔ1 allele. Briefly, in a 50-μl reaction volume, 100 ng of genomic DNA and 20 pmol each of primers A, B, and C were amplified in PCR buffer (Promega, Madison, Wisc.) supplemented with 1.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, and 1.25 U of Taq polymerase (Promega) at 95°C for 3 min and then 35 cycles of 45 s at 94°C, 45 s at 55°C, and 90 s at 72°C, followed by 6 min at 72°C. Twenty microliters of each reaction mixture was separated on a 2.0% agarose gel in 0.5× Tris-acetate-EDTA buffer. Primers A and B amplify a 394-bp fragment from the wild-type allele, and primers A and C amplify a 354-bp fragment from the cul-4AΔ1 allele (Fig. 1D).
Embryos at 7.5, 8.5, and 10.5 dpc were genotyped by PCR analysis of DNA purified from either the yolk sac or the whole embryo. Tissues were digested overnight in 300 μl of lysis buffer (100 mM Tris-HCI [pH 8.5], 5 mM EDTA, 0.2% SDS, 200 mM NaCl, and 200 μg of proteinase K/ml) at 56°C. DNA was then purified by phenol-chloroform extraction, followed by isopropanol precipitation, and then dissolved in 50 μl of 5 mM Tris-HCl (pH 8.0). Two microliters of purified DNA was used for PCR as described above.
Blastocysts and blastocyst outgrowths were genotyped by nested PCR. Outgrowths were washed once with PBS. Then either blastocysts or outgrowths were lysed in 20 μl of PCR lysis buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 2.5 mM MgCl2, 0.1 μg of gelatin/ml, 0.45% NP-40, 0.45% Tween 20, 200 μg of proteinase K/ml) at 56°C for 1 h and then 95°C for 15 min. Two microliters were then amplified in the first PCR with primers C (described above), D (CAACAGTGTTTGCTTGTGCCACTCCCAGGC), and E (TGACAGCTGCCCCCAAAGAAGTGCTCTCAC) (Fig. 1D). The PCR conditions were the same as that described above, except the annealing temperature was 60°C instead of 55°C. Two microliters of the PCR product was used as the template in a subsequent reaction with primers A, B, and C as described for mouse tails and dissected embryos.
RESULTS
Generation and characterization of CUL-4A−/− mice.
To determine the function of CUL-4A during development, mouse ES cell lines with a mutant CUL-4A allele were generated. To construct this allele, a 7.9-kb fragment of the CUL-4A gene that includes its first two coding exons was isolated and subcloned (Fig. 1A) (see Materials and Methods). Because disrupting either CUL-1 or CUL-3 causes an embryonic-lethal phenotype (10, 54, 67), we used the Cre/loxP strategy to generate a CUL-4A deletion allele (described in this report) as well as a conditional disruption allele (K. T. Chun, unpublished data) (reviewed in reference 48). LoxP sites were inserted to flank the first coding exon. The downstream loxP site lies in the downstream intron and is followed by a selectable marker for neomycin resistance and another loxP site (Fig. 1A). In this way, the first coding exon and approximately 1.1 kb of the upstream sequence is floxed. This entire construct was inserted into a plasmid to generate the targeting construct (Materials and Methods).
Mouse ES cells (CCE916) were transfected with the targeting construct, and G418-resistant, ganciclovir-resistant clones were selected to enrich for homologous recombination events as previously described (49, 50). One hundred twenty-seven of these clones were analyzed by Southern analysis to identify five where the targeting construct had undergone homologous recombination at one of the CUL-4A loci (Fig. 1A and B) (see Materials and Methods). Southern analysis with a probe that hybridizes upstream of the targeted region of CUL-4A (E probe; Fig. 1A and B) identifies a 9.5-kb fragment for the wild-type allele and an 8.5-kb fragment for the targeted allele. Analysis with a probe that hybridizes downstream of the targeted region (M probe; Fig. 1A and B) identifies 7.8- and 6.5-kb fragments for the wild-type and targeted alleles, respectively. These analyses confirm the presence of the floxed Neor cassette at the CUL-4A locus. To determine whether the loxP site upstream of the first coding exon is present, PCR and Southern analyses were performed (data not shown). In one clone this site is absent, but it is present in the other four.
These four clones were transiently transfected with a plasmid that encodes the Cre recombinase (pBS185) (51). This plasmid lacks a selectable marker for mammalian cells, so the Cre recombinase was expressed only transiently in transfected cells. After transfection, cells were plated at low density, individual clones were grown and picked, and 153 were genotyped by Southern analysis (H probe; Fig. 1A and C; also see Materials and Methods) to identify those where the targeted locus (corresponding to a 5-kb fragment) had undergone a Cre-mediated deletion. In 11 clones, the first coding exon and the Neor marker were deleted (cul-4AΔ1, identified by a 2-kb fragment). With one of these cell lines, two chimeric male mice that exhibit germ line transmission of cul-4AΔ1 were generated (see Materials and Methods).
Upon Cre-mediated recombination to remove the first coding exon, the resulting deletion mutation should result in a nonfunctional CUL-4A gene. This deletion removes the first 49 codons and the first base pair of the 50th, generating a frameshift mutation as well. Two out-of-frame ATG codons are present before the first in-frame ATG occurs at codon 146. Therefore, even in the unlikely event that translation begins at codon 146, the first 145 amino acids would be absent, resulting in a truncated protein that is only 614 amino acids long instead of the wild-type 759 amino acids. The first 108 amino acids of Cul-2, the first 249 amino acids of Cul-1, and the first 280 amino acids of Cdc53 are required for these cullins to function normally (10, 54, 67). Therefore, it was likely that deleting the first coding exon of CUL-4A would result in a null phenotype.
Offspring hemizygous for the cul-4AΔ1 allele were intercrossed, and 235 weaned pups were genotyped by PCR analysis (Fig. 1A and D; Table 1; also see Materials and Methods). None were homozygous for the deletion allele, indicating that animals lacking functional CUL-4A are not viable (statistically significant at P < 0.001, chi-square test). Surprisingly, 47% of the viable pups were wild type and only 53% were heterozygotes. Assuming a Mendelian ratio of 1:2 for wild-type animals to heterozygotes, twice the number of wild-type animals (111) would be expected to be heterozygotes (222). Therefore, approximately 44% of these expected heterozygotes are also inviable (222 heterozygotes expected minus 124 observed, out of 222 heterozygotes expected), and this deviation from the expected is statistically significant (P < 0.001, chi-square test). However, for the viable heterozygotes, there is no overt phenotypic difference between these and wild-type animals.
TABLE 1.
Genotype of progeny from heterozygous matingsa
| Stage | No. with genotype
|
No. of Empty implantation sites | ||
|---|---|---|---|---|
| +/+ | +/− | −/− | ||
| At weaning | 111 | 124b | 0b | |
| 10.5-dpc embryos | 10 | 15 | 0c | 14 |
| 8.5-dpc embryos | 6 | 12 | 0c | 9 |
| 7.5-dpc embryos | 12 | 20 | 0c | 8 |
| Blastocysts | 10 | 20 | 10 | |
Genotypes were determined by PCR.
Observed frequency departs significantly from the expected ratio of 1:2:1 for +/+:+/−:−/− with a P value of <0.001 (chi-square test).
Observed frequency also departs significantly from the expected ratio with a P value of <0.01 (chi-square test).
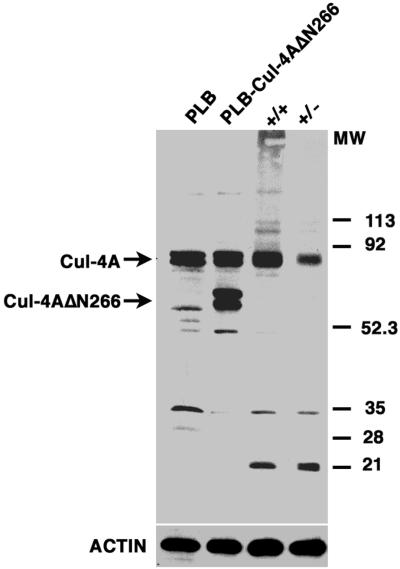
As described above, deletion of the first coding exon left an in-frame ATG at codon 146, so it was possible that a truncated, 614-amino-acid (approximately 68-kDa) CUL-4A protein was expressed instead of the 84-kDa wild-type protein. To examine whether the cul-4AΔ1 allele is null or results in the expression of a truncated CUL-4A protein, thymus total cell lysates from wild type and hemizygotes were analyzed by immunoblot and probed with anti-Cul-4A antiserum (Materials and Methods). As shown in Fig. 2, no truncated forms of Cul-4A were detected. In addition, the amount of Cul-4A expressed in the hemizygote is roughly half that expressed in the wild type. If a truncated gene product had been expressed, this antiserum should have detected it, because all of the portion of Cul-4A used as the immunogen is included in the possible truncated product from the cul-4AΔ1 allele. To confirm that the antiserum would detect this possible cul-4AΔ1 truncated protein, a truncated Cul-4A protein that lacks the first 266 amino acids was expressed in PLB-985 cells (a human cell line; see Materials and Methods). As shown in Fig. 2, the anti-Cul-4A antiserum clearly recognizes this truncated protein. Endogenous, full-length Cul-4A and truncated Cul-4A each appear as doublets, which is consistent with the finding that Cul-4A is modified by the attachment of a ubiquitin-related peptide, Nedd8 (19, 41). Therefore, it is likely that no truncated products are expressed and that cul-4AΔ1 is a null allele.
FIG. 2.
A truncated protein is not expressed from cul-4AΔ1. Protein lysates were prepared from wild-type PLB-985 cells (PLB), PLB-985 cells expressing a truncated CUL-4A protein that lacks its amino-terminal 266 amino acids (PLB-Cul-4AΔN266), thymus total lysate from a wild-type mouse (+/+), and thymus total lysate from a hemizygous littermate (+/−) (see Materials and Methods). Lysates were fractionated on a 7.5% polyacrylamide gel, and an immunoblot was probed with anti-Cul-4A antiserum. Full-length and truncated Cul-4A are each denoted by an arrow, and actin levels were measured to ensure equal loading. MW, molecular weight, given in thousands.
To determine at what stage of development the homozygous mutant embryos die, 7.5-, 8.5-, and 10.5-dpc embryos from heterozygote intercrosses were isolated and genotyped by PCR (Table 1; see Materials and Methods). No homozygous mutant embryos were recovered at each stage, indicating that CUL-4A−/− embryos die prior to 7.5 dpc (P < 0.01). Approximately 20% (8 of 40) of the 7.5-dpc implantation sites appeared empty, half (20 of 40) yielded heterozygous embryos, and similar proportions were observed at 8.5 dpc. However, a slightly smaller proportion than the expected (15 of 39, or 38% instead of 50%) yielded heterozygotes at 10.5 dpc, but this deviation is not statistically significant.
In vitro analysis of early embryos.
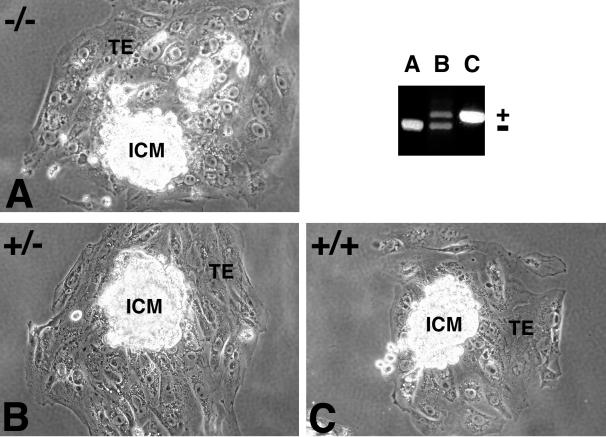
The failure of CUL-4A−/− embryos to develop past 7.5 dpc suggests that CUL-4A may be required for cell viability. To begin to assess CUL-4A's role in this function, 0.5-dpc embryos from heterozygote intercrosses were isolated and allowed to develop for 4 days in vitro, and then viable blastocysts were genotyped by PCR (see Materials and Methods). Twenty-five percent of these were homozygous mutant, 50% were heterozygotes, and 25% were wild type, indicating that CUL-4A is not required for blastocyst formation (Table 1). Similarly prepared blastocysts were cultured on gelatinized petri dishes 4 days longer (in the presence of LIF) and then genotyped. Mutant blastocysts formed an inner cell mass and trophectoderm and were morphologically indistinguishable from wild-type and heterozygous outgrowths (Fig. 3).
FIG. 3.
CUL-4A−/− blastocysts grown in vitro exhibit normal outgrowth characteristics. Blastocysts derived from CUL-4A+/− intercrosses were cultured in vitro for 4 days. The presence of both an inner cell mass (ICM) and trophectoderm (TE) were detected in each outgrowth. The lens objective was ×32. The genotype of each outgrowth was determined by PCR and appears in the upper left corner of each panel. The electrophoresed PCR products for genotyping appear with each lane labeled with the letter of the corresponding outgrowth. PCR products corresponding to the wild-type or deletion allele are labeled by + or −, respectively (see Materials and Methods).
DISCUSSION
To determine the in vivo function of CUL-4A, we generated a mouse line hemizygous for a CUL-4A deletion allele. No viable, homozygous mutant offspring were obtained from heterozygote intercrosses, indicating that CUL-4A is required for normal embryonic development. One quarter of the viable blastocysts (roughly 3.5-dpc embryos) recovered from similar intercrosses were homozygous mutant, and such embryos were able to hatch and form an inner cell mass and trophectoderm, indicating that CUL-4A is not required for development to this stage. However, upon analysis of 7.5- to 10.5-dpc embryos, no CUL-4A−/− embryos were recovered. Approximately one-quarter of the implantation sites observed at these time points appeared empty, indicating that CUL-4A−/− embryos are able to hatch and implant (4.5 dpc) but fail to develop past a point between this stage and 7.5 dpc.
Surprisingly, 44% fewer heterozygous pups were observed than expected by Mendelian genetics, indicating that many of the heterozygotes from these intercrosses are also dying and suggesting that appropriate CUL-4A expression is required for embryonic development. No truncated CUL-4A gene product was detected in lysates from heterozygotes, and the amount of full-length Cul-4A was roughly half that in wild-type littermates, so it is likely that this semidominance results from a reduction in CUL-4A expression and not from a dominant mutant, truncated Cul-4A. Also, the Mendelian proportion of heterozygotes was observed at 10.5 dpc, indicating that the inviable heterozygotes are dying at a stage later than the homozygous mutants. None of the other cullin null alleles generated thus far cause an apparent phenotype in heterozygotes, so the haploinsufficiency of the CUL-4A null allele is novel for this gene family.
Other cullins participate in several different E3 complexes, and each complex often has multiple substrates (reviewed in references 11 and 66). Therefore, it is likely that there are multiple substrates for the Cul-4A-containing E3(s) and that the failure to regulate the degradation of one or more of these substrates results in early embryonic death. A candidate for such a factor is DDB2, which was identified as a likely substrate of Cul-4A-containing E3 (8, 38, 53). DDB2 is mutated in some xeroderma pigmentosum group E individuals, and this gene is required for the p53-dependent activation of the global genomic DNA repair of UV radiation damage (39, 63). A database search for DDB2 cDNAs identified an expressed sequence tag (GenBank accession no. AA516636) from a cDNA that was generated from 6.5-dpc mouse embryos, so it is likely that DDB2 is expressed during the same stage of development when CUL-4A is required. Therefore, dysregulated DDB2 degradation may contribute to the embryonic-lethal phenotype of CUL-4A−/− mice. However, there is no other evidence to indicate a requirement for DDB2 during development. For example, it is not known whether the overexpression of DDB2 by another means (e.g., DDB2 transgenic mice) results in the same phenotype. The cul-4AΔ1 mice provide a suitable system with which to elucidate the molecular mechanism of CUL-4A function during early embryonic development.
As components of ubiquitin ligases that regulate cell cycle regulators, cullins have been shown to be critical for normal cell cycle progression, and this appears to be the case for CUL-4A as well. The observations that CUL-4A is overexpressed in breast cancer and that its mRNA levels are cell cycle regulated suggest a role in regulating cell cycle progression (7, 9). Also, CUL-4A overexpression appears to interfere with the G2/M checkpoint after exposure to ionizing radiation in mammary epithelial cells (15). At 6.5 dpc, there are approximately 500 cells in the mouse embryo, which rapidly increase to over 100,000 cells in the 8.5-dpc embryo, so it is likely that during this rapid proliferation, the proper regulation of cell cycle progression is critical. Our results indicate that CUL-4A is required during roughly this same period of development. Also, CUL-1 and CUL-3 are each required during this period of embryonic development, and each is involved in targeting cell cycle regulators for degradation (10, 54, 67). Apparently, multiple E3s, including one containing Cul-4A, are required to help orchestrate developmental events occurring during this early embryonic stage.
The Cul-4A amino acid sequence is 78% identical and 87% similar to that encoded by its most closely related gene, CUL-4B (GenBank accession no. NM_028288). However, despite this high degree of similarity, disrupting CUL-4A causes a dramatic phenotype, demonstrating that one or more of CUL-4A's functions are distinct from those of CUL-4B. In fact, the predicted amino-terminal amino acid sequence for Cul-4B is different from the first 40 amino acids predicted for Cul-4A, while the remainder of Cul-4A and -4B are 82% identical and 91% similar. In other cullins, the amino terminus is required for binding Skp1 or elongin C and F-box or SOCS-box proteins, which are all involved in substrate recognition (11). Perhaps Cul-4A and Cul-4B bind different adaptor and substrate recognition subunits, which results in their corresponding E3s targeting different substrates for degradation and, in turn, distinct functions. Alternatively, these two genes might exhibit very different expression patterns, resulting in their requirement during different periods of development and/or in different tissues. In fact, we and others observed that in adult human tissues, CUL-4A is most highly expressed in skeletal muscle, cardiac muscle, and the testis (A. W. Roberts and K. T. Chun, unpublished data; 19). CUL-4B is most highly expressed in the spleen, prostate, testis, ovary, and colon and in leukocytes (19). Also, CUL-4B expression was reported to be especially high at day 11 of mouse embryonic development, while CUL-4A expression was not markedly higher at this stage (19).
Studies with mice, C. elegans, plants, and Dictyostelium suggest that cullins, their E3 ligases, and ubiquitin-mediated degradation play critical roles in development (10, 13, 14, 26, 37, 54, 67). For example, the plant hormone auxin is critical for regulating cell division and differentiation, and in Arabidopsis thaliana, mutations in genes encoding SCF subunits (ASK1, related to Skp1, and TIR1, an F-box protein) dramatically alter developmental responses to auxin (14). In Dictyostelium, an apparent E3 containing CulA and FbxA target RegA (required for normal development) for degradation (37). A mutation in either CulA (most closely related to mouse and human CUL-1) or FbxA (encoding an F-box protein) causes a failure to downregulate RegA and defects in cell-type differentiation and development.
The studies described here demonstrate that CUL-4A contributes an essential function for early mammalian development and suggest that as little as a twofold decrease in expression can cause embryonic death. Nearly half of mice heterozygous for the cul-4AΔ1 allele die in utero, apparently during the second half of gestation, thus providing a useful system with which to study the role of CUL-4A in later stages of development. Further study will also be required to determine the mechanism by which CUL-4A is required for early embryonic development. This will include comparing differentiation and cell cycle progression during early embryonic development in CUL-4A-null and wild-type animals. The early death of CUL-4A−/− embryos makes discerning these processes difficult. We are currently generating CUL-4A−/− ES cell lines to examine whether normal differentiation in vitro requires CUL-4A and if so, to identify Cul-4A downstream effectors, perhaps E3 substrates, whose dysregulation ultimately leads to the embryonic-lethal phenotype.
Acknowledgments
This work was supported by an American Heart Association Midwest Affiliate Scientist Development Grant (9930341Z), an Indiana University Cancer Center Development Grant (NIH P30CA82709), a grant from the Showalter Research Trust Fund, an American Cancer Society Indiana University Institutional Research Grant, and an Indiana University School of Medicine Core Centers of Excellence in Molecular Hematology grant (NIH P30DK49218) awarded to K.T.C. and Riley Memorial Association grants awarded separately to J.C.R. and K.T.C.
We thank Dave Skalnik and Merv Yoder for advice and helpful discussions, Loren Field for advice and for expertly isolating 0.5-dpc embryos, the Indiana University Cancer Center knockout mouse core facility for ES cell transfections, blastocyst injections, and generation of chimeric mice, and Stephany Scruggs, Heather Noffsinger, Candice Horn, and Prianto Moeljadi for diligent technical assistance.
REFERENCES
- 1.Aso, T., D. Haque, R. J. Barstead, R. C. Conaway, and J. W. Conaway. 1996. The inducible elongin A elongation activation domain: structure, function and interaction with the elongin BC complex. EMBO J. 15:5557-5566. [PMC free article] [PubMed] [Google Scholar]
- 2.Bai, C., P. Sen, K. Hofmann, L. Ma, M. Goebl, J. W. Harper, and S. J. Elledge. 1996. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-Box. Cell 86:263-274. [DOI] [PubMed] [Google Scholar]
- 3.Borden, K. L., and P. S. Freemont. 1996. The RING finger domain: a recent example of a sequence-structure family. Curr. Opin. Struct. Biol. 6:395-401. [DOI] [PubMed] [Google Scholar]
- 4.Burnatowska-Hledin, M., P. Zhao, B. Capps, A. Poel, K. Parmelee, C. Mungall, A. Sharangpani, and L. Listenberger. 2000. VACM-1, a cullin gene family member, regulates cellular signaling. Am. J. Physiol. Cell Physiol. 279:C266-C273. [DOI] [PubMed] [Google Scholar]
- 5.Byrd, P. J., T. Stankovic, C. M. McConville, A. D. Smith, P. R. Cooper, and A. M. R. Taylor. 1997. Identification and analysis of expression of human VACM-1, a cullin gene family member located on chromosome 11q22-23. Genome Res. 7:71-75. [DOI] [PubMed] [Google Scholar]
- 6.Carrano, A. C., E. Eytan, A. Hershko, and M. Pagano. 1999. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat. Cell Biol. 1:193-199. [DOI] [PubMed] [Google Scholar]
- 7.Chen, L.-C., S. Manjeshwar, Y. Lu, D. Moore, B.-M. Ljung, W.-L. Kuo, S. H. Dairkee, M. Wernick, C. Collins, and H. S. Smith. 1998. The human homologue for the Caenorhabditis elegans cul-4 gene is amplified and overexpressed in primary breast cancers. Cancer Res. 58:3677-3683. [PubMed] [Google Scholar]
- 8.Chen, X., Y. Zhang, L. Douglas, and P. Zhou. 2001. UV-damaged DNA binding proteins are targets of Cul4A-mediated ubiquitination and degradation. J. Biol. Chem. 276:48175-48182. [DOI] [PubMed] [Google Scholar]
- 9.Cho, R. J., M. Huang, M. J. Campbell, H. Dong, L. Steinmetz, L. Sapinoso, G. Hampton, S. J. Elledge, R. W. Davis, and D. J. Lockhart. 2001. Transcriptional regulation and function during the human cell cycle. Nat. Genet. 27:48-54. [DOI] [PubMed] [Google Scholar]
- 10.Dealy, M. J., K. V. T. Nguyen, J. Lo, M. Gstaiger, W. Krek, D. Elson, J. Arbeit, E. T. Kipreos, and R. S. Johnson. 1999. Loss of Cul1 results in early embryonic lethality and dysregulation of cyclin E. Nat. Genet. 23:245-248. [DOI] [PubMed] [Google Scholar]
- 11.Deshaies, R. J. 1999. SCF and Cullin/RING H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15:435-467. [DOI] [PubMed] [Google Scholar]
- 12.Duan, D. R., A. Pause, W. H. Burgess, T. Aso, D. Y. Chen, K. P. Garrett, R. C. Conaway, J. W. Conaway, W. M. Linehan, and R. D. Klausner. 1995. Inhibition of transcription elongation by the VHL tumor suppressor protein. Science 269:1402-1406. [DOI] [PubMed] [Google Scholar]
- 13.Fay, D. S., and M. Han. 2000. Mutations in cye-1, a Caenorhabditis elegans cyclin E homolog, reveal coordination between cell-cycle control and vulval development. Development 127:4049-4060. [DOI] [PubMed] [Google Scholar]
- 14.Gray, W. M., J. C. del Pozo, L. Walker, L. Hobbie, E. Risseeuw, T. Banks, W. L. Crosby, Y. M., H. Ma, and M. Estelle. 1999. Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 13:1678-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta, A., L. X. Yang, and L. Chen. 2002. Study of the G2/M cell cycle checkpoint in irradiated mammary epithelial cells overexpressing Cul-4A gene. Int. J. Radiat. Oncol. Biol. Phys. 52:822-830. [DOI] [PubMed] [Google Scholar]
- 16.Hatakeyama, S., M. Kitagawa, K. Nakayama, M. Shirane, M. Matsumoto, K. Hattori, H. Higashi, H. Nakano, K. Okumura, K. Onoe, R. A. Good, and K. Nakayama. 1999. Ubiquitin-dependent degradation of IκBα is mediated by a ubiquitin ligase Skp1/Cul 1/F-box protein FWD1. Proc. Natl. Acad. Sci. USA 96:3859-3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 18.Hogan, B., R. Beddington, F. Costantini, and E. Lacy. 1994. Manipulating the mouse embryo: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Woodbury, N.Y.
- 19.Hori, T., F. Osaka, T. Chiba, C. Miyamoto, K. Okabayashi, N. Simbara, S. Kato, and K. Tanaka. 1999. Covalent modification of all members of human cullin family proteins by NEDD8. Oncogene 18:6829-6834. [DOI] [PubMed] [Google Scholar]
- 20.Iwai, K., K. Yamanaka, T. Kamura, N. Minato, R. C. Conaway, J. W. Conaway, R. D. Klausner, and A. Pause. 1999. Identification of the von Hippel-Lindau tumor-suppressor protein as part of an active E3 ubiquitin ligase complex. Proc. Natl. Acad. Sci. USA 96:12436-12441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamizono, S., T. Hanada, H. Yasukawa, S. Minoguchi, R. Kato, M. Minoguchi, K. Hattori, S. Hatakeyama, M. Yada, S. Morita, T. Kitamura, H. Kato, K. Nakayama, and A. Yoshimura. 2001. The SOCS box of SOCS-1 accelerates ubiquitin-dependent proteolysis of TEL-JAK2. J. Biol. Chem. 276:12530-12538. [DOI] [PubMed] [Google Scholar]
- 22.Kamura, T., D. Burian, Q. Yan, S. L. Schmidt, W. S. Lane, E. Querido, P. E. Branton, A. Shilatifard, R. C. Conaway, and J. W. Conaway. 2001. Muf1, a novel elongin BC-interacting leucine-rich repeat protein that can assemble with Cul5 and Rbx1 to reconstitute a ubiquitin ligase. J. Biol. Chem. 276:29748-29753. [DOI] [PubMed] [Google Scholar]
- 23.Kamura, T., D. M. Koepp, M. N. Conrad, D. Skowyra, R. J. Moreland, O. Iliopoulos, W. S. Lane, W. G. Kaelin, Jr., S. J. Elledge, R. C. Conaway, J. W. Harper, and J. W. Conaway. 1999. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science 284:657-661. [DOI] [PubMed] [Google Scholar]
- 24.Kamura, T., S. Sato, D. Haque, L. Liu, W. G. Kaelin, R. C. Conaway, and J. W. Conaway. 1998. The elongin BC complex interacts with the conserved SOCS-box motif present in members of the SOCS, ras, WE-40 repeat, and ankyrin repeat families. Genes Dev. 12:3872-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kibel, A., O. Iliopoulos, J. A. DeCaprio, and W. G. Kaelin, Jr. 1995. Binding of the von Hippel-Lindau tumor suppressor protein to elongin B and C. Science 269:1444-1446. [DOI] [PubMed] [Google Scholar]
- 26.Kipreos, E. T., L. E. Lander, J. P. Wing, W. W. He, and E. M. Hedgecock. 1996. cul1-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell 85:829-839. [DOI] [PubMed] [Google Scholar]
- 27.Kroll, M., F. Margottin, A. Kohl, P. Renard, H. Durand, J. P. Concordet, F. Bachelerie, F. Arenzana-Seisdedos, and R. Benarous. 1999. Inducible degradation of IκBα by the proteasome requires interaction with the F-box protein h-βTrCP. J. Biol. Chem. 274:7941-7945. [DOI] [PubMed] [Google Scholar]
- 28.Lacronique, V., A. Boureux, V. D. Valle, H. Poirel, C. T. Quang, M. Mauchauffe, C. Berthou, M. Lessard, R. Berger, J. Ghysdael, and O. A. Bernard. 1997. A TEL-JAK2 fusion protein with constitutive kinase activity in human leukemia. Science 278:1309-1312. [DOI] [PubMed] [Google Scholar]
- 29.Lisztwan, J., G. Imbert, C. Wirbelauer, M. Gstaiger, and W. Krek. 1999. The von Hippel-Lindau tumor suppressor protein is a component of an E3 ubiquitin-protein ligase activity. Genes Dev. 13:1822-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lisztwan, J., A. Marti, H. Sutterlüty, M. Gstaiger, C. Wirbelauer, and W. Krek. 1998. Association of human CUL-1 and ubiquitin-conjugating enzyme CDC34 with the F-box protein p45SKP2: evidence for evolutionary conservation in the subunit composition of the CDC34-SCF pathway. EMBO J. 17:368-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lonergan, K. M., O. Iliopoulos, M. Ohh, T. Kamura, R. C. Conaway, J. W. Conaway, and W. G. Kaelin, Jr. 1998. Regulation of hypoxia-inducible mRNAs by the von Hippel-Lindau tumor suppressor protein requires binding to complexes containing elongins B/C and Cul2. Mol. Cell. Biol. 18:732-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lyapina, S. A., C. C. Correll, E. T. Kipreos, and R. J. Deshaies. 1998. Human CUL1 forms an evolutionarily conserved ubiquitin ligase complex (SCF) with SKP1 and an F-box protein. Proc. Natl. Acad. Sci. USA 95:7451-7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marti, A., C. Wirbelauer, M. Scheffner, and W. Krek. 1999. Interaction between ubiquitin-protein ligase SCFSKP2 and E2F-1 underlies the regulation of E2F-1 degradation. Nat. Cell. Biol. 1:14-19. [DOI] [PubMed] [Google Scholar]
- 34.Mathias, N., S. L. Johnson, M. Winey, A. E. M. Adams, L. Goetsch, J. R. Pringle, B. Byers, and M. G. Goebl. 1996. Cdc53p acts in concert with Cdc4p and Cdc34p to control the G1-to-S phase transition and identifies a conserved family of proteins. Mol. Cell. Biol. 16:6634-6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maxwell, P. H., M. S. Wiesener, G. W. Chang, S. C. Clifford, E. C. Vaux, M. E. Cockman, C. C. Wykoff, C. W. Pugh, E. R. Maher, and P. J. Ratcliffe. 1999. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399:271-275. [DOI] [PubMed] [Google Scholar]
- 36.Moberg, K. H., D. W. Bell, D. C. Wahrer, D. A. Haber, and I. K. Hariharan. 2001. Archipelago regulates Cyclin E levels in Drosophila and is mutated in human cancer cell lines. Nature 413:311-316. [DOI] [PubMed] [Google Scholar]
- 37.Mohanty, S., S. Lee, N. Yadava, M. J. Dealy, R. S. Johnson, and R. A. Firtel. 2001. Regulated protein degradation controls PKA function and cell-type differentiation in Dictyostelium. Genes Dev. 15:1435-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nag, A., T. Bondar, S. Shiv, and P. Raychaudhuri. 2001. The xeroderma pigmentosum group E gene product DDB2 is a specific target of cullin 4A in mammalian cells. Mol. Cell. Biol. 21:6738-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nichols, A. F., P. Ong, and S. Linn. 1996. Mutations specific to the xeroderma pigmentosum group E Ddb− phenotype. J. Biol. Chem. 271:24317-24320. [DOI] [PubMed] [Google Scholar]
- 40.Ohta, T., J. J. Michel, A. J. Schottelius, and Y. Xiong. 1999. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol. Cell 3:535-541. [DOI] [PubMed] [Google Scholar]
- 41.Osaka, F., H. Kawasaki, N. Aida, M. Saeki, T. Chiba, S. Kawashima, K. Tanaka, and S. Kato. 1998. A new NEDD8-ligating system for cullin-4A. Genes Dev. 12:2263-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patton, E. E., A. R. Willems, D. Sa, L. Kuras, D. Thomas, K. L. Craig, and M. Tyers. 1998. Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box protein complexes that regulate cell division and methionine biosynthesis in yeast. Genes Dev. 12:692-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pause, A., S. Lee, K. M. Lonergan, and R. D. Klausner. 1998. The von Hippel-Lindau tumor suppressor gene is required for cell cycle exit upon serum withdrawal. Proc. Natl. Acad. Sci. USA 95:993-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pause, A., S. Lee, R. A. Worrell, D. Y. Chen, W. H. Burgess, W. M. Linehan, and R. D. Klausner. 1997. The von Hippel-Lindau tumor-suppressor gene product forms a stable complex with human CUL-2, a member of the Cdc53 family of proteins. Proc. Natl. Acad. Sci. USA 94:2156-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peeters, P., S. D. Raynaud, J. Cools, I. Wlodarska, J. Grosgeorge, P. Philip, F. Monpoux, L. Van Rompaey, M. Baens, H. Van den Berghe, and P. Marynen. 1997. Fusion of TEL, the ETS-variant gene 6 (ETV6), to the receptor-associated kinase JAK2 as a result of t(9;12) in a lymphoid and t(9;15;12) in a myeloid leukemia. Blood 90:2535-2540. [PubMed] [Google Scholar]
- 46.Pickart, C. M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503-533. [DOI] [PubMed] [Google Scholar]
- 47.Querido, E., P. Blanchette, Q. Yan, T. Kamura, M. Morrison, D. Boivin, W. G. Kaelin, R. C. Conaway, J. W. Conaway, and P. E. Branton. 2001. Degradation of P53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a cullin-containing complex. Genes Dev. 15:3104-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rajewsky, K., H. Gu, R. Kuhn, U. A. K. Betz, W. Muller, J. Roes, and F. Schwenk. 1996. Conditional gene targeting. J. Clin. Investig. 98:600-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roberts, A. W., C. Kim, L. Zhen, J. B. Lowe, R. Kapur, B. Petryniak, A. Spaetti, J. D. Pollock, J. B. Borneo, G. B. Bradford, S. J. Atkinson, M. C. Dinauer, and D. A. Williams. 1999. Deficiency of the hematopoietic cell-specific Rho family GTPase Rac2 is characterized by abnormalities in neutrophil function and host defense. Immunity 10:183-196. [DOI] [PubMed] [Google Scholar]
- 50.Robertson, E., A. Bradley, M. Kuehn, and M. Evans. 1986. Germ-line transmission of genes introduced into cultured pluripotential cells by retroviral vector. Nature 323:445-448. [DOI] [PubMed] [Google Scholar]
- 51.Sauer, B., and N. Henderson. 1990. Targeted insertion of exogenous DNA into the eukaryotic genome by the Cre recombinase. New Biol. 2:441-449. [PubMed] [Google Scholar]
- 52.Seol, J. H., R. M. Feldman, W. Zachariae, A. Shevchenko, C. C. Correll, S. Lyapina, Y. Chi, M. Galova, J. Claypool, S. Sandmeyer, K. Nasmyth, A. Shevchenko, and R. J. Deshaies. 1999. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 13:1614-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shiyanov, P., A. Nag, and P. Raychaudhuri. 1999. Cullin 4A associates with the UV-damaged DNA-binding protein DDB. J. Biol. Chem. 274:35309-35312. [DOI] [PubMed] [Google Scholar]
- 54.Singer, J. D., M. Gurian-West, B. Clurman, and J. M. Roberts. 1999. Cullin-3 targets cyclin E for ubiquitination and controls S phase in mammalian cells. Genes Dev. 13:2375-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Skowyra, D., K. L. Craig, M. Tyers, S. J. Elledge, and J. W. Harper. 1997. F-Box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91:209-219. [DOI] [PubMed] [Google Scholar]
- 56.Skowyra, D., D. M. Koepp, T. Kamura, M. N. Conrad, R. C. Conaway, J. W. Conaway, S. J. Elledge, and J. W. Harper. 1999. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science 284:662-665. [DOI] [PubMed] [Google Scholar]
- 57.Spencer, E., J. Jiang, and Z. J. Chen. 1999. Signal-induced ubiquitination of IκBα by the F-box protein Slimb/β-TrCP. Genes Dev. 13:284-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stebbins, C. E., W. G. Kaelin, and N. P. Pavletich. 1999. Structure of the VHL-elonginC-elonginB complex: implications for VHL tumor suppressor function. Science 284:455-461. [DOI] [PubMed] [Google Scholar]
- 59.Strohmaier, H., C. H. Spruck, P. Kaiser, K. A. Won, O. Sangfelt, and S. I. Reed. 2001. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature 413:316-322. [DOI] [PubMed] [Google Scholar]
- 60.Sutterluty, H., E. Chatelain, A. Marti, C. Wirbelauer, M. Senften, U. Muller, and W. Krek. 1999. p45SKP2 promotes p27Kip1 degradation and induces S phase in quiescent cells. Nat. Cell Biol. 1:207-214. [DOI] [PubMed] [Google Scholar]
- 61.Suzuki, H., T. Chiba, M. Kobayashi, M. Takeuchi, T. Suzuki, A. Ichiyama, T. Ikenoue, M. Omata, K. Furuichi, and K. Tanaka. 1999. IκBα ubiquitination is catalyzed by an SCF-like complex containing Skp1, cullin-1, and two F-box/WD40-repeat proteins, βTrCP1 and βTrCP2. Biochem. Biophys. Res. Commun. 256:127-132. [DOI] [PubMed] [Google Scholar]
- 62.Tan, P., S. Y. Fuchs, A. Chen, K. Wu, C. Gomez, Z. Ronai, and Z. Q. Pan. 1999. Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of IκBα. Mol. Cell 3:527-533. [DOI] [PubMed] [Google Scholar]
- 63.Tang, J. Y., B. J. Hwang, J. M. Ford, P. C. Hanawalt, and G. Chu. 2000. Xeroderma pigmentosum p48 gene enhances global genomic repair and suppresses UV-induced mutagenesis. Mol. Cell 5:737-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsvetkov, L. M., K. H. Yeh, S. J. Lee, H. Sun, and H. Zhang. 1999. p27(Kip1) ubiquitination and degradation is regulated by the SCF(Skp2) complex through phosphorylated Thr187 in p27. Curr. Biol. 9:661-664. [DOI] [PubMed] [Google Scholar]
- 65.Tucker, K. A., M. B. Lilly, L. J. Heck, and T. A. Rado. 1987. Characterization of a new human diploid myeloid leukemia cell line (PLB-985) with granulocytic and monocytic differentiating capacity. Blood 70:372-378. [PubMed] [Google Scholar]
- 66.Tyers, M., and P. Jorgensen. 2000. Proteolysis and the cell cycle: with this RING I do thee destroy. Curr. Opin. Genet. Dev. 10:54-64. [DOI] [PubMed] [Google Scholar]
- 67.Wang, T., S. Penfold, X. Tang, N. Hattori, P. Riley, J. W. Harper, J. C. Cross, and M. Tyers. 1999. Deletion of the Cul1 gene in mice causes arrest in early embryogenesis and accumulation of cyclin E. Curr. Biol. 9:1191-1194. [DOI] [PubMed] [Google Scholar]
- 68.Willems, A. R., S. Lanker, E. E. Patton, K. L. Craig, T. F. Nason, N. Mathias, R. Kobayashi, C. Wittenberg, and M. Tyers. 1996. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell 86:453-463. [DOI] [PubMed] [Google Scholar]
- 69.Winston, J. T., P. Strack, P. Beer-Romero, C. Y. Chu, S. J. Elledge, and J. W. Harper. 1999. The SCFβ-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes Dev. 13:270-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu, K., S. Y. Fuchs, A. Chen, P. Tan, C. Gomez, Z. Ronai, and Z. Q. Pan. 2000. The SCF(HOS/β-TRCP)-ROC1 E3 ubiquitin ligase utilizes two distinct domains within CUL1 for substrate targeting and ubiquitin ligation. Mol. Cell. Biol. 20:1382-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu, L., L. Zhen, and M. C. Dinauer. 1997. Biosynthesis of the phagocyte NADPH oxidase cytochrome b558. Role of heme incorporation and heterodimer formation in maturation and stability of gp91phox and p22phox subunits. J. Biol. Chem. 272:27288-27294. [DOI] [PubMed] [Google Scholar]
- 72.Yu, Z.-K., J. L. M. Gervais, and H. Zhang. 1998. Human CUL-1 associates with the SKP1/SKP2 complex and regulates p21CIP1/WAF1 and cyclin D proteins. Proc. Natl. Acad. Sci. USA 95:11324-11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang, J.-G., A. Farley, S. E. Nicholson, T. A. Willson, L. M. Zugaro, R. J. Simpson, R. L. Moritz, D. Cary, R. Richardson, G. Hausmann, B. J. Kile, S. B. H. Kent, W. S. Alexander, D. Metcalf, D. J. Hilton, N. A. Nicola, and M. Baca. 1999. The conserved SOCS box motif in suppressors of cytokine signaling binds to elongins B and C and may couple bound proteins to proteasomal degradation. Proc. Natl. Acad. Sci. USA 96:2071-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]