Abstract
When viewing the changes in our understanding of inositides over the last 20 years, it is difficult to know whether to be more impressed by the proliferation in the number of inositides themselves (e.g. seven polyphosphoinositol lipids, more than 30 inositol phosphates), or by the number of functions for each. This review will focus on two specific aspects of this diversity: the evolution of the polyphosphoinositides, and the synthesis and functions of the higher inositol phosphates.
Introduction
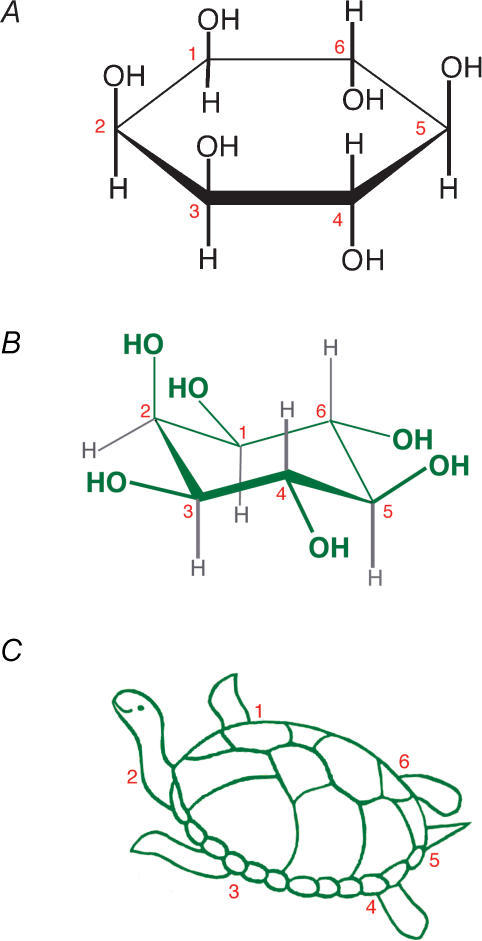
It is the central theme of this lecture and short review to regard myo-inositol as a versatile chemical building block on which some simple modifications, centred on phosphorylation, have generated great diversity. As on previous occasions (e. g. Irvine & Schell 2001) it helps to introduce Agranoff's turtle analogy (Agranoff, 1978). This visualizes myo-inositol as a turtle, with the D-numbering commonly used in biological contexts placing number 1 on its front right flipper and 2 on its head (Fig. 1). Looking at such an analogy prevents confusion with enantiomers (Irvine & Schell, 2001), makes it easier to visualize metabolic and functional links (Fig. 2), and concurrently justifies the title of this review. Previously, when writing a review in a similar context to this one (Irvine, 1995), I sketched out a possible evolutionary pathway of inositide signalling and metabolism as a way of trying to understand their functions. Ten years later, we know very much more about functions. Moreover, we have also learned more about the evolution of the enzymes that make and break inositides, so a revisiting of this topic 10 years on seems reasonable. We can now readdress, but from a much more informed standpoint, the approximate order in which the phosphoinositide pathways evolved. Note that this review will be confined to polyphosphoinositides; as before (Irvine, 1995), I shall not discuss the PtdIns-glycans that anchor proteins to membranes and possibly serve other important functions (see Ferguson, 1994 for review of these). Overall, it will be a brief and generalized approach.
Figure 1. myo-Inositol.
myo-Inositol is depicted as a Haworth projection (A) as a more accurate spatial representation (B), and schematically as a turtle (Agranoff, 1978) (C).
Figure 2. Eukaryotic inositol phosphate metabolism.
Adapted from Irvine & Schell (2001). This is a simplified version, designed to illustrate only specific points discussed in the text. Thus, no phosphatase reactions (other than the conversion of Ins(1,3,4,5)P4 to Ins(1,3,4)P3) are shown, and no pathways from higher plants are illustrated. Thick black arrows indicate reactions universal (as far as we know) in eukaryotes, and thinner coloured arrows illustrate reactions confined to specific groups of organisms (again, as far as we know, and within the limits of the discussion in the text): purple, slime moulds; blue, animals; red, yeast. For fuller versions of these pathways, see Irvine & Schell, 2001; Shears, 2001). Note that the isomeric configuration of (PP)2InsP4 is not yet known for animal cells (Irvine & Schell, 2001).
The evolution of polyphosphoinositides, and of inositol phosphate synthesis
We don't know when or how inositol evolved, nor how it was first synthesized. If this synthesis was the same as it is now in eukaryotes, by the conversion of glucose 6-phosphate firstly to inositol 3-phosphate (Fig. 2) followed by dephosphorylation, then Ins3P would have been the first inositide. The incorporation of inositol into lipids, probably by the reaction that is universal (as far as we know) – the chemical combination of inositol with activated phosphatidic acid (CMP-PtdOH) to form phosphatidylinositol (PtdIns) – must have followed shortly after. We know that some prokaryotes, for example mycobacteria, contain PtdIns (albeit mostly with mannose esterified onto the inositol ring; e.g. Lee & Ballou, 1964), but so far no inositol phosphates derived from lipids have been found in prokaryotes; but then, not many people have looked for them. The route by which prokaryotes break down inositol lipids is not determined, to my knowledge. A phospholipase D activity would generate free inositol from PtdIns (e.g. Becher et al. 1994), but if either a phospholipase C or A is involved in PtdIns catabolism then inositol monophosphate or glycerophosphoinositol will be generated. The latter could in turn also break down to Ins1P or inositol (depending on which side of the phosphorus atom a glycerophosphoinositol phosphodiesterase acts; mammals do both – see for example Dawson & Hemington (1977)versusDawson et al. (1979)). So, with PtdIns and InsP(s) probably present in prokaryotes, what happened next?
Polyphosphoinositol lipids and phosphates have not so far been shown to occur in prokaryotes, so at present we must assume that any further phosphorylation of the inositol ring by kinases evolved as a part of the eukaryotic expansion. But was it a lipid or an inositol phosphate that was phosphorylated first? Inositol phosphate kinases can be traced back to ‘earliest’ eukaryotes, as exemplified by an InsP6 kinase in the genome of Giardia (M. J. Schell & R. F. Irvine, unpublished observations), so perhaps phosphorylation of InsP derived from PtdIns catabolism (see above) followed shortly after it first appeared. Or, phosphorylation of PtdIns itself might have been the starting point. We may never know, and it is intriguing to note here that at least one inositol phosphate ‘multikinase’ is able, at least in vitro, to phosphorylate PtdIns(4,5)P2 to PtdIns(3,4,5)P3 (A. Resnick, S. H. Snyder and A. Saiardi personal communication) – contrast this with the exquisitely specific structure of Ins(1,4,5)P3 3-kinase, which ensures that it never phosphorylates PtdIns(4,5)P2 (Gonzalez et al. 2004; Miller & Hurley, 2004).
PtdIns4P and PtdIns(4,5)P2 are ubiquitous in eukaryotes as far as we know. Moreover, so far all eukaryotic genomes contain a member of the PI-PLC family, which catalyses the enzymic reaction that provides the only certain link between inositol lipids and phosphates. This link is of particular importance for inositol phosphate synthesis in the budding yeast Saccharomyces cerevisiae, as there is clear evidence that in this organism the synthesis of ‘higher’ inositol phosphates (InsP4–8, see below) must start with Ins(1,4,5)P3 that has been generated from PtdIns(4,5)P2 by a PI-PLC (Fig. 2). Deletion mutants of the PLC1 gene make no InsP4–8 (Ongusaha et al. 1998; York et al. 1999), and the enzymes that convert Ins(1,4,5)P3 to Ins(1,4,5,6)P4, then to Ins(1,3,4,5,6)P5, then InsP6 and finally InsP7 (an InsP7 kinase exists, but has not yet been cloned from any organism) have now been identified (Saiardi et al. 1999; York et al. 1999; Odom et al. 2000). Is this the paradigm for the beginning of higher inositol phosphates – the first three phosphates are esterified on the inositol ring when it is part of a lipid, and then the other phosphates are added after liberation of Ins(1,4,5)P3 by PI-PLC?
Or is yeast misleading us, and slime moulds and higher plants actually represent the ‘first stage’? These latter organisms can synthesize InsP6 from inositol without any lipid intermediate being involved. They do this by (a) firstly phosphorylating inositol in the 3-position with an inositol kinase (Loewus et al. 1982; Stephens et al. 1990) (although, because probably all eukaryotes can synthesize Ins3P directly from glucose 6-phosphate (Eisenberg, 1967; Fig. 2), this particular reaction is not essential), and (b) then phosphorylating the Ins3P sequentially up to InsP6 (Stephens & Irvine, 1990; Brearley & Hanke, 1996; Fig. 2– only the slime mould pathway is shown).
Currently it is not absolutely clear how we (mammals) synthesize our InsP6. The generally accepted route is similar in principle to yeast (above), that is, the first three phosphorylations take place in the lipid phase to form PtdIns(4,5)P2, and then Ins(1,4,5)P3 is formed by PI-PLC action. However, the Ins(1,4,5)P3 is apparently converted to InsP6 by a more convoluted route (Fig. 2): Ins(1,4,5)P3 to Ins(1,3,4,5)P4 to Ins(1,3,4)P3 to Ins(1,3,4,6)P4 to Ins(1,3,4,5,6)P5 to InsP6 (Balla et al. 1989; Shears, 1989). There is evidence that this route does happen in intact cells, perhaps the most cogent being that embryos of mice lacking the ‘inositol phosphate multikinase’ (and assuming that in mammals it catalyses the Ins(1,3,4,6)P4 to Ins(1,3,4,5,6)P5 step, as suggested by Chang et al. (2002)) show undetectable incorporation of radiolabelled inositol into InsP6 compared with wild-type animals (J. D. York, personal communication).
However, all these studies relied on radiolabelling, and the quantitative (mass) contribution that this pathway makes to our InsP6 synthesis is still not entirely clear. Two observations suggest that this pathway might not be the only, or even the major, contributor. One is that, as discussed elsewhere (Irvine & Schell, 2001), the incorporation of radiolabelled inositol into InsP6 in mammalian cells in culture is remarkably slower than it is into inositol lipids or the other higher inositol phosphates (e.g. Oliver et al. 1992). Secondly, there is nutritional evidence that rats derive most of their InsP6 from their diet (Grases et al. 2000, 2001), and the data in one of these studies (Grases et al. 2000) also suggest that this is by a direct supply of InsP6 to the body tissues, not by a route involving InsP6 being first dephosphorylated in the gut to inositol and then absorbed and reconverted to InsP6.
These issues remain to be resolved, and indeed they should be resolved, as they have significant medical and physiological relevance. The possibility that plants, slime moulds and the like are making InsP6 for the world is an intriguing one! If they are, then it is arguably a mixed blessing, as it is well known by nutritionists that InsP6 at high levels in the diet can chelate cations, and can thus lead in particular to zinc and iron deficiencies (Cheryan, 1980). Also, perhaps less well known, InsP6 released into the soil as a consequence of animal excretions can be a serious environmental problem (Turner et al. 2002). Whatever the contribution of plants and slime moulds to the world's InsP6, the question as to why slime moulds growing in their axenic phase, which (unlike plants) do not have any obvious need for phosphate storage, make so much InsP6 (Martin et al. 1987; Europe-Finner et al. 1988) so quickly (Stephens & Irvine, 1990), is as puzzling now as it was 15 years ago.
Higher inositol phosphates: form and function
Two extensive reviews have addressed the functions of InsP6 suggested up until 2001 (Irvine & Schell, 2001; Shears, 2001), so I will not expand on these here However, one further aspect of InsP6 biology deserves mention, as it impinges on possible functions. This is the physicochemical form in which InsP6 is found in cells. We don't know exactly how much InsP6 is free in the cytoplasm – certainly in some organisms it is found at high concentrations as a semicrystalline calcium salt, for example in plant seeds (Raboy, 2003), and in the protective cuticle of the parasite Echinococcus granulosus (Irigoin et al. 2002, 2004). Moreover, in the slime mould, where its estimated intracellular concentration is 0.7 mm (Martin et al. 1987), some of this may be in calcium phosphate-rich granules (Marchesini et al. 2002). However, based on equilibrium inositol labelling, the mass of InsP6 in mammalian cells has been estimated to be of the order of 100 μm and most of this appears to be soluble and free in the cytoplasm (Stuart et al. 1994). This is a high concentration for a multiphosphorylated compound which readily forms insoluble salts with polyvalent cations.
A recent detailed quantitative study of the interaction of InsP6 with inorganic cations (Torres et al. 2005) has produced a likely resolution of this conundrum through the surprising finding that InsP6 has an apparently unique relationship with magnesium. All the divalent and trivalent cations form salts with InsP6 that are primarily of the form InsP6: ion in a 1: 1 stoichiometry, with the number of protons dictated by the pH. The 1: 1 ratios are consistent with the unique grouping of the 1,2,3 phosphates (Fig. 1) being primarily responsible for cation interactions (see Hawkins et al. 1993). However, under ionic conditions that approximate to those of the cytoplasm, InsP6 can form a neutral salt with five Mg2+ ions and two protons (its likely full content is Mg5H2InsP6(H2O)22), which is stable and soluble up to a concentration above 100 μm at pH 7.5 (Torres et al. 2005).
This discovery has some functional implications. For example, the interaction of InsP6 with Fe3+ ions, which has been suggested to be of functional significance in protecting cells against Fe3+-catalysed radical formation (Hawkins et al. 1993), probably cannot occur if it is in its Mg5H2InsP6 form (Torres et al. 2005). This refocuses our attention on Ins(1,2,3)P3 as a potential low molecular weight iron carrier in cells (Spiers et al. 1996; Barker et al. 2004). Previously, the higher concentration of InsP6 in cells compared with Ins(1,2,3)P3 would have made the former arguably more likely to fulfil such a role. But if InsP6 is effectively removed from competition by being in the Mg5H2InsP6 form, and as Ins(1,2,3)P3 will more likely form a Mg2+ salt with 1 : 1 stochiometry, Ins(1,2,3)P3 becomes a much more attractive prospect for this iron-carrying role.
Also, we don't yet know if the pyrophosphate-containing inositol phosphates (Fig. 2), PPInsP5 and (PP)2InsP4 (loosely known as InsP7 and InsP8), form a pentavalent salt with Mg2+. If they do not (which is certainly possible, as a pyrophosphate in place of a phosphate might be expected to disrupt the structure of the complex), they will have an ionic interaction with their environment different from that of InsP6 itself, and this may have implications for their functions.
Finally, to turn to new ideas of higher inositol phosphate function, a most provocative potential function for PPInsP5 has emerged recently with the suggestion that it can phosphorylate proteins non-enzymatically (Saiardi et al. 2004). Particularly intriguing is that most of the proteins that it favoured as substrates were nucleolar proteins. If this observation is taken in the context that InsP6 has been suggested to be involved with mRNA export from the nucleus (York et al. 1999), Ins(1,4,5,6)P4 with transcriptional events (Odom et al. 2000), and PPInsP4 with the regulation of telomere length (Saiardi et al. 2005; York et al. 2005), the links of higher inositol phosphates with nuclear functions are burgeoning and growing ever more fascinating. Moreover, to these possibilities must also be added the proposed multiple intranuclear roles of the inositol lipids, especially PtdIns(4,5)P2 (see Irvine, 2003 for review).
So, to close this brief essay on a provocative note, the preceding paragraph, taken together with the likelihood that polyphosphoinositides evolved during the eukaryotic expansion (above), leads me to wonder (again, as we did before; Divecha et al. 1993): did polyphosphoinositol lipids and phosphates first evolve in the nucleus?
Acknowledgments
I would like to acknowledge the debt I owe to Mike Schell for so much of the recent work from my lab on inositol phosphate kinases, especially including the insight we have gained into the evolution of that enzyme family. My work is supported by the Royal Society and the Wellcome Trust.
References
- Agranoff BW. Cyclitol confusion. Trends Biochem Sci. 1978;3:N283–N285. [Google Scholar]
- Balla T, Baukal AJ, Hunyady L, Catt KJ. Agonist-induced regulation of inositol tetrakisphosphate isomers and inositol pentakisphosphate in adrenal glomerulosa cells. J Biol Chem. 1989;264:13605–13611. [PubMed] [Google Scholar]
- Barker CJ, Wright J, Hughes PJ, Kirk CJ, Michell RH. Complex changes in cellular inositol phosphate complement accompany transit through the cell cycle. Biochem J. 2004;380:465–473. doi: 10.1042/BJ20031872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher A, Wissing JB, Wylegalla C, Wagner KG. Phosphatidylinositol-specific isoenzymes of phospholipase D from Catharanthus roseus– purification and chracterization. Plant Sci. 1994;97:143–151. [Google Scholar]
- Brearley CA, Hanke DE. Metabolic evidence for the order of addition of individual phosphate esters in the myo-inositol moiety of inositol hexakisphosphate in the duckweed Spirodela polyrhiza L. Biochem J. 1996;314:227–233. doi: 10.1042/bj3140227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SC, Miller AL, Feng Y, Wente SR, Majerus PW. The human homolog of the rat inositol phosphate multikinase is an inositol 1,3,4,6-tetrakisphosphate 5-kinase. J Biol Chem. 2002;277:43836–43843. doi: 10.1074/jbc.M206134200. [DOI] [PubMed] [Google Scholar]
- Cheryan M. Phytic acid interactions in food systems. Crit Rev Food Sci Nutr. 1980;13:297–335. doi: 10.1080/10408398009527293. [DOI] [PubMed] [Google Scholar]
- Dawson RMC, Hemington N. A phosphodiesterase in rat kidney cortex that hydrolyses glycerylphosphorylinositol. Biochem J. 1977;162:241–245. doi: 10.1042/bj1620241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson RMC, Hemington N, Richards DE, Irvine RF. sn-Glycero(3)phosphoinositol glycerophosphohydrolase. A new phosphodiesterase in rat tissues. Biochem J. 1979;182:39–49. doi: 10.1042/bj1820039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divecha N, Banfic H, Irvine RF. Inositides and the nucleus and inositides in the nucleus. Cell. 1993;74:405–407. doi: 10.1016/0092-8674(93)80041-c. [DOI] [PubMed] [Google Scholar]
- Eisenberg FJ. d-myoinositol 1-phosphate as the product of cyclization of glucose 6-phosphate and substrate for a specific phosphatase in rat testis. J Biol Chem. 1967;242:1375–1382. [PubMed] [Google Scholar]
- Europe-Finner GN, Luderus ME, Small NV, Van Driel R, Reymond CD, Firtel RA, Newell PC. Mutant ras gene induces elevated levels of inositol tris- and hexakisphosphates in Dictyostelium. J Cell Sci. 1988;89:13–20. doi: 10.1242/jcs.89.1.13. [DOI] [PubMed] [Google Scholar]
- Ferguson MA. What can GPI do for you? Parasitol Today. 1994;10:48–52. doi: 10.1016/0169-4758(94)90392-1. [DOI] [PubMed] [Google Scholar]
- Gonzalez B, Schell MJ, Letcher AJ, Veprintsev DB, Irvine RF, Williams RL. Structure of a human inositol 1,4,5-trisphosphate 3-kinase: substrate binding reveals why it is not a phosphoinositide 3-kinase. Mol Cell. 2004;15:689–701. doi: 10.1016/j.molcel.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Grases F, Simonet BM, March JG, Prieto RM. Inositol hexakisphosphate in urine: the relationship between oral intake and urinary excretion. BJU Int. 2000;85:138–142. doi: 10.1046/j.1464-410x.2000.00324.x. [DOI] [PubMed] [Google Scholar]
- Grases F, Simonet BM, Prieto RM, March JG. Variation of InsP4,InsP5 and InsP6 levels in tissues and biological fluids depending on dietary phytate. J Nutr Biochem. 2001;12:595–601. doi: 10.1016/s0955-2863(01)00178-4. [DOI] [PubMed] [Google Scholar]
- Hawkins PT, Poyner DR, Jackson TR, Letcher AJ, Lander DA, Irvine RF. Inhibition of iron-catalysed hydroxyl radical formation by inositol polyphosphates: a possible physiological function for myo-inositol hexakisphosphate. Biochem J. 1993;294:929–934. doi: 10.1042/bj2940929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irigoin F, Casaravilla C, Iborra F, Sim RB, Ferreira F, Diaz A. Unique precipitation and exocytosis of a calcium salt of myo-inositol hexakisphosphate in larval Echinococcus granulosus. J Cell Biochem. 2004;93:1272–12781. doi: 10.1002/jcb.20262. [DOI] [PubMed] [Google Scholar]
- Irigoin F, Ferreira F, Fernandez C, Sim RB, Diaz A. myo-Inositol hexakisphosphate is a major component of an extracellular structure in the parasitic cestode Echinococcus granulosus. Biochem J. 2002;362:297–304. doi: 10.1042/0264-6021:3620297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine RF. Inositide evolution: what can it tell us about functions? Biochem Soc Trans. 1995;23:27–35. doi: 10.1042/bst0230027. [DOI] [PubMed] [Google Scholar]
- Irvine RF. Nuclear lipid signalling. Nat Rev Mol Cell Biol. 2003;4:349–360. doi: 10.1038/nrm1100. [DOI] [PubMed] [Google Scholar]
- Irvine RF, Schell MJ. Back in the water: the return of the inositol phosphates. Nat Rev Mol Cell Bio. 2001;2:327–338. doi: 10.1038/35073015. [DOI] [PubMed] [Google Scholar]
- Lee YC, Ballou CE. Sructural studies on the myo-inositol mannosides from the glycolipids of Mycobacterium tuberculosis and Mycobaterium phlei. J Biol Chem. 1964;239:1316–1327. [PubMed] [Google Scholar]
- Loewus MW, Sasaki K, Leavitt AL, Muscell L, Sherman WR, Loewus FA. Enanantiomeric form of myo-inositol 1-phosphate produced by myo-inositol 1-phosphate synthetase and myo-inositol kinase in higher plants. Plant Physiol. 1982;70:1661–1663. doi: 10.1104/pp.70.6.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchesini N, Ruiz FA, Vieira M, Docampo R. Acidocalcisomes are functionally linked to the contractile vacuole of Dictyostelium discoideum. J Biol Chem. 2002;277:8146–8153. doi: 10.1074/jbc.M111130200. [DOI] [PubMed] [Google Scholar]
- Martin JB, Foray MF, Klein G, Satre M. Identification of inositol hexaphosphate in 31P-NMR spectra of Dictyostelium discoideum amoebae. Relevance to intracellular pH determination. Biochim Biophys Acta. 1987;931:16–25. doi: 10.1016/0167-4889(87)90045-0. [DOI] [PubMed] [Google Scholar]
- Miller GJ, Hurley JH. Crystal structure of the catalytic core of inositol 1,4,5-trisphosphate 3-kinase. Mol Cell. 2004;15:703–711. doi: 10.1016/j.molcel.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Odom AR, Stahlberg A, Wente SR, York JD. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science. 2000;287:2026–2029. doi: 10.1126/science.287.5460.2026. [DOI] [PubMed] [Google Scholar]
- Oliver KG, Putney JW, Jr, Obie JF, Shears SB. The interconversion of inositol 1,3,4,5,6-pentakisphosphate and inositol tetrakisphosphates in AR4-2J cells. J Biol Chem. 1992;267:21528–21534. [PubMed] [Google Scholar]
- Ongusaha PP, Hughes PJ, Davey J, Michell RH. Inositol hexakisphosphate in Schizosaccharomyces pombe: synthesis from Ins(1,4,5),P3 and osmotic regulation. Biochem J. 1998;335:671–679. doi: 10.1042/bj3350671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raboy V. myo-Inositol-1,2,3,4,5,6-hexakisphosphate. Phytochemistry. 2003;64:1033–1043. doi: 10.1016/s0031-9422(03)00446-1. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Bhandari R, Resnick AC, Snowman AM, Snyder SH. Phosphorylation of proteins by inositol pyrophosphates. Science. 2004;306:2101–2105. doi: 10.1126/science.1103344. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Erdjument-Bromage H, Snowman AM, Tempst P, Snyder SH. Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr Biol. 1999;9:1323–1326. doi: 10.1016/s0960-9822(00)80055-x. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Resnick AC, Snowman AM, Wendland B, Snyder SH. Inositol pyrophosphates regulate cell death and telomere length through phosphoinositide 3-kinase-related protein kinases. Proc Natl Acad Sci U S A. 2005;102:1911–1914. doi: 10.1073/pnas.0409322102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shears SB. The pathway of myo-inositol 1,3,4-trisphosphate phosphorylation in liver. Identification of myo-inositol 1,3,4-trisphosphate 6-kinase, myo-inositol 1,3,4-trisphosphate 5-kinase, and myo-inositol 1,3,4,6-tetrakisphosphate 5-kinase. J Biol Chem. 1989;264:19879–19886. [PubMed] [Google Scholar]
- Shears SB. Assessing the functional omnipotence of inositol hexakisphophosphate. Cell Sig. 2001;13:151–158. doi: 10.1016/s0898-6568(01)00129-2. [DOI] [PubMed] [Google Scholar]
- Spiers ID, Barker CJ, Chung SK, Chang YT, Freeman S, Gardiner JM, Hirst PH, Lambert PA, Michell RH, Poyner DR, Schwalbe CH, Smith AW, Solomons KR. Synthesis and iron binding studies of myo-inositol 1,2,3-trisphosphate and (±)-myo-inositol 1,2-bisphosphate, and iron binding studies of all myo-inositol tetrakisphosphates. Carbohydr Res. 1996;282:81–99. doi: 10.1016/0008-6215(95)00361-4. [DOI] [PubMed] [Google Scholar]
- Stephens LR, Irvine RF. Stepwise phosphorylation of myo-inositol leading to myo-inositol hexakisphosphate in Dictyostelium. Nature. 1990;346:580–583. doi: 10.1038/346580a0. [DOI] [PubMed] [Google Scholar]
- Stephens LR, Kay RR, Irvine RF. A myo-inositol D-3 hydroxykinase activity in Dictyostelium. Biochem J. 1990;272:201–210. doi: 10.1042/bj2720201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart JA, Anderson KL, French PJ, Kirk CJ, Michell RH. The intracellular distribution of inositol polyphosphates in HL60 promyeloid cells. Biochem J. 1994;303:517–525. doi: 10.1042/bj3030517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres J, Dominguez S, Cerda MF, Obal G, Mederos A, Irvine RF, Diaz A, Kremer C. Solution behaviour of myo-inositol hexakisphosphate in the presence of multivalent cations. Prediction of a neutral pentamagnesium species under cytosolic/nuclear conditions. J Inorg Biochem. 2005;99:828–840. doi: 10.1016/j.jinorgbio.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Turner BL, Paphazy MJ, Haygarth PM, McKelvie ID. Inositol phosphates in the environment. Philos Trans R Soc Lond B Biol Sci. 2002;357:449–469. doi: 10.1098/rstb.2001.0837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York SJ, Armbruster BN, Greenwell P, Petes TD, York JD. Inositol diphosphate signaling regulates telomere length. J Biol Chem. 2005;280:4264–4269. doi: 10.1074/jbc.M412070200. [DOI] [PubMed] [Google Scholar]
- York JD, Odom AR, Murphy R, Ives EB, Wente SR. A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science. 1999;285:96–100. doi: 10.1126/science.285.5424.96. [DOI] [PubMed] [Google Scholar]