Abstract
Human activating signal cointegrator 1 (hASC-1) was originally isolated as a transcriptional coactivator of nuclear receptors. Here we report that ASC-1 exists as a steady-state complex associated with three polypeptides, P200, P100, and P50, in HeLa nuclei; stimulates transactivation by serum response factor (SRF), activating protein 1 (AP-1), and nuclear factor κB (NF-κB) through direct binding to SRF, c-Jun, p50, and p65; and relieves the previously described transrepression between nuclear receptors and either AP-1 or NF-κB. Interestingly, ectopic expression of Caenorhabditis elegans ASC-1 (ceASC-1), an ASC-1 homologue that binds P200 and P100, like hASC-1, while weakly interacting only with p65, in HeLa cells appears to replace endogenous hASC-1 from the hASC-1 complex and exerts potent dominant-negative effects on AP-1, NF-κB, and SRF transactivation. In addition, neutralization of endogenous P50 by single-cell microinjection of a P50 antibody inhibits AP-1 transactivation; the inhibition is relieved by coexpression of wild-type P50, but not of P50ΔKH, a mutant form that does not interact with P200. Overall, these results suggest that the endogenous hASC-1 complex appears to play an essential role in AP-1, SRF, and NF-κB transactivation and to mediate the transrepression between nuclear receptors and either AP-1 or NF-κB in vivo.
Transcription coactivators bridge transcription factors and the components of the basal transcriptional apparatus and/or remodel the chromatin structures (reviewed in reference 18). Notably, many transcription coactivators function in the context of multiprotein complexes in vivo. These include TRAP/DRIP/ARC (6, 22, 25), P/CAF (28), steroid receptor coactivator 1 (SRC-1), and CREB binding protein (CBP) complexes (19). CBP and its functional homologue p300, SRC-1 and its family members, and activating signal cointegrator 2 (ASC-2) have been shown to be essential for the activation of transcription by a large number of regulated transcription factors, including serum response factor (SRF), activating protein 1 (AP-1), and nuclear factor κB (NF-κB) (reviewed in references 13 and 18). These transcription factors are known to control a surprisingly diverse set of genes. SRF, along with ternary complex factor (TCF), binds to and activates the serum response element present in the upstream regulatory sequences of a number of myogenic and immediate-early genes (reviewed in reference 27). SRF belongs to the MADS box family of proteins and recognizes a CArG box in the serum response element, whereas TCF does not bind autonomously to the element but rather requires the assistance of SRF to efficiently contact the DNA. The AP-1 complex, which controls the expression of immediate-early response genes, consists of a heterodimer of a Fos family member and a Jun family member (reviewed in reference 10). This complex binds the consensus DNA sequence TGAGTCA (termed an AP-1 site) found in a variety of promoters. NF-κB, composed of homo- and heterodimeric complexes of members of the Rel (NF-κB) family of polypeptides, regulates genes involved with various immunological processes (reviewed in reference 3). In vertebrates, this family consists of p50, p65 (RelA), c-Rel, p52, and RelB. In the majority of cells, NF-κB exists in an inactive form in the cytoplasm, bound to the inhibitory IκB proteins. In response to various inducers, IκB proteins are rapidly degraded and the NF-κB dimers spared from degradation translocate to the nucleus to activate gene transcription.
ASC-1, originally referred to TRIP4 (14), is a novel transcription coactivator molecule of nuclear receptors (11). ASC-1 harbors an autonomous transactivation domain that contains a putative zinc finger motif, which also serves as a binding site for TATA-binding protein (TBP), TFIIA, SRC-1, CBP/p300, and nuclear receptors (11). Strikingly, ASC-1, at least when overexpressed in single-cell microinjections, was found in the cytoplasm under serum-deprived conditions but remained in the nucleus when serum starved in the presence of a ligand or coexpressed CBP or SRC-1 (11). Thus, it was suggested that ASC-1 plays an important role in establishing distinct coactivator complexes under various different cellular conditions (11).
In this work, we report the purification of a steady-state ASC-1 complex from HeLa nuclei and identify SRF, AP-1, and NF-κB as new target transcription factors of ASC-1. We also present experimental results that support the importance of the endogenous ASC-1 complex for AP-1, NF-κB, and SRF transactivation in vivo.
MATERIALS AND METHODS
Plasmids.
PCR fragments encoding P200, P200 deletion mutants, P100, P100s, P50, and P50ΔKH, as well as Caenorhabditis elegans ASC-1 (ceASC-1), ceASC-1 residues 1 to 182 fused to human ASC-1 (hASC-1) residues 325 to 581 (chASC-1), hASC-1 residues 1 to 324 fused to ceASC-1 residues 183 to 425 (hcASC-1), and hASC-1 deletion mutants, were cloned into EcoRI and XhoI/SalI restriction sites of LexA fusion vector pEG202PL, B42 fusion vector pJG4-5, Gal4 fusion vector pCMX-Gal4-N, hemagglutinin (HA) tagging vector pJ3H, and pcDNA3 (Invitrogen). The C. elegans expressed sequence tag (EST) clone yk282d8 corresponding to the full-length ceASC-1 was obtained from Yuji Kohara (National Institute of Genetics, Mishima, Japan). Similarly, PCR fragments encoding SRFΔC and SRFΔN were cloned into EcoRI and XhoI restriction site of pGEX4T (Pharmacia). Glutathione S-transferase (GST) fusion vectors encoding SRF, ASC-1, c-Jun, c-Fos, p50, and p65, LexA, B42 fusion vectors for ASC-1, SRF, p50, p65, c-Jun, and c-Fos, mammalian expression/T7 in vitro transcription pcDNA3 vectors for SRF, ASC-1, p300, SRC-1, c-Fos, c-Jun, p50, and p65, transfection indicator construct pRSV-β-gal, and reporter constructs SRE-LUC, AP1-LUC, κB-LUC, Gal4-LUC, and AP1-lacZ were as previously described (11, 14-16).
GST pull-down and yeast β-galactosidase assays, cell culture, and transfections.
The GST pull-down and yeast β-galactosidase assays as well as cell cultures, transfections, and luciferase assays were done as previously described (11, 14-16). For luciferase assays, normalized luciferase expression from triplicate samples relative to LacZ expression was calculated and the results were expressed as fold activation over the value obtained with a reporter alone. The data are representative of three similar experiments.
Purification of the ASC-1 complex, peptide microsequencing, and antibody generation.
HeLa nuclear extract (500 mg of protein) (4) was loaded onto a HiTrap heparin column (Phamarcia) equilibrated with G buffer (20 mM HEPES-KOH [pH 7.9], 0.5 mM EDTA, 0.05% NP-40, 10% glycerol, 1 mM dithiothreitol [DTT], protease inhibitors [21]) containing 100 mM KCl. The bound proteins were eluted with 60 ml of a linear salt gradient (100 to 600 mM KCl in G buffer). The fractions immunoreactive with anti-ASC-1 (11) were pooled (350 mM KCl, 200 mg of protein) and dialyzed in Q buffer (20 mM Tris-HCl [pH 7.8], 0.5 mM EDTA, 20% glycerol, 1 mM DTT, protease inhibitors) containing 80 mM KCl for 2 h. The samples were loaded onto a HiTrap Q column (Pharmacia) equilibrated with Q buffer containing 100 mM KCl, and the bound proteins were eluted with 60 ml of a salt gradient from 100 to 500 mM KCl in Q buffer. The fractions containing ASC-1 were pooled (300 mM KCl, 15 mg of protein) and mixed with 200 μl of protein G-agarose conjugated with an affinity-purified anti-ASC-1 antibody (11) at 4°C for 12 h. The beads were recovered and washed three times with 1 ml of IP-150 buffer (20 mM Tris-HCl [pH 7.8], 0.1 mM EDTA, 0.2% NP-40, 10% glycerol, 1 mM DTT, protease inhibitors, 150 mM potassium acetate). Bound proteins were eluted twice with 200 μl of 100 mM glycine (pH 3.0), precipitated with 10% trichloroacetate, and visualized by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and Coomassie staining. Peptide microsequencing was performed at the Harvard Microchemistry Facility by microcapillary reverse-phase HPLC nano-electrospray tandem mass spectrometry (μLC/MS/MS) on a Finnegan LCQ quadrupole ion trap mass spectrometer and corresponding cDNA clones for P200, P100, and P50 were isolated from a HeLa cDNA library. Anti-P200 antisera were generated in rats by using, as antigens, a recombinant P200 fragment (P200 residues 1588 to 1917) expressed in Escherichia coli. Similarly, anti-P100 and anti-P50 antisera were generated in rats by using full-length P100 and P50 residues 120 to 356 as antigens, respectively. Antibody coupling to protein G-agarose and immunoprecipitation were performed as previously described (17).
Microinjection of P50 antibody.
Insulin-responsive Rat-1 fibroblasts, made quiescent by incubation in serum-free medium for 24 h, were microinjected with either preimmune immunoglobulin G (IgG) or affinity-purified anti-P50 IgG along with empty, p50, or p50ΔKH expression vectors and either the AP-1-lacZ or Gal4-lacZ reporter construct (25 μg/ml each). About 1 h after injection, cells were stimulated with 0.1 μM tetradecanoyl phorbol acetate (TPA). After 4 h of incubation, cells were fixed and stained to detect injected IgG by using fluorescein isothiocyanate-conjugated antibodies and examined for β-galactosidase expression as described previously (15). The image was photographed with Zeiss AxioplanII microscope equipped with a PIXERA camera.
Isolation of a ceASC-1-containing complex from HeLa cells.
HeLa cells (5 × 106) grown in Dulbecco's modified Eagle medium plus 10% fetal bovine serum were transfected with 50 μg of pJ3H-HA-ceASC-1 by the calcium phosphate method and incubated for 36 h at 37°C. The nuclear extracts (2.7 mg of protein) prepared from these cells (4) were loaded onto a 1-ml HiTrap Q column (Pharmacia) equilibrated with Q buffer containing 100 mM KCl, and bound proteins were eluted with 6 ml of a linear salt gradient (150 to 500 mM KCl in Q buffer). Each fraction was subjected to immunoblot analyses with affinity-purified anti-ASC-1 and anti-P100 antibodies and, for ceASC-1, an anti-HA monoclonal antibody (Boehringer Mannheim). Immunoreactive fractions were pooled and subjected to either a Superose 6 sizing column (Pharmacia) equilibrated with G buffer containing 300 mM KCl or immunoprecipitation with antibodies as previously described (2).
Nucleotide sequence accession numbers.
The GenBank accession numbers for ceASC-1, mouse ASC-1 (mASC-1), P200, P100, and P50 genes are AF197575, AF197574, AY013288, AY013289, and AY013290, respectively.
RESULTS
Identification of SRF, AP-1, and NF-κB as novel targets for ASC-1.
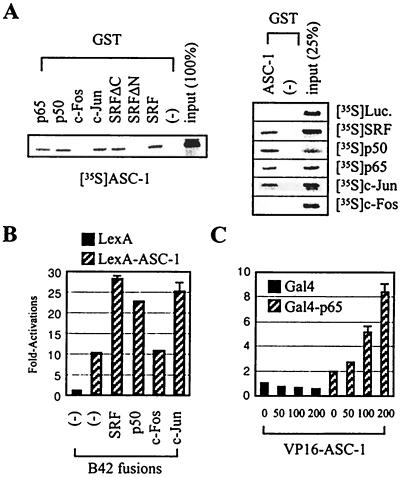
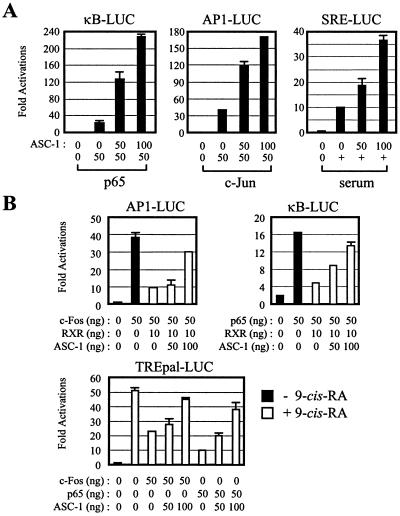
We have examined whether ASC-1, originally isolated as a novel transcriptional coactivator of nuclear receptors (11), is a multifunctional cointegrator molecule, like SRC-1, ASC-2, and CBP/p300 (13, 18). GST alone and various GST fusion proteins were expressed, purified, and tested for interaction with in vitro-translated luciferase, ASC-1, SRF, p50, p65, c-Jun, and c-Fos. As shown in Fig. 1A, the radiolabeled ASC-1 readily interacted with fusions of GST to p65, p50, c-Jun, and SRF but not with GST alone or a fusion of GST to c-Fos. Interestingly, ASC-1 interacted with GST-SRFΔC (SRF residues 1 to 52) but not with GST-SRFΔN (SRF residues 53 to 504), localizing the ASC-1 interaction interface to the N-terminal region of SRF. In reciprocal experiments, GST-ASC-1 exhibited identical interaction patterns (Fig. 1A, right). Similar results were also obtained in the yeast and mammalian two-hybrid tests (Fig. 1B and C). From these results, we concluded that ASC-1 directly binds to SRF, NF-κB components p50 and p65, and AP-1 component c-Jun, in addition to a series of nuclear receptors as we previously reported (11). Confirming the functionality of these interactions, cotransfected ASC-1 stimulated transactivation mediated by reporter constructs κB-LUC, AP1-LUC, and SRE-LUC (16) in a dose-dependent manner (Fig. 2A). ASC-1 also relieved the previously described transrepression between retinoid X receptor and either AP-1 or NF-κB (8, 26) (Fig. 2B). From these results, we concluded that ASC-1 is clearly a multifunctional transcriptional integrator molecule.
FIG. 1.
Interaction of ASC-1 with SRF, p50, p65, and c-Jun. (A) Indicated proteins were labeled with [35S]methionine by in vitro translation and incubated with glutathione beads containing GST alone (−) or fusions of GST to c-Jun, c-Fos, p50, p65, SRFΔC, SRFΔN, SRF, and ASC-1. Beads were washed, and specifically bound material was eluted with reduced glutathione and resolved by SDS-polyacrylamide gel electrophoresis. (B) The indicated B42 and LexA plasmids were transformed into a yeast strain containing an appropriate lacZ reporter gene, as described previously (2). (C) HeLa cells were transfected with a lacZ expression vector, a Gal4-LUC reporter construct, and an increasing amount of VP16-ASC-1 expression vector (in nanograms).
FIG. 2.
ASC-1 as a coactivator of AP-1, NF-κB, and SRF (A) and as a mediator of transrepression between nuclear receptor and either AP-1 or NF-κB (B). HeLa cells were transfected with the lacZ expression vector and increasing amounts of the ASC-1 expression vector along with the indicated reporter genes. For activation, the expression vector for p65 or c-Jun or serum shock (i.e., addition of 20% fetal bovine serum to cells deprived of serum) was applied, as indicated (in nanograms). Solid and open bars (B), absence and presence, respectively of 100 nM 9-cis-retinoic acid (RA). RXR, retinoic X receptor.
Purification of an ASC-1 complex from HeLa nuclei.
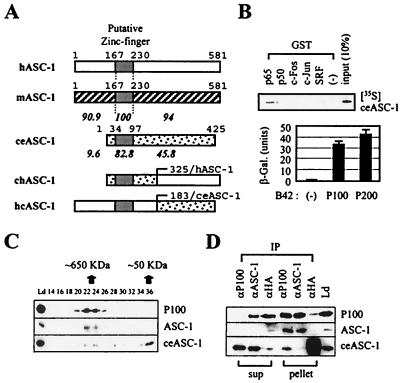
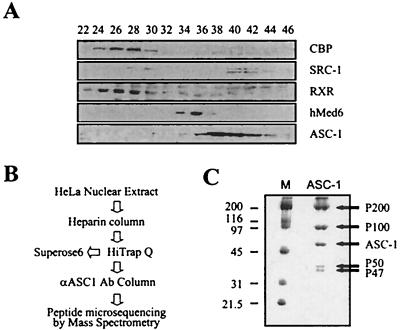
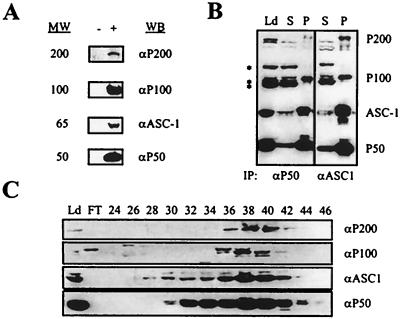
Many transcription coactivators exist as steady-state complexes in vivo (6, 19, 22, 25, 28). Interestingly, 65-kDa protein ASC-1 eluted as an approximately 650-kDa complex from a Superose 6 sizing column (see Fig. 8C). In addition, ASC-1 did not copurify with other coactivator proteins including CBP, SRC-1, and hMed6 (17), as demonstrated by HiTrap Q and other column fractionations (Fig. 3A and data not shown). Thus, we subjected HeLa nuclear extract to a series of biochemical fractionations and immunoaffinity purification with an anti-ASC-1 antibody (Fig. 3B), which ultimately resulted in five distinct protein bands (Fig. 3C). On the basis of their molecular weights, we named these proteins P200, P100, P65, P50, and P47. Peptide microsequencing of these bands by μLc/MS/MS analysis revealed that the P65 protein was ASC-1 as expected and that P47 was the proteolytic degradation product of the P50 protein. Derived peptide sequences of P200, P100, and P50 proteins were used to search the GenBank databases with the BLAST algorithm (1) and were matched to a partial cDNA clone (AJ223948), several EST clones, and a full-length cDNA clone (AF132952.1), respectively. A full-length cDNA clone for P50 was directly isolated from a HeLa cDNA library by using PCR, whereas the P200 and P100 partial sequences in databases were used as probes to screen for the full-length clones. Despite our repeated efforts, however, we failed to recover the full-length P200 clone (thus, all the P200 clones described as P200 in this paper are the longest partial clones encoding 1,917 residues; see Fig. 5A). Based on a comparison of the sizes of the in vitro-translated product of this partial P200 cDNA clone and the native protein by column fraction, at least 100 amino acids appear to be missing from the N terminus (data not shown). Importantly, antibodies raised against the recombinant proteins derived from these cDNAs specifically recognized each cognate band in the original purification (Fig. 4A), confirming the authenticity of these cDNA clones. Immunoprecipitation of ASC-1-containing HiTrap Q column fractions with anti-P50, anti-P100, and anti-ASC-1 antibodies yielded four identical protein bands (Fig. 4B and data not shown). In addition, the association of these polypeptides with hASC-1 was confirmed by their cofractionation throughout the purification procedures (Fig. 4C and data not shown).
FIG. 8.
Interaction profiles of ceASC-1. (A) Schematic representations of hASC-1, mASC-1, and ceASC-1 as well as two chimeric ASC-1 constructs. The putative zinc finger domain and the amino acid numbers for each construct are indicated. The relative percentages of similarity to hASC-1 for the central zinc finger domain and the N- and C-terminal domains are shown. (B) In the GST pull-down assays, radiolabeled ceASC-1 was incubated with GST alone or fusions of GST to p65, p50, c-Fos, c-Jun, and SRF. The fusion of LexA to ceASC-1 was cotransformed into yeast cells along with B42 alone or B42 fusion proteins, as indicated. β-Gal., β-galactosidase. (C) Transiently expressed ceASC-1 was copurified with other subunits of the hASC-1 complex. The HiTrap Q column fraction containing HA-tagged ceASC-1 (see Materials and Methods) was resolved in a Superose 6 sizing column, and the fractions were analyzed by immunoblotting with antibodies to the proteins indicated at the right (anti-HA for ceASC-1). (D) Coimmunoprecipitation of ceASC-1 with other ASC-1 complex subunits, but not with hASC-1. The HA-ceASC-1-containing column fraction (HiTrap Q) was immunoprecipitated with an anti-100, anti-ASC-1, or anti-HA antibody. Twenty percent of the load (Ld) and supernatant (sup) and 100% of the pellet from the immunoprecipitation (IP) were blotted and probed with antibodies to the proteins indicated at the right.
FIG. 3.
Purification of the ASC-1 complex in vivo. (A) HiTrap Q column fractionations reveal distinct elution profiles for different factors as indicated. RXR, retinoic X receptor. (B) Overall purification schemes for the ASC-1 complex summarized. Ab, antibody. (C) An immunoaffinity-purified ASC-1 complex from HeLa cells was resolved in SDS-polyacrylamide gel and stained with Coomassie blue. The sizes of marker proteins (M) are in kilodaltons.
FIG. 5.
(A, C, and D) Amino acid sequences of P200, P100, and P50. The peptide sequences originally obtained from mass spectrometry are underlined. The italicized boldface residues within P100 and P50 represent residues deleted in P100s and a KH domain (24), respectively. (B) Duplicated RNA helicase domains in P200. The RNA helicase consensus sequences are as described previously (29). h, hydrophobic residue; Nt, N terminus; Ct, C terminus.
FIG. 4.
Confirmations for the authenticity of the isolated cDNAs. (A) Antibodies were generated against polypeptides encoded by cDNAs which were isolated based on the peptide sequencing data of the purified proteins. + and −, HiTrap Q fractions containing the ASC-1 complex from HeLa nuclear extracts and unrelated fractions, respectively. Each antibody recognized a protein with the expected molecular weight (MW; in thousands). (B) The ASC-1 complex fraction (HiTrap Q) was immunoprecipitated with anti-P50 or anti-ASC-1 antibody. Equivalent amounts of load (Ld), supernatant (S), and pellet (P) from the immunoprecipitation (IP) were blotted and probed with antibodies to the proteins indicated at the right. Asterisks, protein bands nonspecifically immunoreactive with the anti-P100 antibody. (C) HiTrap Q fractions were subjected to Western blotting (WB) using the antibodies indicated. P200, P100, and P50 were cofractionated in this and other column fractionations (data not shown). FT, flowthrough.
Identities of the ASC-1 complex components and their interactions.
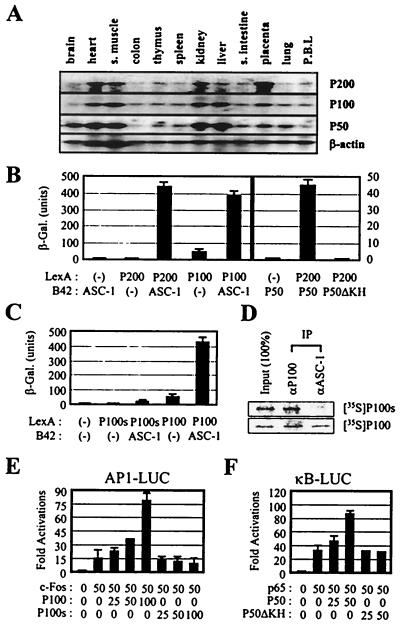
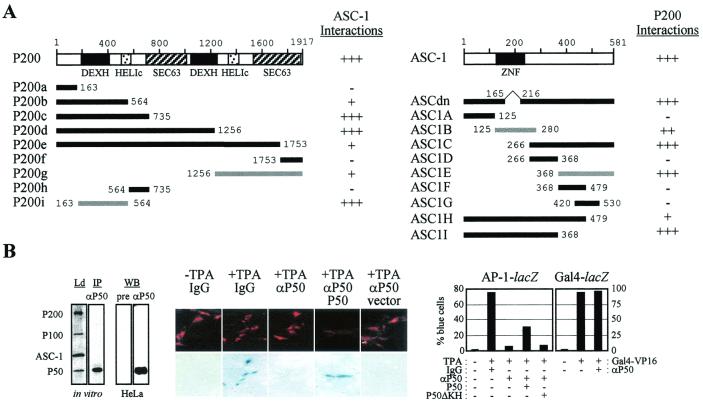
P200 encoded a protein that shows a strong homology to a group of proteins known as the DExH type RNA helicases (29) (Fig. 5A). Among the RNA helicase consensus patterns (29), domains I (ATP-binding domain; -A-G-GKT), II, III, IV, and VI (basic RNA-binding domain, -GR-GR-) are well conserved and duplicated (Fig. 5B). Intriguingly, P50, a protein of 357 amino acids, also contains an RNA-binding motif called KH (24) (Fig. 5D). P100, a novel protein without significant homology to any known protein in databases, consists of 756 amino acids (Fig. 5C). Interestingly, while we screened for the full-length P100 cDNA, we also isolated cDNAs encoding a novel isoform of P100 (i.e., P100s) which has an internal deletion of 25 residues (Fig. 5C). Northern analyses indicated that mRNAs for P200, P100, and P50 (approximately 10, 2.8, and 2.1 kb, respectively) are ubiquitously expressed (Fig. 6A), consistent with the expression pattern of ASC-1 mRNA (11). As shown in Fig. 6B, P200 strongly interacted with ASC-1 and P50 and P100 showed an additional interaction with ASC-1 in yeast. However, a deletion of P50 residues 40 to 162, containing the KH motif (24) (i.e., P50ΔKH), completely abolished the interaction with P200. Similar results were also obtained in coimmunoprecipitation experiments with in vitro-translated proteins as well as in GST pull-down assays (data not shown). In contrast to P100, P100s did not show any significant interaction with ASC-1 in yeast (Fig. 6C), although similar levels of protein expression were observed for P100 and P100s (data not shown). Corroborating these results, P100 but not P100s was copurified with ASC-1 when the two were translated together in vitro and immunoprecipitated by anti-ASC-1 (Fig. 6D). Although the function of this P100 isoform is currently unclear, cotransfection of P100 but not P100s significantly enhanced NF-κB and AP-1 transactivation, consistent with their interaction profile (Fig. 6E and data not shown). Likewise, P50 but not P50ΔKH enhanced NF-κB and AP-1 transactivation (Fig. 6F and data not shown). Next, we localized the interaction interfaces between P200 and ASC-1 in the yeast two hybrid systems using a series of deletion constructs (Fig. 7A) in which a region consisting of P200 residues 163 to 564 (i.e., P200i) was mapped as a major binding site for ASC-1, although an additional, weak interaction site within the C-terminal region of P200 was found. Efficient integration of P200 into the whole ASC-1 complex may require both of these ASC-1 interaction interfaces, along with the P50 interaction interface, based on the fact that P200i alone had no significant effects on AP-1, NF-κB, and SRF transactivation in cotransfections (data not shown). Similarly, the P200 binding site was mapped to two distinct regions of ASC-1, one of which encompasses the central zinc finger region (Fig. 7A). Consistent with these results, ASCdn, in which a part of the ASC-1 zinc finger region was deleted, was previously shown to function as a dominant negative mutant in the transactivation by various nuclear receptors (11). Overall, these results suggest that the molecular mechanisms for the ASC-1 action may involve interactions with RNA molecules and that formation of the ASC-1 complex results from extensive interactions among its different constituents.
FIG. 6.
Interactions among the ASC-1 complex components. (A) Northern blot analyses indicate that P200, P100, and P50 are ubiquitously expressed as previously described for ASC-1 (11). s. muscle, smooth muscle; s. intestine, small intestine; P.B.L, peripheral blood lymphocytes. (B, C) Plasmids encoding the indicated B42 and LexA proteins were transformed into a yeast strain containing an appropriate lacZ reporter gene, as described previously (2). The Western analysis with the anti-P100 antibody showed comparable levels of expression of P100 and P100s in yeast (data not shown). β-Gal., β-galactosidase. (D) ASC-1 was cotranslated in vitro with either P100 or P100s in the presence of [35S]methionine and immunoprecipitated with an anti-P100 or anti-ASC-1 antibody. (E and F) HeLa cells were transfected with the lacZ expression vector and increasing amounts of P100, P100s, P50, and P50ΔKH expression vector along with indicated reporter genes. For activation, expression vectors for p65 and c-Fos were also cotransfected, as indicated (in nanograms).
FIG. 7.
P200-ASC-1 interaction interfaces and essentiality of P50 in AP-1 transactivation. (A) A combination of fusions of B42-ASC-1 and LexA to wild-type P200 or P200 deletion mutants (left panel) and fusions of B42 to the wild-type ASC-1 or ASC-1 deletion mutants and LexA-P200 (right panel) were transformed into a yeast strain containing an appropriate lacZ reporter gene, as described previously (2). None of the LexA-P200 constructs exhibited autonomous transactivation function (data not shown). +++, strongly blue colonies after 2 days of incubation; ++, light blue colonies after 2 days of incubation; +, light blue colonies after more than 2 days of incubation; −, white colonies. Gray bars, deduced interaction domains. Two copies of DExH-type helicase domains, the helicase superfamily C-terminal domains (HELIc), and SEC63 domains of unknown function within P200 as well as the zinc finger (ZNF) domain of ASC-1 are indicated. (B) (Left) Specificity of the P50 antibody was demonstrated by specific immunoprecipitation (IP) of P50 from a mixture of in vitro-translated and radiolabeled constituents of the ASC-1 complex as well as specific detection of P50 from HeLa nuclear extract in Western analysis (WB). Ld and pre, 20% of the reaction mixture and preserum, respectively. (Middle) Photographs of FITC-stained injected cells (top) and the corresponding pattern of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-thiogalactopyranoside) staining (bottom) with microinjection of either control IgG or anti-P50 IgG. The presence or absence of 0.1 μM TPA is indicated. (Right) The number of cells that express lacZ relative to the total number of FITC-positive cells microinjected with either control IgG or anti-P50 IgG. The presence or absence of TPA (0.1 μM) and a P50, P50ΔKH, or Gal4-VP16 expression vector is indicated. The reporter constructs were AP-1-lacZ and Gal4-lacZ. Experiments were repeated twice with similar results (error range, 5 to 10%), with >200 cells injected.
P50 as an essential component for AP-1 transactivation in vivo.
To investigate the function of the ASC-1 complex in vivo, we utilized microinjection techniques (15, 16). The lacZ reporter gene was placed under the control of a simian virus 40 minimal promoter containing TPA-responsive AP-1 sites (16). Given the putative importance of RNA recognition in the function of the ASC-1 complex, we targeted the KH motif-containing P50. The affinity-purified anti-P50 IgG utilized in these experiments was highly specific, as demonstrated by its ability to immunoprecipitate only P50 from a mixture of the in vitro-translated four constituents of the ASC-1 complex and to specifically recognize P50 from HeLa nuclear extract (Fig. 7B). Remarkably, microinjection of this anti-P50 IgG almost completely prevented TPA from activating an AP-1-dependent transcription unit. The percentage of cells that expressed the lacZ reporter (i.e., blue cells) was negligible among cells microinjected with control IgG in the absence of TPA but increased to approximately 75% in the presence of TPA (Fig. 7B, right). However, only approximately 5% of cells turned blue, even in the presence of TPA, when microinjected with anti-P50 IgG. When anti-P50 IgG was coinjected with the P50 expression vector, the percentage of blue cells increased to approximately 32%, while coinjection of anti-P50 IgG with the P50ΔKH expression vector was without any significant effect. In contrast, microinjection of the anti-P50 IgG had no effect on transactivation mediated by Gal4-lacZ and Gal4-VP16. From these results, taken together with the transient transfection data (Fig. 2), we concluded that P50 is essential for AP-1 transactivation in vivo, in which the KH domain may play an indispensable role. Experiments extending these results to other transcription factors, including nuclear receptors, NF-κB, and SRF, are in progress.
The dominant-negative phenotype of ceASC-1 in mammalian cells.
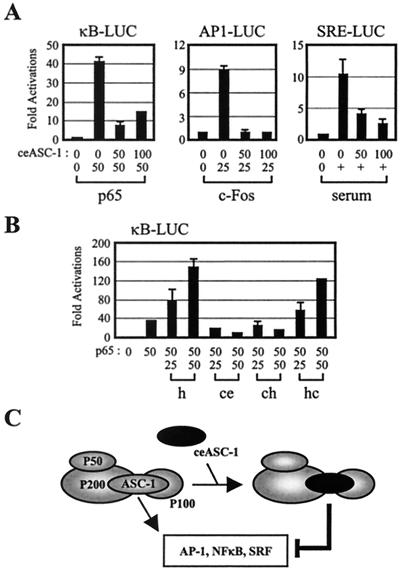
We have isolated a mouse ASC-1 homologue from a mouse liver cDNA library and also obtained an EST clone encoding the full-length C. elegans ASC-1 homologue. These two proteins, mASC-1 and ceASC-1, showed approximately 93 and 31% overall identity to the original human form, respectively. Similar proteins were also identified from the Drosophila melanogaster and Xenopus laevis cDNA databases (data not shown), suggesting their structural and functional conservation throughout evolution. Notably, the central putative zinc finger domain, in which ceASC-1 showed 82.8% similarity to the human form, was conserved the most, whereas the C-terminal domain of ceASC-1 was moderately conserved (approximately 45.8% similarity) (Fig. 8A). Interestingly, ceASC-1 lacked the N-terminal 122 amino acids, which are highly conserved among mammalian ASC-1s. Importantly, ceASC-1 interacted very weakly with NF-κB component p65, but not with SRF, p50, c-Fos, and c-Jun, in the GST pull-down assays (Fig. 8B; note that only a small portion of input protein was bound to GST/p65, relative to the amount pulled down in Fig. 1A). Similarly, the p65-ceASC-1 interactions in the mammalian two-hybrid tests were also very weak relative to the p65-hASC-1 interactions (Fig. 1C) (data not shown). Interestingly, a fusion of LexA to the full-length ceASC-1 (i.e., LexA-ceASC-1) did not show an autonomous transactivation activity in yeast (Fig. 8B, bottom), in contrast to its human homologue (11); the significance of this difference is not currently understood. Surprisingly, however, ceASC-1 interacted with P200 and P100 (Fig. 8B, bottom). Consistent with these results, a part of HA-tagged ceASC-1 eluted at approximately the 650-kDa position from a Superose 6 sizing column, analogous to endogenous hASC-1 and P100, when transfected into HeLa cells (Fig. 8C). Notably, the appearance of ceASC-1 in this 650-kDa position coincided with the disappearance or significant decrease of the endogenous ASC-1 protein compared to the stoichiometric amount of ASC-1 in the original endogenous complex (Fig. 3C). A significant amount of cotransfected ceASC-1 was also visible around its native-molecular-mass position (i.e., approximately 50 kDa), which represents a free or dissociated form of overexpressed ceASC-1 during purification. Consistent with these results, when HiTrap Q column fractions representing the ASC-1/ceASC-1 complexes were pooled and immunoprecipitated by an anti-HA antibody, the precipitates contained only ceASC-1, not hASC-1 (Fig. 8D). Conversely, the hASC-1 immunoprecipitates included only hASC-1, not ceASC-1, whereas the P100 precipitates contained both ASC-1 and ceASC-1. Thus, we concluded that at least a part of the transfected ceASC-1 competes with the endogenous ASC-1 to form a distinct complex containing ceASC-1 but not hASC-1. These results, along with the inability of ceASC-1 to properly interact with SRF, AP-1, and NF-κB (Fig. 8B), led us to predict that overexpressed ceASC-1 will likely show a potent dominant-negative phenotype with these transcription factors in mammalian cells. This prediction was indeed confirmed in cotransfection studies with SRE-LUC, κB-LUC, and AP1-LUC (Fig. 9A). However, ceASC-1 was without any significant effect on transactivation directed by either pRSV-β-gal or Gal4-VP16 (data not shown). To elucidate the ceASC-1 region responsible for the dominant-negative effects, we constructed two chimeras (Fig. 8A); one contains ceASC-1 residues 1 to 182 fused to hASC-1 residues 325 to 581 (i.e., chASC-1), and the other contains hASC-1 residues 1 to 324 fused to ceASC-1 residues 183 to 425 (i.e., hcASC-1). While hcASC-1 was capable of stimulating NF-κB and AP-1 transactivation as efficiently as the wild-type hASC-1, chASC-1 exhibited a dominant-negative phenotype, like ceASC-1 (Fig. 9B and data not shown). These results indicate that the N-terminal region containing the zinc finger region is responsible for the functional difference between ceASC-1 and hASC-1. Consistent with the strong sequence similarity, however, mASC-1 was functionally indistinguishable from the human form (data not shown). Overall, these results indirectly but clearly demonstrate that the endogenous ASC-1 complex is essential for AP-1, NF-κB, and SRF transactivation in vivo.
FIG. 9.
Dominant-negative phenotype of ceASC-1. (A and B) HeLa cells were transfected with the lacZ expression vector and increasing amounts of ceASC-1, hASC-1, chASC-1, and hcASC-1 expression vectors along with the indicated reporter genes. For activation, expression vectors for p65 and c-Fos were also cotransfected, as indicated. +, serum shock; h, ce, ch, and hc, hASC-1, ceASC-1, chASC-1, and hcASC-1, respectively. (C) Schematic representation of the ASC-1 complex. P200 binds both ASC-1 and P50, and ASC-1 also binds P100. The N-terminal region of P200 has not been isolated yet; it may make additional contacts with other components. Cotransfected ceASC-1 in HeLa cells appears to replace the endogenous ASC-1 from the complex (Fig. 8C and D) and acts as a dominant-negative mutant, likely due to the inability of ceASC-1 to properly interact with target transcription factors (see the text for further discussion).
DISCUSSION
CBP/p300, SRC-1, and ASC-2 define a distinct group of transcriptional coactivator molecules that directly bind and coactivate a wide spectrum of different transcription factors and thus designated transcription integrators or cointegrators (13, 18). In this report, we added ASC-1, originally isolated as a coactivator of nuclear receptors (11), to the list of multifunctional transcription integrators. ASC-1 is a coactivator of AP-1, NF-κB, and SRF (Fig. 1 and 2A) and is involved in the previously described transrepression between nuclear receptors and either AP-1 or NF-κB (reviewed in references 8 and 26), based on the observation that coexpressed ASC-1 effectively relieved the repression (Fig. 2B). As with many transcription coactivators (6, 19, 22, 25, 28), ASC-1 was found to stably associate with three additional polypeptides, P200, P100, and P50 (Fig. 3C). Unfortunately, our numerous attempts to identify the missing N-terminal region of P200 were without any success; this region was estimated to consist of approximately 100 amino acids based on the size differences between the in vitro-translated product of the longest cDNA we isolated and the P200 protein in the original purification (results not shown). The annotated human genome sequence at the University of California, Santa Cruz, predicts that the N-terminal P200 sequence we obtained (underlined portion of MIMDFCLKFLLFILNRLFELLGPEGLELIEL…) has only 25 fewer residues than the product of one gene on chromosome 6 (Affymetrix prediction) but has as many as 215 fewer residues than the product of another gene predicted by another program. Interestingly, homologues of ASC-1, P200, and P50 are also found in mice, Drosophila, Xenopus, and C. elegans, whereas a P100 homologue was not found in C. elegans, suggesting that P100 may have integrated into the complex later in evolution. Thus, it was intriguing that P200 and P50, but not P100, were also found in the cytoplasm (our unpublished results), like ASC-1 (11). Interestingly, both P200 and P50 have RNA helicase and RNA-binding motifs (24, 29) (Fig. 5), suggesting that the function of the ASC-1 complex may require RNA binding. Notably, p68 RNA helicase was recently isolated as a transcriptional coactivator specific to N-terminal activating function 1 of estrogen receptor α (5). RNA helicase A was found to mediate association of CBP with RNA polymerase II (23). In addition, novel transcriptional coactivator p52 also interacted with essential splicing factor ASF/SF2 both in vitro and in vivo to modulate ASF/SF2-mediated pre-mRNA splicing (7). More recently, PGC-1 was shown to mediate a direct coupling of transcription initiation and mRNA processing in vivo (20). It is important that posttranscriptional mRNA processing such as 5′ capping, splicing, and polyadenylation can take place cotranscriptionally in vivo (reviewed in reference 9). Thus, these transcriptional coactivator proteins, including the ASC-1 complex, may also act as adapter molecules to coordinate various events of pre-mRNA processing and transcriptional initiation of class II genes. Interestingly, a point-mutated version of P200 was recently identified as an immunodominant antigen recognized by autologous cytolytic T lymphocytes of human melanoma (J.-F. Baurain, personal communications), studies of which may provide an important insight into the transactivation function of the ASC-1 complex in vivo.
As summarized in the model (Fig. 9C), multiple intersubunit interactions occur within the ASC-1 complex, including the interactions of ASC-1 with P100 and P200, as well as the additional P50-P200 interactions (Fig. 6B and 7A). In addition, the missing N-terminal region of P200 could also provide an additional binding interface for P100 within the complex, such that P200 acts as a scaffold protein to enhance the integrity of the whole complex in vivo. Interestingly, the combined molecular masses of all the ASC-1 complex components (i.e., 415 kDa) was different from the apparent molecular mass from the Superose 6 column (i.e., approximately 650 kDa) (Fig. 8C), raising the possibility that some of its components could be multimeric within the complex. However, this possibility is not likely because the four bands isolated appeared to be highly stoichiometric (Fig. 3C). Alternatively, a component loosely attached to the core ASC-1 complex or a nonprotein component such as SRA, a putative RNA component of the SRC-1 complex (12), could have been lost during our purification procedures.
Our preliminary results suggested that P200, P100, and P50 differentially enhanced transactivation by various nuclear receptors, AP-1, NF-κB, and SRF in cotransfections (Fig. 6 and data not shown). In particular, P50 appeared to be an efficient stimulator of transactivation by thyroid hormone receptor, whereas, with AP-1 and NF-κB transactivation, P100 was a much better coactivator than either ASC-1 or P50. Although the exact mechanisms by which cotransfected ASC-1, P50, P100, and P200 differentially coactivate various transcription factors are not clear, these individually overexpressed proteins may help assemble a maximum number of the complexes within the cell, likely through mobilizing excess amounts of the other components being expressed. Confirming the importance of the complex itself, the P100 isoform (i.e., P100s) and P50ΔKH, which were not capable of interacting with ASC-1 and P200, respectively, did not stimulate AP-1 and NF-κB transactivation (Fig. 6 and data not shown). In addition, transfected ceASC-1, replacing the endogenous hASC-1 and incorporating itself into a steady-state complex, caused a concomitant decrease in the amount of the functional hASC-1-containing complex (Fig. 8C and D). However, it is interesting that ceASC-1 complex containing P200, P100, and P50 may not be stable enough to survive the column purification procedures, based on the relative abundance of free form of ceASC-1 (Fig. 8C). It is also possible that the stability of the complex itself may require multiple, cryptic interaction interfaces among the complex constituents in addition to the P200/P50, P200/ASC-1, and ASC-1/P100 interactions we already defined (Fig. 6B and 7A). These cryptic interactions could be missing in ceASC-1, resulting in instability of the ceASC-1 complex (Fig. 8C). Importantly, coexpressed ceASC-1 acted as a potent dominant-negative mutant in AP-1, NF-κB, and SRF transactivation in mammalian cells (Fig. 9A). These results suggest that ceASC-1 expressed in mammalian cells, although capable of forming a complex similar to the LASC-1 complex, is not functional with these transcription factors, likely due to its lack of proper interactions with target transcription factors (Fig. 8B). Alternatively, the negative effects may have simply reflected the decrease of the functional hASC-1 complex. In any event, the endogenous ASC-1 complex is likely to be essential for transactivation by at least a subset of transcription factors in vivo. Notably, ASC-1 interacts with basal transcription factors TBP and TFIIA as well as with integrators CBP/p300 and SRC-1 (11). Interestingly, ceASC-1 did not show any autonomous transactivation activity in yeast (Fig. 8B), as opposed to hASC-1 (11), and ceASC-1 showed a dominant-negative phenotype with transactivation by retinoic acid receptor but not thyroid hormone receptor, although both receptors appear to bind ceASC-1 (our unpublished results). Thus, the dominant-negative phenotype of ceASC-1 (Fig. 9A) may also involve its impaired interactions with other effector molecules, such as TBP, TFIIA, CBP/p300, and SRC-1.
Interestingly, our extensive effort to localize the interaction interfaces of ASC-1 for each of its target transcription factors or to overexpress it for structural studies has been largely unsuccessful because of the consistent difficulty of expressing full-length ASC-1 or fragments of ASC-1 in various expression systems. Similar difficulties were observed with P200, P100, and P50. It is possible that each constituent of the complex alone may not be stable enough in solution; stability may only be adequate when constituents are coexpressed together. Consistently, the displaced hASC-1 in ceASC-1-overexpressing cells was not visible as a free polypeptide when biochemically fractionated (Fig. 8C). The N-terminal subregion of ASC-1, which is not present in ceASC-1 and which is not essential for the ASC-1 function, at least in AP-1, SRF, and NF-κB transactivation (data not shown), could be responsible for this instability, because ceASC-1 was readily detectable as a free form (Fig. 8C).
In conclusion, we have identified AP-1, NF-κB, and SRF as new target molecules of ASC-1 and purified the steady-state ASC-1 complex. Our results suggest that the hASC-1 complex plays a pivotal role in transactivation by these newly identified ASC-1 target transcription factors in mammalian cells. Further studies of the ASC-1 complex should provide important insights into multifactorial regulatory circuits of gene expression controls as well as cross-talk among different signaling pathways.
Acknowledgments
We thank Yuji Kohara for the ceASC-1 plasmid and Bill Lane for peptide microsequencing.
D.-J. Jung and H.-S. Sung contributed equally to this work.
This work was supported by grants 2000-2-20900-006-1 from the Basic Research Program of the Korea Science and Engineering Foundation (Y.C.L) and the Basic Science Research Institute of Pohang University of Science and Technology and GenoCheck, Inc. (J.W.L.).
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.) 1995. Current protocols in molecular biology. Greene Publishing Associates, New York, N.Y.
- 3.Baldwin, A. S. 1996. The NF-kappa B and I kappa B proteins: new discoveries and insights. Annu. Rev. Immunol. 14:649-681. [DOI] [PubMed] [Google Scholar]
- 4.Dignam, J. D., P. L. Martin, B. S. Shastry, and R. G. Roeder. 1983. Eukaryotic gene transcription with purified components. Methods Enzymol. 101:582-598. [DOI] [PubMed] [Google Scholar]
- 5.Endoh, H., K. Maruyama, Y. Masuhiro, Y. Kobayashi, M. Goto, H. Tai, J. Yanagisawa, D. Metzger, S. Hashimoto, and S. Kato. 1999. Purification and identification of p68 RNA helicase acting as a transcriptional coactivator specific for the activation function 1 of human estrogen receptor α. Mol. Cell. Biol. 19:5363-5372. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Fondell, J. D., H. Ge, and R. G. Roeder. 1996. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc. Natl. Acad. Sci. USA 93:8329-8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ge, H., Y. Si, and A. P. Wolffe. 1998. A novel transcriptional coactivator, p52, functionally interacts with the essential splicing factor ASF/SF2. Mol. Cell 2:751-759. [DOI] [PubMed] [Google Scholar]
- 8.Gottlicher, M., S. Heck, and P. Herrlich. 1998. Transcriptional cross-talk, the second mode of steroid hormone receptor action. J. Mol. Med. 76:480-489. [DOI] [PubMed] [Google Scholar]
- 9.Hirose, Y., and J. L. Manley. 2000. RNA polymerase II and the integration of nuclear events. Genes Dev. 14:1415-1429. [PubMed] [Google Scholar]
- 10.Karin, M., Z. Liu, and E. Zandi. 1997. AP-1 function and regulation. Curr. Opin. Cell Biol. 9:240-246. [DOI] [PubMed] [Google Scholar]
- 11.Kim, H.-J., J.-Y. Yi, H.-S. Sung, D. D. Moore, B. H. Jhun, Y. C. Lee, and J. W. Lee. 1999. Activating signal cointegrator 1, a novel transcription coactivator of nuclear receptors, and its cytosolic localization under conditions of serum deprivation. Mol. Cell. Biol. 19:6323-6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lanz, R. B., N. J. McKenna, S. A. Onate, U. Albrecht, J. Wong, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1999. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell 97:17-27. [DOI] [PubMed] [Google Scholar]
- 13.Lee, J. W., Y. C. Lee, S.-Y. Na, D.-J. Jung, and S.-K. Lee. 2001. Transcriptional coregulators of the nuclear receptor superfamily: coactivators and corepressors. Cell. Mol. Life Sci. 58:289-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee, J. W., H. S. Choi, J. Gyuris, R. Brent, and D. D. Moore. 1995. Two classes of proteins dependent on either the presence or absence of thyroid hormone for interaction with the thyroid hormone receptor. Mol. Endocrinol. 9:243-254. [DOI] [PubMed] [Google Scholar]
- 15.Lee, S.-K., S. L. Anzick, J.-E. Choi, L. Bubendorf, X.-Y. Guan, Y.-K. Jung, O.-P. Kallioniemi, J. Kononen, J. M. Trent, D. Azorsa, B. H. Jhun, J. H. Cheong, Y. C. Lee, P. S. Meltzer, and J. W. Lee. 1999. A nuclear factor, ASC-2, as a cancer-amplified transcriptional coactivator essential for ligand-dependent transactivation by nuclear receptors in vivo. J. Biol. Chem. 274:34283-34293. [DOI] [PubMed] [Google Scholar]
- 16.Lee, S.-K., S.-Y. Na, S.-Y. Jung, J.-E. Choi, B. H. Jhun, J. Cheong, P. S. Meltzer, Y. C. Lee, and J. W. Lee. 2000. Activating protein-1, nuclear factor-kappaB, and serum response factor as novel target molecules of the cancer-amplified transcription coactivator ASC-2. Mol. Endocrinol. 14:915-925. [DOI] [PubMed] [Google Scholar]
- 17.Lee, Y. C., and Y.-J. Kim. 1998. Requirement for a functional interaction between mediator components Med6 and Srb4 in RNA polymerase II transcription. Mol. Cell. Biol. 18:5364-5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKenna, N. J., R. B. Lanz, and B. W. O'Malley. 1999. Nuclear receptor coregulators: cellular and molecular biology. Endocr. Rev. 20:321-344. [DOI] [PubMed] [Google Scholar]
- 19.McKenna, N. J., Z. Nawaz, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1998. Distinct steady-state nuclear receptor coregulator complexes exist in vivo. Proc. Natl. Acad. Sci. USA 95:11697-11702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monsalve, M., Z. Wu, G. Adelmant, P. Puigserver, M. Fan, and B. M. Spiegelman. 2000. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol. Cell 6:307-316. [DOI] [PubMed] [Google Scholar]
- 21.Myers, L. C., K. Leuther, D. A. Bushnell, C. M. Gustafsson, and R. D. Kornberg. 1997. Yeast RNA polymerase II transcription reconstituted with purified proteins. Methods 12:212-216. [DOI] [PubMed] [Google Scholar]
- 22.Naar, A. M., P. A. Beaurang, S. Zhou, S. Abraham, W. Solomon, and R. Tjian. 1999. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature 398:828-832. [DOI] [PubMed] [Google Scholar]
- 23.Nakajima, T., C. Uchida, S. F. Anderson, C. G. Lee, J. Hurwitz, J. D. Parvin, and M. Montminy. 1997. RNA helicase A mediates association of CBP with RNA polymerase II. Cell 90:1107-1112. [DOI] [PubMed] [Google Scholar]
- 24.Ostareck-Lederer, A., D. H. Ostareck, and M. W. Hentze. 1998. Cytoplasmic regulatory functions of the KH-domain proteins hnRNPs K and E1/E2. Trends Biochem. Sci. 23:409-411. [DOI] [PubMed] [Google Scholar]
- 25.Rachez, C., Z. Suldan, J. Ward, C. P. Chang, D. Burakov, H. Erdjument-Bromage, P. Tempst, and L. P. Freedman. 1998. A novel protein complex that interacts with the vitamin D3 receptor in a ligand-dependent manner and enhances VDR transactivation in a cell-free system. Genes Dev. 12:1787-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saatcioglu, F., F. X. Claret, and M. Karin. 1994. Negative transcriptional regulation by nuclear receptors. Semin. Cancer Biol. 5:347-359. [PubMed] [Google Scholar]
- 27.Treisman, R. 1994. Ternary complex factors: growth factor regulated transcriptional activators. Curr. Opin. Genet. Dev. 4:96-101. [DOI] [PubMed] [Google Scholar]
- 28.Vassilev, A., J. Yamauchi, T. Kotani, C. Prives, M. L. Avantaggiati, J. Qin, and Y. Nakatani. 1998. The 400 kDa subunit of the PCAF histone acetylase complex belongs to the ATM superfamily. Mol. Cell 2:869-875. [DOI] [PubMed] [Google Scholar]
- 29.Widner, W. R., and R. B. Wickner. 1993. Evidence that the SKI antiviral system of Saccharomyces cerevisiae acts by blocking expression of viral mRNA. Mol. Cell. Biol. 13:4331-4341. [DOI] [PMC free article] [PubMed] [Google Scholar]