Abstract
We have investigated the role that S259 phosphorylation, S621 phosphorylation, and 14-3-3 binding play in regulating Raf-1 activity. We show that 14-3-3 binding, rather than Raf-1 phosphorylation, is required for the correct regulation of kinase activity. Phosphorylation of S621 is not required for activity, but 14-3-3 binding is essential. When 14-3-3 binding to conserved region 2 (CR2) was disrupted, Raf-1 basal kinase activity was elevated and it could be further activated by V12,G37Ras, V23TC21, and V38R-Ras. Disruption of 14-3-3 binding at CR2 did not recover binding of Raf-1 to V12,G37Ras but allowed more efficient recruitment of Raf-1 to the plasma membrane and stimulated its phosphorylation on S338. Finally, V12Ras, but not V12,G37Ras, displaced 14-3-3 from full-length Raf-1 and the Raf-1 bound to Ras. GTP was still phosphorylated on S259. Our data suggest that stable association of Raf-1 with the plasma membrane requires Ras-mediated displacement of 14-3-3 from CR2. Small G proteins that cannot displace 14-3-3 fail to recruit Raf-1 to the membrane efficiently and so fail to stimulate kinase activity.
The Ras/Raf/MEK/ERK signaling pathway is a conserved signaling module that regulates complex cellular functions such as proliferation, differentiation, cell death, and T-cell activation (for reviews, see references 37 and 49). This pathway consists of a kinase cascade that is activated in a Ras-dependent manner. The first kinases in the cascade are the Raf serine/threonine-specific protein kinases. Active Raf proteins phosphorylate and activate the dual-specificity MEK protein kinases, which in turn phosphorylate and activate the serine/threonine-specific extracellular signal-regulated kinase (ERK) mitogen-activated protein kinases. ERKs phosphorylate and regulate the activity of proteins in the cytosol and the nucleus. The Raf proteins therefore couple Ras signaling to ERK activation. In mammals there are three Raf genes (Raf-1, A-Raf, and B-Raf), and comparison of their protein sequences reveals three conserved regions, conserved region 1 (CR1), CR2, and CR3 (22, 35). CR1 and CR2, found within the N-terminal half of the proteins, appear to be regulatory, while the kinase domain, encapsulated within CR3, is in the C-terminal half of the proteins.
Raf regulation has been the subject of intense scrutiny (for reviews, see references 3, 27, and 43). Raf proteins bind to activated Ras (Ras-GTP) with high affinity through a region called the Ras binding domain (RBD) which is within CR1. Ras is anchored to the inner surface of the plasma membrane and in resting cells, Raf proteins are cytosolic. However, in the presence of Ras-GTP, the Raf proteins are recruited to the plasma membrane, where activation occurs through a mechanism involving phosphorylation, dimerization, binding to other proteins and lipid interactions. Two amino acids whose phosphorylation at the plasma membrane is critical for Raf-1 activity are serine 338 (S338) and tyrosine 341 (Y341), which are in a region that we call the N region (for negative-charge regulatory region) (15, 17, 38).
One family of proteins that interact with Raf-1 are the 14-3-3 adaptor-scaffold proteins. These are highly conserved acidic proteins with molecular masses of ∼30 kDa that bind to a large number and variety of client proteins (see references 2 and 46). The binding of 14-3-3 to client proteins occurs through short peptide motifs. For some peptides, binding occurs only if a specific serine within the motif is phosphorylated, but binding to other motifs is phosphorylation independent (45, 47, 61). An emerging concept in 14-3-3 biology is that they bind to and sequester client proteins into inappropriate subcellular compartments, thereby suppressing client protein activity (see reference 46). For example, phosphorylation dependent binding of 14-3-3 to the transcription factor forkhead blocks its ability to repress transcription by removing it from the nucleus (7, 48). Similarly, phosphorylation-dependent binding of 14-3-3 to the proapoptotic protein BAD blocks apoptosis by displacing BAD from mitochondria (11).
The role that 14-3-3 binding plays in regulating Raf-1 is controversial. Many studies suggest that 14-3-3 binding is essential for kinase activity (19, 20, 23, 31, 39, 52, 56, 62), while others suggest that it is not (21, 40, 55). In part, the confusion stems from the fact that there are two 14-3-3 binding sites on Raf-1 that appear to play opposing roles. Both sites conform to the consensus sequence RSXpSXP (single amino acid code [24]: pS, phosphorylated serine; X, any amino acid) (Fig. 1) with binding being dependent on phosphorylation of the central serine (45, 61). One motif is in CR2 and requires phosphorylation of serine 259 (S259) (Fig. 1). The other, in CR3, is at the C-terminal end of the kinase domain and requires phosphorylation of serine 621 (S621) (Fig. 1). Binding of 14-3-3 to CR2 appears to suppress Raf-1 activity, whereas binding to CR3 appears to be essential. Thus, since 14-3-3 proteins are dimeric and can simultaneously bind to two peptides (47, 61), one model suggests that one 14-3-3 dimer binds to both CR2 and CR3 to keep Raf-1 in a closed, inactive conformation (51, 57). Activation requires release of CR2, and 14-3-3 can then bind to a third, unidentified site to maintain the active conformation (see reference 3). Recent studies have shown that protein kinase B can suppress Raf-1 activity by directly phosphorylating S259 (50, 63), and it has also been suggested that protein phosphatase 1 (PP1) and 2A (PP2A) mediate S259 dephosphorylation as a prerequisite for Raf-1 activation in growth factor-stimulated cells (1, 26). It has also been suggested that 14-3-3 dimers bridge Raf-1 to substrates or to other signaling molecules (6, 10).
FIG. 1.
14-3-3 binding motifs in Raf-1. The amino acid sequences surrounding the 14-3-3 binding motif in CR2 and CR3 of Raf-1 are shown, together with the consensus motif (45). The single amino acid code is used (X, any amino acid; pS, phosphorylated serine), and amino acids whose positions cannot be altered are underlined in the consensus sequence. Vertical lines indicate conserved residues, and the numbers 256, 261, 618, and 623 refer to amino acid positions in Raf-1.
There also appears to be competition between Ras and 14-3-3 for binding to Raf-1. Both Ras and 14-3-3 have secondary binding sites within the cysteine-rich domain (CRD), which is also in CR1 and C terminal to the RBD (9, 40, 60). In vitro, Ras-GTP displaces 14-3-3 from the isolated N terminus of Raf-1 (51), possibly due to competition for this second site (12). Genetic evidence from yeast two-hybrid analysis using Saccharomyces cerevisiae also suggests competitive binding. Raf-1 binds to oncogenic Ras (V12Ras) in yeast cells, but binding is disrupted when glutamate 37 (E37) in Ras is replaced with glycine (V12,G37Ras) (59). Intriguingly, mutations to arginine 256 (R256) or serine 257 (S257) of Raf-1, both of which are within the RSXpSXP motif at CR2, restored Raf-1 binding to V12,G37Ras in yeast (59) but not in solution (25). These mutations would be expected to disrupt 14-3-3 binding to CR2, but this has not been tested. The ability of V12,G37Ras to activate Raf-1 in which S257 was replaced with leucine has not been tested directly, but when coexpressed, these proteins activate ERK in cells and stimulate reporter gene expression, something that they cannot do when expressed alone (25, 59).
Finally, it is difficult to analyze the role that 14-3-3 binding to CR3 plays, because S621 mutations are inactivating (4, 44). In one study, it was suggested that Raf-1 activation requires 14-3-3 displacement from CR3 and S621 dephosphorylation (42). It was suggested that S621 mutations are inactivating because the dephosphorylated serine performs a specific function(s) that is required for activity. This model also suggests that 14-3-3 is completely displaced from active Raf-1, and in some experimental conditions, this appears to be the case (52). However, a number of other studies argue that binding of 14-3-3 to CR3 is essential for activity, with the strongest evidence coming from peptide displacement studies. Active Raf-1 can be inactivated by phosphopeptides that displace 14-3-3; Raf-1 is reactivated by subsequent addition of recombinant 14-3-3 (39, 56, 57). Importantly, this approach also works with the isolated kinase domain, supporting binding to CR3 as being essential for activity (62). Furthermore, it has been shown that 14-3-3 binding to CR3 protects S621 from dephosphorylation to maintain Raf-1 activity (56) and also that 14-3-3 protects active Raf-1 from PP1- and PP2A-mediated inactivation (13).
In this study, we have further investigated the complex roles played by S259 phosphorylation, S621 phosphorylation, and 14-3-3 binding in regulating Raf-1 activity. We show that the binding of 14-3-3 to Raf-1, rather than S259 or S621 phosphorylation, is the essential event in Raf-1 regulation. We demonstrate that V12,G37Ras directly activates Raf-1 when 14-3-3 binding to CR2 is disrupted and that this activation requires Ras binding, membrane localization, and S338 phosphorylation. We show that V12Ras, but not V12,G37Ras, displaces 14-3-3 from Raf-1 but that the Raf-1 that is bound to Ras-GTP is still phosphorylated on S259. We also demonstrate that TC21 and R-Ras can only activate Raf-1 when 14-3-3 binding to CR2 is disrupted. Our data suggest that 14-3-3 antagonizes Raf-1 recruitment to the plasma membrane to ensure that Raf-1 is not activated in resting cells and cannot be activated by all Ras-related small G proteins.
MATERIALS AND METHODS
Expression vectors.
All cloning steps were performed by standard techniques (53). The myc-epitope-tagged Raf-1 (mRaf-1) expression vector (pEFm/Raf-1.6) has been described previously (8). It uses the elongation factor 1α (EF1α) promoter for efficient protein expression and fuses a myc epitope tag (MEQKLISEEDLGS) onto the Raf-1 N terminus. This tag is recognized by the mouse monoclonal antibody 9E10 (16) and by the rat monoclonal antibody JAC/6 (K. Maycroft, unpublished data). All the point mutations in mRaf-1, as indicated in the present work, were introduced into this vector by using the PCR with sequence verification by automated dideoxy sequencing procedures. The R-18 sequence was also introduced into this vector by PCR. To introduce the R-18 peptide into CR3, codons 621 to 648 of Raf-1 were replaced with codons encoding R-18 (PHCVPRDLSWLDLEANMCLP). To introduce R-18 into CR2, codons 257 to 268 of mRaf-1 were deleted and R-18 codons were fused between codons 256 and 269. The cDNAs for activated TC21 (V23TC21) and activated R-Ras (V38R-Ras) were cloned into pEFplink.6, which also uses the EF1α promoter but does not incorporate a tag onto the N terminus of the protein. The cDNAs for 14-3-3ζ, V12H-Ras, and V12,G37H-Ras were cloned into the expression vector pEFHA/Plink.6, which also uses the EF1α promoter but fuses an N-terminal hemagglutinin (HA) epitope tag (MDYPYDVPDYAGS) to the expressed proteins. This tag is recognized by the mouse monoclonal antibody 12CA5.
Cell culture and biochemical techniques.
COS cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. Transfections were performed using Lipofectamine (Gibco-BRL Life Technologies). Cell extractions and Raf-1 kinase assays were performed as described previously (33, 34). Western blotting for the myc tag and HA tag was performed as described previously using the monoclonal antibodies 9E10 and 12CA5, respectively (33, 34); Western blotting for ERK was performed using polyclonal antibody 122 (29) or monoclonal antibody to phospho-ERK (catalog no. M8159; Sigma). The S338 phospho-specific antibody has been described previously (38), and phospho-Raf (S259) antibody was obtained commercially (catalog no. 9421; New England Biolabs), as was the monoclonal Raf-1 antibody (catalog no. R19120; Transduction Labs). The analysis for complexes between Raf-1 and Ras and between Raf-1 and 14-3-3 was performed essentially as described previously (32) by probing the Western blots for the appropriate proteins as described in the text. Microinjection studies in MDCK cells were performed essentially as described previously (33). Metabolic labeling with [32P]orthophosphate and transfer of radiolabeled mRaf-1 to Immobilon P membranes (Millipore) were performed as described previously (33). The radiolabeled bands were excised and digested with trypsin (catalog no. v5111; Promega), and the peptides were eluted and resolved on crystalline cellulose thin-layer chromatography plates as described previously (36).
RESULTS
14-3-3 binding to CR3, rather than S621 phosphorylation, is required for Raf-1 activity.
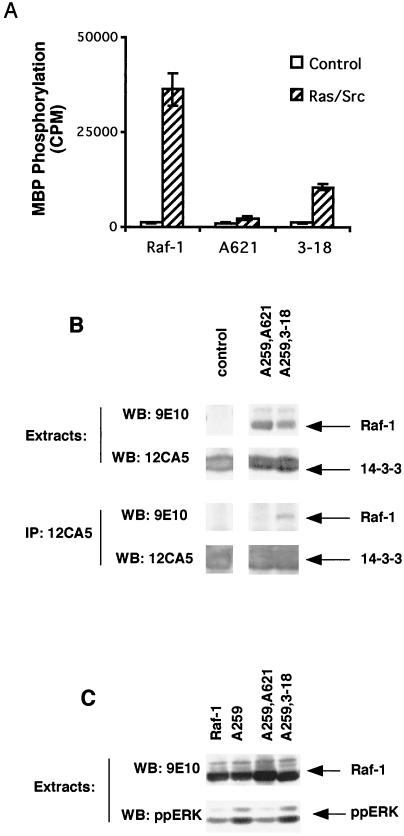
We first investigated whether S621 phosphorylation or 14-3-3 binding to CR3 was required for Raf-1 activity. For these studies, we transiently expressed mRaf-1 in COS cells together with an HA-epitope-tagged oncogenic version of H-Ras (HAV12Ras) and activated Src (F527Src). Raf-1 kinase activity was measured in an immunoprecipitation-kinase cascade assay using GST-MEK, GST-ERK, and myelin basic proteins as sequential substrates. In agreement with our previously determined data (33, 34), wild-type mRaf-1 had low basal kinase activity and was strongly activated by HAV12Ras and F527Src (Fig. 2A). However, when S621 was replaced with alanine (mA621Raf-1) to prevent phosphorylation, Raf-1 was inactive (Fig. 2A).
FIG. 2.
Binding of 14-3-3 to CR3 of Raf-1 is required for kinase activity. (A) Raf-1 kinase activity. mRaf-1 (Raf), mA621Raf-1 (A621), or m3-18Raf-1 (3-18) was expressed in COS cells together with V12Ras (Ras) and F527Src (Src) as indicated, and the cells were serum starved for 24 h prior to extraction. The mRaf-1 proteins were immunoprecipitated for Raf kinase activity determination. The results are from one experiment assayed in triplicate, with error bars representing standard deviations from the means. The background counts (∼1,000 cpm) were subtracted, and similar results were obtained in three independent assays. (B) 14-3-3 binding. mA259,A621Raf-1 (A259,A621) or mA259,3-18Raf-1 (A259,3-18) was expressed in COS cells, together with HA14-3-3. The results for levels of individual protein expression are shown in the upper four panels. HA14-3-3 was immunoprecipitated (IP) using 12CA5, and the samples were resolved on sodium dodecyl sulfate (SDS)-7% polyacrylamide gels and probed by Western blotting (WB) with 9E10 for mRaf-1 and 12CA5 for HA14-3-3 as indicated (lower four panels). Similar results were obtained in three independent assays. (C) Endogenous ERK activation. mRaf-1 (Raf-1), mA259Raf-1 (A259), mA259,A621Raf-1 (A259,A621), and mA259,3-18Raf-1 (A259,3-18) were expressed in COS cells, together with HA14-3-3. Cell extracts were resolved on SDS-7% polyacrylamide gels and probed by Western blotting (WB) for myc-tagged Raf-1 by using 9E10 and for activated endogenous ERK by using a phospho-specific antibody. Similar results were obtained in two independent assays.
In order to determine whether the activity was lost because S621 phosphorylation was necessary or was lost due to the consequent loss of 14-3-3 binding, we replaced the RSXpSXP 14-3-3 binding motif in CR3 with a phosphorylation-independent binding motif. The peptide R-18, which has been identified in phage display libraries and contains the sequence WLDLE, has been shown to bind to 14-3-3 in a phosphorylation-independent manner (47, 58). We therefore deleted amino acids 621 to 648 of Raf-1 and fused the R-18 peptide to amino acid 620, creating m3-18Raf-1. First, we tested whether the fusion of R-18 to mRaf-1 recovered binding of 14-3-3 to CR3. However, the results of Raf-1-14-3-3 binding studies can be indeterminate because of the two 14-3-3 binding sites in Raf-1. Therefore, these studies were performed in the background of a serine-for-alanine substitution at position 259, which abolishes binding of 14-3-3 to the CR2 motif (45, 61). mRaf-1 constructs were coexpressed in COS cells with HA-epitope-tagged 14-3-3 (HA14-3-3), and the HA14-3-3 was immunoprecipitated with the 12CA5 monoclonal antibody. The immune complexes were examined for bound Raf-1 by Western blotting for the myc epitope. As expected, mRaf-1 in which both S259 and S621 were replaced with alanine (mA259,A621Raf-1) did not bind to 14-3-3 (Fig. 2B). By contrast, 14-3-3 did bind to mA259,3-18Raf-1 (Fig. 2B), indicating that the R-18 peptide did direct 14-3-3 binding to CR3. The R-18 peptide also partially restored (∼30%) mRaf-1 kinase activity (Fig. 2A) and also restored Raf-1 activity in vivo, as shown by its effect on endogenous ERK. As described previously, Raf-1 in which S259 is replaced with alanine (mA259Raf-1) has elevated basal kinase activity (1, 63) (also see below). We used the extracts from the 14-3-3 binding experiments to examine the activation of endogenous ERK in COS cells, using an antibody that binds only to the dually phosphorylated, active form of ERK1/2. As shown (Fig. 2C), mA259Raf-1 overexpression led to elevated ERK phosphorylation, a result which was not seen with wild-type mRaf-1. By contrast, mA259,A621Raf-1 failed to stimulate ERK phosphorylation under these conditions, whereas mA259,3-18Raf-1 did (Fig. 2C). These data demonstrate that Raf-1 can be activated in the absence of S621 phosphorylation, provided that 14-3-3 binds to CR3.
S259 is phosphorylated in L257Raf-1, but 14-3-3 binding is lost.
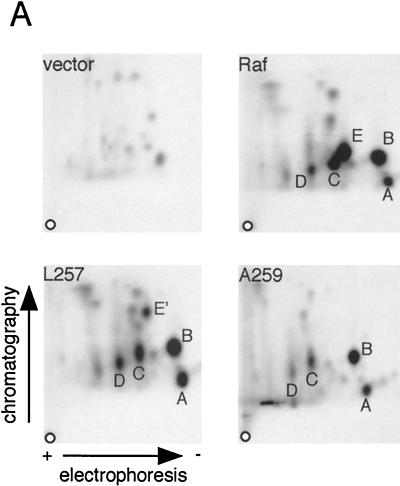
Using similar approaches, we also examined the role of 14-3-3 binding to CR2. For these studies, we compared wild-type mRaf-1 to two versions of mRaf-1 in which the CR2 motif is mutated: mA259Raf-1 (as described above) and an mRaf-1 with a leucine-for-serine substitution at position 257 (mL257Raf-1). In L257Raf-1, the first serine in the RSXpSXP motif is mutated, which restores binding of Raf-1 to V12,G37Ras (59). We first examined how these mutations affected phosphorylation of Raf-1 on S259. COS cells expressing mRaf-1 proteins were metabolically labeled with [32P]orthophosphate, and the radiolabeled mRaf-1 was subjected to two-dimensional tryptic phosphopeptide mapping. Wild-type mRaf-1 produced five major radiolabeled peptides (peptides A to E) (Fig. 3A). In agreement with previously determined data (44), peptide B was absent from maps of mA621Raf-1 and peptide A was absent from maps in which S43 was substituted for alanine (unpublished data); the origin of peptides C and D is unknown. Peptide E was absent from the maps generated from mL257Raf-1 and also from the maps generated from mA259Raf-1 (Fig. 3A). However, the mL257Raf-1 map contained an additional peptide, E', which was absent from all of the other maps (Fig. 3A). E' and E appear to have similar charges, because they migrate the same distance in the electrophoretic dimension, but E' appears to be the more hydrophobic of the two, as it was more mobile in the ascending chromatography (Fig. 3A). These data suggest that E' may be derived from phosphorylation of S259 in mL257Raf-1. To test this directly, we used a phospho-specific antibody that only binds to Raf-1 when S259 is phosphorylated. In Western blot analysis, the pS259 antibody bound to wild-type mRaf-1 and mA621Raf-1 but not to mA259Raf-1 (Fig. 3B). It also bound to mL257Raf-1 (albeit at lower levels) but not to mL257,A259Raf-1 (Fig. 3B). These data show that mL257Raf-1 is phosphorylated on S259, although the levels of phosphorylation may be reduced (see Discussion).
FIG. 3.
Binding of 14-3-3 to CR2. (A) Phosphopeptide analysis. COS cells expressing mRaf-1 (Raf), mL257Raf-1 (L257), mA259Raf-1 (A259), or vector control (vector) were metabolically labeled with [32P]orthophosphate, and the radiolabeled mRaf-1 proteins were analyzed by two-dimensional tryptic phosphopeptide mapping. The positions of the five major phosphopeptides are indicated by letters A to E. The directions of the electrophoretic and chromatographic dimensions are indicated. o, origin. (B) Phosphorylation of S259. mRaf-1 (Raf-1), mL257Raf-1 (L257), mA259Raf-1 (A259), mL257,A259Raf-1 (L257,A259), or mA621Raf-1 (A621) was expressed in COS cells. The results for levels of individual proteins are shown in the upper panel. mRaf-1 was immunoprecipitated (IP) using 9E10, and the samples were resolved on SDS-7% polyacrylamide gels and probed by Western blotting (WB) for phosphorylation of Raf-1 S259 by using a phospho-specific antibody (lower panel). Similar results were obtained in two independent assays. (C) 14-3-3 binding. Shown are COS cells expressing mA621Raf-1 (A621), mL257,A621Raf-1 (L257,A621), or mA259,A621Raf-1 (A259,A621) with (+) or without (−) HA14-3-3 (14-3-3) as indicated and processed for 14-3-3 binding as described in the Fig. 2C legend. Results for levels of protein expression are shown in the upper two panels and those for the levels of mRaf-1 associated with HA14-3-3 in the lower panel.
We next tested whether these mRaf-1 proteins bound to 14-3-3 in vivo. Once again, in order to simplify the analysis, these studies were performed in the mA621Raf-1 background to prevent confusion arising due to 14-3-3 binding to CR3. MA621Raf-1 efficiently coimmunoprecipitated with HA14-3-3, whereas mL257,A621Raf-1 and mA259,A621Raf-1 did not (Fig. 3C). Thus, when S259 phosphorylation was prevented as in A259Raf-1, 14-3-3 binding to CR2 was disrupted. Despite the fact that S259 was phosphorylated in mL257Raf-1, albeit to lower levels, we did not detect any 14-3-3 binding to this mutant (Fig. 3C).
14-3-3 binding to CR2 suppresses the activation of Raf-1 by V12,G37Ras.
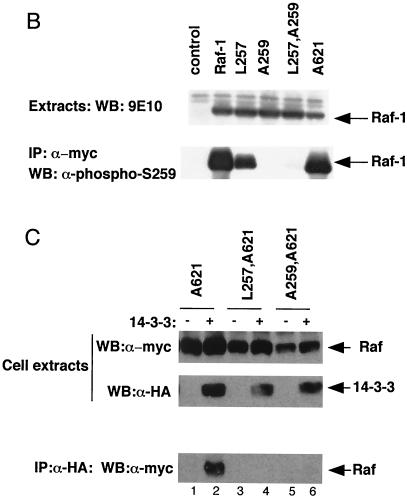
We next tested whether these CR2 mutations affected Ras-mediated Raf-1 activation. Since V12Ras activates Raf-1 more weakly in the absence of F527Src (33), the protein expression levels and kinase assays were adjusted to allow accurate measurement of lower levels of Raf kinase activity. The basal kinase activity of mRaf-1 was still low in serum-starved cells, but it was strongly activated by HAV12Ras (Fig. 4A, lanes 1 and 2). The level of basal kinase activity of both mL257Raf-1 and mA259Raf-1 was elevated compared to that of wild-type mRaf-1 but was still higher in the presence of HAV12Ras (Fig. 4A, lanes 4, 5, 7, and 8). Next, we tested whether a HAV12Ras in which the glutamate at position 37 was replaced by glycine (HAV12,G37Ras) could activate mRaf-1. HAV12,G37Ras did not activate wild-type mRaf-1 but activated both mL257Raf-1 and mA259Raf-1, although to lower levels than those achieved by HAV12Ras (Fig. 4A, lanes 3, 6, and 9). The ability of HAV12,G37Ras to activate mL257Raf-1 and mA259Raf-1 in vivo was confirmed by their effects on endogenous ERK. HAV12,G37Ras with wild-type mRaf-1 only stimulated very low levels of endogenous ERK phosphorylation, but when HAV12,G37Ras was coexpressed with mL257Raf-1 or mA259Raf-1, ERK was strongly phosphorylated (Fig. 4B). (Note that the stimulation of ERK phosphorylation by mA259Raf-1 alone in this experiment appears lower than that seen in Fig. 2C. This is because the sensitivity of the assay was adjusted to allow us to analyze ERK phosphorylation stimulated by V12,G37Ras in the presence of the Raf-1 mutants.)
FIG. 4.
Activation of Raf-1 by V12,G37Ras. mRaf-1 (Raf-1), mL257Raf-1 (L257), mA259Raf-1 (A259), m2-18Raf-1 (2-18), mA621Raf-1 (A621), mL257,A621Raf-1 (L257,A621), mA259,A621Raf-1 (A259,A621), or m2-18,A621Raf-1 (2-18,A621) was expressed in COS cells alone (control), with HAV12Ras (Ras), with HAV12,G37Ras (G37), or with HA14-3-3 (+) as indicated. (A) Raf-1 kinase activity. Raf-1 kinase assays were performed, and the results are from one experiment assayed in triplicate, with error bars to represent deviations from the means; background counts (∼1,445 cpm) were subtracted, and similar results were obtained in three independent experiments. (B) ERK activation. Cell extracts were resolved in SDS-7% polyacrylamide gels and subjected to Western blotting (WB) for activation of endogenous ERKs by using a phospho-specific antibody. Similar results were obtained in three independent assays. (C) 14-3-3 binding. Binding of Raf-1 to HA14-3-3 was determined as described in the Fig. 2C legend. The results for levels of mRaf-1 proteins are shown in the upper panel, and those for levels of the mRaf-1 proteins associated with HA14-3-3 are shown in the middle panel. The results for efficiency of the 12CA5 immunoprecipitate (IP) are shown in the lower panel.
These data show that activation of Raf-1 by V12,G37Ras correlates with loss of 14-3-3 binding rather than loss of S259 phosphorylation. To test this model further, we replaced the RSXpSXP motif in CR2 with the WLDLE motif containing the R-18 peptide (creating m2-18Raf-1) so that 14-3-3 binding to CR2 would be independent of S259 phosphorylation. First, we tested whether 14-3-3 would bind to the CR2 region of m2-18Raf-1, performing these experiments in the context of mA621Raf-1 to simplify the analysis. Whereas mL257,A621Raf-1 and mA259,A621Raf-1 did not bind to HA14-3-3, m2-18,A621 Raf-1 bound with similar affinity to that of mA621Raf-1 (Fig. 4C). Thus, the replacement of the RSXpSXP motif in CR2 by the WLDLE motif containing the R-18 peptide recovered 14-3-3 binding to CR2. What is more, the basal kinase activity of m2-18Raf-1 was similar to that of wild-type mRaf-1, and the ability of HAV12,G37Ras to activate m2-18Raf-1 was strongly suppressed compared to its ability to activate mL257Raf-1 and mA259Raf-1 (Fig. 4A, lanes 1 to 3 and 10 to 12). Finally, m2-18Raf-1 was less efficient than either mL257Raf-1 or mA259Raf-1 at cooperating with HAV12,G37Ras to stimulate activation of endogenous ERK (Fig. 4B).
Activation of L257Raf-1 requires N-region phosphorylation and membrane recruitment.
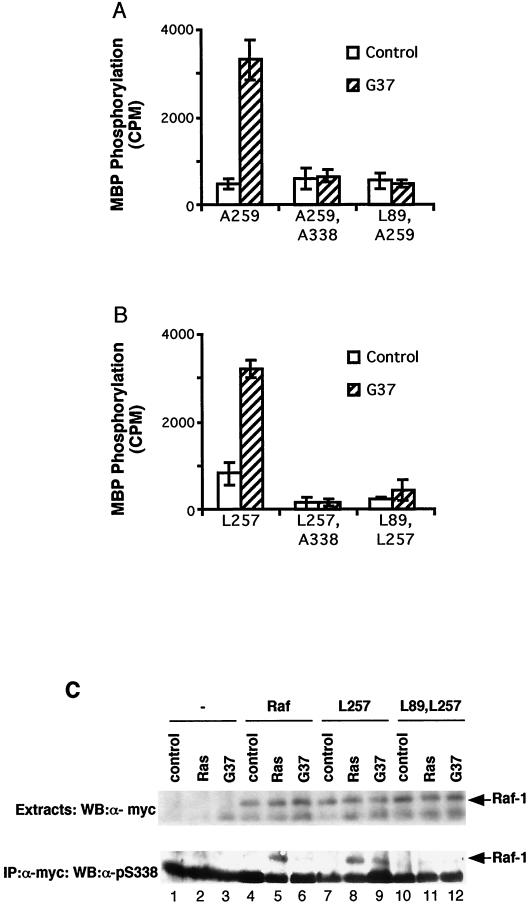
The above data show that V12,G37Ras was able to activate Raf-1 when 14-3-3 binding to CR2 was disrupted. We wished to establish how V12,G37Ras activated these Raf-1 mutants. First, we tested whether S338 phosphorylation was required. HAV12,G37Ras did not activate an mA259Raf-1 or mL257Raf-1 in which S338 was replaced by alanine (mA259,A338Raf-1 and mL257,A338Raf-1, respectively) (Fig. 5A and B). Furthermore, using an S338 phospho-specific antibody (38), we did not detect any S338 phosphorylation on mL257Raf-1 in serum-starved cells, and whereas HAV12,G37Ras did not stimulate S338 phosphorylation on wild-type mRaf-1, it did stimulate S338 phosphorylation on mL257Raf-1 (Fig. 5C, lanes 6, 7, and 9).
FIG. 5.
Raf-1 activity requires S338 phosphorylation. mRaf-1 (Raf), mL257Raf-1 (L257), mA259Raf-1 (A259), mL89,L257Raf-1 (L89,L257), mL89,A259Raf-1 (L89,A259), mA259,A338Raf-1 (A259,A338) or mL257,A338Raf-1 (L257,A338) was expressed in COS cells alone (control), with HAV12Ras (Ras), or with HAV12,G37Ras (G37) as indicated. (A and B) Raf-1 kinase activity. Raf-1 kinase assays were performed, and the data are shown for one experiment assayed in triplicate with background counts (1,100 cpm) subtracted and error bars to represent standard deviations from the means. Similar results were obtained in three independent assays. (C) S338 phosphorylation. The mRaf-1 proteins were immunoprecipitated (IP), and the levels of S338 phosphorylation were determined using a phospho-specific antibody. The upper panel shows results for the levels of mRaf-1 protein expression, and the lower panel those for the corresponding levels of S338 phosphorylation. WB, Western blotting.
The above data show that V12,G37Ras-mediated activation of L257Raf-1 and A259Raf-1 requires Ras-mediated recruitment of the Raf-1 proteins to the plasma membrane and S338 phosphorylation, as previously described for wild-type Raf-1 (38). We therefore tested whether L257Raf-1 and A259Raf-1 bound directly to HAV12,G37Ras. HAV12,G37Ras and mRaf-1 were expressed in COS cells, and the Ras was immunoprecipitated with monoclonal antibody Y13-238. The complexes were examined for coprecipitated mRaf-1 by Western blotting for the myc epitope antibody tag. mRaf-1, mL257Raf-1, mA259Raf-1, and m2-18Raf-1 all bound to HAV12Ras strongly (Fig. 6A). However, we did not detect binding of any of these mRaf-1 proteins to HAV12,G37Ras (Fig. 6A), suggesting that there was no direct binding to V12,G37Ras. To test the dependence of direct binding to Ras, we examined the activation of these mutations in a Raf-1 double mutant in which arginine 89 (R89) was also replaced by leucine (mL89,L257Raf-1 and mL89,A259Raf-1, respectively). R89 is in the Raf-1 RBD, and the replacement of arginine by leucine blocks binding to RasGTP, preventing plasma membrane recruitment, S338 phosphorylation, and Raf-1 activation (18, 33, 38). HAV12,G37Ras did not activate mL89,A259Raf-1 or mL89,L257Raf-1 (Fig. 5A and B) and did not stimulate S338 phosphorylation on mL89,L257Raf-1 (Fig. 5C, lanes 10 to 12).
FIG. 6.
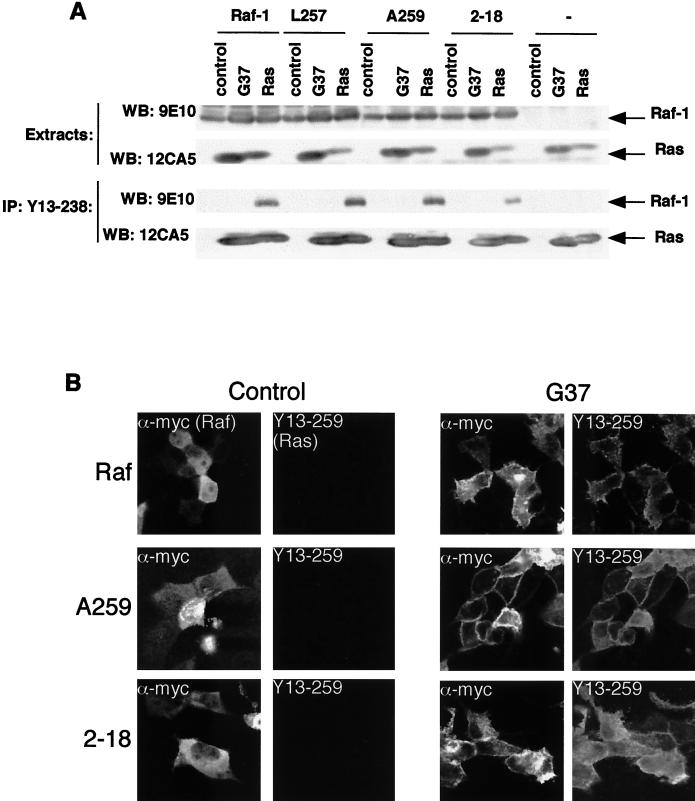
Membrane recruitment of Raf-1. (A) Ras-Raf binding. COS cells were transfected with expression constructs for mRaf-1 (Raf), mL257Raf-1 (L257), mA259Raf-1 (A259), or m2-18Raf-1 (2-18) alone (control), with HAV12Ras (Ras), or with HAV12,G37Ras (G37) as indicated, and the results for levels of protein expression are shown in the upper two panels, as revealed by Western blotting (WB) with 9E10 for mRaf-1 proteins or 12CA5 for Ras. The Ras proteins were immunoprecipitated (IP) with the Y13-238 antibody and the complexes were resolved on SDS-7% polyacrylamide gels, as shown in the lower two panels. Western blotting (WB) with 12CA5 as shown in the bottom panel revealed the efficiency of the Ras precipitation, and the bound mRaf-1 proteins were revealed by Western blotting with 9E10, shown in the lower middle panel. Similar results were obtained in at least three independent assays. (B) Membrane recruitment. MDCK cells were microinjected with expression constructs for mRaf-1 (Raf; top row), mA259Raf-1 (A259; middle row) or m2-18Raf-1 (2-18; bottom row) alone (control; left two columns) or with HAV12,G37Ras (G37; right two columns) as indicated. For each sample, the same field of cells, stained for mRaf-1 protein expression (α-myc; leftmost and middle right columns) or for Ras expression (Y13-259; rightmost and middle left columns), is shown. Similar results were obtained in three independent assays.
Finally, we tested whether HAV12,G37Ras could recruit mL257Raf-1 and mA259Raf-1 to the plasma membrane. MDCK cells were microinjected with Ras and Raf-1 expression constructs and the cells were immunostained for Raf-1 with 9E10 or for Ras with Y13-259. In resting cells, both mRaf-1 and mA259Raf-1 were cytosolic, and whereas wild-type mRaf-1 was recruited to the plasma membrane very poorly by HAV12,G37Ras, mA259Raf-1 recruitment was very efficient (Fig. 6B). ML257Raf-1 behaved in a manner similar to that of mA259Raf-1; it was cytosolic in resting cells and was efficiently recruited to the plasma membrane by HAV12,G37Ras (data not shown), whereas m2-18Raf-1 behaved like wild-type mRaf-1, since its recruitment was very inefficient (Fig. 6B). We did observe that when the cells were incubated for an extended time or the expression levels were high, wild-type Raf-1 was recruited to the plasma membrane by V12,G37Ras (data not shown), but we cannot confirm that this was due to a direct effect. Taken together, these data show that although disruption of 14-3-3 binding to CR2 does not allow efficient binding of Raf-1 to V12,G37Ras, V12,G37Ras was able to recruit Raf-1 to the plasma membrane for S338 phosphorylation and activation when 14-3-3 binding to CR2 was disrupted.
Ras-GTP displaces 14-3-3 from Raf-1, but S259 is still phosphorylated.
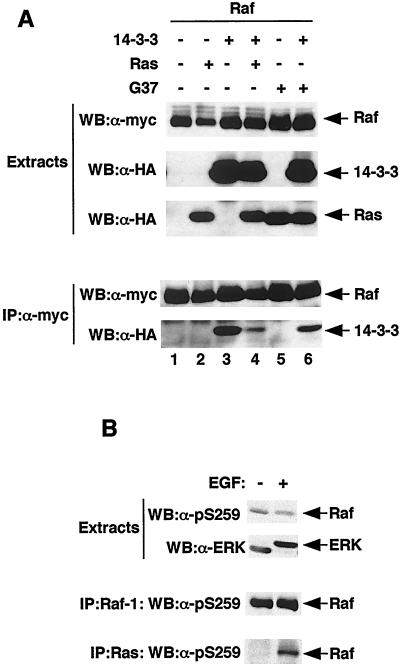
These data suggest that 14-3-3 binding to CR2 antagonizes Ras-mediated Raf-1 recruitment to the plasma membrane and that wild-type Ras-GTP must therefore displace 14-3-3 from CR2 in order to recruit Raf-1 to the membrane. To test this model, mRaf-1 was immunoprecipitated from COS cells by using the rat anti-myc antibody JAC/6 and the binding of HA14-3-3 was examined by Western blotting with 12CA5. In resting cells, HA14-3-3 was efficiently immunoprecipitated by JAC/6 in the presence of mRaf-1, indicating strong binding between these proteins (Fig. 7A, lanes 1 and 3). In the presence of HAV12Ras, however, 14-3-3 binding was significantly reduced, whereas in the presence of HAV12,G37Ras, it was not (Fig. 7A, lanes 3, 4, and 6). In similar experiments using mA259Raf-1 or mL89Raf-1, no reduction in binding to HA14-3-3 was observed on coexpression of HAV12Ras (data not shown). Thus, as Ras-GTP did displace 14-3-3 from CR2 in Raf-1, we tested whether the binding of Raf-1 to Ras-GTP was accompanied by S259 dephosphorylation. These experiments were performed on endogenous proteins in COS cells treated with epidermal growth factor (EGF), which activates Ras and stimulates the formation of Ras-Raf-1 complexes (32). These complexes were immunoprecipitated with the Ras monoclonal antibody Y13-238, and the Raf-1 in the complexes was subjected to Western blotting with the pS259 phospho-specific antibody. S259 was phosphorylated in resting COS cells, and EGF treatment did not significantly alter these levels of phosphorylation (Fig. 7B). Furthermore, the Raf-1 that was bound to Ras was recognized by the S259 phospho-specific antibody, showing that the binding of Raf-1 to Ras-GTP does not require dephosphorylation of S259 (Fig. 7B).
FIG. 7.
Displacement of 14-3-3 from Raf-1 by Ras does not require S259 dephosphorylation. (A) 14-3-3 binding. COS cells were transfected with expression constructs for mRaf-1 (Raf), HA14-3-3 (14-3-3), HAV12Ras (Ras), or HAV12,G37Ras (G37) as indicated. The levels of protein expression were revealed by Western blotting (WB) for mRaf-1 with 9E10 (top panel of top three) and for HA14-3-3 and HA-tagged Ras protein by Western blotting with 12CA5 (middle and bottom panels of top three, respectively). The mRaf-1 was immunoprecipitated (IP) using the rat monoclonal antibody JAC/6, and the complexes were resolved on SDS-7% polyacrylamide gels and probed for mRaf-1 by using 9E10 (upper panel of bottom two) or for HA14-3-3 by using 12CA5 (lower panel of bottom two). Similar results were obtained in two independent experiments. (B) S259 phosphorylation. COS cells were treated with EGF (10 ng, 20 min), and the levels of pS259 on Raf-1 were examined by Western blotting (WB) with a phospho-specific antibody, either in extracts (upper panel of top two) or following immunoprecipitation (IP) of Raf-1 by using a Raf-1 monoclonal antibody (upper panel of bottom two). As a loading control and to ensure that EGF had activated this pathway, the mobility of endogenous ERK2 in SDS gels was examined using antibody 122 (lower panel of top two). Finally, endogenous Ras was immunoprecipitated, and the levels of S259 phosphorylation on the coprecipitated Raf-1 were determined using the phospho-specific antibody (lower panel of bottom two). Similar results were obtained in three independent experiments.
TC21 and R-Ras activate A259Raf-1 but not wild-type Raf-1.
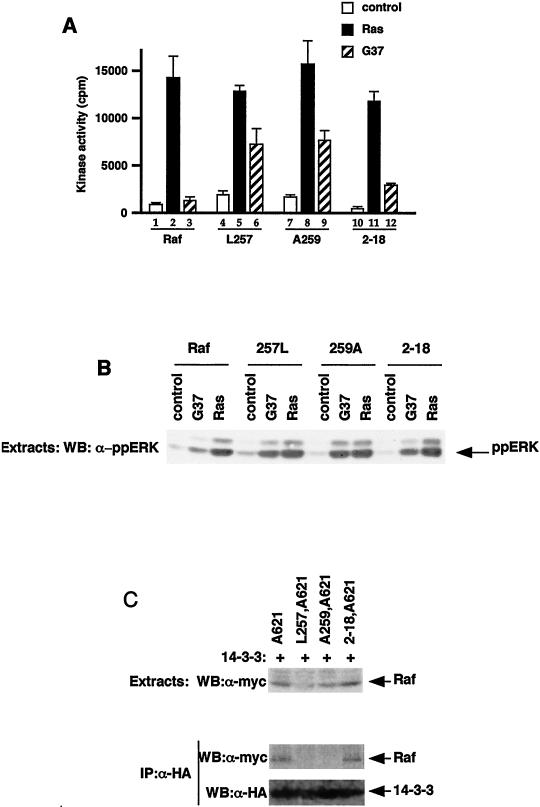
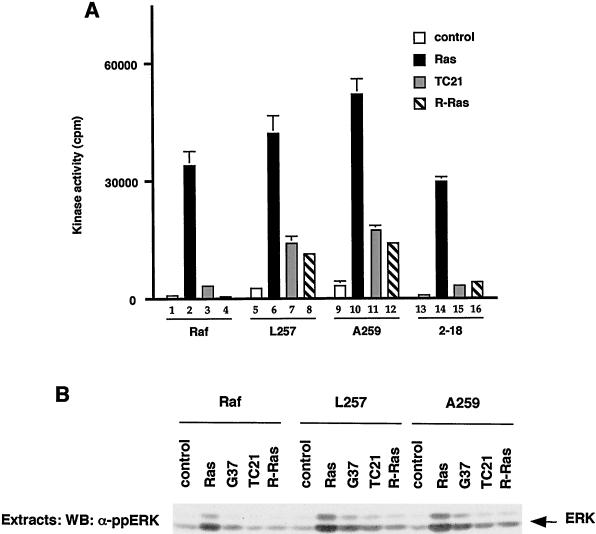
Finally, we wished to examine the biological consequences of 14-3-3 binding and Raf-1 activation by small G proteins. A number of Ras family members have a very similar effector domain to Ras and bind to Raf-1 in vitro but do not activate Raf-1 (see reference 5). We therefore tested whether these proteins could activate the mutant Raf-1 proteins characterized above. Activated versions of Rap1a, Rap2a, and Rap2b failed to activate any of the mRaf-1 proteins (data not shown). By contrast, whereas activated versions of TC21 (V23TC21) and R-Ras (V38R-Ras) did not activate wild-type mRaf-1, they did activate both mL257Raf-1 and mA259Raf-1, albeit less efficiently than HAV12Ras (Fig. 8A). We also tested the in vivo activity of these proteins by examining their effects on endogenous ERK. When expressed with wild-type mRaf-1, V23TC21 and V38R-Ras failed to stimulate endogenous ERK phosphorylation, but they did stimulate ERK phosphorylation when expressed with mL257Raf-1 and mA259Raf-1 (Fig. 8B), indicating that the latter proteins were also activated in vivo. Finally, V23TC21 and V38R-Ras failed to activate m2-18Raf-1 (Fig. 8A). Thus, V23TC21 and V38R-Ras could activate Raf-1 only when 14-3-3 binding to CR2 was disrupted.
FIG. 8.
Activation of Raf-1 by TC21 and R-Ras. mRaf-1 (Raf-1), mL257Raf-1 (L257), mA259Raf-1 (A259), or m2-18Raf-1 (2-18) was expressed in COS cells alone (control) or with HAV12Ras (Ras), HAV12,G37Ras (G37), V23TC21 (TC21), or V38R-Ras (R-Ras) as indicated. (A) Raf-1 kinase activity. Raf-1 kinase assays were performed, and the results are from one experiment assayed in triplicate, with error bars to represent deviations from the means; background counts (3,164 cpm) were subtracted, and similar results were obtained in two independent experiments. (B) ERK activation. COS cell extracts were resolved on SDS-7% polyacrylamide gels and probed by Western blotting (WB) for activated ERK by using a phospho-specific antibody. Similar results were obtained in two independent experiments.
DISCUSSION
Previously it had not been possible to analyze the role played by 14-3-3 binding to CR3 in Raf-1 regulation, because mutations to S621 are inactivating. However, we show that Raf-1 activation can occur in the absence of S621 phosphorylation, provided that 14-3-3 binding to CR3 is maintained. The R-18 peptide provided phosphorylation-independent 14-3-3 binding to CR3 and supported Raf-1 activity in vitro and in vivo, even though S621 was completely absent. We also observed that V12Ras did not completely displace 14-3-3 from full-length Raf-1 (Fig. 7A), which is a result consistent with the data of others who have suggested that the residual binding is likely to occur at CR3 (28, 39, 51). While our data do not prove that 14-3-3 is bound to CR3 in active Raf-1, they do show that S621 is not absolutely required for activity and so argue that this phosphorylation does not perform an alternative function in the active conformation, as has been suggested (42). Rather, we support the model proposed for peptide displacement and protection from phosphatase-mediated inactivation studies (13, 56, 57, 62), in which 14-3-3 remains bound to CR3 in active Raf-1, where it performs an essential role.
We note that the recovery of activity in 3-18Raf-1 was only ∼30% of that of wild-type Raf-1 (Fig. 2A), suggesting that the R-18 peptide does not replace all functions of the RSXpSXP motif. This may be because 14-3-3 binds to these motif peptides in different ways or because the spacing between 14-3-3 and the kinase domain is slightly compromised in the R-18 fusion. We are presently examining these issues, but nevertheless, our data clearly show that Raf-1 activity can be maintained in the absence of S621 phosphorylation, and this is the first report that demonstrates that it is possible to replace phosphorylation-dependent 14-3-3 binding sites with phosphorylation-independent sites and maintain activity in vitro and in vivo. This may therefore be a useful technique for the study of the role played by 14-3-3 binding in other client proteins, since binding can be made independent of the kinase and phosphatase activity. Studies are ongoing to further examine the role played by 14-3-3 binding to CR3 in Raf kinase activity.
We also examined how 14-3-3 binding to CR2 affects Raf-1 activation mediated by V12,G37Ras. For those studies, we used two Raf-1 proteins that have mutations in CR2, L257Raf-1, and A259Raf-1. The interaction between 14-3-3 and L257Raf-1 has not been examined before, although it has been suggested that V12,G37Ras activates L257Raf-1 because these proteins interact at the plasma membrane, resulting in a conformation change in the Raf-1 (41). For those studies, however, Raf-1 was anchored to the plasma membrane by fusion of the lipid anchor from Ras (RafCAAX) (30, 54), which leads to constitutive phosphorylation on S338 (an event that occurs at the plasma membrane) (38), so it was not possible to assess the role played by Ras-mediated membrane recruitment or S338 phosphorylation in the activation process. Furthermore, for those studies, reporter gene assays and ERK activation were used as surrogate measures of Raf-1 activity and direct measurements were not made (25, 59).
We showed here that V12,G37Ras activates L257Raf-1 and A259Raf-1 in vitro and in vivo (Fig. 4A) and stimulates S338 phosphorylation on L257Raf-1. However, V12,G37Ras did not activate L89,L257Raf-1 or L89,A259Raf-1 and did not stimulate S338 phosphorylation on L89,L257Raf-1 (Fig. 5). Thus, when the RBD of these mutants was disrupted, S338 phosphorylation and activation were blocked, suggesting that they must still interact with V12,G37Ras and be recruited to the plasma membrane for S338 phosphorylation and activation. Our unpublished data also suggest that L257Raf-1 and A259Raf-1 associate with the membrane more readily than wild-type Raf-1. We performed membrane-cytosol fractionation studies and found that even in resting cells, small proportions of L257Raf-1 and A259Raf-1 were in the membrane fraction, whereas those of the R89L versions were not (data not shown). Unfortunately, in our hands the results of these experiments were somewhat variable, for reasons that are unclear, but they are consistent with results published while this paper was being reviewed. Dhillon et al. demonstrated that in their hands, a small amount of A259Raf-1 was in the membrane fraction of resting cells and that V12Ras recruited A259Raf-1 to the membrane more efficiently than it recruited wild-type Raf-1 (14). They also demonstrated that L89,A259Raf-1 is not recruited to the plasma membrane and is not activated.
Together, these studies suggest that L257Raf-1 and A259Raf-1 partition to the plasma membrane more readily than wild-type Raf-1 but that this association still requires Ras binding, which is presumably provided by low levels of Ras signaling present in resting cells. Both wild-type Raf-1 and L257Raf-1 bind to G37Ras-GTP in vitro with similar but very low affinities (25), whereas in yeast L257Raf-1 binds to V12,G37Ras with higher affinity than wild-type Raf-1 (59). We also found a distinction between the in vivo and in vitro results. Wild-type Raf-1, L257Raf-1, and A259Raf-1 failed to bind to V12,G37Ras in extracts (Fig. 6A), but A259Raf-1 was still efficiently recruited to the plasma membrane by V12,G37Ras whereas wild-type Raf-1 was not (Fig. 6B). Thus, the differences in the binding of L257Raf-1 and wild-type Raf-1 to V12,G37Ras appear to manifest themselves only in vivo.
We interpret these data with the following model. We propose that Raf-1 recruitment to the plasma membrane requires at least two events. One is the direct binding of Raf-1 to Ras, and the other is neutralization of a negative function mediated by CR2. Wild-type Ras-GTP both binds to Raf-1 and neutralizes the CR2 function. G37Ras-GTP, by contrast, can bind to Raf-1 but cannot neutralize CR2 and so does not stabilize Raf-1 interaction with the membrane. However, the CR2 function is already neutralized in the L257Raf-1 and A259Raf-1 mutants, so that even though the binding to G37Ras-GTP is weak, a stable association with the membrane can form, allowing S338 phosphorylation and activation. This model predicts that these mutants are very sensitive even to weak Ras signals and so suggests that unlike wild-type Raf-1, the mutants are able to respond to the weak Ras signals in resting cells, which explains why A259Raf-1 weakly associates with the plasma membrane in these cells (14) and why L257Raf-1 and A259Raf-1 have elevated basal activity (Fig. 4). It also explains why the R89L and S338A versions do not have so-called elevated basal activity (Fig. 5A and B), because in our model, the basal activity requires Ras interaction and S338 phosphorylation.
We propose that the function at CR2 that Ras must overcome is 14-3-3 binding, and we propose that Ras-GTP must displace 14-3-3 from CR2 for Raf-1 to form a stable interaction with the plasma membrane, allowing S338 phosphorylation and Raf-1 activation. When the RSXpSXP motif at CR2 was replaced by R-18, the resulting protein was more similar to wild-type Raf-1 than to the CR2 mutants. First, 14-3-3 binding to CR2 was restored (Fig. 4C). Second, the basal activity of Raf-1 was suppressed (Fig. 4A). Third, V12,G37Ras activation of Raf-1 was strongly suppressed (Fig. 4A). Fourth, the ability of Raf-1 to couple to V12,G37Ras in vivo and activate endogenous ERK was suppressed (Fig. 4B). Fifth, the ability of V12,G37Ras to recruit Raf-1 to the plasma membrane was strongly suppressed (Fig. 6B), and sixth, we show that V12,G37Ras could not displace 14-3-3 from full-length Raf-1, whereas V12Ras could.
We note that the R-18 peptide replacement was not perfect and that 2-18Raf-1 had properties that are intermediate between those of wild-type Raf-1 and L257Raf-1/A259Raf-1; 2-18Raf-1 was activated by V12,G37Ras more strongly than wild-type Raf-1, and it coupled to V12,G37Ras to activate endogenous ERK more strongly than Raf-1. However, the data demonstrate that the binding of 14-3-3 to CR2 suppresses the ability of V12,G37Ras to be able to achieve efficient recruitment of Raf-1 to the plasma membrane, such that the loss of 14-3-3 binding at this region plays a crucial role in the subsequent activation of Raf-1 and ERK. It has been suggested that Ras and 14-3-3 compete with each other for binding to the CRD of Raf-1, which flanks the RBD and separates it from CR2 (for a review, see reference 3). It is possible that this region affects Ras-mediated 14-3-3 displacement, since both proteins have secondary binding sites in the CRD. A theme growing in importance in 14-3-3 biology is that 14-3-3 proteins regulate the subcellular distribution of client proteins (see reference 46), and our model suggests that 14-3-3 prevents Raf-1 association with the plasma membrane in the presence of weak Ras signals.
Our data also suggest that it is not the phosphorylation of S259 that mediates the negative effect at CR2. We demonstrate that S259 was still phosphorylated in L257Raf-1, albeit to lower levels (although this may reflect a reduction in antibody binding due to S257 forming part of the antibody epitope), but that the properties of L257Raf-1 and A259Raf-1 were indistinguishable. Also, S259 was absent in 2-18Raf-1, so it could not be responsible for mediating the effects on membrane association and activation. Finally, we show that the Raf-1 bound to RasGTP is still phosphorylated on S259. While we cannot determine the stoichiometry of this phosphorylation, it clearly shows that S259 dephosphorylation is not necessary for Raf-1 to form a stable interaction with Ras. It has recently been suggested that S259 dephosphorylation is necessary to allow Raf-1 to bind to Ras-GTP (1, 26). However, this suggestion is counterintuitive, because the phosphate on the serine of peptides bound to 14-3-3 is deeply buried and would be inaccessible to phosphatases (61). Therefore, 14-3-3 displacement must precede dephosphorylation, and we propose that Ras-GTP performs this displacement, a model which is in agreement with the demonstration that Ras-GTP can displace 14-3-3 from the isolated N terminus of Raf-1 (51). Subsequent dephosphorylation of S259 may play a role in sustaining Raf-1 activity under some circumstances.
Finally, our model proposes that one of the important biological consequences of 14-3-3 binding to CR2 is that it prevents Raf-1 from being activated by all Ras family members. We show that activated versions of TC21 and R-Ras, despite having the same effector binding domain as Ras, do not activate wild-type Raf-1, but do activate L257Raf-1 and A259Raf-1 (Fig. 8). Again, we attribute this difference in activation levels to 14-3-3 binding, since these proteins were largely unable to activate 2-18Raf-1. It will be interesting to determine which aspects of TC21 and R-Ras prevent them from being able to activate wild-type Raf-1.
In conclusion, we propose that 14-3-3 binding to CR2 of Raf-1 prevents it from being activated by weak Ras signals in resting cells and from being inappropriately activated by all members of the Ras superfamily, despite the fact that these proteins share very similar effector binding domains. We propose that Ras must displace 14-3-3 from CR2 in order to be able to stabilize Raf-1 association with the plasma membrane and allow phosphorylation and activation. Finally, we suggest that S259 dephosphorylation is not required for the displacement of 14-3-3, although it may have a role in regulating activation under other circumstances.
Acknowledgments
We thank C. J. Marshall, D. Barford, and members of the Signal Transduction Team for their helpful comments on the work in progress and for comments on the manuscript.
This work was funded by Cancer Research UK and The Institute of Cancer Research.
REFERENCES
- 1.Abraham, D., K. Podar, M. Pacher, M. Kubicek, N. Welzel, B. A. Hemmings, S. M. Dilworth, H. Mischak, W. Kolch, and M. Baccarini. 2000. Raf-1-associated protein phosphatase 2A as a positive regulator of kinase activation. J. Biol. Chem. 275:22300-22304. [DOI] [PubMed] [Google Scholar]
- 2.Aitken, A. 1995. 14-3-3 proteins on the MAP. Trends Biochem. Sci. 20:95-97. [DOI] [PubMed] [Google Scholar]
- 3.Avruch, J., A. Khokhlatchev, J. M. Kyriakis, Z. Luo, G. Tzivion, D. Vavvas, and X. F. Zhang. 2001. Ras activation of the Raf kinase: tyrosine kinase recruitment of the MAP kinase cascade. Recent Prog. Horm. Res. 56:127-155. [DOI] [PubMed] [Google Scholar]
- 4.Baek, K. H., J. R. Fabian, F. Sprenger, D. K. Morrison, and L. Ambrosio. 1996. The activity of D-raf in torso signal transduction is altered by serine substitution, N-terminal deletion, and membrane targeting. Dev. Biol. 175:191-204. [DOI] [PubMed] [Google Scholar]
- 5.Bos, J. 1998. All in the family? New insights and questions regarding interconnectivity of Ras, Rap1 and Ral. EMBO J. 17:6776-6782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braselmann, S., and F. McCormick. 1995. BCR and RAF form a complex in vivo via 14-3-3 proteins. EMBO J. 14:4839-4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunet, A., A. Bonni, M. J. Zigmond, M. Z. Lin, P. Juo, L. S. Hu, M. J. Anderson, K. C. Arden, J. Blenis, and M. E. Greenberg. 1999. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96:857-868. [DOI] [PubMed] [Google Scholar]
- 8.Chiloeches, A., C. S. Mason, and R. Marais. 2001. S338 phosphorylation of Raf-1 is independent of phosphatidylinositol 3-kinase and Pak3. Mol. Cell. Biol. 21:2423-2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark, G. J., J. K. Drugan, K. L. Rossman, J. W. Carpenter, K. Rogers-Graham, H. Fu, C. J. Der, and S. L. Campbell. 1997. 14-3-3 zeta negatively regulates raf-1 activity by interactions with the Raf-1 cysteine-rich domain. J. Biol. Chem. 272:20990-20993. [DOI] [PubMed] [Google Scholar]
- 10.Conklin, D. S., K. Galaktionov, and D. Beach. 1995. 14-3-3 proteins associate with cdc25 phosphatases. Proc. Natl. Acad. Sci. USA 92:7892-7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Datta, S. R., A. Katsov, L. Hu, A. Petros, S. W. Fesik, M. B. Yaffe, and M. E. Greenberg. 2000. 14-3-3 proteins and survival kinases cooperate to inactivate BAD by BH3 domain phosphorylation. Mol. Cell 6:41-51. [PubMed] [Google Scholar]
- 12.Daub, M., J. Jockel, T. Quack, C. K. Weber, F. Schmitz, U. R. Rapp, A. Wittinghofer, and C. Block. 1998. The RafC1 cysteine-rich domain contains multiple distinct regulatory epitopes which control Ras-dependent Raf activation. Mol. Cell. Biol. 18:6698-6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dent, P., T. Jelinek, D. K. Morrison, M. Weber, and T. W. Sturgill. 1995. Reversal of Raf-1 activation by purified and membrane-associated protein phosphatases. Science 269:1902-1906. [DOI] [PubMed] [Google Scholar]
- 14.Dhillon, A. S., S. Meikle, Z. Yazici, M. Eulitz, and W. Kolch. 2002. Regulation of Raf-1 activation and signalling by dephosphorylation. EMBO J. 21:64-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diaz, B., D. Barnard, A. Filson, S. MacDonald, A. King, and M. Marshall. 1997. Phosphorylation of Raf-1 serine 338-serine 339 is an essential regulatory event for Ras-dependent activation and biological signaling. Mol. Cell. Biol. 17:4509-4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evan, G. I., G. K. Lewis, G. Ramsay, and J. M. Bishop. 1985. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol. Cell. Biol. 5:3610-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fabian, J. R., I. O. Daar, and D. K. Morrison. 1993. Critical tyrosine residues regulate the enzymatic and biological activity of Raf-1 kinase. Mol. Cell. Biol. 13:7170-7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fabian, J. R., A. B. Vojtek, J. A. Cooper, and D. K. Morrison. 1994. A single amino acid change in Raf-1 inhibits Ras binding and alters Raf-1 function. Proc. Natl. Acad. Sci. USA 91:5982-5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fantl, W. J., A. J. Muslin, A. Kikuchi, J. A. Martin, A. M. MacNicol, R. W. Gross, and L. T. Williams. 1994. Activation of Raf-1 by 14-3-3 proteins. Nature 371:612-614. [DOI] [PubMed] [Google Scholar]
- 20.Freed, E., M. Symons, S. G. Macdonald, F. McCormick, and R. Ruggieri. 1994. Binding of 14-3-3 proteins to the protein kinase Raf and effects on its activation. Science 265:1713-1716. [DOI] [PubMed] [Google Scholar]
- 21.Fu, H., K. Xia, D. C. Pallas, C. Cui, K. Conroy, R. P. Narsimhan, H. Mamon, R. J. Collier, and T. M. Roberts. 1994. Interaction of the protein kinase Raf-1 with 14-3-3 proteins. Science 266:126-129. [DOI] [PubMed] [Google Scholar]
- 22.Hagemann, C., and U. R. Rapp. 1999. Isotype-specific functions of Raf kinases. Exp. Cell Res. 253:34-46. [DOI] [PubMed] [Google Scholar]
- 23.Irie, K., Y. Gotoh, B. M. Yashar, B. Errede, E. Nishida, and K. Matsumoto. 1994. Stimulatory effects of yeast and mammalian 14-3-3 proteins on the Raf protein kinase. Science 265:1716-1719. [DOI] [PubMed] [Google Scholar]
- 24.IUPAC-IUB Commission on Biochemical Nomenclature. 1969. A one-letter notation for amino acid sequences. Tentative rules. Biochem. J. 113:1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaitner, B. K., J. Becker, T. Linnemann, C. Herrmann, A. Wittinghofer, and C. Block. 1997. Discrimination of amino acids mediating Ras binding from noninteracting residues affecting raf activation by double mutant analysis. J. Biol. Chem. 272:29927-29933. [DOI] [PubMed] [Google Scholar]
- 26.Jaumot, M., and J. F. Hancock. 2001. Protein phosphatases 1 and 2A promote Raf-1 activation by regulating 14-3-3 interactions. Oncogene 20:3949-3958. [DOI] [PubMed] [Google Scholar]
- 27.Kolch, W. 2000. Meaningful relationships: the regulation of the Ras/Raf/MEK/ERK pathway by protein interactions. Biochem. J. 351:289-305. [PMC free article] [PubMed] [Google Scholar]
- 28.Koyama, S., L. T. Williams, and A. Kikuchi. 1995. Characterization of the interaction of Raf-1 with ras p21 or 14-3-3 protein in intact cells. FEBS Lett. 368:321-325. [DOI] [PubMed] [Google Scholar]
- 29.Leevers, S. J., and C. J. Marshall. 1992. Activation of extracellular signal-regulated kinase, ERK2, by p21ras oncoprotein. EMBO J. 11:569-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leevers, S. J., H. F. Paterson, and C. J. Marshall. 1994. Requirement for Ras in Raf activation is overcome by targeting Raf to the plasma membrane. Nature 369:411-414. [DOI] [PubMed] [Google Scholar]
- 31.Li, S., P. Janosch, M. Tanji, G. C. Rosenfeld, J. C. Waymire, H. Mischak, W. Kolch, and J. M. Sedivy. 1995. Regulation of Raf-1 kinase activity by the 14-3-3 family of proteins. EMBO J. 14:685-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marais, R., Y. Light, C. Mason, H. Paterson, M. Olson, and C. J. Marshall. 1998. Requirement of Ras-GTP-Raf complexes for activation of Raf-1 by protein kinase C. Science 280:109-112. [DOI] [PubMed] [Google Scholar]
- 33.Marais, R., Y. Light, H. F. Paterson, and C. J. Marshall. 1995. Ras recruits Raf-1 to the plasma membrane for activation by tyrosine phosphorylation. EMBO J. 14:3136-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marais, R., Y. Light, H. F. Paterson, C. S. Mason, and C. J. Marshall. 1997. Differential regulation of Raf-1, A-Raf and B-Raf by oncogenic Ras and tyrosine kinases. J. Biol. Chem. 272:4378-4383. [DOI] [PubMed] [Google Scholar]
- 35.Marais, R., and C. J. Marshall. 1996. Control of the ERK MAP kinase cascade by Ras and Raf, p. 101-125. In P. J. Parker and T. Pawson (ed.), Cell signalling, vol. 27. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 36.Marais, R., J. Wynne, and R. Treismann. 1993. The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell 73:381-393. [DOI] [PubMed] [Google Scholar]
- 37.Marshall, C. J. 1995. Specificity of receptor tyrosine kinase signalling: transient versus sustained extracellular signal-regulated kinase activation. Cell 80:179-185. [DOI] [PubMed] [Google Scholar]
- 38.Mason, C. S., C. Springer, R. G. Cooper, G. Superti-Furga, C. J. Marshall, and R. Marais. 1999. Serine and tyrosine phosphorylations cooperate in Raf-1, but not B-Raf activation. EMBO J. 18:2137-2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McPherson, R. A., A. Harding, S. Roy, A. Lane, and J. F. Hancock. 1999. Interactions of c-Raf-1 with phosphatidylserine and 14-3-3. Oncogene 18:3862-3869. [DOI] [PubMed] [Google Scholar]
- 40.Michaud, N. R., J. R. Fabian, K. D. Mathes, and D. K. Morrison. 1995. 14-3-3 is not essential for Raf-1 function: identification of Raf-1 proteins that are biologically activated in a 14-3-3- and Ras-independent manner. Mol. Cell. Biol. 15:3390-3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mineo, C., R. G. W. Anderson, and M. A. White. 1997. Physical interaction with Ras enhances activation of membrane-bound Raf (RafCAAX). J. Biol. Chem. 272:10345-10348. [DOI] [PubMed] [Google Scholar]
- 42.Mischak, H., T. Seitz, P. Janosh, M. Eulitz, H. Steen, M. Schellerer, A. Philipp, and W. Kolch. 1996. Negative regulation of Raf-1 by phosphorylation of serine 621. Mol. Cell. Biol. 16:5409-5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morrison, D. K., and R. E. J. Cutler. 1997. The complexity of Raf-1 regulation. Curr. Opin. Cell Biol. 9:174-179. [DOI] [PubMed] [Google Scholar]
- 44.Morrison, D. K., G. Heidecker, U. R. Rapp, and T. D. Copeland. 1993. Identification of the major phosphorylation sites of the Raf-1 kinase. J. Biol. Chem. 268:17309-17316. [PubMed] [Google Scholar]
- 45.Muslin, A. J., J. W. Tanner, P. M. Allen, and A. S. Shaw. 1996. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 84:889-897. [DOI] [PubMed] [Google Scholar]
- 46.Muslin, A. J., and H. Xing. 2000. 14-3-3 proteins: regulation of subcellular localization by molecular interference. Cell. Signal. 12:703-709. [DOI] [PubMed] [Google Scholar]
- 47.Petosa, C., S. C. Masters, L. A. Bankston, J. Pohl, B. Wang, H. Fu, and R. C. Liddington. 1998. 14-3-3ζ binds a phosphorylated Raf peptide and an unphosphorylated peptide via its conserved amphipathic groove. J. Biol. Chem. 273:16305-16310. [DOI] [PubMed] [Google Scholar]
- 48.Rittinger, K., J. Budman, J. Xu, S. Volinia, L. C. Cantley, S. J. Smerdon, S. J. Gamblin, and M. B. Yaffe. 1999. Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol. Cell 4:153-166. [DOI] [PubMed] [Google Scholar]
- 49.Robinson, M. J., and M. H. Cobb. 1997. Mitogen-activated protein kinase pathways. Curr. Opin. Cell Biol. 9:180-186. [DOI] [PubMed] [Google Scholar]
- 50.Rommel, C., B. A. Clarke, S. Zimmermann, L. Nunez, R. Rossman, K. Reid, K. Moelling, G. D. Yancopoulos, and D. J. Glass. 1999. Differentiation stage-specific inhibition of the Raf-MEK-ERK pathway by Akt. Science 286:1738-1741. [DOI] [PubMed] [Google Scholar]
- 51.Rommel, C., G. Radziwill, J. Lovric, J. Noeldeke, T. Heinicke, D. Jones, A. Aitken, and K. Moelling. 1996. Activated Ras displaces 14-3-3 proteins from the amino terminus of c-Raf-1. Oncogene 12:609-619. [PubMed] [Google Scholar]
- 52.Roy, S., R. A. McPherson, A. Apolloni, J. Yan, A. Lane, J. Clyde-Smith, and J. F. Hancock. 1998. 14-3-3 facilitates Ras-dependent Raf-1 activation in vitro and in vivo. Mol. Cell. Biol. 18:3947-3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd edition. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 54.Stokoe, D., S. G. Macdonald, K. Cadwallader, M. Symons, and J. F. Hancock. 1994. Activation of Raf as a result of recruitment to the plasma membrane. Science 264:1463-1467. [DOI] [PubMed] [Google Scholar]
- 55.Suen, K. L., X. R. Bustelo, and M. Barbacid. 1995. Lack of evidence for the activation of the Ras/Raf mitogenic pathway by 14-3-3 proteins in mammalian cells. Oncogene 11:825-831. [PubMed] [Google Scholar]
- 56.Thorson, J. A., L. W. Yu, A. L. Hsu, N. Y. Shih, P. R. Graves, J. W. Tanner, P. M. Allen, H. Piwnica-Worms, and A. S. Shaw. 1998. 14-3-3 proteins are required for maintenance of Raf-1 phosphorylation and kinase activity. Mol. Cell Biol. 18:5229-5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tzivion, G., Z. Lou, and J. Avruch. 1998. A dimeric 14-3-3 protein is an essential cofactor for Raf kinase activity. Nature 394:536-539. [DOI] [PubMed] [Google Scholar]
- 58.Wang, B., H. Yang, Y.-C. Liu, T. Jelinek, L. Zhang, E. Ruoslahti, and H. Fu. 1999. Isolation of high-affinity peptide antagonists of 14-3-3 proteins by phage display. Biochemistry 38:12499-12504. [DOI] [PubMed] [Google Scholar]
- 59.White, M. A., C. Nicolette, A. Minden, A. Polverino, A. L. Van, M. Karin, and M. H. Wigler. 1995. Multiple Ras functions can contribute to mammalian cell transformation. Cell 80:533-541. [DOI] [PubMed] [Google Scholar]
- 60.Williams, J. G., J. K. Drugan, G. S. Yi, G. J. Clark, C. J. Der, and S. L. Campbell. 2000. Elucidation of binding determinants and functional consequences of Ras/Raf-cysteine-rich domain interactions. J. Biol. Chem. 275:22172-22179. [DOI] [PubMed] [Google Scholar]
- 61.Yaffe, M. B., K. Rittinger, S. Volinia, P. R. Caron, A. Aitken, H. Leffers, S. J. Gamblin, S. J. Smerdon, and L. C. Cantley. 1997. The structural basis for 14-3-3:phosphopeptide binding specificity. Cell 91:961-971. [DOI] [PubMed] [Google Scholar]
- 62.Yip-Schneider, M. T., W. Miao, A. Lin, D. S. Barnard, G. Tzivion, and M. S. Marshall. 2000. Regulation of the Raf-1 kinase domain by phosphorylation and 14-3-3 association. Biochem. J. 351:151-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zimmermann, S., and K. Moelling. 1999. Phosphorylation and regulation of Raf by Akt (protein kinase B). Science 286:1741-1744. [DOI] [PubMed] [Google Scholar]