Abstract
Drosophila melanogaster DNA replication-related element (DRE) factor (dDREF) is a transcriptional regulatory factor required for the expression of genes carrying the 5′-TATCGATA DRE. dDREF has been reported to bind to a sequence in the chromatin boundary element, and thus, dDREF may play a part in regulating insulator activity. To generate further insights into dDREF function, we carried out a Saccharomyces cerevisiae two-hybrid screening with DREF polypeptide as bait and identified Mi-2 as a DREF-interacting protein. Biochemical analyses revealed that the C-terminal region of Drosophila Mi-2 (dMi-2) specifically binds to the DNA-binding domain of dDREF. Electrophoretic mobility shift assays showed that dMi-2 thereby inhibits the DNA-binding activity of dDREF. Ectopic expression of dDREF and dMi-2 in eye imaginal discs resulted in severe and mild rough-eye phenotypes, respectively, whereas flies simultaneously expressing both proteins exhibited almost-normal eye phenotypes. Half-dose reduction of the dMi-2 gene enhanced the DREF-induced rough-eye phenotype. Immunostaining of polytene chromosomes of salivary glands showed that dDREF and dMi-2 bind in mutually exclusive ways. These lines of evidence define a novel function of dMi-2 in the negative regulation of dDREF by its DNA-binding activity. Finally, we postulated that dDREF and dMi-2 may demonstrate reciprocal regulation of their functions.
The promoters of Drosophila melanogaster genes related to DNA replication, such as those for the 180-kDa catalytic subunits and 73-kDa regulatory subunits of DNA polymerase α, as well as PCNA, contain a common 8-bp palindromic sequence (5′-TATCGATA) named the DNA replication-related element (DRE) (10), in addition to E2F-binding sites (4, 29). The DRE requirement for promoter activation has been confirmed in both cultured cells and transgenic flies (11, 26, 28). Studies using the latter have further shown that DRE is required for the function of the PCNA promoter throughout development, except in adult females.
We have found a specific DRE-binding factor (DREF), Drosophila DREF (dDREF), consisting of an 80-kDa polypeptide homodimer, and molecular cloning of its cDNA allowed confirmation that dDREF is a transactivator for DRE-containing genes (11). Ectopic expression of the dominant-negative form of dDREF by the GAL4 upstream activation sequence (UAS) system resulted in inhibited endo-replication of larval salivary gland cells and significantly reduced DNA replication in the second mitotic wave in eye imaginal discs (12). Furthermore, ectopic expression of full-length dDREF in eye imaginal discs caused ectopic DNA synthesis and apoptosis in normally postmitotic cells, inhibited differentiation of photoreceptor cells, and resulted in a severe rough-eye phenotype in the adult flies (9). These results indicate that dDREF is involved in the regulation of DNA replication in both mitotic and endo cell cycles as well as in differentiation processes.
Another important role of dDREF as an antagonist of the boundary element-associated factor (BEAF), which is involved in the boundary activity of the scs′ region of the Drosophila hsp70 gene, has been proposed (7, 8). Staining of polytene chromosomes with anti-dDREF and anti-BEAF antibodies revealed that about 50% of signals from the two proteins overlapped. Furthermore, by using a chromatin precipitation method, others have demonstrated that dDREF can bind to the same sequences as BEAF in the scs′ special chromatin domain present in the hsp70 locus. From their findings, they concluded that competition of binding between dDREF and BEAF is important for the regulation of activity at the chromatin boundary.
To generate further insight into the molecular mechanisms of DRE and dDREF regulation, we screened DREF-interacting proteins with a Saccharomyces cerevisiae two-hybrid system by using dDREF or a recently characterized human DREF homologue (hDREF/KIAA0785) (F. Hirose and N. Ohshima, unpublished data) as bait and identified Mi-2 as one of the DREF-interacting proteins. The Mi-2 protein was initially characterized as an autoantigen in patients with dermatomyositis (6) and belongs to the CHD (chromo-helicase and ATPase-DNA-binding) family related to SWI2/SNF2 (15). Four CHD proteins have been identified in vertebrates: CHD1, CHD2, CHD3 (Mi-2 α), and CHD4 (Mi-2 β) (1, 24). CHD1 and CHD2 have a DNA-binding domain, whereas Mi-2 α and Mi-2 β lack an obvious DNA-binding domain but contain two PHD fingers and a truncated helix-turn-helix motif resembling the DNA-binding domain of c-myb. Although physiological functions of each CHD protein have yet to be clarified, Mi-2 α and/or Mi-2 β has been identified as a subunit of an ATP-dependent chromatin-remodeling complex with histone deacetylase activity. Furthermore, biochemical analyses of NRD (nucleosome remodeling and deacetylation) showed that Drosophila Mi-2 (dMi-2) is a nucleosome-stimulated ATPase that uses the energy from ATP hydrolysis both to mobilize histone octamers relative to DNA and to facilitate the deacetylation of nucleosomal histones in certain situations (3). Recent work demonstrated that Mi-2 complexes contain MBD3 in addition to Rpd3-like deacetylase and RbAp48/p46 histone-binding proteins, indicating that they may be involved in coupling DNA methylation to chromatin remodeling and histone deacetylation (23, 32). Thus, the Mi-2 complexes are believed to be recruited by protein-protein interaction with a specific transcription factor and to repress transcription through chromatin remodeling and deacetylation. However, target genes of the Mi-2 complexes, functional differences or redundancy between Mi-2 α and Mi-2 β, and physiological regulation of Mi-2 complexes are just beginning to be characterized.
In Drosophila, a single gene (dMi-2) encoding a vertebrate Mi-2 α and Mi-2 β homologue has been identified. Kehle et al. clearly demonstrated that dMi-2 interacts physically and genetically with hunchback, a repressor of HOX gene expression, by use of several lines of dMi-2 mutant flies (13). In the present study, we provide several lines of evidence indicating that dMi-2 specifically binds to the DNA-binding domain of dDREF and thereby inhibits its activity as a transcriptional activator. From the results, we propose a novel function of Mi-2, with repression of gene expression by direct interaction with a transcription factor. Furthermore, we describe the possible biological significance for interactions between dDREF and dMi-2.
MATERIALS AND METHODS
Cell culture.
Schneider cells derived from Drosophila melanogaster embryos were grown at 25°C in M(3)BF medium supplemented with 10% fetal calf serum in the presence of 5% CO2.
Fly strains.
Fly stocks were maintained at 25°C on standard food. The Canton S fly was used as the wild-type strain.
The dMi-21, dMi-24, and dMi-26 alleles described previously (13) were kindly supplied by Jurg Muller. The dMi-2j3D4 allele was obtained from Bloomington, Stock Center, Bloomington, Ind. The UAS-dDREF fly was described earlier (12), as was the transgenic fly line (line number 16) carrying pGMR-GAL4 on the X chromosome (20). The GAL4 enhancer trap lines carrying homozygous hs-GAL4 in the third chromosome and P[Sg-GAL4](l)/Binsinscy were provided by Brand and Perrimon (2). Transgenic flies carrying −119PCNAlacZW8HS, −86P3NPCNAlacZW8HS, and −168mutΔ6PCNAlacZW8HS were described previously (27).
Establishment of transgenic flies.
P-element-mediated germ line transformation was carried out as described previously (19), F1 transformants being selected on the basis of white color rescue (17). Six and five independent lines were established for the plasmids pUAS-Flag-dDREF and pUAS-HA-dMi-2, respectively, and subjected to immunoblot analysis to confirm the expression of HA-dMi-2 or Flag-dDREF in the transgenic flies.
Oligonucleotides.
HA and Flag oligonucleotides were synthesized for construction of the plasmids pUAS-HA-dMi-2 and pUAS-Flag-dDREF, respectively. HA oligonucleotide, 5′-GGCCGCATGGCTTACCCATACGATGTTCCAGATTACGCTG and 5′-GATCCAGCGTAATCTGGAACATCGTATGGGTATGCCATGC. Sequences corresponding to the region encoding the HA epitope are shown in boldface, and sequences compatible with NotI and BamHI sites are underlined.
Flag oligonucleotide, 5′-GATCTATGGACTACAAGGATGACGATGACAAGGCGGCCGCACA and 5′-TATGTGCGGCCGCCTTGTCATCGTCATCCTTATAGTCCATA. Sequences corresponding to the region encoding the Flag epitope are shown in boldface, and sequences that are compatible with BglII and NdeI sites are underlined. The oligonucleotide DRE-P used as a probe for an electrophoretic mobility shift assay (EMSA) was described previously (10).
Plasmids.
For construction of hDREF/pAS2 as a bait plasmid for two-hybrid screening of DREF, a full-length cDNA of hDREF was cut out from hDREF/pcDNA3-HA (Hirose et al., unpublished) with NdeI and XhoI digestion and inserted into the NdeI-SalI sites of the pAS2 vector. The series of plasmids expressing glutathione S-transferase (GST)-dDREF (GST-dDREF1-708, GST-dDREF16-607, GST-dDREF16-251, GST-dDREF16-205, GST-dDREF16-185, GST-dDREF16-165, GST-dDREF16-145, GST-dDREF16-125, GST-dDREF16-105, GST-dDREF16-95, GST-dDREF32-230, GST-dDREF52-230, GST-dDREF72-230, GST-dDREF69-157, GST-dDREF157-242, and GST-dDREF230-608) were as described previously (11). The dMi-24-1982/pT7-l template plasmid for in vitro transcription and translation reactions was a gift from Jurg Muller. A series of deletion mutants of dMi-2/pT7-l (amino acids [aa] 935-1982, 935-1514, 4-439/935-1982, 4-439/1503-1982, 1503-1982, 1758-1982, 1503-1758 and 4-935/1503-1982) were generated by partially digesting dMi-24-1982/pT7-l with EcoRI and self-ligating plasmids. Plasmid 203-1982 was constructed by BamHI digestion and self-ligation of plasmid dMi-24-1982/pT7-l.
To construct the plasmid pUAS-HA-dMi-2 containing HA-tagged dMi-2, the cDNA plasmid dMi-24-1982/pT7-l was digested with XbaI and then partially digested with BamHI. The resultant dMi-2 cDNA fragments (4 to 1,982 residues) were separated by agarose gel electrophoresis, isolated from the gel, and inserted into the NotI and XbaI sites of the pUAST vector with the HA oligonucleotide as an adapter.
To construct the plasmid pUAS-Flag-dDREF, dDREF cDNA was isolated from dDREF/pAS-2 by digestion with NdeI and SalI and inserted into the BglII and XhoI sites of the pUAST vector with the Flag oligonucleotide as an adapter.
To construct −168DPCNAluc and −168mutΔ6PCNAluc for the luciferase transient expression assay, plasmids p5′-168PCNACAT and p5′-168mutΔ6PCNACAT (26) were digested with SacI and blunt-ended. Then the fragments were digested with SalI, and the DNA fragments containing the PCNA gene promoter were isolated and inserted between the blunt-ended BglII and XhoI sites of the plasmid pGVB.
Two-hybrid screening.
The yeast two-hybrid screen was performed by using the MATCHMAKER pretransformed library system (Clontech). The cells of Saccharomyces strain PJ69-2A (MATa trp1 leu2 ura3 his3 gal4 gal80 LYS2::GAL1UAS-GAL1TATA-HIS3 GAL2UAS-GAL2TATA-ADE2) transformed with hDREF/pAS2 were mated with Y187 cells (MATα ura3 his3 ade2 trp1 leu2 gal4 gal80 met− URA3::GAL1UAS-GAL1TATA-lacZ) pretransformed with human fetal brain cDNA/pACT2. Mating efficiency was calculated as 3.8%. Diploid his+ ade+ trp+ leu+ transformants were selected on minimal medium lacking histidine, adenine, tryptophan, and leucine but supplemented with 2% glucose. Transformants were further confirmed to express β-galactosidase with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) as a substrate. The plasmid DNAs, isolated from the yeast candidate clones, were transfected to transform Escherichia coli KC8 carrying the hisB, leuB, and trpC mutations. KC8 cells transformed with pACT2 were grown on M9 minimal medium supplemented with essential amino acids and thiamine but lacking leucine.
GST pull-down assay.
In vitro transcription and translation reactions were carried out with the TNT-coupled reticulocyte lysate system (Promega) in the presence of [35S]methionine. The sizes and amounts of the in vitro translation products were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
GST-dDREF fusion proteins were produced in E. coli XL1-blue as described previously (11), and the amounts of each of the GST-dDREF proteins were estimated by SDS-PAGE. Glutathione beads (10 μl) were equilibrated in binding buffer (50 mM Tris-HCl [pH 8.0], 5 mM MgCl2, 10% glycerol, 0.1 mM EDTA, 150 mM NaCl, 0.1% NP-40, 0.1% Triton X-100, 5 mM β-mercaptoethanol, 1.5 mg of bovine serum albumin (BSA)/ml, 1 mM phenylmethylsulfonyl fluoride, 5 μg of leupeptin/ml, 2 μg of pepstatin/ml, 1 μg of aprotinin/ml), and 200-μl aliquots were incubated for 1 h at 4°C with E. coli extracts containing approximately 1 μg of GST or GST-dDREF recombinant protein. After washing five times with the same buffer, we added a 5-μl aliquot of the in vitro-translated dMi-2 polypeptide and incubated the mixture at room temperature for 1 h. After washing six times with 200 μl of the same buffer, we eluted bound proteins by incubating them for 1 h at 4°C in 20 μl of the binding buffer supplemented with 5 mM reduced glutathione and separated the proteins by SDS-PAGE. After Coomassie brilliant blue staining to verify that equal amounts of the fusion proteins were loaded, the gel was dried and 35S-labeled proteins were detected with a BAS2000 imaging analyzer (Fuji Photo Film) or by autoradiography.
EMSA.
EMSAs were performed as described previously (10), with minor modifications. A reaction mixture containing 20 mM HEPES [pH 7.6], 100 mM KCl, 0.1 mM EDTA, 0.5 mM dithiothreitol, 10% glycerol, and 1 μg of salmon sperm DNA was used with Kc cell nuclear extracts. 32P-labeled probes (10,000 cpm) were incubated in 15 μl of the reaction mixture. To test for the effects of dMi-2 on the DNA-binding activity of dDREF, Kc cell nuclear extracts were incubated with E. coli extracts containing GST or GST-dMi-2 proteins for 15 min at 4°C before or after addition of the probe. DNA-protein complexes were electrophoretically resolved on 4% polyacrylamide gels in 100 mM Tris-borate [pH 8.3]-2 mM EDTA containing 2.5% glycerol at 25°C. The gels were dried and then autoradiographed.
Scanning electron microscopy.
Adult flies were anesthetized, mounted on stages, and observed in a Hitachi S-100 scanning electron microscope in the low-vacuum mode.
Antibodies.
Polyclonal anti-dMi-2 rabbit serum (αdMi-2-C) was a gift from Jerg Muller (3). Anti-dDREF polyclonal and monoclonal antibodies were as described previously (11).
Immunoprecipitation.
Immunoprecipitation was carried out in the same buffer used for the GST pull-down assay. Ten microliters of supernatant of hybridoma-producing anti-dDREF monoclonal antibody no. 1 was added to 300 μl of crude nuclear extract from Drosophila embryos (approximately 150 μg of protein) and incubated for 3 h at 4°C. Ten microliters of protein G-Sepharose beads (Roche) was added, and incubation was continued for 1 h. Protein G-antibody complexes were collected by centrifugation, washed six times in 0.5 ml of the binding buffer, resuspended in loading buffer for SDS-PAGE, and boiled for 3 min. Polypeptides in the immunoprecipitates were separated by SDS-PAGE, transferred to Hybond membranes, and probed with relevant antibodies.
DNA transfection and luciferase assay.
DNA transfection of various DNA mixtures in Schneider cells was performed with the CellFectin reagent (Life Technologies, Inc.). Schneider cells (1.5 × 105/well) were grown in 24-well culture plates for 24 h and cotransfected with 50 ng of reporter plasmid, 50 to 400 ng of pUAS-HA-dMi-2 and 100 ng of pAct-GAL4 as effector plasmids, and 1 ng of pRL-actin5C (18) as an internal control plasmid. The total amount of DNA in the transfection mixture was adjusted to 1 μg by addition of pGEM3. Cells were harvested 48 h after transfection. The luciferase assay was carried out by means of the Dual-Glo Luciferase Reporter Assay System (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity. Transfections were performed three times with at least two independent plasmid preparations.
Immunocytochemical detection of β-galactosidase.
Expression patterns of lacZ in brain lobes were detected by immunohistochemistry as described previously (28). Third-instar larva were dissected in Drosophila Ringer's solution, and imaginal discs were fixed in 4% paraformaldehyde-phosphate-buffered saline (PBS) for 20 min at room temperature. After washing with PBS-0.3% Triton X-100 (PBS-T), the samples were blocked with PBS-T containing 10% normal goat serum for 30 min at room temperature. Samples were then incubated with mouse anti-β-galactosidase monoclonal antibody (Promega) at a 1:1,000 dilution at 4°C for 16 h. After extensive washing with PBS-T, the brain lobes were incubated with alkaline phosphatase-conjugated goat anti-mouse immunoglobulin G (IgG; Promega) at a 1:2,000 dilution for 2 h at room temperature and washed with PBS-T. Color was developed with a solution containing 100 mM Tris-HCl (pH 9.5), 100 mM NaCl, 5 mM MgCl2, 0.34 mg of nitroblue tetrazolium salts/ml, and 0.175 mg of 5-bromo-4-chloro-3-indolyl phosphate toluidinium salts (X-phosphate)/ml. The tissues were washed with PBS and mounted in 90% glycerol-PBS for microscopic observation.
Immunostaining of polytene chromosomes.
Polytene chromosome spreads were made according to the protocol of Zink et al. from Canton S wild-type wandering third-instar larvae (33). Squashes were stored in PBS-0.05% Tween 20-1% BSA at 4°C overnight and then incubated with rabbit polyclonal antibody specific for dMi-2 at a 1:400 dilution or with culture supernatant of hybridoma producing anti-DREF monoclonal antibody at a 1:200 dilution at room temperature for 1 h. After extensive washing with PBS-0.05% Tween 20-1% BSA, samples were incubated at room temperature for 1 h with anti-mouse IgG conjugated with Alexa594 (Molecular Probes) and anti-rabbit IgG conjugated with Alexa488 (Molecular Probes), each at a 1:400 dilution. After placing coverslips over the samples and mounting them in 90% glycerol-PBS, the preparations were viewed under an Olympus laser scanning confocal microscope.
RESULTS
In vivo association of dMi-2 with dDREF.
To generate further insight into the molecular mechanisms of DRE/DREF regulation, we here screened for DREF-interacting proteins with the yeast two-hybrid system by using dDREF or the recently identified human DREF (hDREF/KIAA0785) as bait (Hirose et al., unpublished). With hDREF/KIAA0785 we isolated 40 independent clones containing 14 different genes encoding DREF-interacting proteins. Ten of the 40 clones encoded 300 aa residues of the Mi-2 α polypeptide C terminus. In interaction tests using β-galactosidase reporter in yeast cells, this C-terminal region exhibited the strongest interaction with hDREF/KIAA0785 among the clones (Hirose et al., unpublished). To clarify the biological meaning of the interaction between DREF and Mi-2, we decided to use the Drosophila system for two reasons. The first was that the gene encoding the Drosophila Mi-2 polypeptide exists as a single copy and that mutant flies were available. Second, our laboratory had transgenic flies ectopically expressing dDREF (9, 12, 20) as well as transgenic flies carrying PCNA-lacZ (26, 28), which were useful for the elucidation of in vivo DREF activity.
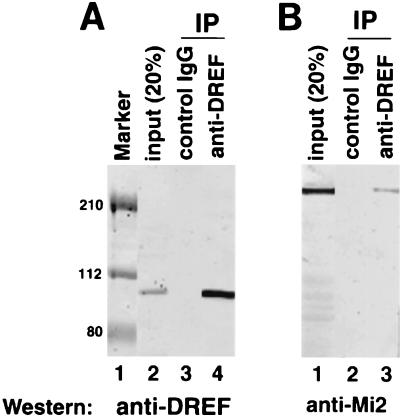
In order to test interactions between dMi-2 and dDREF, immunoprecipitation experiments were performed. Using the anti-dDREF polyclonal antibody (11) and crude nuclear extracts from Drosophila embryos, we detected dMi-2 polypeptide with the anti-dMi-2 polyclonal antibody αdMi-2-C (3). Anti-dDREF antibodies coimmunoprecipitated the 220-kDa dMi-2 polypeptide (Fig. 1B, lane 3), whereas normal IgG did not (Fig. 1B, lane 2), indicating that a dDREF/dMi-2 complex exists in vivo. Under these experimental conditions, most dDREF polypeptides in extracts were immunoprecipitated (Fig. 1A, lanes 2 and 4). Thus, we can estimate that 5% of the total dMi-2 polypeptide was immunoprecipitated with dDREF.
FIG. 1.
dDREF and dMi-2 interact in vivo. Ten microliters of supernatant of a hybridoma producing anti-dDREF monoclonal antibody no. 1 were added to 300 μl of crude nuclear extract from Drosophila embryos (approximately 150 μg of protein) and incubated for 3 h at 4°C. Ten microliters of protein G-Sepharose beads (Roche) were then added to the mixture, followed by incubation for 1 h. After extensive washing, proteins bound to the beads were separated by SDS-PAGE, transferred to a Hybond membrane, and probed with (A) anti-dDREF antibody or (B) anti-dMi-2 antibody. (A) Lane 1, prestained molecular size markers; lane 2, 20% of input protein used for immunoprecipitation (IP); lane 3, immunoprecipitation with normal rabbit IgG as a negative control; lane 4, immunoprecipitation with the anti-DREF antibody. (B) Lane 1, 20% of input protein used for immunoprecipitation; lane 2, immunoprecipitation without normal rabbit IgG as a negative control; lane 3, immunoprecipitation with anti-DREF antibody.
Determination of domains involved in dMi2-dDREF interactions.
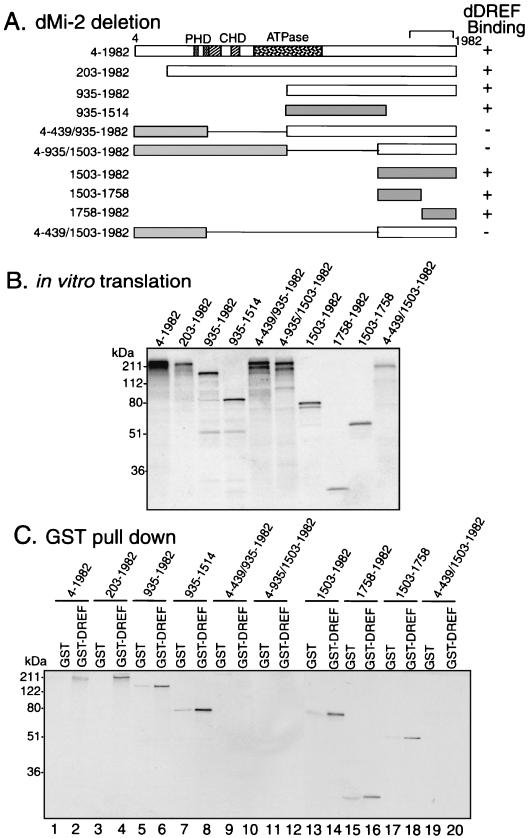
Next, we determined the domains involved in interactions between dMi-2 and dDREF by GST pull-down assay (Fig. 2 and 3). Deletion analysis of GST-dMi-2 fusion constructs revealed that the C-terminal half of the dMi-2 polypeptide is essential for binding (Fig. 2C, lane 6) and three subregions (aa 935 to 1514, 1503 to 1758, and 1758 to 1982) of the C-terminal half are sufficient to bind the dDREF polypeptide (Fig. 2C, lanes 8, 14, and 16). In addition, we found the N-terminal region of the dMi-2 polypeptide to have an inhibitory effect on binding, since deletion of the first 203 aa enhanced the binding affinity for the dDREF polypeptide (aa 203 to 1982) (Fig. 2C, lane 4). Furthermore, addition of the N-terminal region (aa 4 to 439) to the C-terminal binding subdomain conversely decreased binding affinity for the dDREF polypeptide (aa 4 to 439/935 to 1982 and aa 4 to 439/1503 to 1982) (Fig. 2C, lanes 10 and 20). These findings for the dMi-2 domain involved in the interaction with dDREF are consistent with the results obtained with yeast two-hybrid analysis (data not shown).
FIG. 2.
Mapping of the dMi-2 domain interacting with dDREF by GST pull-down assay. (A) Schematic representation of deletions for the dMi-2 polypeptide. (B) In vitro translation products of deleted dMi-2 forms used for the GST pull-down assay. (C) GST pull-down assay using GST-dDREF and in vitro-translated 35S-dMi-2. Odd lanes, GSTs were immobilized on glutathione-Sepharose beads and incubated with either 35S-dMi-2 polypeptide as indicated; even lanes, GST-dDREF (aa 1 to 708) was immobilized on glutathione-Sepharose beads and incubated with either 35S-dMi-2 polypeptide as indicated. Protein complexes were washed and resolved by SDS-PAGE, and bound proteins were detected by autoradiography.
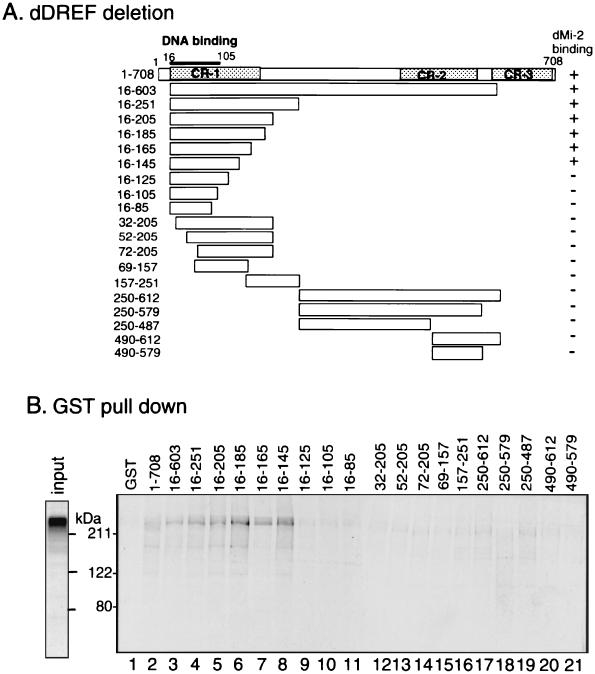
FIG. 3.
Mapping of the dDREF domain interacting with dMi-2 by GST pull-down assay. (A) Schematic representation of deletions for the dDREF polypeptide. These deletion polypeptides were expressed as fusion proteins with GST. (B) GST pull-down assay results with GST-dDREF and in vitro-translated 35S-dMi-2. GST (lane 1) or GST-dDREF polypeptides (lanes 2 through 21) were immobilized on glutathione-Sepharose beads and incubated with either 35S-dMi-2 polypeptide (aa residues 4 through 1982). Protein complexes were washed and resolved by SDS-PAGE, and bound dMi-2 was detected by autoradiography. The input gel on the left of the panel shows an aliquot of 35S-dMi-2 polypeptide used for each binding reaction.
We also determined the interaction domain in the dDREF polypeptide. As shown in Fig. 3, 130 aa residues (aa 16 to 145) near the N-terminal domain of dDREF polypeptide are responsible. This region is mapped within the region (CR-1) (Fig. 3A) exhibiting the highest conservation among DREFs of Drosophila melanogaster, Drosophila virilis (20), and humans (Hirose et al., unpublished).
dMi-2 inhibits DRE-binding activity of DREF.
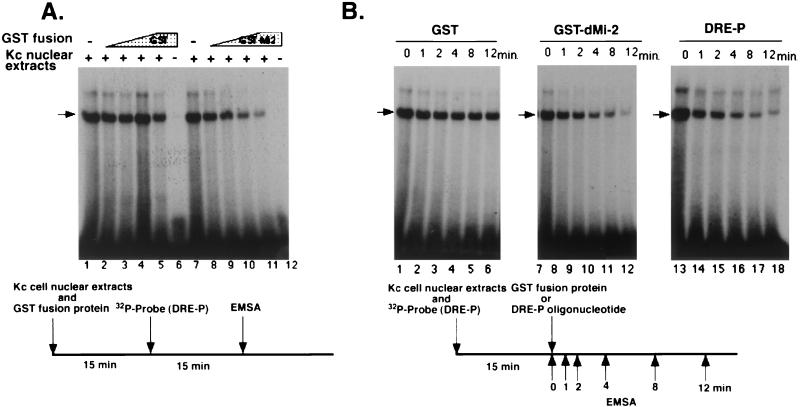
The amino acid region of dDREF involved in the interaction with dMi-2 (aa 16 to 145) includes an amino acid sequence responsible for its DNA-binding (aa 16 to 105), suggesting that dMi-2 might impede DREF function by masking the DNA-binding domain. Thus, we examined the effect of dMi-2 protein on the DNA-binding activity of dDREF by EMSA with the radiolabeled oligonucleotide DRE-P containing the DRE sequence from the PCNA gene promoter as a probe. As shown in Fig. 4A, a shifted band was observed with this probe and Kc cell nuclear extracts (lanes 1 and 7). It has already been confirmed by use of anti-dDREF monoclonal antibodies that this shift band represents the dDREF/DRE complex (11). Neither GST nor GST-dMi-2 fusion proteins bound DRE oligonucleotides (Fig. 4A, lanes 6 and 12). Preincubation of Kc cell nuclear extract with extracts of E. coli expressing the GST-dMi-2 fusion protein reduced the signals in a dose-dependent manner (Fig. 4A, lanes 8 through 11), whereas preincubation with extracts of E. coli expressing GST did not (Fig. 4A, lanes 2 through 5). Similar results were obtained in EMSA with purified GST-dDREF protein in place of Kc cell nuclear extract (data not shown), indicating that the dMi-2 protein might directly inhibit DNA-binding of dDREF.
FIG. 4.
dMi-2 inhibits DNA-binding activity of dDREF. (A) Nuclear extracts from Kc cells were preincubated with E. coli extracts containing GST (lanes 2 through 5) or GST-dMi-2 fusion protein (lanes 8 through 11), and then 32P-labeled DRE-P oligonucleotides were added. After an additional 15-min incubation, complexes of DRE/dDREF were analyzed by EMSA. (B) Nuclear extracts from Kc cells were incubated with 32P-labeled DRE-P oligonucleotides for 15 min; then E. coli extracts containing GST (lanes 1 through 6), E.coli extracts containing GST-dMi-2 fusion protein (lanes 7 through 12), or a 200-fold molar excess of unlabeled DRE-P oligonucleotide as a competitor (lanes 13 through 18) were added to reactions. Aliquots of reaction mixtures were subjected to electrophoresis at the indicated time points after addition of E. coli extracts or competitor oligonucleotide.
Next, we examined whether dMi-2 protein was able to dissociate dDREF from DNA/dDREF complexes. To test this possibility, labeled DRE-P oligonucleotides were preincubated with Kc cell nuclear extracts for 15 min, and then E. coli extracts containing GST-dMi-2 were added to the reaction mixtures and aliquots were subjected to EMSA at different time points (Fig. 4B, lanes 7 through 12). In a parallel experiment to estimate the rate of dissociation of DRE/dDREF complexes, excess amounts of DRE-P oligonucleotide were added to the reaction mixture instead of E. coli extract (Fig. 4B, lanes 13 through 18). The time required for half of the dDREF molecules to become dissociated from the labeled DRE-P probe was approximately 40 s, and addition of GST-dMi-2 extracts did not change the dissociation rate. Furthermore, addition of GST-dMi-2 did not change the mobility of DRE/dDREF complexes. It can thus be concluded that dMi-2 interacts with free dDREF and thereby inhibits its DNA-binding activity but cannot bind to the DNA-bound form of dDREF.
dMi-2 genetically interacts with dDREF.
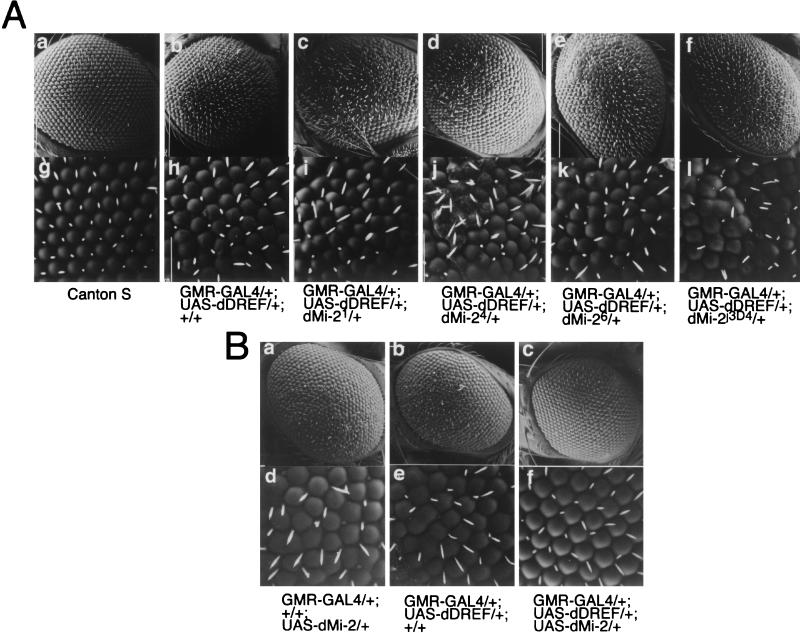
Next, a genetic approach was adopted to confirm interactions between dMi-2 and dDREF in vivo. It was shown previously that dDREF in eye imaginal discs was overexpressed using the GAL4-UAS targeted expression system, the results being induction of ectopic DNA synthesis in the cells behind the morphogenetic furrow that are normally postmitotic, abolition of photoreceptor specification, apoptosis in the imaginal disk cells, and a rough-eye phenotype in adult flies (9). This fly is very useful for genetic interaction testing and was therefore employed for the present study. A half-dose reduction of dMi-2 was achieved by crossing GMR-GAL4/UAS-DREF flies with loss-of-function mutants for dMi-2 (dMi-21, dMi-24, dMi-26, and dMi-2j3D4) (Fig. 5A, panels c through l). As shown in Fig. 5A, the dDREF-induced rough-eye phenotype was enhanced by a half-dose reduction of dMi-2. Immunohistochemical and immuno-Western blot analysis revealed that a half-dose reduction of dMi-2 did not change dDREF amounts in the imaginal disk cells (data not shown). Thus, the data imply that dMi-2 negatively regulates dDREF function in vivo. In order to further evaluate this repressive function of dMi-2 for dDREF, we established several transgenic lines carrying UAS-HA-dMi-2 and attempted coexpression experiments. Ectopic expression of dMi-2 in eye imaginal disk cells resulted in a mild rough-eye phenotype in adult flies (Fig. 5B, panels a and d). As noted previously, adult flies expressing dDREF in eye imaginal disk cells exhibited a more severe rough-eye phenotype than dMi-2- expressing flies (Fig. 5B, panels b and e). However, the eyes of adult flies simultaneously expressing dDREF and dMi-2 appeared to be normal (Fig. 5B, panels c and f). This means that the dDREF-induced rough-eye phenotype and the dMi-2-induced rough-eye phenotype were suppressed by the expression of dMi-2 and dDREF, respectively. From these results, we concluded that dMi-2 is a negative regulator of dDREF. Furthermore, we hypothesized that dDREF and dMi-2 may negatively regulate each other's functions in vivo.
FIG. 5.
dMi-2 genetically interacts with dDREF. (A) Half-dose reduction of the dMi-2 gene enhances the dDREF rough-eye phenotype. All alleles of dMi-2 are balanced with TM6C or TM6B balancer chromosomes. Female flies expressing dDREF (GMR-GAL4/GMR-GAL4, UAS-dDREF/UAS-dDREF, +/+) were crossed with males carrying dMi-21, dMi-24, dMi-26, and dMi-2j3D4 alleles. F1 progeny developed at 25°C without a balancer chromosome were used for analysis of eye phenotype by scanning electron microscopy. (a and g) Wild-type eye; (b and h) GMR-GAL4/+, UAS-dDREF/+, and +/+; (c and i) GMR-GAL4/+, UAS-dDREF/+, and dMi-21/+; (d and j) GMR-GAL4/+, UAS-dDREF/+, and dMi-24/+; (e and k) GMR-GAL4/+, UAS-dDREF/+, and dMi-26/+; (f and l) GMR-GAL4/+, UAS-dDREF/+, and dMi-2j3D4/+. (B) Expression of dMi-2 suppresses the dDREF-induced rough-eye phenotype. Scanning electron micrographs of adult compound eyes. (a and d) GMR-GAL4/+, +/+, and UAS-HA-dMi-2/+; (b and e) GMR-GAL4/+, UAS-Flag-dDREF/+, and +/+; (c and f) GMR-GAL4/+, UAS-Flag-dDREF/+, and UAS-HA-dMi-2/+. Note that the transgenic fly simultaneously expressing dDREF and dMi-2 in eye imaginal discs exhibits a normal eye morphology.
dMi-2 negatively regulates the promoter activity of the PCNA gene.
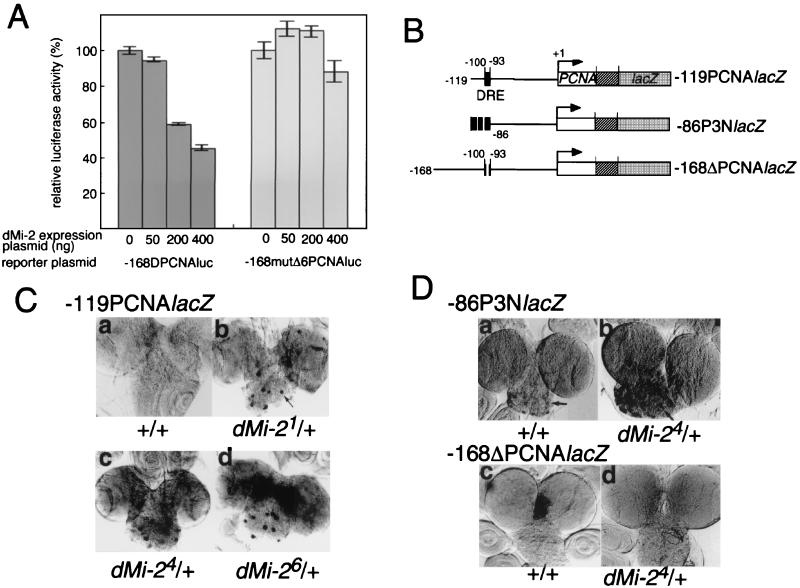
To investigate whether expression of dMi-2 represses promoter activity containing DRE, we transfected Schneider cells with −168DPCNAluc, in which expression of the luciferase gene is controlled by the promoter region from −168 to +23 of the PCNA gene and by increasing amounts of the dMi-2 expression plasmid. The DRE is located from −100 to −93 of the PCNA gene promoter as a single copy (26). Expression of dMi-2 reduced significantly the PCNA gene promoter-directed luciferase activity (Fig. 6A). In contrast, no repression by dMi-2 was observed with −168mutΔ6PCNAluc, a 6-bp deletion derivative of −168DPCNAluc. These results show that dMi-2 expression can repress expression of the PCNA gene promoter via a DRE sequence.
FIG. 6.
dMi-2 negatively regulates the PCNA gene promoter. (A) Effect of cotransfecting dMi-2-expressing plasmids on luciferase activity directed by the PCNA gene promoter. Schneider cells (1.5 × 105/well) were cotransfected with 50 ng of reporter plasmid (either −168DPCNAluc or −168mutΔ6PCNAluc), 50 to 400 ng of pUAS-HA-dMi-2 and 100 ng of pAct-GAL4 as effector plasmids, and 1 ng of pRL-actin5C as an internal control plasmid. The total amount of DNA in the transfection mixture was adjusted to 1 μg by the addition of pGEM3. Cells were harvested 48 h after transfection. The luciferase assay was carried out by means of the Dual-Glo Luciferase Reporter Assay System (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity. Averaged values obtained from three independent transfections are shown. (B) Constructs of PCNA-lacZ fusion genes used to establish the transgenic lines analyzed in panels C and D. (C) Half-dose reduction of dMi-2 activates the PCNA gene promoter depending on the presence of DRE. Detection of expression of β-galactosidase in brain lobes from transgenic flies carrying the PCNA-lacZ reporter gene. Male transgenic flies carrying −119PCNAlacZ on the second chromosome were crossed with females with dMi-21, dMi-24, and dMi-26 alleles balanced with the TM6C chromosome. Brain lobes were dissected from the third-instar larvae of F1 progeny, fixed, and immunostained with anti-β-galactosidase antibody. (D) Male transgenic flies carrying either −86P3NlacZ or −168ΔPCNAlacZ reporter genes on the second chromosome were crossed with females with the dMi-24 allele balanced with the TM6C chromosome. Brain lobes were dissected from the third-instar larvae of F1 progeny, fixed, and immunostained with anti-β-galactosidase antibody.
To address whether dMi-2 expression in cells represses the PCNA gene promoter activity in living flies, we examined the effects of a half-dose reduction of the dMi-2 gene on the PCNA gene promoter activity by using transgenic flies carrying PCNA (−119 to +137) and lacZ fusion genes (−119PCNAlacZ) (26). It has already been demonstrated that both anti-dDREF and anti-PCNA antibodies stained the nuclei of similar sets of cells containing proliferative neuroblasts in brain lobes (27) and that three copies of the DRE sequence ensure a high level of activity of the PCNA gene minimal promoter (−86 to +137) in this tissue (28). As shown in Fig. 6C, half-dose reduction of functional dMi-2 protein by crossing −119PCNAlacZ male flies with females carrying dMi-2 loss-of-function alleles (dMi-21, dMi-24, and dMi-26) resulted in an increase in lacZ expression (Fig. 6C, panels b through d). In particular, we observed a high level of ectopic expression of lacZ in proliferative neuroblasts, progenitor cells of neurons, as indicated by arrows in the figure. To confirm that the activation of the PCNA promoter by half-dose reduction of the dMi-2 is due to activation of dDREF, we investigated the effects of the dMi-2 gene dose on other PCNA-lacZ in vivo reporters, as illustrated in Fig. 6B. LacZ expression in the brain lobes of transgenic flies carrying the −86P3NlacZ construct with three tandemly aligned 30-bp oligonucleotides containing the DRE sequence upstream of the PCNA gene minimal promoter (−86 to +137) was significantly enhanced by half-dose reduction of the dMi-2 (Fig. 6D, panel b). In contrast, lacZ expression of the transgenic fly carrying −168Δ6PCNAlacZ with a 6-bp deletion in DRE did not change (Fig. 6D, panel d). These results indicate that the DRE sequence is responsible for up-regulation of the PCNA gene promoter by half-dose reduction of the dMi-2, so that dMi-2 can negatively regulate genes under the control of the DRE/dDREF system.
dMi-2 and dDREF bind to polytene chromosomes in a mutually exclusive manner.
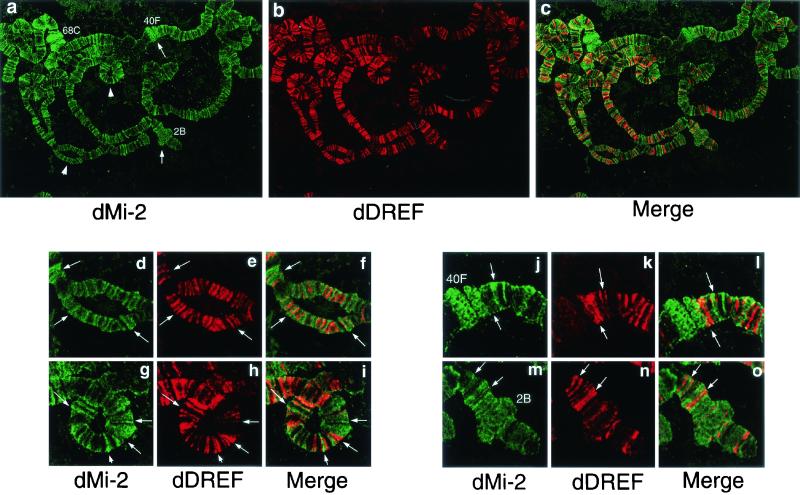
We examined the distribution of dMi-2 and dDREF proteins on salivary gland polytene chromosomes in third-instar larvae by immunofluorescence microscopy. dDREF was found to bind strongly to a number of discrete sites as described earlier (Fig. 7b) (8). We detected several hundred signals for dMi-2 in interband regions and readily recognizable puffs (Fig. 7a), such as 2B on the X chromosome (Fig. 7m) and 68C on the left arm of the third chromosome (Fig. 7j). Neither anti-dDREF antibody nor anti-dMi-2 antibody stained chromocenters known to correspond to the proximal heterochromatin of the X, second, third, and fourth chromosomes. Merging of images for dDREF and dMi-2 revealed no colocalization of the two proteins, although we observed that dMi-2 and dDREF appeared to be in contact with each other at the border in many loci. Enlarged confocal images of the signals showed mutually exclusive binding to the chromosomes (Fig. 7d through i). A similar binding pattern was observed when polytene chromosomes of transgenic flies expressing HA-tagged dMi-2 and Flag-tagged dDREF were double-stained with anti-HA and anti-Flag antibodies (data not shown), excluding the possibility that association of dDREF and dMi-2 hinders epitopes for anti-dDREF and/or anti-dMi-2 antibodies. These data suggest that the chromatin-bound form of dMi-2 polypeptide cannot interact with the chromatin-bound form of dDREF. The molecular mechanism of the exclusive binding property of dMi-2 and dDREF is discussed below, taking into consideration the results indicating that dMi-2 cannot interact with DNA-bound dDREF.
FIG. 7.
Immunostaining of polytene chromosomes with anti-dMi-2 and anti-dDREF antibodies. (a through c) Polytene chromosomes from the third-instar larvae of wild-type flies were spread and double-stained with rabbit anti-dMi-2 antiserum (green) and mouse anti-dDREF monoclonal antibodies (red). Arrows indicate puffs. Arrowheads indicate regions shown as enlarged images in panels d through o. (d through i) Enlarged images of the localization patterns of dMi-2 and dDREF. Note that they bind DNA in a mutually exclusive manner. Typical features of localization of the two proteins are highlighted by arrows. (j through o) Enlarged images of the localization patterns of dMi-2 and dDREF in puffed regions.
DISCUSSION
Promoters of Drosophila genes related to DNA replication contain the DRE sequence (10) in addition to E2F-binding sites. We have isolated and characterized dDREF as a DREF and demonstrated its requirement for DNA replication in both endo and mitotic cell cycles by using transgenic flies (12). We have further speculated that dDREF may play an additional role in regulating gene expression in cooperation with other interacting factors for the following reasons. Firstly, a pattern search (Berkeley Drosophila Genome Project) of the total Drosophila genome sequence with the 5′-TATCGATA DRE sequence as a query hit 620 loci containing a considerable variety of genes in specific chromosome loci, such as subtelomeric heterochromatin repeats flanking naturally occurring regulatory P elements inserted at the X chromosome telomere. Secondly, Hart et al. demonstrated that dDREF can compete for binding to the scs′ region of the hsp70 gene and function as an antagonist of BEAF (8). Thirdly, we found that dDREF genetically interacts with some Polycomb/trithorax-group genes involved in determining chromatin structure or chromatin remodeling (brahma, moira, osa, and Distal-less) (9). To elucidate novel functions of DREF in vivo and interacting proteins, we therefore performed the present yeast two-hybrid screen, which demonstrated a novel interaction between dDREF and dMi-2.
The Mi-2 polypeptide is a member of the SWI/SNF2 family of DNA-stimulated ATPases and has been purified in the form of NuRD/Mi-2 chromatin-remodeling complexes (21, 22, 25, 31). The NuRD/Mi-2 chromatin-remodeling complexes contain seven major polypeptides, including Mi-2, MTA2, MBD3, and histone deacetylases HDAC1/2 and RbAp46/48, and are believed to repress transcription of target genes through their remodeling and deacetylation activities in a targeted manner. Recent work revealed that recruitment of NuRD/Mi-2 complexes to distinct promoter regions is conducted via interactions between Mi-2 complexes and sequence-specific DNA-binding proteins, such as the Drosophila hunchback protein (13) p53 (16) and Ikaros (14), or via recognition of methylated DNA by its MBD3 subunit (23, 32). These findings indicate that association of NuRD/Mi-2 complexes with specific DNA-binding factors might be essential in achieving repression of transcription of the target genes.
In this report, we propose a novel mechanism whereby dMi-2 is involved in repressing transcription of DRE-containing genes by inhibiting the DNA binding of dDREF. The observations point to a first example of a member of the SWI/SNF2 family of DNA-stimulated ATPases directly interacting with a transcription factor to attenuate its activity. Although the present biochemical and genetic analyses clearly indicated direct interaction between dDREF and dMi-2, it is uncertain whether the dMi-2 polypeptide alone or in association with another subunit of the chromatin-remodeling complex, such as HDAC, binds to dDREF in vivo. It is worth noting that treatment of Drosophila cultured cells with trichostatin A, a microbial metabolite generally used as an inhibitor of HAT (30), did not affect the PCNA promoter activity, whereas cotransfection of a dMi-2-expressing plasmid with reporter plasmid significantly decreased PCNA promoter activity depending on the presence of the DRE sequence (Hirose et al., unpublished). This indicates that accompanying histone acetyltransferase activity might not be involved in repression by dMi-2 (or the dMi-2 complex). However, we cannot rule out a requirement for other subunits. Although previous studies on mammalian Mi-2 (Mi-2 β, CHD4) complexes characterized the Mi-2 polypeptide as a major component, biological functions of separate components have not been examined. Importantly, several different strategies resulted in the purification of slightly different Mi-2 complexes. In the case of the original NRD complex, the purification was performed by pursuing HDAC activity by conventional chromatography, followed by affinity chromatography for Mi-2 β (CHD4) (25). With this purification method, the bulk of the tightly associated NRD core complex does not contain sequence-specific DNA-binding protein. Recently, Feng and Zhang purified MeCP1 complexes to homogeneity and demonstrated the presence of the core polypeptide of the known NRD complex (5), indicating that several kinds of complexes, including the Mi-2 polypeptide, might exist in cells. Considering that only 5% of the total dMi-2 polypeptide was estimated to be associated with dDREF by immunoprecipitation experiments, we hypothesize that binding with dDREF in vivo may also be limited. Furthermore, it is interesting to note that the amino-terminal region of dMi-2 exhibits inhibitory effects on its binding to dDREF, suggesting a possible regulation by change in the structure of the molecule. To assess this possibility, a challenge for the future will be the determination of the three-dimensional structure of Mi-2 (or the Mi-2 complex) that binds and modulates dDREF activity. We have established transgenic fly lines expressing HA epitope-tagged dMi-2 and Flag epitope-tagged dDREF by using the GAL4-UAS system. These flies should be powerful tools for the purification of dDREF/dMi-2 complexes.
We here observed that dMi-2 protein is localized at several hundred loci of the polytene chromosomes of salivary glands. Kehle et al. proposed a model of dMi-2 protein function featuring repression of transcription by binding to a Polycomb group protein in the form of a hunchback-dMi-2 complex, with consequent recruitment to DNA (13). However, we observed dMi-2 in interbands and regions associated with high transcriptional activity (puffs), suggesting an ability to enhance as well as to repress gene expression. To address this question, it is important that genes that are positively regulated by dMi-2 be identified.
Another important finding of our immunostaining is that dDREF and dMi-2 bind to polytene chromosomes in a mutually exclusive manner. This seems contrary to the results of immunoprecipitation and in vitro binding experiments but can be explained as follows. Since the DNA-binding domain and the Mi-2-binding domain of DREF overlap, dMi-2 cannot interact with dDREF bound to DNA. On the other hand, dMi-2 presumably has access to free dDREF. If dMi-2 cannot disrupt dDREF/DNA complexes, genes adjacent to dDREF binding sites will be kept in a transcriptionally active state. Furthermore, we demonstrated that overexpression of dDREF or dMi-2 in eye imaginal discs induces a rough-eye phenotype although the eyes of transgenic flies simultaneously expressing dDREF and dMi-2 appear normal. These results suggest that dDREF and dMi-2 negatively regulate each other's functions. To date, although there is no evidence that molecules recruit dMi-2 to specific loci of polytene chromosomes, it can be speculated that dDREF could be involved in the regulation of such dMi-2 recruitment. If this is the case, an important mechanism for the maintenance of epigenetic activation (or silencing) of genes can be envisaged. This idea is not contradictory to the model postulated by Hart et al. (8), in which dDREF contributes to the cancellation of chromatin boundary function by displacing BEAF from its binding sites.
In summary, we here have provided a line of evidence of a novel function of dMi-2 in repressing transcription of DRE-containing genes by attenuating the DRE-binding activity of dDREF. In addition, we hypothesize that dDREF and dMi-2 may demonstrate reciprocal regulation of their functions. To probe this possibility, efforts to isolate dDREF mutant flies and determine the dMi-2 (complex) structure in association with dDREF are necessary in the future.
Acknowledgments
We are grateful to Jurg Muller for his kind gifts of dMi-2 mutant flies and anti-dMi-2 antibody, N. Perrimon for pUAST, and M. Moore for critical reading of the manuscript.
This work was supported in part by grants-in-aid from the Ministry of Education, Science, Sports, Culture, and Technology of Japan.
REFERENCES
- 1.Aubry, F., M.-G. Mattei, and F. Galibert. 1998. Identification of a human 17p-located cDNA encoding a protein of the Snf2-like helicase family. Eur. J. Biochem. 254:558-564. [DOI] [PubMed] [Google Scholar]
- 2.Brand, A. H., and N. Perrimon. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401-415. [DOI] [PubMed] [Google Scholar]
- 3.Brehm, A., G. Langst, J. Kehle, C. R. Clapier, A. Imhof, A. Eberharter, J. Muller, and P. B. Becker. 2000. dMi-2 and ISWI chromatin remodelling factors have distinct nucleosome binding and mobilization properties. EMBO J. 19:4332-4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duronio, R. J., P. H. O'Farrell, J. E. Xie, A. Brook, and N. Dyson. 1995. The transcription factor E2F is required for S phase during Drosophila embryogenesis. Genes Dev. 9:1445-1455. [DOI] [PubMed] [Google Scholar]
- 5.Feng, Q., and Y. Zhang. 2001. The MeCP1 complex represses transcription through preferential binding, remodeling, and deacetylating methylated nucleosome. Genes Dev. 15:827-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ge, Q., D. S. Nilasena, C. A. O'Brien, M. B. Frank, and I. N. Targoff. 1995. Molecular analysis of a major antigenic region of the 240-kD protein of Mi-2 autoantigen. J. Clin. Investig. 96:1730-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hart, C. M., K. Zhao, and U. K. Laemmli. 1997. The scs" boundary element: characterization of boundary element-associated factors. Mol. Cell. Biol. 17:999-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hart, C. M., O. Cuvier, and U. K. Laemmli. 1999. Evidence for an antagonistic relationship between the boundary element-associated factor BEAF and the transcription factor DREF. Chromosoma 108:375-383. [DOI] [PubMed] [Google Scholar]
- 9.Hirose, F., N. Ohshima, M. Shiraki, Y. H. Inoue, O. Taguchi, Y. Nishi, A. Matsukage, and M. Yamaguchi. 2001. Ectopic expression of DREF induces DNA syntheses, apoptosis, and unusual morphogenesis in the Drosophila eye imaginal disc: possible interaction with Polycomb and trithorax group proteins. Mol. Cell. Biol. 21:7231-7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirose, F., M. Yamaguchi, H. Handa, Y. Inomata, and A. Matsukage. 1993. Novel 8-base pair sequence (Drosophila DNA replication-related element) and specific binding factor involved in the expression of Drosophila genes for DNA polymerase α and proliferating cell nuclear antigen. J. Biol. Chem. 268:2092-2099. [PubMed] [Google Scholar]
- 11.Hirose, F., M. Yamaguchi, K. Kuroda, A. Omori, T. Hachiya, M. Ikeda, Y. Nishimoto, and A. Matsukage. 1996. Isolation and characterization of cDNA for DREF, a promoter-activating factor for Drosophila DNA replication-related genes. J. Biol. Chem. 271:3930-3937. [DOI] [PubMed] [Google Scholar]
- 12.Hirose, F., M. Yamaguchi, and A. Matsukage. 1999. Targeted expression of the DNA binding domain of DRE-binding factor, a Drosophila transcription factor, attenuates DNA replication of the salivary gland and eye imaginal disc. Mol. Cell. Biol. 19:6020-6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kehle, J., D. Beuchle, S. Treuheit, B. Christen, J. A. Kennison, M. Bienz, and J. Muller. 1998. dMi-2, a hunchback-interacting protein that functions in polycomb repression. Science 282:1897-1900. [DOI] [PubMed] [Google Scholar]
- 14.Kim, J., S. Sif, B. Jones, A. Jackson, J. Koipally, E. Heller, S. Winandy, A. Viel, A. Sawyer, T. Ikeda, R. Kingston, and K. Georgopoulos. 1999. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity 10:345-355. [DOI] [PubMed] [Google Scholar]
- 15.Kingston, R. E., and G. J. Narlikar. 1999. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13:2339-2352. [DOI] [PubMed] [Google Scholar]
- 16.Luo, J., F. Su, D. Chen, A. Shiloh, and W. Gu. 2000. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature 408:377-381. [DOI] [PubMed] [Google Scholar]
- 17.Robertson, H. M., C. R. Preston, R. W. Philips, D. M. Johnson-Schlitz, W. K. Benz, and W. R. Engels. 1988. A stable genomic source of P-element transposase in Drosophila melanogaster. Genetics 118:461-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sawado, T., F. Hirose, Y. Takahashi, T., Sasaki, T., Shinomiya, K. Sakaguchi, A. Matsukage, and M. Yamaguchi. 1998. The DNA replication-related element (DRE)/DRE-binding factor system is a transcriptional regulator of the Drosophila E2F gene. J. Biol. Chem. 273:26042-26051. [DOI] [PubMed] [Google Scholar]
- 19.Spradling, A. C. 1986. P-element-mediated transformation, p.175-197. In D. B. Roberts (ed.), Drosophila: a practical approach. IRL Press, Oxford, England.
- 20.Takahashi, Y., F. Hirose, A. Matsukage, and M. Yamaguchi. 1999. Identification of three conserved regions in the DREF transcription factors from Drosophila melanogaster and Drosophila virilis. Nucleic Acids Res. 27:510-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tong, J. K., C. A. Hassig, G. R. Schnitzler, R. E. Kingston, and S. L. Schreiber. 1998. Chromatin deacetylation by an ATP-dependent nucleosome remodeling complex. Nature 395:917-921. [DOI] [PubMed] [Google Scholar]
- 22.Wade, P. A., P. L. Jones, D. Vermaak, and A. Wolffe. 1998. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr. Biol. 8:843-846. [DOI] [PubMed] [Google Scholar]
- 23.Wade, P. A., A. Gegonne, P. L. Jones, E. Ballestar, F. Aubry, and A. Wolffe. 1999. Mi-2 complex couples DNA methylation to chromatin remodeling and histone deacetylation. Nature Genet. 23:62-66. [DOI] [PubMed] [Google Scholar]
- 24.Woodage, T., M. Basrai, A. D. Baxevanis, P. Hieter, and F. S. Collins. 1997. Characterization of the CHD family of proteins. Proc. Natl. Acad. Sci. USA 94:11472-11477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xue, Y., J. Wong, G. T. Moreno, M. K. Young, J. Cote, and W. Wang. 1998. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell. 2:851-861. [DOI] [PubMed] [Google Scholar]
- 26.Yamaguchi, M., Y. Hayashi, Y. Nishimoto, F. Hirose, and A. Matsukage. 1995. A nucleotide sequence essential for the function of DRE, a common promoter element for Drosophila DNA replication-related genes. J. Biol. Chem. 270:15808-15814. [DOI] [PubMed] [Google Scholar]
- 27.Yamaguchi, M., F. Hirose, Y. Nishimoto, T. Naruge, M. Ikeda, T. Hachiya, K. Tamai, K. Kuroda, and A. Matsukage. 1995. Expression patterns of DNA replication enzymes and the regulatory factor DREF during Drosophila development analyzed with specific antibodies. Biol. Cell 85:147-155. [DOI] [PubMed] [Google Scholar]
- 28.Yamaguchi, M., F. Hirose, and A. Matsukage. 1996. Roles of multiple promoter elements of the proliferating cell nuclear antigen gene during Drosophila development. Genes Cells 1:47-58. [DOI] [PubMed] [Google Scholar]
- 29.Yamaguchi, M., Y. Hayashi, and A. Matsukage. 1995. Essential role of E2F recognition sites in regulation of the proliferating cell nuclear antigen gene promoter during Drosophila development. J. Biol. Chem. 270:25159-25165. [DOI] [PubMed] [Google Scholar]
- 30.Yoshida, M., S. Horinouchi, and T. Beppu. 1995. Trichostatin A and trapoxin: novel chemical probes for the role of histone acetylation in chromatin structure and function. Bioessays 17:423-430. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, Y., G. LeRoy, H. P. Seelig, W. S. Lane, and D. Reinberg. 1998. The dermatomyositis-specific autoantigen Mi-2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95:279-289. [DOI] [PubMed] [Google Scholar]
- 32.Zhang, Y., H.-H. Ng, H. Erdjument-Bromage, P. Tempst, A. Bird, and D. Reinberg. 1999. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 13:1924-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zink, B., Y. Engstrom, W. J. Gehring, and R. Paro. 1991. Direct interaction of the Polycomb protein with Antennapedia regulatory sequences in polytene chromosomes of Drosophila melanogaster. EMBO J. 10:153-162. [DOI] [PMC free article] [PubMed] [Google Scholar]