Abstract
The mammalian ING1 gene encodes a tumor suppressor required for the function of p53. In this study we report a novel function for YNG1, a yeast homolog of ING1. Yng1p is a stable component of the NuA3 histone acetyltransferase complex, which contains Sas3p, the yeast homolog of the mammalian MOZ proto-oncogene product, as its catalytic subunit. Yng1p is required for NuA3 function in vivo but surprisingly is not required for the integrity of the complex. Instead, we find that Yng1p mediates the interaction of Sas3p with nucleosomes and is thus required for the ability of NuA3 to modify histone tails. These data, and the observations that other ING1 homologs are found in additional yeast complexes that posttranslationally modify histones, suggest that members of the ING1 class of proteins may have broad roles in enhancing or modifying the activities of chromatin-modifying complexes, thereby regulating their activities in transcription control.
Eukaryotic DNA is packaged into a nucleoprotein structure known as chromatin, consisting of DNA, histones, and nonhistone proteins. This structure is able to modulate access of the cellular machinery to DNA, thus regulating processes like transcription, replication, repair, and recombination. Numerous multiprotein complexes capable of modifying chromatin exist within a cell. The most studied of these complexes include the ATP-dependent chromatin remodeling complexes, which use the energy of ATP hydrolysis to alter the association of DNA with histones, and the histone modifying complexes, which posttranslationally modify histones.
The best-characterized class of histone-modifying complexes is the histone acetyltransferases (HATs), which use acetyl-coenzyme A as a substrate to acetylate lysine residues within the amino-terminal tails of histones. In the yeast Saccharomyces cerevisiae these include the GCN5-dependent SAGA, ADA, and HAT-A2 complexes, the SAS3-dependent NuA3 complex, the ESA1-dependent NuA4 complex, the ELP3-dependent Elongator complex, and the Nut1p-containing Mediator complex (28). These complexes differ substantially with regard to their components, substrate specificity, and suspected functions in the cell.
Interestingly, the substrate specificity of many HAT complexes is dictated not by the catalytic subunit but rather by the additional protein components within the complex. For example, recombinant Esa1p, Gcn5p, and Sas3p can acetylate both histones H3 and H4 when assayed on free histones, but they lack the ability to acetylate nucleosomal histones (10, 16, 26, 29). When incorporated into complexes, these proteins gain the ability to acetylate nucleosomes and become limited to primarily acetylating histone H3 in the case of Gcn5p and Sas3p or H4 for Esa1p (2, 10, 15). How the noncatalytic proteins within HAT complexes serve to alter the substrate specificity of the catalytic subunit is the focus of much research.
The SAS3-dependent NuA3 complex is approximately 450 kDa and consists of at least five subunits (15). In addition to Sas3p, the only known subunit of the complex is TAF30. Loss of TAF30 has no effect on the complex in terms of structure or substrate specificity, and thus the function of this protein within the complex is not known (15). We wished to identify novel subunits within the NuA3 complex, and therefore we purified the native NuA3 complex from yeast and subjected it to mass spectrometry analysis. By using this approach we identified Yng1p as a novel component of this complex. YNG1 is a homolog of the mammalian ING1 (inhibitor of growth 1) gene. Other yeast homologs of ING1 include Yng2p and Pho23p, which are components of the NuA4 HAT complex, and an Rpd3p histone deacetylase complex, respectively (18-20). ING1 encodes a tumor suppressor that physically interacts with p53 and is necessary for p53-dependent transcription activation in vivo (9). We were unable to detect a similar role for NuA3 in this regard, but instead showed that Yng1p is specifically required for the ability of the NuA3 complex to bind to and acetylate histones. These results may shed light on the function of ING1 and other homologs in mediating transcriptional activation by p53.
MATERIALS AND METHODS
Yeast strains, plasmids, and genetic methods.
Strains used in this study are listed in Table 1 and were either published previously or were created for this study by using standard yeast manipulations (3). Mating assays were performed as described by others (27). Plasmids pJW217, pJW218, and pJW219 contain the ANC1, GCN5, and YNG1 open reading frames (ORFs), respectively, and also contain ∼1,000 bp of upstream sequences, cloned into the URA3 centromeric vector, YCplac33, fused to a FLAG-CYC cassette (15). Plasmids pJW220, pJW221, and pJW222 contain the SAS3, YNG1, and YNG1ΔPHD ORFs, respectively, and also contain ∼1,000 bp of upstream sequences, cloned into the CEN/ARS TRP1 vector pRS314 fused to a triple hemagglutinin (HA)-CYC cassette (8). Plasmids pJW223 and pJW224 are CEN/ARS TRP1 plasmids bearing amino-terminal HA-tagged, full-length, and carboxyl terminally truncated SAS3 under the control of a GAL promoter (15). The plasmids pSas3FLG, pJW214, and pJW215 have been described previously (13, 15).
TABLE 1.
Strains used in this study
| Strain | Genotype |
|---|---|
| JRY2069 | MATα HMRa-e** ade2-101 his3 lys2 tyr1 ura3-52 |
| JRY2726 | MATahis4 |
| YJW120 | MATaade2-1 his3-11 leu2-3,112 trp1-1 ura3-1 can1-100 sas3::URA3 pJW224 |
| YJW134 | MATahis3Δ200 leu2Δr1 ura3-52 trp1Δ63 gcn5::HIS3 sas3::HIS3MX6 pJW214 |
| YJW207 | MATaade2-1 his3-11 leu2-3,112 trp1-1 ura3-1 can1-100 ESA1-3HA::HISMX6 |
| YJW534 | MATaade2-1 his3-11 leu2-3,112 trp1-1 ura3-1 can1-100 YCplac33 |
| YJW535 | MATaade2-1 his3-11 leu2-3,112 trp1-1 ura3-1 can1-100 pJW219 |
| YJW536 | MATaade2-1 his3-11 leu2-3,112 trp1-1 ura3-1 can1-100 sas3::HISMX6 pJW219 |
| YJW537 | MATahis3Δ200 leu2Δr1 lys2-128δ ura3-52 trp1Δ63 sas3::HISMX6 pJW220 |
| YJW538 | MATahis3Δ200 leu2Δr1 lys2-128δ ura3-52 trp1Δ63 sas3::HISMX6 pJW219 pJW220 |
| YJW539 | MATahis3Δ200 leu2Δr1 lys2-128δ ura3-52 trp1Δ63 yng1::HISMX6 pJW221 |
| YJW540 | MATahis3Δ200 leu2Δr1 lys2-128δ ura3-52 trp1Δ63 sas3::HISMX6 pJW217 pJW220 |
| YJW541 | MATahis3Δ200 leu2Δr1 lys2-128δ ura3-52 trp1Δ63 sas3::HISMX6 yng1::HISMX6 pJW217 pJW220 |
| YJW542 | MATahis3Δ200 leu2Δr1 lys2-128δ ura3-52 trp1Δ63 gcn5::HIS3 yng1::HISMX6 pJW218 |
| YJW543 | MATahis3Δ200 leu2Δr1 lys2-128δ ura3-52 trp1Δ63 yng1::HISMX6 pJW217 pJW221 |
| YJW544 | MATahis3Δ200 leu2Δr1 lys2-128δ ura3-52 trp1Δ63 sas3::HISMX6 yng1::HISMX6 pJW217 pJW221 |
| YJW545 | MATahis3Δ200 leu2Δr1 lys2-128δ ura3-52 trp1Δ63 sas3::HISMX6 pSas3FLG |
| YJW546 | MATahis3Δ200 leu2Δr1 lys2-128δ ura3-52 trp1Δ63 sas3::HISMX6 yng1::HISMX6 pSas3FLG |
| YJW547 | MATα HMRa-e** ade2-101 his3 lys2 tyr1 ura3-52 sas3::HISMX6 |
| YJW548 | MATα HMRa-e** ade2-101 his3 lys2 tyr1 ura3-52 yng1::HISMX6 |
| YJW549 | MATahis3Δ200 leu2Δr1 lys2-128δ ura3-52 trp1Δ63 sas3::HISMX6 pJW223 |
| YJW550 | MATahis3Δ200 leu2Δr1 lys2-128δ ura3-52 trp1Δ63 sas3::HISMX6 yng1::HISMX6 pJW223 |
Purification of the NuA3 HAT complex.
The NuA3 HAT complex was purified from yeast strains YJW545 and YJW546 by Ni2+-nitrilotriacetic acid (NTA) agarose (Qiagen, Valencia, Calif.) and MonoQ ion-exchange chromatography as previously described (7). Fractions containing the NuA3 complex were pooled and incubated with αFLAG M2 resin (100 μl of resin for 12 liters of starting culture; Sigma, St. Louis, Mo.) overnight at 4°C with rotation. The beads were washed three times with 10 bed volumes of a solution containing 50 mM Tris (pH 8.0), 350 mM NaCl, 10% glycerol, 0.1% Tween 20, 1 mM phenylmethylsulfonyl (PMSF), 2 μg of leupeptin/ml, and 2 μg of pepstatin/ml. The complex was eluted from the beads by washing with 1 bed volume of a solution containing 50 mM Tris (pH 8.0), 350 mM NaCl, 10% glycerol, 0.1% Tween 20, and 0.1 mg of FLAG peptide/ml (Sigma). The complex was trichloroacetic acid precipitated, and the proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and subjected to mass spectrometry as described previously (11).
Preparation of yeast whole-cell extracts, immunoprecipitations, and glutathione S-transferase (GST) pull-down assays.
For immunoprecipitations of FLAG epitopes, yeast whole-cell extracts were prepared in a solution containing 50 mM Tris (pH 8.0), 350 mM NaCl, 10% glycerol, 0.1% Tween 20, 1 mM PMSF, 2 μg of leupeptin/ml, and 2 μg of pepstatin/ml. Approximately 5 mg of extract was rotated at 4°C overnight with 10 μl of washed, packed αFLAG M2 resin (Sigma). The beads were washed three times with a solution containing 50 mM Tris (pH 8.0), 350 mM NaCl, 10% glycerol, 0.1% Tween 20, 1 mM PMSF, 2 μg of leupeptin/ml, and 2 μg of pepstatin/ml. The proteins were either eluted from the beads by boiling in SDS-PAGE loading buffer or were assayed for HAT activity as described previously (7). Immunoprecipitation of Spt16p was performed as described previously (15).
For GST pull-down assays, yeast whole-cell extracts and GST fusion proteins were prepared as described previously (23, 30). Approximately 2 mg of extract was rotated at 4°C for 2 h with 10 μl of washed, packed beads. The beads were washed three times with 50 volumes of a solution containing 50 mM HEPES (pH 7.4), 150 mM NaCl, 16 mM magnesium acetate, 1 mM EGTA, 0.1% Nonidet P-40, 0.5 mM dithiothreitol, 10% glycerol, 1 mM PMSF, 2 μg of leupeptin/ml, and 2 μg of pepstatin/ml prior to boiling in SDS sample buffer.
Size exclusion chromatography.
Yeast whole-cell extracts were prepared in a solution containing 50 mM Tris (pH 8.0), 500 mM NaCl, 10% glycerol, 0.1% Tween 20, 1 mM PMSF, 2 μg of leupeptin/ml, and 2 μg of pepstatin/ml. Five milligrams of extract was resolved on a Superose 6 HR 10/30 column (Pharmacia, Piscataway, N.J.) equilibrated with the same buffer.
Immobilized template pull-down assay.
Immobilized oligonucleosome templates were prepared from the G5E4-5S fragment as previously described (12). Approximately 200 ng of array was incubated in 100 μl of a solution containing 10 mM HEPES (pH 7.8), 150 ml NaCl, 5 mM dithiothreitol, 0.5 mM PMSF, 5% glycerol, and 0.25 mg of bovine serum albumin/ml, with 40 μl of NuA3 purified by Ni2+-NTA agarose (Qiagen) and MonoQ ion-exchange chromatography. The samples were rotated at 37°C for 1 h, and the beads were washed two times with 200 μl of a solution containing 10 mM HEPES (pH 7.8), 150 ml NaCl, 5 mM dithiothreitol, 0.5 mM PMSF, 5% glycerol, and 0.25 mg of bovine serum albumin/ml. The beads were boiled in SDS-PAGE sample buffer, and the bound proteins were analyzed by Western blotting.
RESULTS
Yng1p is a component of the NuA3 complex.
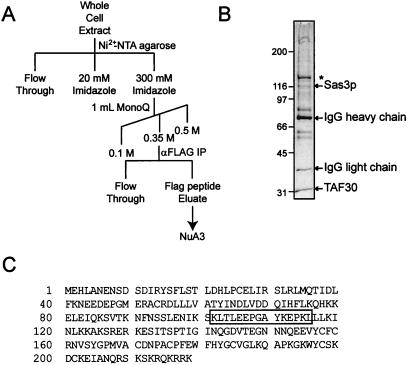
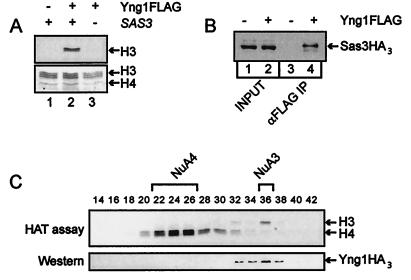
To understand the function of the SAS3-dependent S. cerevisiae NuA3 HAT complex, we attempted to identify novel protein subunits within this complex. To this end, we purified NuA3 from a FLAG epitope-tagged Sas3p strain (15) by using the scheme shown in Fig. 1A and subjected the resulting proteins to mass spectrophotometer analysis (Fig. 1B). The band corresponding to TAF30 was used as a positive control, but surprisingly this sample generated a peptide encoded by the YNG1/YOR064C ORF (Fig. 1C). Immunoprecipitation of Yng1p from whole-cell extracts has been previously shown to precipitate histone H3-specific HAT activity (18), thus making this protein a likely candidate for a subunit of NuA3. To confirm this, the YNG1 ORF and its endogenous promoter were fused to a carboxyl-terminal FLAG epitope tag and were cloned into an ARS/CEN plasmid. The resulting plasmid was introduced into a wild-type yeast strain, and whole-cell extracts from the resulting strain were subjected to immunoprecipitation with an αFLAG antibody. As shown previously, histone H3-specific HAT activity was coimmunoprecipitated from a tagged Yng1p strain but not from an untagged strain (Fig. 2A, compare lanes 1 and 2). However, when the same experiment was performed with a sas3Δ strain, Yng1p failed to coimmunoprecipitate HAT activity, suggesting that the acetyltransferase activity associated with Yng1p is that of the NuA3 complex (Fig. 2A, compare lanes 2 and 3).
FIG. 1.
Yng1p is associated with purified NuA3 HAT complex. (A) Scheme used to generate purified NuA3. (B) Silver stained, SDS-PAGE (4 to 15% gradient) of the purified NuA3 complex. Bands corresponding to the verified subunits, Sas3p and TAF30, are indicated. An asterisk indicates a contaminating band. (C) Complete amino acid sequence for Yng1p. The peptide sequence obtained by mass spectrometry from purified NuA3 is shown boxed. IP, immunoprecipitation.
FIG. 2.
Yng1p is a stable component of the NuA3 HAT complex. (A and B) Whole-cell extracts from the indicated strains were immunoprecipitated (IP) with αFLAG resin and either were assayed for HAT activity on free HeLa histones (panel A; the fluorogram and Coomassie brilliant blue stain of an SDS-PAGE of these histones are shown) or were Western blotted and probed for Sas3HA3 (B). (C) Whole-cell extracts from a strain expressing HA-tagged Yng1p was purified by Ni2+-NTA agarose and MonoQ ion-exchange chromatography. The MonoQ fractions were assayed for HAT activity on free histones by SDS-PAGE and fluorography and for the presence of Yng1HA3 by Western blotting.
To provide further support for an interaction between Yng1p and Sas3p, the Yng1-FLAG tag construct was introduced into cells expressing triple HA-tagged Sas3p. Figure 2B shows that an αFLAG antibody coimmunoprecipitated Sas3p from a FLAG-tagged Yng1p strain but not from an untagged strain. We also show that, when fused with a triple HA tag, Yng1p copurified with NuA3 HAT activity by our standard purification technique, demonstrating that Yng1p is a stable component of the NuA3 complex (Fig. 2C).
Loss of YNG1 is not disruptive to the NuA3 HAT complex.
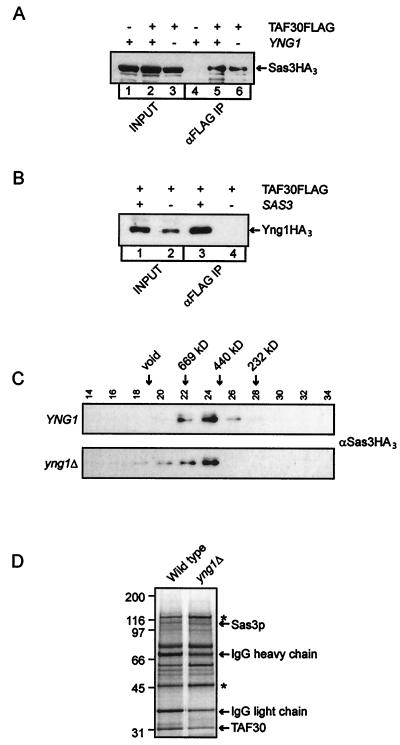
To understand the function of Yng1p within the NuA3 complex, we first asked whether loss of this protein results in disruption of the NuA3 complex. Currently the known subunits of NuA3 are Sas3p and TAF30. To determine whether these two proteins still exist in a complex in yng1Δ cells, we introduced FLAG-tagged TAF30 and triple HA-tagged Sas3p expressing plasmids into wild-type and yng1Δ strains. Whole-cell extracts were prepared from these strains and were subjected to immunoprecipitation for the FLAG epitope. As shown previously, immunoprecipitation of TAF30 coprecipitated Sas3p (Fig. 3A, compare lanes 4 and 5) (15), but loss of YNG1 did not affect this interaction (Fig. 3A, compare lanes 5 and 6). In a reciprocal experiment, deletion of SAS3 resulted in a failure to coimmunoprecipitate Yng1HA3 with TAF30FLAG (Fig. 3B, compare lanes 3 and 4), demonstrating that, unlike Sas3p, Yng1p is not required for the integrity of the NuA3 complex. To provide further support that NuA3 is still intact in yng1Δ cells, whole-cell extracts from Sas3HA3-expressing wild-type and yng1Δ cells were subjected to Superose 6 size exclusion chromatography. NuA3 purified from wild-type cells elutes in the 400- to 500-kDa size range from this column (Fig. 3C) (15). Loss of YNG1 does not alter this elution to a significant extent, suggesting that although Yng1p is an integral component of NuA3, a stable NuA3 complex still exists in cells lacking Yng1p. As a final confirmation, we purified NuA3 from both wild-type and yng1Δ cells by using the scheme depicted in Fig. 1A. The proteins of the resulting complexes were separated by SDS-PAGE and were visualized by silver staining. As is shown in Fig. 3D, all protein bands originating from wild-type NuA3 are also present in NuA3 isolated from the yng1Δ strain, further demonstrating that a stable NuA3 complex can exist in the absence of Yng1p.
FIG. 3.
Loss of YNG1 does not disrupt the NuA3 HAT complex. (A and B) Whole-cell extracts from the indicated strains were immunoprecipitated with αFLAG resin. The input whole-cell extracts and immunoprecipitates (IP) were Western blotted and probed for HA. (C) Whole-cell extracts from the indicated strains were resolved on a Superose 6 HR 10/30 column, and the fractions were assayed for the presence of Sas3HA3 by Western blotting. (D) Silver stained, SDS-PAGE (4% to 15% gradient) of NuA3 complexes purified from wild-type (YJW545) and yng1Δ (YJW546) strains. Bands corresponding to the verified subunits Sas3p and TAF30 are indicated. Contaminating bands are indicated by asterisks. IgG, immunoglobulin G.
Loss of YNG1 disrupts NuA3 function.
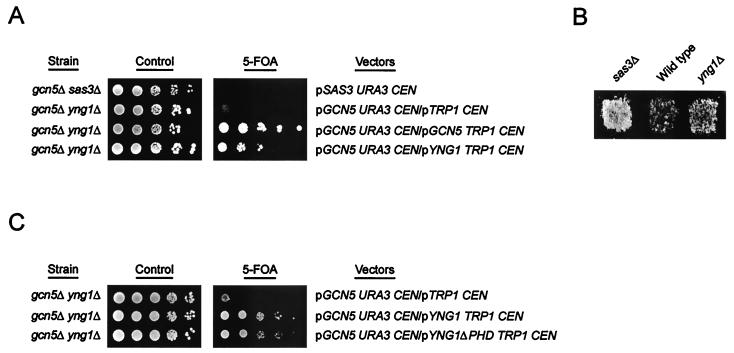
The loss of SAS3 results in only very minor phenotypes (22), suggesting that the NuA3 complex is not essential for normal cell growth. However, simultaneous loss of SAS3 and GCN5, the catalytic subunits of numerous HAT complexes, is synthetically lethal, suggesting that NuA3 has overlapping functions with other HAT complexes that are essential for viability (13). To determine whether YNG1 is required for the function of the NuA3 complex, we asked whether gcn5Δ yng1Δ strains are viable. To this end, a diploid strain with single deletions of GCN5 and YNG1 was transformed with a URA3-based plasmid carrying wild-type GCN5. The transformed strain was sporulated and dissected, and haploid spore products with disruptions of both GCN5 and YNG1 but carrying the GCN5 plasmid were identified. These double mutant strains grew extremely poorly when subjected to negative selection for the URA3-based plasmid on medium containing 5-fluoroorotic acid (5-FOA) (Fig. 4A). However, if simultaneously transformed with non-URA3-based plasmids carrying wild-type copies of GCN5 or YNG1, the double mutants grew well on 5-FOA. These results demonstrate that although gcn5Δ yng1Δ strains are viable, they are severely compromised for growth.
FIG. 4.
Loss of YNG1 disrupts NuA3 function. (A) gcn5Δ yng1Δ strains are synthetically sick. Yeast strain YJW542 (MATa his3Δ200 leu2Δ1 ura3-52 trp1Δ63 gcn5::HIS3 yng1::HISMX6 pJW218) was transformed with a CEN/ARS TRP1 plasmid alone (pRS314) or with CEN/ARS TRP plasmids encoding Gcn5p or Yng1p (pJW215 and pJW221, respectively), and the resulting transformants were plated on either synthetic complete medium (control) or synthetic complete medium with 5-FOA. Strain YJW134 (MATa his3Δ200 leu2Δ1 ura3-52 trp1Δ63 gcn5::HIS3 sas3::HISMX6 pJW214) was used as a control. (B) Deletion of YNG1 restores silencing to strains with mutations in HMR-E. Wild-type (JRY2069; MATα HMRa-e∗∗ ade2-101 his3 lys2 tyr1 ura3-52) and yng1Δ (YJW548; MATα HMRa-e∗∗ ade2-101 his3 lys2 tyr1 ura3-52 yng1::HISMX6) strains were assayed for mating efficiency, with JRY2726 (MATa his4) as a tester strain. Strain YJW547 (MATα HMRa-e∗∗ ade2-101 his3 lys2 tyr1 ura3-52 sas3::HISMX6) was used as a positive control. (C) Yeast strains YJW542 (MATa his3Δ200 leu2Δ1 ura3-52 trp1Δ63 gcn5::HIS3 yng1::HISMX6 pJW218) was transformed with a CEN/ARS TRP1 plasmid alone (pRS314) or with CEN/ARS TRP1 plasmids encoding Yng1p or Yng1ΔPHD (pJW221 and pJW222, respectively), and the resulting transformants were plated on either synthetic complete medium (control) or synthetic complete medium with 5-FOA.
An interesting characteristic of the gcn5Δ sasΔ synthetic lethality is that it is not due to loss of the Gcn5p-dependent HAT complexes, since loss of SAS3 is compatible with deletions of ADA2 or ADA3, mutations which disrupt all known Gcn5p-dependent HAT complexes (10, 13, 24). We find a similar pattern in yng1Δ strains; although gcn5Δ yng1Δ strains are extremely sick, the ada2Δ yng1Δ strains are not (data not shown). In addition, we find that deletion of YNG1 results in other sas3Δ-specific phenotypes, including the rescue of silencing defects of an HMR locus with mutations in the Rap1p and Abf1p binding sites (Fig. 4B). The fact that deletion of YNG1 recapitulates loss of SAS3 suggests that Yng1p is required for the function of the NuA3 complex.
Members of the ING1 family, including the yeast proteins Yng1p, Yng2p, and Pho23p, all contain a PHD finger domain within their C-terminal region (18). This zinc finger-like motif occurs in various proteins thought to be involved in chromatin-mediated gene regulation (1). Despite the intriguing possibility that this motif mediates the interaction of NuA3 with chromatin, we find that Yng1p lacking the PHD domain is still able to rescue the gcn5Δ yng1Δ growth defect (Fig. 4C), suggesting that this region of the protein is not required for the function of the NuA3 complex. This is consistent with work by others showing that Yng2p lacking the PHD finger is still able to rescue growth defects of yng2Δ strains as well as NuA4 HAT activity (20).
Yng1p is required for NuA3 HAT activity.
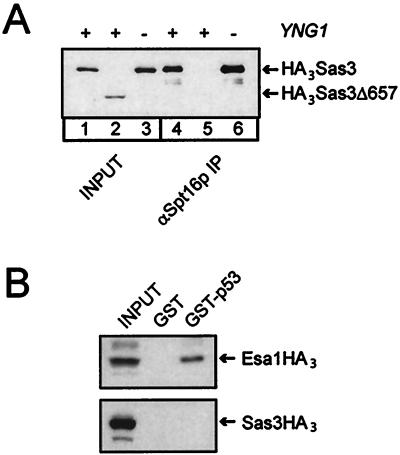
YNG1 is required for the function, but not the integrity, of the NuA3 complex, suggesting that this protein is necessary for either the proper targeting or the catalytic activity of this complex. It has been previously shown that the NuA3 complex can interact with Spt16p both in vivo and in vitro (15). Spt16p can also interact directly with histones, which may provide a mechanism for targeting this HAT complex to its substrate (21). However, we find that we are still able to coimmunoprecipitate Sas3p with Spt16p in the absence of Yng1p, suggesting that Yng1p does not play a role in this interaction (Fig. 5A, compare lanes 4 and 6). A strain expressing HA3Sas3Δ657, which lacks the region of Sas3p required for interaction of Sas3p with Spt16p (15), was included as a negative control.
FIG. 5.
Yng1p does not mediate interactions of Spt16p or p53 with NuA3. (A) Spt16p was immunoprecipitated (IP) from yeast whole-cell extracts from YNG1 (YJW549) and yng1Δ (YJW550) strains expressing an amino-terminal-tagged Sas3p. A YNG1 strain expressing a carboxyl-terminal-truncated version of HA3Sas3p (YJW120) was used as a negative control. The input whole-cell extracts and the bead fractions were assayed for the presence of HA3Sas3p by Western blotting. (B) Yeast whole-cells extracts from strains expressing HA-tagged Sas3p (YJW537) or Esa1p (YJW207) were incubated with either GST or GST-p53 fusion proteins bound to glutathione Sepharose beads. The input whole-cell extracts and the bead fractions were assayed for the presence of Esa1HA3 and Sas3HA3 by Western blotting.
A mammalian homolog of Yng1p, the candidate tumor suppressor ING1, can coimmunoprecipitate with p53 and modulates p53-dependent transcriptional activation (9). Moreover, the yeast NuA4 HAT complex, which contains the ING1 homolog Yng2p, has also been shown to interact with p53 and is required for p53-dependent transcription activation in yeast (20). Although NuA4 lacking Yng2p is still able to interact with p53, the strength of the interaction is diminished compared to that of NuA4 from wild-type strains. These data suggest that conserved motifs within the ING1 and YNG2 gene products may serve as important interaction partners for p53. To test this hypothesis, we asked whether the Yng1p-containing NuA3 complex was also capable of interacting with p53. Yeast whole-cell extracts from cells expressing triple HA-tagged Sas3p were incubated with full-length p53 immobilized on Sepharose beads. We used triple HA-tagged Esa1p, the catalytic subunit of the NuA4 complex, as a positive control. Although we were able to reproduce the interaction between the NuA4 complex and p53 (Fig. 5B), we found that the NuA3 complex was unable to interact with p53, suggesting that the presence of an ING1 homolog alone is not sufficient to mediate interaction with p53. This result, however, must be interpreted with some caution, since it is possible that the recombinant p53 is lacking some modification required for the interaction between p53 and the ING1 family proteins.
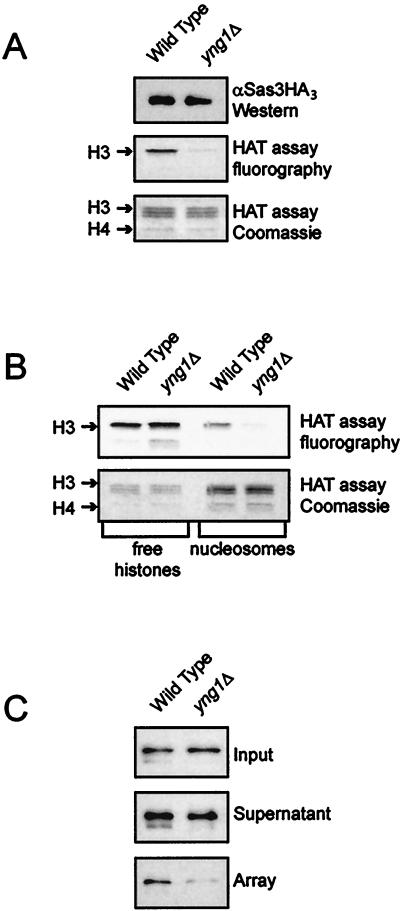
We next asked whether Yng1p is required for the catalytic activity of the NuA3 complex. NuA3 affinity purified by using FLAG-tagged TAF30 from both wild-type and yng1Δ strains was subjected to HAT assays with free histones as a substrate. The reactions were normalized for levels of Sas3p. Figure 6A shows that loss of Yng1p severely compromises Sas3p HAT activity. When the assays were normalized for levels of HAT activity on free histones, we found that lack of Yng1p has an even greater effect on nucleosomal HAT activity, since in the absence of Yng1p NuA3 is no longer capable of acetylating nucleosomes (Fig. 6B).
FIG. 6.
Yng1p is required for NuA3 HAT activity. (A) Wild-type (YJW540) and yng1Δ (YJW541) TAF30 affinity-purified NuA3, normalized for levels of Sas3HA3 by Western blotting, were assayed for HAT activity on free HeLa histones. The fluorogram and Coomassie brilliant blue stain of an SDS-PAGE of these histones are shown. (B) NuA3 purified from strains YJW540 and YJW541, normalized for levels of HAT activity on free histones, was assayed for HAT activity on HeLa nucleosomes. (C) Yng1p mediates interaction of NuA3 with nucleosomal arrays. Biotinylated G5E4-5S nucleosome arrays were bound to paramagnetic beads (Dynabeads) coupled to streptavidin. NuA3 purified from either wild-type or yng1Δ strains was added to the array template where indicated. The template beads were concentrated with a magnet, and the presence of Sas3p in the input, supernatants, and beads was analyzed by Western blotting.
One possible explanation for the loss of NuA3 HAT activity in yng1Δ strains is that Yng1p mediates the interaction of Sas3p with histones. To test this hypothesis, we reconstituted an array of nucleosomes bound to paramagnetic beads. The immobilized array was incubated with partially purified NuA3 isolated from either wild-type or yng1Δ strains. The beads were washed and the proteins that remained bound to the arrays were eluted in SDS-PAGE sample buffer. Western blot analysis revealed that in the absence of Yng1p, the ability of Sas3p to interact with nucleosomes is severely decreased (Fig. 6C). These results clearly demonstrate that a biological role of Yng1p is to facilitate interaction between the NuA3 complex and chromatin.
DISCUSSION
A valuable technique for deciphering the function of multiprotein complexes in yeast is to identify the protein subunits of these complexes and the phenotypes associated with their loss. In this study we have demonstrated that Yng1p, a homolog of the human candidate tumor suppressor ING1, is a stable component of the NuA3 HAT complex and is required for the interaction and acetylation of histones by NuA3.
The requirement of Yng1p for Sas3p HAT activity is reminiscent of the relationship between Ada2p and Ada3p with Gcn5p. Bacterially expressed Gcn5p can acetylate histones well, but it exhibits poor nucleosomal HAT activity (10, 17). However, when incorporated into a complex with Ada2p and Ada3p, Gcn5p acetylates nucleosomes well (4, 24). Thus, one of the functions of the additional proteins within the ADA and SAGA complexes is to allow Gcn5p to access nucleosomal histones. Our results suggest that Yng1p might serve a similar function in NuA3 by enhancing the access of Sas3p to histone tails in both free and nucleosomal histones. The ability of an ING1 homolog to facilitate the use of histones as substrates may not be limited to the NuA3 HAT complex. Two other yeast homologs of ING1, Yng2p and Pho23p, have also been found to be associated with HAT activity (18, 20). Moreover, Pho23p and the ING1 gene product, p33ING1b, are found functionally associated with histone deacetylase complexes (19, 25). Thus, these proteins may serve general roles in permitting various enzymatic activities to modify histone tails.
A homolog of Yng1p, ING1/p33, has been shown to interact with the tumor suppressor p53 by coimmunoprecipitation (9). ING1 is also important for p53 function in vivo (9). In contrast, despite a high degree of conservation with ING1, Yng1p is not able to interact with p53 in the context of NuA3. These data may suggest that although ING1 and YNG1 share common ancestors, their functions have diverged. An alternative hypothesis is that other proteins mediate the interaction of ING1 with p53 and that ING1 has an important role in transcriptional activation by p53, which is distinct from directly interacting with this protein.
A clue to the answer to this question comes from work with the Yng2p-containing histone H4-specific HAT complex, NuA4. This complex has also been shown to interact with p53 and to mediate p53-dependent transcription in vivo (20). However, this interaction is not strictly dependent on Yng2p, as NuA4 isolated from yng2Δ cells is still capable of some interaction with p53. Furthermore, NuA4 contains the protein Tra1p, which has been shown to interact with multiple acidic trans-activation domains and thus is a likely candidate for mediating the interaction of NuA4 with p53 (5). In light of this data, it is conceivable that the interaction of ING1 with p53 in mammalian cells is mediated by its incorporation into a complex homologous to NuA4, such as the Tip60 complex (14). If ING1 is not directly required for recruitment of the Tip60 complex by p53, then it must be required for some other function of this complex. Considering the data presented in this manuscript, one possibility is to potentiate the HAT activity of the recruited complex. This hypothesis is also supported by experiments with NuA4, as it was found that Yng2p is required for both the abundance and the HAT activity of the NuA4 complex in yeast (6, 20). Thus, although ING1/Yng2p has important functions in mediating p53-dependent transcription, this may only be in the context of the complex associated with these proteins. Taken together, the ING1/Yng2p/Yng1p proteins may have a broader role in enhancing or modulating the activity of their respective HAT complexes.
Acknowledgments
Support for this work was provided by grant GM47867 from the National Institute of General Medical Sciences to J.L.W and by the NCRR Yeast Center grant RR11823. L.H. is supported by a postdoctoral fellowship from the Canadian Institutes of Health Research. T.K. is a postdoctoral associate of the Howard Hughes Medical Institute. J.L.W. is an associate investigator of the Howard Hughes Medical Institute.
We gratefully acknowledge the valuable comments provided by members of the Workman, Côté, Simpson, Tan, and Reese laboratories. We are also grateful to J. Côté, J. Rine, and T. Formosa for providing strains and reagents.
REFERENCES
- 1.Aasland, R., T. J. Gibson, and A. F. Stewart. 1995. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 20:56-59. [DOI] [PubMed] [Google Scholar]
- 2.Allard, S., R. T. Utley, J. Savard, A. Clarke, P. Grant, C. J. Brandl, L. Pillus, J. L. Workman, and J. Cote. 1999. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 18:5108-5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M. 1987. Current protocols in molecular biology. J. Wiley, New York, N.Y.
- 4.Balasubramanian, R., M. G. Pray-Grant, W. Selleck, P. A. Grant, and S. Tan. 2002. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J. Biol. Chem. 277:7989-7995. [DOI] [PubMed] [Google Scholar]
- 5.Brown, C. E., L. Howe, K. Sousa, S. C. Alley, M. J. Carrozza, S. Tan, and J. L. Workman. 2001. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 292:2333-2337. [DOI] [PubMed] [Google Scholar]
- 6.Choy, J. S., B. T. Tobe, J. H. Huh, and S. J. Kron. 2001. Yng2p-dependent NuA4 Histone H4 acetylation activity is required for mitotic and meiotic progression. J. Biol. Chem. 276:43653-43662. [DOI] [PubMed] [Google Scholar]
- 7.Eberharter, A., S. John, P. A. Grant, R. T. Utley, and J. L. Workman. 1998. Identification and analysis of yeast nucleosomal histone acetyltransferase complexes. Methods 15:315-321. [DOI] [PubMed] [Google Scholar]
- 8.Eberharter, A., D. E. Sterner, D. Schieltz, A. Hassan, J. R. Yates III, S. L. Berger, and J. L. Workman.1999. The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:6621-6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garkavtsev, I., I. A. Grigorian, V. S. Ossovskaya, M. V. Chernov, P. M. Chumakov, and A. V. Gudkov. 1998. The candidate tumour suppressor p33ING1 cooperates with p53 in cell growth control. Nature 391:295-298. [DOI] [PubMed] [Google Scholar]
- 10.Grant, P. A., L. Duggan, J. Côté, S. M. Roberts, J. E. Brownell, R. Candau, R. Ohba, T. Owen-Hughes, C. D. Allis, F. Winston, S. L. Berger, and J. L. Workman. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11:1640-1650. [DOI] [PubMed] [Google Scholar]
- 11.Grant, P. A., D. Schieltz, M. G. Pray-Grant, J. R. Yates III, and J. L. Workman. 1998. The ATM-related cofactor Tra1 is a component of the purified SAGA complex. Mol. Cell. 2:863-867. [DOI] [PubMed] [Google Scholar]
- 12.Hassan, A. H., K. E. Neely, and J. L. Workman. 2001. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell 104:817-827. [DOI] [PubMed] [Google Scholar]
- 13.Howe, L., G. Auston, P. A. Grant, S. John, R. G. Cook, J. L. Workman, and L. Pillus. 2001. Histone H3 specific acetyltransferases are essential for cell cycle progression. Genes Dev. 15:3144-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikura, T., V. V. Ogryzko, M. Grigoriev, R. Groisman, J. Wang, M. Horikoshi, R. Scully, J. Qin, and Y. Nakatani. 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102:463-473. [DOI] [PubMed] [Google Scholar]
- 15.John, S., L. Howe, S. T. Tafrov, P. A. Grant, R. Sternglanz, and J. L. Workman. 2000. The something about silencing protein, Sas3, is the catalytic subunit of NuA3, a yTAF(II)30-containing HAT complex that interacts with the Spt16 subunit of the yeast CP (Cdc68/Pob3)-FACT complex. Genes Dev. 14:1196-1208. [PMC free article] [PubMed] [Google Scholar]
- 16.Kuo, M. H., J. E. Brownell, R. E. Sobel, T. A. Ranalli, R. G. Cook, D. G. Edmondson, S. Y. Roth, and C. D. Allis. 1996. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature 383:269-272. [DOI] [PubMed] [Google Scholar]
- 17.Kuo, M.-H., J. Zhou, P. Jambeck, M. E. A. Churchill, and C. D. Allis. 1998. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 12:627-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loewith, R., M. Meijer, S. P. Lees-Miller, K. Riabowol, and D. Young. 2000. Three yeast proteins related to the human candidate tumor suppressor p33(ING1) are associated with histone acetyltransferase activities. Mol. Cell. Biol. 20:3807-3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loewith, R., J. S. Smith, M. Meijer, T. J. Williams, N. Bachman, J. D. Boeke, and D. Young. 2001. Pho23 is associated with the Rpd3 histone deacetylase and is required for its normal function in regulation of gene expression and silencing in Saccharomyces cerevisiae. J. Biol. Chem. 276:24068-24074. [DOI] [PubMed] [Google Scholar]
- 20.Nourani, A., Y. Doyon, R. T. Utley, S. Allard, W. S. Lane, and J. Cote. 2001. Role of an ING1 growth regulator in transcriptional activation and targeted histone acetylation by the NuA4 complex. Mol. Cell. Biol. 21:7629-7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orphanides, G., W. H. Wu, W. S. Lane, M. Hampsey, and D. Reinberg. 1999. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature 400:284-288. [DOI] [PubMed] [Google Scholar]
- 22.Reifsnyder, C., J. Lowell, A. Clarke, and L. Pillus. 1996. Yeast SAS silencing genes and human genes associated with AML and HIV-1 Tat interactions are homologous with acetyltransferases. Nat. Gen. 14:42-49. [DOI] [PubMed] [Google Scholar]
- 23.Saleh, A., V. Lang, R. Cook, and C. J. Brandl. 1997. Identification of native complexes containing the yeast coactivator/repressor proteins NGG1/ADA3 and ADA2. J. Biol. Chem. 272:5571-5578. [DOI] [PubMed] [Google Scholar]
- 24.Sendra, R., C. Tse, and J. C. Hansen. 2000. The yeast histone acetyltransferase A2 complex, but not free Gcn5p, binds stably to nucleosomal arrays. J. Biol. Chem. 275:24928-24934. [DOI] [PubMed] [Google Scholar]
- 25.Skowyra, D., M. Zeremski, N. Neznanov, M. Li, Y. Choi, M. Uesugi, C. A. Hauser, W. Gu, A. V. Gudkov, and J. Qin. 2001. Differential association of products of alternative transcripts of the candidate tumor suppressor ING1 with the mSin3/HDAC1 transcriptional corepressor complex. J. Biol. Chem. 276:8734-8739. [DOI] [PubMed] [Google Scholar]
- 26.Smith, E. R., A. Eisen, W. Gu, M. Sattah, A. Pannuti, J. Zhou, R. G. Cook, J. C. Lucchesi, and C. D. Allis. 1998. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc. Natl. Acad. Sci. USA 95:3561-3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sprague, G. F., Jr. 1991. Assay of yeast mating reaction. Methods Enzymol. 194:77-93. [DOI] [PubMed] [Google Scholar]
- 28.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64:435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takechi, S., and T. Nakayama. 1999. Sas3 is a histone acetyltransferase and requires a zinc finger motif. Biochem. Biophys. Res. Commun. 266:405-410. [DOI] [PubMed] [Google Scholar]
- 30.Utley, R. T., K. Ikeda, P. A. Grant, J. Côté, D. J. Steger, A. Eberharter, S. John, and J. L. Workman. 1998. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature 394:498-502. [DOI] [PubMed] [Google Scholar]