Abstract
Background
The number of species within the Malagasy genus Lepilemur and their phylogenetic relationships is disputed and controversial. In order to establish their evolutionary relationships, a comparative cytogenetic and molecular study was performed. We sequenced the complete mitochondrial cytochrome b gene (1140 bp) from 68 individuals representing all eight sportive lemur species and most major populations, and compared the results with those obtained from cytogenetic studies derived from 99 specimens.
Results
Interspecific genetic variation, diagnostic characters and significantly supported phylogenetic relationships were obtained from the mitochondrial sequence data and are in agreement with cytogenetic information. The results confirm the distinctiveness of Lepilemur ankaranensis, L. dorsalis, L. edwardsi, L. leucopus, L. microdon, L. mustelinus, L. ruficaudatus and L. septentrionalis on species level. Additionally, within L. ruficaudatus large genetic differences were observed among different geographic populations. L. dorsalis from Sahamalaza Peninsula and from the Ambanja/Nosy Be region are paraphyletic, with the latter forming a sister group to L. ankaranensis.
Conclusion
Our results support the classification of the eight major sportive lemur taxa as independent species. Moreover, our data indicate further cryptic speciation events within L. ruficaudatus and L. dorsalis. Based on molecular data we propose to recognize the sportive lemur populations from north of the Tsiribihina River, south of the Betsiboka River, and from the Sahamalaza Peninsula, as distinct species.
Background
Sportive lemurs, genus Lepilemur, are small nocturnal primates endemic to the island of Madagascar. They are amongst the most widely distributed lemurs, occurring in almost all natural evergreen or deciduous forest formations on the island (Fig. 1). Because pelage colouration and other external characteristics are inconspicuous in sportive lemurs, the early classifications [1,2] based on these features were disputed until a comprehensive cytogenetic approach allowed the recognition of six species [3-6]: L. dorsalis, L. edwardsi, L. leucopus, L. mustelinus L. ruficaudatus and L. septentrionalis. For a seventh, L. microdon, the karyotype remained unknown. Recently, L. septentrionalis has been split into two separate species, L. septentrionalis and L. ankaranensis [7,8] and the karyotype of L. microdon has been established [9], so that there are now eight cytogenetically recognized species.
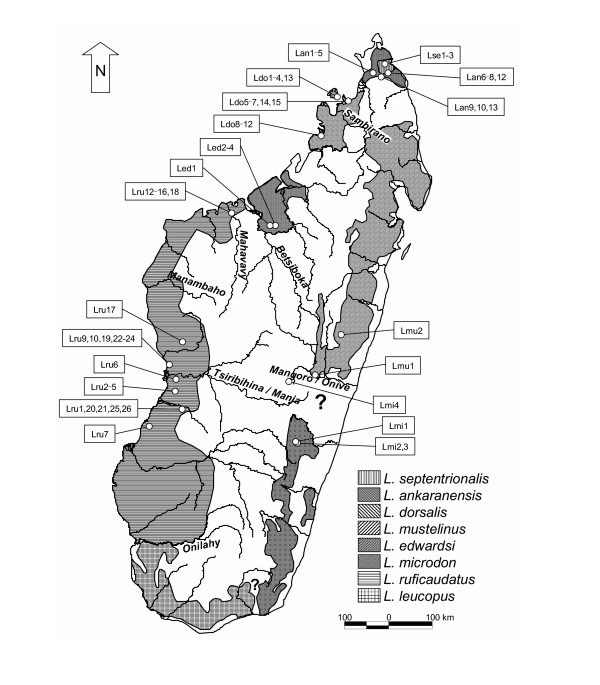
Figure 1.
Distribution of Lepilemur species (based on [25]). Circles indicate origin of analysed individuals (abbreviations are listed in additional file 1). Labelled rivers represent possible biogeographical barriers. Question marks indicate areas with ambiguous or missing information on Lepilemur distribution.
Molecular studies, especially the sequencing of mitochondrial genes, have expanded enormously during the last decade and have helped to characterise biodiversity and biogeographic patterns of Malagasy lemurs [10-19]. Such studies have been particularly fruitful among the family Cheirogaleidae, in which several new mouse lemur species were described and others elevated from synonymy [11,18,20], despite the fact that no chromosomal differences were detected within the subfamily Cheirogaleinae [21-23].
The application of mitochondrial sequence data to the reconstruction of phylogenetic relationships within the genus Lepilemur is still rare. However, pioneering studies using this method have helped to solve taxonomic issues. Some of the results have confirmed previous conclusions such as the species status of L. septentrionalis and L. ankaranensis [24], while other studies revealed unexpected results such as the paraphyly of L. edwardsi from south and north of the Betsiboka River [12,17], suggesting the existence of two different species [6,12,17]. Within L. ruficaudatus, Roos [25] also detected a paraphyly, with specimens from Andramasay, north of the Tsiribihina River, being more closely related to L. edwardsi from south of the Betsiboka River than they are to L. ruficaudatus from Kirindy, south of the Tsiribihina. The classification of specimens from south of the Betsiboka as L. edwardsi was, however, based on erroneous information on the extension of the species' range as far south as the Tsiribihina [3,5], despite the report of the existence of L. ruficaudatus north of the Tsiribihina in the Antsalova region [4]. Later, sportive lemurs south of the Betsiboka were cytogenetically classified as L. ruficaudatus [26].
In order to obtain a more complete picture of the sportive lemur's evolution, diversity and biogeography, we conducted a comparative molecular study by combining mitochondrial sequence data with cytogenetic information from all currently recognized species. We sequenced the complete mitochondrial cytochrome b gene from 68 individuals from 21 geographic locations, and compared the obtained results with conclusions from cytogenetic data derived from 99 individuals.
Results
Cytogenetics
Karyograms were established for 99 individuals during studies performed between 1975 and 2005, with one to 28 specimens from each study population (see additional files 1 and 2). As shown in additional file 2, the diploid chromosome number within the genus ranges from 20 to 38, and one to 19 chromosomal rearrangements were detected between species. Among the eight recognized species, karyotypes are species specific. Among the different populations of the traditional L. ruficaudatus and L. dorsalis no differences in diploid chromosome number and in the R-banding pattern were detected (see additional file 2, Fig. 2, 3, 4). Only one L. ruficaudatus from Anjahamena was analyzed by Giemsa staining and this specimen showed a karyotype identical to that of other L. ruficaudatus (Fig. 4). Further details about diploid chromosome numbers for all species and populations, and rearrangements among them, are given in additional file 2.
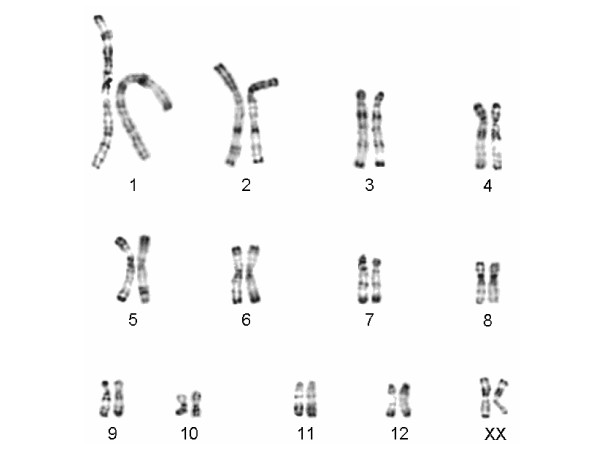
Figure 2.
Comparison of the half karyotypes of Lepilemur dorsalis from Sahamalaza Peninsula (left) and Ambanja (right). No differences in the R-banding pattern were detected.
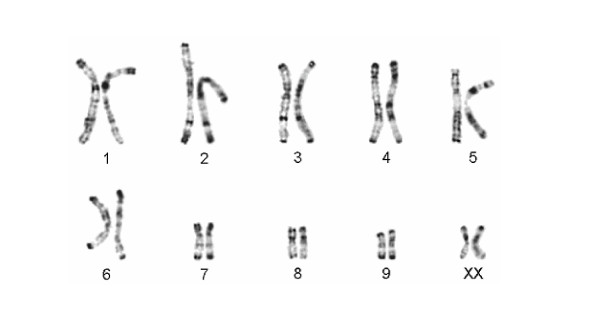
Figure 3.
Comparison of the half karyotypes of Lepilemur ruficaudatus from Andramasay (left) and Kirindy/CFPF (right). No differences in the R-banding pattern were detected.
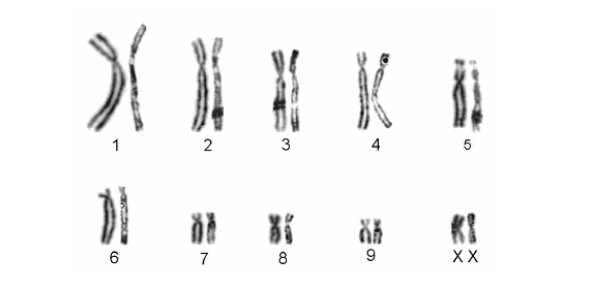
Figure 4.
Comparison of the half karyotypes of Lepilemur ruficaudatus from Kirindy/CFPF (left) and Anjahamena (right). No differences in the Giemsa-stained karyotypes were detected.
Molecular genetics
Complete mitochondrial cytochrome b gene sequences (1140 bp) were generated from 68 individuals representing all currently recognized species. Among the 68 sequences we detected 39 haplotypes. Uncorrected pairwise differences within the genus range from 0.00–16.82%, with overlapping intra- and inter-specific differences (intra-specific: 0.00–7.63%, inter-specific: 2.90–16.82%). Although most inter-specific differences are in the range of 7.37–16.82%, there are also two exceptions which show that differences between species can be much lower (L. ankaranensis – L. dorsalis from Sahamalaza Peninsula: 4.56–5.35%; L. ankaranensis – L. dorsalis from Ambanja/Nosy Be: 2.90–3.60%). Intra-specific differences range from 0.00–7.63%. Interestingly, the observed differences of 5.88–7.63% between the three major populations of L. ruficaudatus, which are separated by wide rivers or large distances, and those found between L. dorsalis from Sahamalaza Peninsula and from Ambanja/Nosy Be (5.18–5.88%) exceed those detected between L. ankaranensis and the two L. dorsalis populations, indicating speciation events in L. ruficaudatus and L. dorsalis. Further details about pairwise differences within and between species are reported in additional file 3.
Based on the 1140 bp of the mitochondrial cytochrome b gene, a population aggregation analysis was performed with 373 positions serving as diagnostic characters (see additional file 4). With 163–183 diagnostic characters, L. mustelinus is clearly separated from the other species. The remaining species differ in 32–146 diagnostic characters, with the lowest detected between L. ankaranensis and L. dorsalis from the Ambanja/Nosy Be population. The three L. ruficaudatus populations are separated by 64–78 diagnostic characters, comparable to those distinguishing the two L. dorsalis populations (57 characters), or to those detected between the two widely recognized species L. septentrionalis and L. ankaranensis (82 characters).
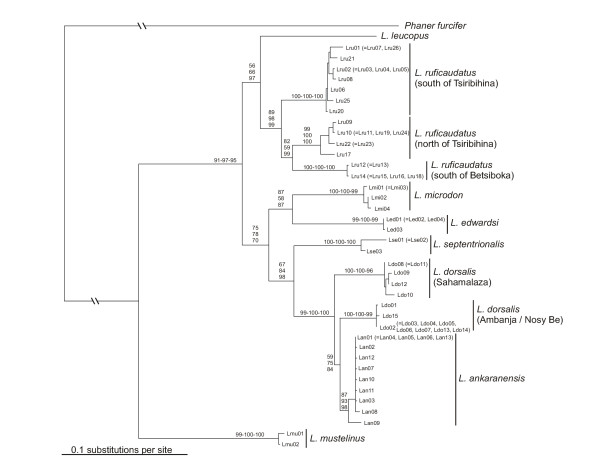
Phylogenetic trees reconstructed on the basis of different algorithms and models of sequence evolution produced identical tree topologies, with mainly significantly supported branching patterns (Fig. 5). Based on inferred phylogenetic relationships, L. mustelinus was the first species to split off. The remaining species diverged into two subgroups, of which one contained L. ankaranensis, L. dorsalis, L. edwardsi, L. microdon and L. septentrionalis, and the other L. leucopus and L. ruficaudatus. Within L. ruficaudatus three very distinct clades were observed, corresponding to different geographic locations. In the other subgroup a major split occurred between L. microdon/L. edwardsi and the remaining species, which later separated into L. septentrionalis and a group consisting of L. dorsalis and L. ankaranensis. L. dorsalis is further divided into two subclades, with one containing individuals from Ambanja and Nosy Be, and the other those from Sahamalaza. Moreover, L. dorsalis emerges as paraphyletic, with specimens from Nosy Be/Ambanja forming a sister clade to L. ankaranensis.
Figure 5.
Phylogenetic relationships as obtained from complete mitochondrial cytochrome b gene sequences (39 haplotypes), with branch lengths drawn according to those estimated by the NJ algorithm and by applying the TVM + I + Γ model of sequence evolution. Abbreviations refer to those listed in additional file 1 and numbers on nodes indicate support values for internal branches (first: NJ, second: MP, third: ML) based on 1,000 bootstrap replicates (NJ, MP) or 10,000 quartet puzzling steps (ML).
Discussion
As in many nocturnal mammals, pelage colouration among sportive lemurs is not a suitable characteristic for distinguishing among taxa, since it is not well defined and consistent within taxa. In contrast, cytogenetics or molecular methods such as sequencing of marker genes offer powerful tools that lead to important insights into the diversity and phylogeny of several Malagasy lemur genera [17,19,28,29]. In order to determine the diversity and phylogenetic relationships among sportive lemur taxa we conducted a comparative molecular study by combining mitochondrial sequence information and cytogenetic data from all currently recognized Lepilemur species.
Based on cytogenetic data, the genus Lepilemur can be divided into the eight traditionally recognized species, L. ankaranensis, L. dorsalis, L. edwardsi, L. leucopus, L. microdon, L. mustelinus, L. ruficaudatus and L. septentrionalis. These differ by several chromosomal rearrangements (3 to 19), with the exception of L. septentrionalis and L. ankaranensis that only one rearrangement separates [7-9] (see additional file 2). Nevertheless, the phylogenetic positions as well the genetic distances separating these two latter species suggest a separation on species level [24]. Among the three L. ruficaudatus populations and the two L. dorsalis populations, no chromosomal differences were detected.
The mitochondrial sequence data also confirm the specific status of the eight species mentioned above. The genetic differences detected among them are in the range of those observed among species of other lemur genera such as Mirza [11], Microcebus [11,14,18], Hapalemur [10,16] and Propithecus [13,30]. Interestingly, the distance of 14.47–16.82% observed between L. mustelinus and the remaining Lepilemur species is the largest detected among lemur species to date, and exceeds those observed among mouse lemur species [11,18].
Besides the high genetic differences observed among the eight Lepilemur species, further large differences were detected within L. ruficaudatus and L. dorsalis. The molecular data strongly indicate that the three L. ruficaudatus populations that are distributed in distinct subclades and separated by high genetic differences of 5.88–7.63% represent different taxa, confirming previous suggestions of a separate taxon status for specimens from Kirindy, Andramasay and Anjahamena [6,12,17,25,31]. Although no chromosomal differences were detected among the three L. ruficaudatus populations, the genetic differences observed among them are higher than those detected among some other Lepilemur species (e.g. L. dorsalis – L. ankaranensis: 2.90–3.60%), as well as from comparable data from other lemur genera, e.g. Microcebus [11,18]. Thus a separation of the three populations at the species level is proposed, which is additionally supported by the fact that these populations occupy ranges between major biogeographical barriers, such as Madagascar's large rivers (Fig. 1). These rivers are barriers also for a number of other species (Tsiribihina: Geogale aurita, Echinops telfairi, Hypogeomys antimena, Mungotictis decemlineata, Propithecus verreauxi verreauxi, Microcebus berthae; Mahavavy: Eulemur fulvus rufus, Galida elegans occidentalis; Betsiboka: Oryzorictes talpoides, Avahi occidentalis; Sambirano: Phaner furcifer parienti and possibly Avahi unicolor).
The L. dorsalis populations of Ambanja and Nosy Be are closely related and form one clade, indicating both their common origin and a recent isolation of the island of Nosy Be. The Nosy Be/Ambanja L. dorsalis and L. ankaranensis together form a sister group to L. dorsalis from Sahamalaza, indicating a paraphyly of L. dorsalis. These phylogenetic and biogeographic patterns are also confirmed by RAPD analysis [32], and are similar to patterns obtained for the genus Propithecus (Indriidae) in that P. v. coquereli appears to cluster with P. tattersalli instead with other subspecies of P. verreauxi [13,17,25,30]. Although ten chromosomal rearrangements were detected between L. ankaranensis and Nosy Be/Ambaja L. dorsalis (see additional file 2), genetic differences are, at 2.90–3.60%, the lowest detected among all Lepilemur species (see additional file 3). L. dorsalis from Sahamalaza differs from the Nosy Be/Ambanja L. dorsalis and L. ankaranensis at 4.56–5.88%, which is comparable with differences among other Lepilemur species. Taking together the paraphyly of L. dorsalis and the large genetic differences among analysed populations, we propose to divide the traditional L. dorsalis into two distinct species.
Furthermore, the distribution of diagnostic characters derived from a PAA also suggests a split of the L. ruficaudatus population into three different taxa and the L. dorsalis population into two taxa. This pattern is congruent with the results of the pairwise distance comparison and the phylogenetic tree reconstruction.
Since the type locality of L. ruficaudatus and its synonym L. pallidicauda is simply "Morondava" and no other synonyms are available, the populations north of the Tsiribihina River and between the rivers Betsiboka and Mahavavy need to be named. Questionable, however, is the classification of another new species announced by Edward Louis from Mitsinjo [33], a village located on the south side of the Mahavavy du Sud River. Whether this species is the same as the one we describe from the region between the Betsiboka and Mahavavy rivers needs to be evaluated by further studies. The locality of the L. dorsalis type specimen is very imprecisely labelled as "Madagascar". The only possible synonym for L. dorsalis is "Lepidolemur" grandidieri, with the similarly imprecise origin "North-western Madagascar". Most museum specimens are from the Ambanja region (see also [11]), and hence we presume that this is also the case for the L. dorsalis and L. grandidieri holotypes. Therefore, the population from the Sahamalaza Peninsula needs to be named. Below, we describe the sportive lemurs from the regions between the Betsiboka and Mahavavy rivers, north of the Tsiribihina River, and the Sahamalaza Peninsula, as three new species:
Lepilemur aeeclis sp. nov. (Fig. 6)
Figure 6.
L. aeeclis from Anjahamena during the day (a) and night (b) (Photographs by U. Thalmann).
Type Series
Skull (UM 2003-Lem-100, see additional file 5) stored at the University of Mahajanga. The individual was found dead at the type locality. Hair and DNA samples from an additional five different individuals are stored at the University Louis Pasteur Strasbourg, France and the Gene Bank of Primates, German Primate Centre, Germany (GBP 1028-1032). Measurements for individuals are given in additional file 6.
Type locality
Antafia (approx. 16°03.057'S, 45°54.522'E), north-east side of the Mahavavy du Sud River, Fokotany Ambatomahavavy, Firaisana Antongomena-Bevary, Fivondronona Mitsinjo, Province Mahajanga, Madagascar.
Description
Pelage coloration is considerably variable in the expression of the colours, possibly as a function of the age of individuals. Depending on light conditions (daylight or flashlight at night) the impression of colours may change subjectively. However, some constant characters are present, though variably expressed. The face is essentially grey and the ears are protruding and rounded. Sometimes there is the impression of a "facemask", in that there may be a darker diffuse, patch of hair in the middle of the forehead. Above the eyes darker coloured but diffuse stripes may run upwards to join in the middle of the head. These confluent stripes continue as one darker and distinct stripe along the back. The stripe is especially well expressed until it reaches the middle of the back, and then continues less prominently to the tail. On the back, the animals are essentially grey and reddish grey. The middle part of the back, especially, may show considerable reddish colouration that extends onto both shoulders and the upper and lower arms. The thigh and lower limb in general are less reddish than the upper part of the body. The ventral pelage is light to darker grey. The tail is variably coloured between grey with some red influence to deep rusty red with negligible grey influence.
Diagnosis
Differs, with the exception of L. randrianasoli and L. ruficaudatus, from all other sportive lemurs in karyotype (2N = 20, Fig. 4, see additional file 2). In the complete mitochondrial cytochrome b gene, L. aeeclis differs from its closest relative, L. randrianasoli by 5.88–6.75% and in 64 diagnostic characters. It is slightly larger than L. randrianasoli but is similar in size to L. ruficaudatus (see additional file 6). Head measurements (length and width) are more similar to L. ruficaudatus than to L. randrianasoli. However, hind foot length is more similar to L. randrianasoli than to L. ruficaudatus, which has the shortest hind foot.
Etymology
L. aeeclis is named in honour of the Association Européenne pour l'Etude et la Conservation des Lémuriens (A.E.E.C.L.), which has supported our fieldwork for 12 years.
Distribution
The taxon occurs between the Betsiboka and Mahavavy du Sud rivers. The southern extension of the taxon across the Mahavavy du Sud River is unknown, and needs further research.
Lepilemur randrianasoli sp. nov. (Fig. 7)
Figure 7.
L. randrianasoli from the southern bank of Manambolo River, approx. 35 km NE of type location (a) and from Ambalarano (b) (Photographs by U. Thalmann (a) and N. Andriaholinirina (b)).
Holotype
Tissue and DNA from one individual stored at the Gene Bank of Primates, German Primate Centre, Germany (GBP 941).
Type locality
Andramasay (approx. 44°29'E, 19°28'S), Province Toliary, Madagascar.
Description
Measurements of five males and four females from the type locality Andramasay are listed in additional file 6.
Diagnosis
Differs, with the exception of L. aeeclis and L. ruficaudatus, from all other sportive lemurs in karyotype (2N = 20, Fig. 3, see additional file 2). In the complete mitochondrial cytochrome b gene, L. randrianasoli differs from its closest relative, L. aeeclis, by 5.88–6.75% and in 64 diagnostic characters. The species is slightly smaller than L. aeeclis and L. ruficaudatus (see additional file 5). It differs from these species by having a narrower but slightly longer head. These differences in head size are most pronounced in males. Hind feet are of similar length to L. aeeclis, but longer than in L. ruficaudatus. Tail length is similar in all three species.
Etymology
L. randrianasoli is named in honour of our late colleague, Georges Randrianasolo, who worked from 1970 to 1977 to sample the sportive lemurs necessary for the first taxonomic revision based on cytogenetic studies, and who walked for two weeks to obtain data and samples from the L. ruficaudatus from Antsalova.
Distribution
Currently, L. randrianasoli is restricted to the type locality and Bemaraha. The Tsiribihina River is most likely the southern limit of the species' range. Further fieldwork is necessary to determine the northern limit. A possible northern barrier could be one of the major rivers Manambaho or the Mahavavy du Sud.
Lepilemur sahamalazensis sp. nov. (Fig. 8)
Figure 8.
L. sahamalazensis at its type locality Sahamalaza Peninsula (Photographs by T. Hahn (a) and R. Hilgartner (b)).
Type Series
Tissue and DNA samples from 4 individuals stored at the University Louis Pasteur Strasbourg, France (Ldo158s, Ldo153s, Ldo40, Lepi205Ak99).
Type locality
Sahamalaza Peninsula (approx. 47°58'E, 14°16'S), Province Mahajanga, Madagascar. Hypodigm: 6 animals from Ankarafa (47°45'E, 14°22'S), 25 km southeast of the type locality, for which morphometric measurements have been taken.
Description
Pelage coloration is variable, possibly also depending on age of individuals. Depending on light conditions (daylight or flashlight at night) the impression of colours may change subjectively. However, some constant characters are present, though variably expressed. The face is essentially grey. The forehead and the hairline around the ears are red-brown with sometimes darker diffuse patches. A dark diffuse line runs from the middle of the upper skull down the spine, ending in the middle or at the lower part of the back, but is never present on the tail. The dorsal pelage, including shoulders and the upper and lower arms, is predominantly red-brown, whereas the thigh and lower limbs in general are less reddish than the upper part of the body. The ventral pelage is generally grey to creamy. The coloration of the tail is red-brown to deep brown. Measurements of 2 males and of 4 females from Ankarafa are listed in additional file 7.
Diagnosis
Differs, with the exception of L. dorsalis and L. leucopus, from all other sportive lemurs in diploid chromosome number (2N = 26 Fig. 2, see additional file 2). The karyotypes of L. leucopus and L. sahamalazensis differ in six chromosomal rearrangements, whereas none exists between the latter and L. dorsalis. In the complete mitochondrial cytochrome b gene, L. sahamalazensis differs from its closest relatives, L. dorsalis and L. ankaranensis, in 5.18–5.88% and 4.56–5.35% and by 57 and 50 diagnostic characters, respectively. The few morphometric data which are available at the moment indicate that L. sahamalazensis is smaller and lighter than L. dorsalis. The tibia of L. sahamalazensis seems to be longer than in L. dorsalis, although tarsus length does not differ.
Etymology
L. sahamalazensis is named after the type locality, the Sahamalaza Peninsula.
Distribution
The species is restricted to the type locality of the Sahamalaza Peninsula, with the Sambirano River most likely as its northern boundary. Further field studies are required to determine the exact distribution range.
Based on our analyses, the classification of sportive lemurs is now as in additional file 8.
Conclusion
The combination of cytogenetic and molecular approaches reported here revealed important information about the diversity and evolution of the sportive lemurs. Phylogenetic relationships obtained from the mitochondrial sequence data are completely resolved and significantly supported, so that they most likely display the real evolutionary relationships among the analysed species and populations. Both methods were able to confirm the species status of the eight traditionally recognized species, while mitochondrial sequencing indicated in addition further cryptic speciation events in L. dorsalis and L. ruficaudatus. Especially in the light of less pronounced differences in pelage colouration and other external characteristics in nocturnal species compared to diurnal taxa, molecular studies are imperative. However, no chromosomal differences were detected between the three new proposed species and their respective sister taxa. Moreover, only a relatively small number of specimens per locality and only one mitochondrial locus were analysed in this study, so that additional (genetic) data, e.g. from nuclear DNA loci, are required to confirm the distinct species status of the populations. Furthermore, behavioural and morphological studies will provide independent data to delimit the species status of the three new forms. The discovery of three new possible primate species however, shows how diverse Madagascar's fauna is and how limited our knowledge currently still is. Ongoing work in the field and in the laboratory is urgently required to describe the complete diversity of sportive lemurs, especially of the different populations representing L. leucopus, L. microdon and L. mustelinus, and of other Malagasy lemurs. Greater knowledge of ecological, evolutionary and biogeographic patterns and processes will provide the necessary basis for protecting Madagascar's unique biodiversity.
Methods
Fieldwork
Samples from 68 individuals representing all currently recognized Lepilemur species were collected during field surveys in Madagascar (Fig. 1). Skin biopsies were taken under general anaesthesia with a 2 mg/kg injection of ketamine solution (Ketalar® Parke-Davis) or GM2 [34]. A part of each sample was directly frozen in liquid nitrogen, while the other part was preserved with a cryoprotector (DMSO 10%) with the aim of growing fibroblast cultures. Other tissue samples were stored in 75% ethanol, or in Queen's lysis buffer [35]. From some animals, 0.5 ml blood was also taken in heparinized tubes for further lymphoblast cultures. From most animals, standard morphometric measurements, including body mass, head length and width, head-body length, tail length and hind foot length, were collected according to [36]. For morphometric comparisons, we included only individuals with a body mass of at least 660 g, because this is the lower limit of adult body mass for L. ruficaudatus in the Kirindy forest. However, morphometric data were collected by different people under various conditions, and hence we expect low inter-observer reliability. We therefore did not use morphometric data for species delimitation, but rather use them as a tool to present a picture of the taxa. Immediately after recovery from anaesthesia, animals were released in their respective capture areas.
Cytogenetics
A total of 99 sportive lemur individuals with at least one specimen per locality was cytogenetically analysed during studies performed from 1975 till 2005 [8,9,26,37-39]. Chromosomes were prepared either from lymphoblast or fibroblast cultures, following classical methods [8,9,26,37-39]. Giemsa staining was performed for all 99 individuals. With the exception of one population, R-banded and C-banded chromosomes were prepared for at least one individual (see additional file 2).
Molecular genetics
DNA from the biopsies was extracted using a standard proteinase K digestion, followed by a phenol chloroform extraction [40] with minor modifications [41], or isolated with the QIAamp DNA Mini Kit as recommended by the supplier.
The complete mitochondrial cytochrome b gene was amplified via PCR using the oligonucleotide primers CYT-LEP-L: 5'-AATGATATGAAAAACCATCGTTGTA-3' and CYT-LEP-H: 5'-GGCTTACAAGGCCGGGGTAA-3'. Standard, wax-mediated hot-start PCRs were carried out for 40 cycles, each with a denaturation step at 94°C for 60 s, annealing at 60°C for 60 s, and extension at 72°C for 90 s, followed by a final extension step at 72°C for 5 min. Aliquots of the PCR amplifications were checked by agarose gel electrophoresis. Subsequently, PCR products were cleaned using the Qiagen PCR Purification Kit and sequenced on an ABI 3100-Avant sequencer using the BigDye Terminator Cycle Sequencing Kit (Applied Biosystems), primers as indicated above and the internal primers CYT-LEP-L400: 5'-TGAGGACAAATATCATTCTGAGG-3' and CYT-LEP-H545: 5'-TGGAGTGCGAAGAATCGGGT-3'. The respective sequences were deposited in GenBank and are available under the accession numbers DQ108990-DQ109034 and DQ234881-DQ234900.
Sequences were easily aligned by eye due to the lack of insertions or deletions, and were checked for their potential to be correctly transcribed in order to eliminate data set contaminations with pseudogenes. For a comprehensive evaluation of the sequence data, we expanded our data set with orthologous sequences, already deposited at GenBank, from two L. ruficaudatus and one L. dorsalis. As outgroup for phylogenetic tree reconstructions, we selected Phaner furcifer because it displays the most similar orthologous sequence of all Malagasy lemurs to Lepilemur [29]. Further details about analysed individuals and sequences are summarized in Figure 1 and additional file 1.
Uncorrected pairwise differences within and between species and major populations were calculated with PAUP 4.0b10 [42] and DnaSP 3.52 [43].
A population aggregation analysis (PAA) was performed according to the diagnostic character framework described in [44]. Accordingly, fixed nucleotide characters provide the unit for which aggregation of taxonomic units occurs. For diagnosis, attributes whose fixed unique states unite a group (populations, species), to the exclusion of other groups, are considered characters. Polymorphic attributes, or traits, are indicative of population frequency differences. To identify diagnostic sites, sequences were imported into MacClade 3.0 [45].
Phylogenetic tree reconstructions were carried out with the maximum-parsimony (MP), neighbor-joining (NJ) and maximum-likelihood (ML) algorithms as implemented in PAUP or TREEPUZZLE 5.0 [46]. For MP analyses, all characters were treated as unordered and equally weighted throughout. A heuristic search was performed with the maximum number of trees set to 100. NJ and ML trees were constructed with the TVM + I (= 0.5156) + Γ (= 2.2387) model of sequence evolution as it was selected as best-fitting model with MODELTEST 3.06 [47], as well as with standard models. Relative support of internal nodes was performed by bootstrap analyses with 1,000 replications (MP, NJ), or by the quartet puzzling support values on the basis of 10,000 puzzling steps (ML).
Authors' contributions
NA participated in the design of the study and the concept of the revision work on Lepilemur. She did the field work for L. microdon, L. mustelinus, L. edwardsi and L. aeeclis, took morphometric measurements and did a part of the cytogenetic study in the Institut Pasteur de Madagascar (IPM) in Antananarivo.
JLF designed Lepilemur specific primers, generated several sequences, performed the statistical analysis of the phylogenetic data and wrote a draft version of the MS in collaboration with YR.
ChR designed Lepilemur specific primers, generated several sequences, performed the statistical analysis of the phylogenetic data and wrote the draft and final version of the MS in collaboration with DZ.
DZ organised, lead and conducted the expedition to collect data and samples of L. ruficaudatus south of the Morondava River. He also sampled L. ruficaudatus in the Kirindy Forest and took morphometric measurements in collaboration with RH. DZ generated sequences from these individuals, did the statistical analysis of the morphometric data and wrote the draft and final version of the MS in collaboration with ChR.
UT conceived, planned, organised, lead and conducted the expeditions to collect the samples, data and pictures of L. aeeclis sp. nov. in the field, participated in the design and drafting of the MS, critically revised it several times and approved the final version.
ClR participated in the design of the study and the concept of the revision work on Lepilemur, in collaboration with YR and NA. He organized and conceptualized the field work of NA, participated in the collection of samples of L. edwardsi and participated in a revision of a draft version of the MS.
IR participated in the design of the study and the concept of the revision work on Lepilemur. She did the field work for L. septentrionalis, L. ankaranensis, L. dorsalis in Ambanja and part of Nosy Be.
JG sampled L. randrianasoli and L. ruficaudatus and provided morphometric data and tissue samples.
BM organized financed and conceptualized part of the field work on on Nosy Be (with CL), in Sahamalaza Peninsula (with TH and IR), on the ridge of Andrafiamena (with IR) and in Ankarana (with IR). At several of these sites, he collected samples and animal measurements.
RH sampled L. ruficaudatus south of the Tsiribihina River and took morphometric measurements in collaboration with DZ and he took photographs of L. sahamalazensis sp. nov. in the field.
LW designed Lepilemur specific primers and generated several sequences.
AZ participated in the expeditions to collect data on L. aeeclis sp. nov., and found the skull of the animal of the type series. He considerably facilitated field work and data collection.
CL did field work on Nosy Be.
TH did field work on Sahamalaza Peninsula.
EZ organized, financed and conceptualized part of the field work and revised the MS.
UR organized, financed and conceptualized part of the field work and revised the MS.
MC conducted part of the field work including capturing, measuring and sampling the Sahamalaza population, and revised the MS.
JT participated in the design of the study and the concept of the revision work on Lepilemur.
IT provided systematic input and checked the MS.
YR participated in the design of the study and the concept of the revision work on Lepilemur, wrote a draft version of the MS, critically revised it several times and approved its final version.
Supplementary Material
A table showing details on studied sportive lemur individuals.
A table showing diploid number (2N) and chromosomal rearrangements among species and populations [9].
A table showing minimum and maximum uncorrected pairwise genetic differences (in %) within and among analysed species and populations based on complete mitochondrial cytochrome b sequence data.
A table showing number of diagnostic characters as obtained from population aggregation analysis (PAA).
A table showing morphometric measurements for the L. aeeclis syntype skull
A table showing morphometric measurements of L. randrianasoli and L. aeeclis in comparison with L. ruficaudatus.
A table showing morphometric measurements for L. sahamalazensis in comparison to L. dorsalis.
A table showing classification of sportive lemurs.
Acknowledgments
Acknowledgements
Our thanks go to all the following persons and entities: the Direction des Eaux et Forêts of Antananarivo and Mahajanga, the Association Nationale pour la Gestion des Aires Protégées of Antananarivo and Mahajanga (ANGAP) for their permission to capture the animals, to take samples and for delivery CITES. Under an agreement between the University Louis Pasteur of Strasbourg and the Institut Pasteur of Antananarivo, the latter allowed N.A. to do lymphoblast cultures. We thank the Faculty of Sciences, University of Antananarivo, Madagascar, and the CFPF Morondava, for support.
We are grateful to J. Cuisin and M. Tranier from the Muséum National d'Histoire Naturelle de Paris who graciously provided a biological sample of L. microdon (MNHN CG 2002.2 coll.nr 48) and helped with the systematic nomenclature. Furthermore, we thank B. Rakotosamimanana, C. Schwitzer and N. Schwitzer for logistic help. For valuable support in the field we thank L. Razafimanantsoa, R. Rasoloarison and G. Olivieri, and we thank D. Montagnon, K. Platner, C. Overdieck and C. Schwarz for technical assistance with the genetic analyses. We also thank A. Petter-Rousseaux for her help in the determination of museum specimens, M. Hauwy for his cytogenetic work, M. Lavaux for her secretarial assistance and A. Yoder for her helpful comments on molecular results.
This study was financially supported by various organizations. Y. Rumpler's and J Fausser's work was partly supported by the Association Européenne pour l'Etude et la Conservation des Lémuriens (A.E.E.C.L.), the I.G.B.M.C. (Pf P. Chambon) and the DREIF-MENESR (EA3428). The work of U. Thalmann was funded by the Margot Marsh Biodiversity Foundation and the A.H. Schultz-Foundation. The work of R. Hilgartner and D. Zinner was supported by the German Science Council (DFG KA 1082/6-1, 2) and the German Primate Center, DPZ. The work of J.U. Ganzhorn was supported by the German Science Council (DFG GA 342/3-1,2). The work of E. Zimmermann, U. Radespiel and M. Craul was supported by the Volkswagen Foundation (VW I, Aktenzeichen I/76828, Zimmermann) and the „Gesellschaft der Freunde der Tierärztlichen Hochschule Hannover e.V.", Germany.
Contributor Information
Nicole Andriaholinirina, Email: nicole-ludes@netcourrier.com.
Jean-Luc Fausser, Email: jean-luc.Fausser@medecine.u-strasbg.fr.
Christian Roos, Email: croos@dpz.gwdg.de.
Dietmar Zinner, Email: dzinner@gwdg.de.
Urs Thalmann, Email: uthal@aim.unizh.ch.
Clément Rabarivola, Email: cjrabary@hotmail.com.
Iary Ravoarimanana, Email: iary.ravaoarimanana@botschaft-madagaskar.de.
Jörg U Ganzhorn, Email: Ganzhorn@zoologie.uni-hamburg.de.
Bernhard Meier, Email: bmlegr@t-online.de.
Roland Hilgartner, Email: hilgartner@gmx.de.
Lutz Walter, Email: lwalter@gwdg.de.
Alphonse Zaramody, Email: zaramody@univ-mahajanga.mg.
Christoph Langer, Email: christoph-langer@gmx.de.
Thomas Hahn, Email: thahn@mpimf-heidelberg.mpg.de.
Elke Zimmermann, Email: elke.zimmermann@tiho-hannover.de.
Ute Radespiel, Email: ute.radespiel@tiho-hannover.de.
Mathias Craul, Email: mathias.craul@tiho-hannover.de.
Jürgen Tomiuk, Email: juergen.tomiuk@uni-tuebingen.de.
Ian Tattersall, Email: iant@amnh.org.
Yves Rumpler, Email: marguerite.lavaux@embryo-ulp.u-strasbg.fr.
References
- Petit G. Le genre Lepidolemur et sa répartition géographique. C R Soc Biogéogr. 1933;10:33–37. [Google Scholar]
- Petter JJ, Petter-Rousseaux A. Remarque sur la systématique du genre Lepilemur. Mammalia. 1960;24:76–86. [Google Scholar]
- Petter JJ, Albignac R, Rumpler Y. Faune de Madagascar. Vol. 44. Paris: C.N.R.S./O.R.S.T.O.M; 1977. Mammifères lémuriens (Primates prosimiens) pp. 1–513. [Google Scholar]
- Rumpler Y, Albignac R. Chromosomes studies of the Lepilemur, an endemic Malagasy genus of lemurs: Contribution of the cytogenetics to their taxonomy. J Hum Evol. 1978;7:191–196. [Google Scholar]
- Tattersall I. The Primates of Madagascar. New York: Columbia University Press; 1982. [Google Scholar]
- Thalmann U, Ganzhorn JU. Lepilemur, Sportive Lemur. In: Goodmann SM, Benstead JP, editor. The Natural History of Madagascar. Chicago: The University of Chicago Press; 2003. pp. 1336–1340. [Google Scholar]
- Groves C. Primate Taxonomy. Washington: Smithsonian Institution Press; 2001. [Google Scholar]
- Rumpler Y, Ravaoarimanana B, Hauwy M, Warter S. Cytogenetic arguments in favour of taxonomic revision of Lepilemur septentrionalis species. Folia Primatol. 2001;72:308–315. doi: 10.1159/000052747. [DOI] [PubMed] [Google Scholar]
- Andriaholinirina N, Rabarivola C, Hauwy M, Rumpler Y. Cytogenetic study of Lepilemur microdon. Folia Primatol. 2005;76:238–241. doi: 10.1159/000086027. [DOI] [PubMed] [Google Scholar]
- Fausser JL, Prosper P, Donati G, Ramanamanjato JB, Rumpler Y. Phylogenetic relationships between Hapalemur species and subspecies based on mitochondrial DNA sequences. BMC Evol Biol. 2002;2:1. doi: 10.1186/1471-2148-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappeler PM, Rasoloarison RM, Razafimanantsoa L, Walter L, Roos C. Morphology, behaviour and molecular evolution of giant mouse lemurs (Mirza spp.) Gray, with description of a new species. Primate Report. 1870;71:3–26. [Google Scholar]
- Pastorini J. PhD thesis. University of Zürich; 2000. Molecular systematics of lemurs. [Google Scholar]
- Pastorini J, Forstner MRJ, Martin RD. Phylogenetic history of Sifakas (Propithecus: Lemuriformes) derived from mtDNA sequences. Am J Primatol. 2001;53:1–17. doi: 10.1002/1098-2345(200101)53:1<1::AID-AJP1>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Pastorini J, Martin RD, Ehresmann P, Zimmermann E, Forstner MRJ. Molecular phylogeny of the lemur family Cheirogaleidae (Primates) based on mitochondrial DNA sequences. Mol Phylogenet Evol. 2001;19:45–56. doi: 10.1006/mpev.2000.0904. [DOI] [PubMed] [Google Scholar]
- Pastorini J, Forstner MRJ, Martin RD. Phylogenetic relationships among Lemuridae (Primates): evidence from mtDNA. J Hum Evol. 2002;43:463–478. [PubMed] [Google Scholar]
- Pastorini J, Forstner MRJ, Martin RD. Phylogenetic relationships of the gentle lemurs (Hapalemur) Evol Anthropol. 2002;11:150–154. doi: 10.1002/evan.10079. [DOI] [Google Scholar]
- Pastorini J, Thalmann U, Martin RD. A molecular approach to comparative phylogeography of extant Malagasy lemurs. Proc Natl Acad Sci USA. 2003;100:5879–5884. doi: 10.1073/pnas.1031673100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder AD, Rasoloarison RM, Goodman SM, Irwin JA, Atsalis S, Ravosa MJ, Ganzhorn JU. Remarkable species diversity in Malagasy mouse lemurs (Primates, Microcebus) Proc Natl Acad Sci USA. 2000;97:11325–11330. doi: 10.1073/pnas.200121897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder AD, Olson LE, Hanley C, Heckman KL, Rasoloarison RM, Russell AL, Ranivo J, Soarimalala V, Karanth KP, Raselimanana AP, Goodman SM. A multidimensional approach for detecting species patterns in Malagasy vertebrates. Proc Nat Acad Sci USA. 2005;102:6587–6594. doi: 10.1073/pnas.0502092102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasoloarison RM, Goodman SM, Ganzhorn JU. A taxonomic revision of mouse lemurs (Microcebus) occurring in the western portions of Madagascar. Int J Primatol. 2000;21:963–1019. doi: 10.1023/A:1005511129475. [DOI] [Google Scholar]
- Rumpler Y, Albignac R. Cytogenetic study of the endemic Malagasy lemurs subfamily Cheirogaleinae (Gregory 1915) Am J Phys Anthropol. 1973;38:261–264. doi: 10.1002/ajpa.1330380220. [DOI] [PubMed] [Google Scholar]
- Rumpler Y, Warter S, Hauwy M, Meier B, Peyrieras A, Albignac R, Petter JJ, Dutrillaux B. Cytogenetic study of Allocebus trichotis, a Malagasy prosimian. Am J Primatol. 1995;36:239–244. doi: 10.1002/ajp.1350360307. [DOI] [PubMed] [Google Scholar]
- Rumpler Y, Ganzhorn JU, Tomiuk J, Leipoldt M, Warter S. A cytogenetic study of Microcebus myoxinus. Folia Primatol. 1998;69:307–311. doi: 10.1159/000021644. [DOI] [PubMed] [Google Scholar]
- Ravaoarimanana B, Tiedemann R, Montagnon D, Rumpler Y. Molecular and cytogenetic evidence for cryptic speciation within a rare endemic Malagasy lemur, the Northern Sportive Lemur (Lepilemur septentrionalis) Mol Phylogenet Evol. 2004;31:440–448. doi: 10.1016/j.ympev.2003.08.020. [DOI] [PubMed] [Google Scholar]
- Roos C. PhD thesis. Technical University München; 2003. Molekulare Phylogenie der Halbaffen, Schlankaffen und Gibbons. [Google Scholar]
- Zaramody A, Andriaholinirina N, Dominique R, Rabarivola C. Nouvelle répartition respective de Lepilemur microdon et L. mustelinus, et de L. ruficaudatus et L. edwardsi. Lemur News. 2005;10:19–20. [Google Scholar]
- Rumpler Y. What cytogenetic studies may tell us about species diversity and speciation of lemurs. Int J Primatol. 2000;21:865–881. doi: 10.1023/A:1005598726749. [DOI] [Google Scholar]
- Rumpler Y, Dutrillaux B. Chromosomal evolution and speciation in Primates. In: University of the Basque Country, editor. Revisiones Sobre Biologia Celular. Vol. 23. Berlin/Heidelberg/New York: Springer-Verlag; 1990. pp. 1–112. [PubMed] [Google Scholar]
- Roos C, Schmitz J, Zischler H. Primate jumping genes elucidate strepsirrhine phylogeny. Proc Nat Acad Sci USA. 2004;101:10650–10654. doi: 10.1073/pnas.0403852101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor MI, Sommer JA, Houck M, Zaonarivelo J, Wright P, Ingram C, Engel S, Louis EE., Jr Specific status of Propithecus spp. Int J Primatol. 2004;25:875–900. doi: 10.1023/B:IJOP.0000029127.31190.e9. [DOI] [Google Scholar]
- Bachmann L, Rumpler Y, Ganzhorn JU, Tomiuk J. Genetic differentiation among natural populations of Lepilemur ruficaudatus. Int J Primatol. 2000;21:853–864. doi: 10.1023/A:1005546709911. [DOI] [Google Scholar]
- Ravaoarimanana B, Fausser JL, Rumpler Y. Genetic comparison of wild populations of Lepilemur septentrionalis and Lepilemur dorsalis using RAPD markers. Primates. 2001;42:221–231. [Google Scholar]
- Anonymous Named in honor of Ulie Seal, this newly discovered lemur inhabits the rain forest of eastern Madagascar. CBSG News. 2005;16:40. [Google Scholar]
- Rensing S. Immobilization and anesthesia of nonhuman primates. Primate Report. 1999;55:33–38. [Google Scholar]
- Seutin G, White BN, Boag PT. Preservation of avian blood and tissue samples for DNA analysis. Can J Zool. 1991;69:82–90. [Google Scholar]
- Rasoloharijaona S, Rakotosamimanana B, Randrianambinina B, Zimmermann E. Pair-specific usage of sleeping sites and their implications for social organization in a nocturnal Malagasy primate, the Milne Edwards' sportive lemur (Lepilemur edwardsi) Am J Phys Anthropol. 2003;122:251–258. doi: 10.1002/ajpa.10281. [DOI] [PubMed] [Google Scholar]
- Rumpler Y, Albignac R. Intraspecific chromosome variability in a lemur from the north of Madagascar: Lepilemur septentrionalis, species nova. Am J Phys Anthropol. 1975;42:425–429. doi: 10.1002/ajpa.1330420308. [DOI] [PubMed] [Google Scholar]
- Rumpler Y, Ishak B, Warter S, Dutrillaux B. Chromosomal evolution in Malagasy lemurs. VIII. Chromosome banding studies of Lepilemur ruficaudatus, L leucopus, and L septentrionalis. Cytogenet Cell Genet. 1985;39:194–199. doi: 10.1159/000132134. [DOI] [PubMed] [Google Scholar]
- Rumpler Y, Ishak B, Dutrillaux B, Warter S, Ratsirarson J. Chromosomal evolution in Malagasy lemurs. IX. Chromosomal banding studies of Lepilemur mustelinus, L. dorsalis, and L. edwardsi. Cytogenet Cell Genet. 1986;42:164–168. doi: 10.1159/000132270. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2. Cold Spring Harbor: Cold Spring Harbor Press; 1989. [Google Scholar]
- Fausser JL, Rabarivola C, Meier B, Hahn T, Rumpler Y. Genetic comparison between different populations of Eulemur macaco flavifrons in northwest Madagascar using RAPD markers. Am J Primatol. 2000;51:249–255. doi: 10.1002/1098-2345(200008)51:4<249::AID-AJP4>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: Phylogenetic analysis using parsimony (*and other methods), version v40b10. Sunderland, MA, Sinauer Associates; 2002. [Google Scholar]
- Rozas J, Rozas R. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics. 1998;15:174–175. doi: 10.1093/bioinformatics/15.2.174. [DOI] [PubMed] [Google Scholar]
- Davis JI, Nixon KC. Populations, genetic variation, and the delimitation of phylogenetic species. Syst Biol. 1992;41:421–435. [Google Scholar]
- Maddison WP, Maddison DR. MacClade, version 3. Sunderland, MA, Sinauer Associates; 1992. [Google Scholar]
- Strimmer K, von Haeseler A. Quartet puzzling: A maximum likelihood method for reconstructing tree topologies. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitutions. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A table showing details on studied sportive lemur individuals.
A table showing diploid number (2N) and chromosomal rearrangements among species and populations [9].
A table showing minimum and maximum uncorrected pairwise genetic differences (in %) within and among analysed species and populations based on complete mitochondrial cytochrome b sequence data.
A table showing number of diagnostic characters as obtained from population aggregation analysis (PAA).
A table showing morphometric measurements for the L. aeeclis syntype skull
A table showing morphometric measurements of L. randrianasoli and L. aeeclis in comparison with L. ruficaudatus.
A table showing morphometric measurements for L. sahamalazensis in comparison to L. dorsalis.
A table showing classification of sportive lemurs.