Abstract
We demonstrate that transformation-transactivation domain-associated protein (TRRAP) binding and the recruitment of histone H3 and H4 acetyltransferase activities are required for the transactivation of a silent telomerase reverse transcriptase (TERT) gene in exponentially growing human fibroblasts by c-Myc or N-Myc protein. However, recruitment of TRRAP by c- or N-Myc is dispensable for the partial induction of several basally expressed genes in exponentially growing primary and immortalized fibroblasts. Furthermore, recruitment of TRRAP is required for c-Myc- or N-Myc-mediated oncogenic transformation but not for the partial restoration of the growth defect in myc-null fibroblasts. A segment of the adenovirus E1A protein fused to a transformation-defective N-Myc protein carrying a small deletion in the transactivation domain specifically restores interaction with TRRAP, activates the silent TERT gene, induces acetylation of histones H3 and H4 at the TERT promoter, and transforms primary cells. Accordingly, wild-type L-Myc is much less efficient in TRRAP binding, activation of the silent TERT gene, and transformation of primary fibroblasts. Nevertheless, L-Myc is a potent activator of several basally expressed genes and can fully restore the growth defect of myc-null cells. These results suggest a differential requirement for TRRAP for several Myc-mediated activities.
Myc overexpression has long been associated with a number of human malignancies and with the acquisition of the transformed phenotype in many animal models (19, 27). In addition, basal expression of Myc is required for progression through the cell cycle and for normal embryonic development (14). Myc is a transcriptional activator when it binds as a heterodimer with Max to the consensus site CACGTG. Only the C-terminal 100 amino acids of Myc are required for Max dimerization and DNA binding, whereas the remaining N-terminal domain contains segments capable of transcriptional transactivation and nuclear cofactor binding. The relationship between transactivation, the recruitment of nuclear cofactors, and Myc biological activity is complex and unclear. On one hand, there is a general dependence on transactivation for many Myc biological activities; however, a naturally occurring shorter form of Myc (MycS) that is defective in transactivation can still rescue the growth defect in c-myc-null cells (51). On the other hand, small deletions of evolutionarily conserved domains (called Myc boxes I and II [MBI and MBII]) abolish Myc oncogenic activity in primary cells, the induction of apoptosis, and the ability to block differentiation (18, 22, 46). These same mutants retain the ability to transactivate most promoters in transient-expression assays (5).
A growing number of genes have been ascribed as downstream targets of Myc regulation; however, only a small number have been shown to be direct targets in vivo (12, 13). The criteria for considering a given gene as a Myc target vary between studies, and Myc may regulate different sets of genes dependent on Myc level and other cellular factors (12). However, the most compelling evidence for direct Myc-dependent regulation is the ability to cross-link Myc to a specific chromosomal consensus binding site. We were interested in determining mechanisms of activation of specific cellular promoters in the context of their native chromosomal location. We focused on four promoters, CAD, CDK4, HSP60, and telomerase reverse transcriptase (TERT), that have functional binding sites for Myc/Max heterodimers and/or that can be directly cross-linked to Myc in vivo (9, 16, 21, 28). However, TERT regulation differs dramatically from the other genes in exponentially growing cells, since TERT is silent in almost all somatic cells (25), whereas the other genes are ubiquitously expressed. Interestingly, Myc is the only transcription factor capable of interacting with the TERT promoter and inducing its expression from the chromosomal locus (49). Proteins like E2F or E1A, which in many in vivo assays demonstrate activities similar to that of Myc, fail to activate TERT (49). Since TERT expression is up-regulated in the vast majority of human cancer cells, it was of particular interest to address how this normally silenced gene can be activated by Myc.
An important aspect of gene regulation is the interplay between basal transcription factors and local chromatin structure. The packaging of DNA into an ordered array of nucleosomes is a general repressor of transcription, and many genetic and biochemical studies demonstrate that gene regulation is dependent on factors that remodel or modify the local chromatin structure surrounding specific promoters (reviewed in reference 47). The discovery of the transformation-transactivation domain-associated protein (TRRAP) and TIP49/TIP48 as cofactors essential for Myc-induced transformation provided an important link between chromatin modification and Myc-dependent phenotypes (35, 36, 50). Interestingly, recruitment of these cofactors by Myc occurs via interactions with the MBII domain. A series of recent studies have attributed TRRAP and TIP49/TIP48 to a number of complexes. First was a human ortholog of the yeast SAGA complex, which contains TRRAP, the human homologue of GCN5 or PCAF, but no TIP49/TIP48 (37). This complex possesses H3-histone acetyltransferase activity (HAT). Another TRRAP-containing complex has the TIP60 H2/H4 HAT as well as TIP49/TIP48 (29). This complex shares many subunits with a p400 complex (23), but the latter complex lacks HAT activity. The TIP49/TIP48 proteins are also involved in other distinct complexes (39), and the majority of cellular TIP49/TIP48 exists in complexes that do not contain TRRAP (G. LeRoy and G. Wang, unpublished data).
The role of histone acetylation in transcriptional activation by Myc is not clear. On one hand, a number of studies reported that activation of the Myc target genes CAD and TERT occurs without concomitant increases in histone H3 or H4 acetylation (16). On the other hand, Myc recruits HAT activity (36), and the recruitment of TRRAP to the promoter of several Myc-responsive genes following serum stimulation was associated with induction of H4 and/or H3 acetylation (8, 21).
In the present study, we were interested in determining the mechanism of activation of specific cellular promoters in the context of their native chromosomal location in exponentially growing primary and immortalized cells. We found that the activation of a silent TERT gene in exponentially growing primary human fibroblasts requires TRRAP recruitment and is accompanied by both H3 and H4 acetylation. On the other hand, TERT regulation differs dramatically from that of other genes in exponentially growing cells. We also demonstrate that Myc mutants that lose their ability to interact with TRRAP fail to activate TERT in primary cells but are still able to activate transcription of basally expressed genes in both primary cells and myc-null fibroblasts (34). An analysis of wild-type L-Myc protein and N-Myc mutants demonstrated that TRRAP binding was largely dispensable for normal cell proliferation and the induction of basally expressed genes.
MATERIALS AND METHODS
Vectors.
Expression plasmids were created by standard methods in a cytomegalovirus promoter-driven vector containing a FLAG epitope tag or in the retroviral expression vector pLXSH and were verified by sequence analysis. Details of individual constructs are available upon request.
Cell culture, transfection, and retroviral infection.
Primary human fibroblasts (IMR90), rat embryo fibroblasts, rat c-myc-null fibroblasts (HO15.19), the retroviral producer PhoeNX cell line, and HEK 293 cells were cultured in Dulbecco's modified Eagle medium supplemented with 10% calf serum.
Retroviral infection of IMR90 and HO15.19 cells was performed according to the method described in reference 40, using PhoeNX cells. PhoeNX cells were transfected via the calcium phosphate method. To obtain a transduced population, after completion of infection IMR90 or HO15.19 cells were selected for resistance to hygromycin (150 μg/ml) for 7 days.
Reverse transcription-PCR.
Total RNA was isolated from infected cells using TRIzol reagent (GIBCO-BRL). Reverse transcription of 1 to 3 μg of RNA was performed using the SuperScript First-Strand Synthesis System (GIBCO-BRL), according to the manufacturer's instructions. PCR was performed on cDNA using the following sets of oligonucleotides: human TERT-specific oligonucleotides, HT1 (CGGAAGAGTGTCTGGAGCAA) and HT2 (GGATGAAGCGGAGTCTGGA); human glyceraldehyde-3-phosphate dehydrogenase (GAPDH)-specific oligonucleotides, HG1 (CTCAG-ACACCATGGGGAAGGTGA) and HG2 (ATGATCTTGAGGCTGTTGTCATA); human CDK4-specific oligonucleotides, HCD1 (GTGGACATGTGGAGTGTTGG) and HCD2 (GCCCTCTCAGTGTCCAGAAG); human HSP60-specific oligonucleotides, HHSP1 (ACAAGTGATGTTGAAGTGAATG) and HHSP2 (ATTGCTGGAATTTTGAGTGTTC); rat TERT-specific oligonucleotides, RT1 (CGTAAGAGTGTGTGGAGCAA) and RT2 (GGATGAAGCGCAGTCTGCA); rat GAPDH-specific oligonucleotides, RG1 (AACGGATTTGGCCGTATTGGCCG) and RG2 (GACAATCTTGAGGGAGTTGTCATA); rat CDK4-specific oligonucleotides, RCD1 (TGGGTGCGGTGCCTATGGGA) and RCD2 (CGCATCTGGTAGCTGTAGATTC); and rat HSP60-specific oligonucleotides, RHSP1 (ACAAGTGATGTTGAAGTGAATG) and RHSP2 (ATTGCAGGAATTTTAAGTGCTC). Oligonucleotides specific for human and rat CAD cDNA were CAD1 (GGTGGATCTGGAGCATGAGTGGA) and CAD2 (AGATGGAAGCGGCCATCAGGAAG). For the quantitative analysis of PCR products, oligonucleotides HT1, HG1, HCD1, HHSP1, RT1, RG1, RCD1, RSP1, and CAD1 were end labeled with [γ-32P]ATP. Amplification was performed in a T3 Thermocycler (Biometra) for 31 cycles (TERT), 19 cycles (GAPDH), and 21 cycles (CAD) at 94°C for 45 s, 60°C for 45 s, and 72°C for 60 s, followed by a final extension step at 72°C for 5 min. The optimal number of cycles for exponential amplification was determined by kinetic analysis. All quantitative PCRs were repeated at least two times. PCR products were resolved on a 6% polyacrylamide gel and were quantitated using the Molecular Dynamics Phosphor Imaging System.
Immunoprecipitation.
HEK 293 cells were transfected by the calcium phosphate method. Cells were lysed using F buffer (44) 48 h after transfection. For immunoprecipitations, lysates were incubated with anti-FLAG antibodies with constant rocking at 4°C overnight, followed by several 1-h washes in F buffer and/or F buffer supplemented with 0.3 M NaCl. Elution was carried out overnight at 4°C in F buffer supplemented with FLAG peptide (100 μg/ml). To determine levels of protein production, proteins eluted from beads were resolved on sodium dodecyl sulfate (SDS) polyacrylamide gels and were Western blotted to a nitrocellulose membrane. Membranes were probed with anti-FLAG antibodies (M2, Sigma), anti-c-Myc (N262), anti-N-myc (C19), anti-TRRAP (T17), and anti-GCN5 (N18), all from Santa Cruz. Antibodies against TIP49 and BAF53 were previously described (39, 50).
Chromatin immunoprecipitation.
Retrovirally infected IMR90 cells were cross-linked by adding formaldehyde (final concentration, 1%; Fisher Scientific) directly to the cells on a rocking tissue culture dish for 10 min at room temperature. Cross-linking was terminated by addition of glycine to a final concentration of 125 mM. Fixed cells were washed with phosphate-buffered saline containing 0.5 mM phenylmethylsulfonyl fluoride (PMSF) followed by scraping cells in swelling buffer [5 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (pH 8.0), 85 mM KCl, 0.5% NP-40, 0.5 mM PMSF, and 100 ng of leupeptin and aprotinin per ml]. Cells were incubated on ice for 20 min and were then Dounce homogenized. Nuclei were harvested by centrifugation (4,500 × g) following resuspension in sonication buffer (0.1% SDS, F buffer, 0.5 mM PMSF, and 100 ng of leupeptin and aprotinin per ml) and were sonicated on ice to obtain 1,000- to 2,000-bp-long DNA fragments. After sonication, lysates were cleared by microcentrifugation at 10,000 × g. Lysates were normalized according to their optical density measurements. Each lysate was diluted six times with dilution buffer (F buffer, 0.5 mM PMSF, and 100 ng of leupeptin and aprotinin per ml) and were normalized by dilution in immunoprecipitation buffer (0.015% SDS, F buffer, 0.5 mM PMSF, and 100 ng of leupeptin and aprotinin per ml). The normalized chromatin lysates were precleared overnight with preblocked protein A/G beads. Protein A/G beads were preblocked by incubation in immunoprecipitation buffer containing 1 μg of salmon sperm DNA per ml and 1 μg of bovine serum albumin per ml for 3 h at 4°C. Precleared lysates were incubated with 1.5 μg or 5 μl of anti-acetylated histone H3 or H4 antibodies (Upstate Biotechnology), respectively, at 4°C overnight. Beads were washed and eluted, followed by reversing cross-linking and DNA isolation using the Qiagen PCR purification kit. Eluted material was subjected to quantitative PCR amplification. PCR was performed using human TERT-promoter specific primers SC-9 (AGTGGATTCGCGGGCACAGA) and SC-10 (AGCACCTCGCGGTAGTGGCT). To normalize samples by the amount of nonspecific DNA, we amplified a region in the third intron of the human β-globin gene using the following primers: SC-46 (ATCTTCCTCCCACAGCTCCT) and SC-47 (TTTGCAGCCTCACCTTCTTT). Oligonucleotides SC-9 and SC-46 were end labeled with [γ-32P]ATP. The optimal number of cycles for exponential amplification was determined by kinetic analysis. PCR products were resolved on a 4 or 6% polyacrylamide gel. PCR products were quantitated and normalized using the Molecular Dynamics Phosphor Imaging System.
HAT assay.
Complexes containing FLAG-tagged proteins were eluted from the FLAG-conjugated beads by addition of excess FLAG peptide (100 μg/ml) in HAT assay buffer (50 mM Tris-HCl at pH 8.0, 10% glycerol, 50 mM KCl, 0.1 mM EDTA, 1 mM dithiothreitol, 1 mM PMSF, and 10 mM butyric acid). Core histones from HeLa cells (3 μg) along with [14C]acetyl coenzyme A were incubated with each eluate at 30°C for 30 min.
RESULTS
Considerable effort has been expended to define a collection of genes that mediate Myc function, but few studies have addressed the mechanism by which Myc activates individual target genes. Mutations that disrupt the evolutionarily conserved MBII region in the transactivation domain of c-Myc have generally been found to have unaltered transactivation in transient assays (5, 30), yet these same mutants are defective in cell transformation assays (10, 46). On the other hand, the L-Myc protein, which possesses an intact MBII domain, is also severely defective for oncogenesis in most assays (6). Gene knockouts of L-myc are viable, unlike the knockouts of c-myc and N-myc (14, 26, 43, 45). We were interested in determining if chromatin-modifying factors interacting with the Myc family proteins were required for Myc-dependent activation of specific cellular promoters in the context of their native chromosomal location in normally growing cells and if different Myc family proteins exhibited differential gene activation. We focused on several promoters (TERT, CAD, HSP60, and CDK4) that have functional binding sites for Myc/Max heterodimers and that can be directly cross-linked to Myc in vivo.
MBII is required for activation of the TERT gene in IMR90 cells.
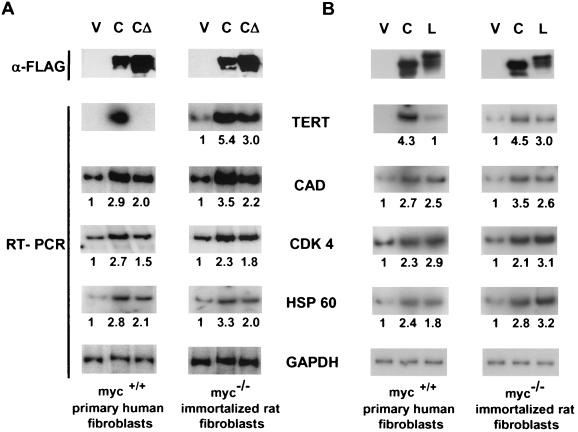
As an initial assay for the activation of Myc regulated genes, we tested gene expression in response to wild-type c-Myc and the c-Myc(Δ129-145) mutant (c-MycMBIIΔ hereafter) that is defective for oncogenic activity. The TERT gene is silent in IMR90 cells, and ectopic expression of wild-type c-Myc activates expression as described previously (49). In contrast, the c-MycMBIIΔ mutant was completely defective in TERT activation (Fig. 1A). Unlike the case for TERT, expression of the CAD, CDK4, and HSP60 genes is readily detectable in IMR90 cells, and expression is enhanced approximately threefold by ectopic wild-type c-Myc. Interestingly, the c-Myc-MBIIΔ protein also induced CAD, CDK4, and HSP60 expression, although to a lesser extent (Fig. 1A). These data demonstrate that the ability of c-Myc to activate a silent gene like TERT is completely dependent on MBII, whereas other genes exhibit only a modest MBII dependence.
FIG. 1.
Activation of endogenous genes by c-Myc, c-MycΔ, and L-Myc. IMR90 and HO15.19 cells were infected with empty vector or retroviral vectors expressing FLAG-tagged c-myc or c-mycΔ cDNAs (A) or empty vector and c-myc or L-myc cDNA (B). To determine protein expression, normalized cell lysates were immunoprecipitated with anti-FLAG antibodies and were resolved on an SDS-8% polyacrylamide gel followed by Western blotting with anti-FLAG specific antibodies. Quantitative PCR was performed with cDNA synthesized on total RNA isolated from infected cells using radiolabeled oligonucleotides specific for human or rat genes indicated in the center. Quantitation of PCR products was performed using the Molecular Dynamics Phosphor Imaging System. Numbers below a panel indicate the induction (n-fold) determined by dividing a TERT-, CDK4-, HSP60-, or CAD-specific signal by a corresponding GAPDH-specific signal and by the ratio of these signals in the vector (V) lane. RT-PCR, reverse transcriptase PCR.
We next examined expression of all four genes in an established fibroblast cell line in which the c-myc genes were knocked out by homologous recombination (34). Unlike what is found in IMR90 cells, the TERT gene is expressed even in the absence of endogenous c-Myc in c-myc-null fibroblasts (Fig. 1A). TERT is induced by reconstitution with either wild-type c-Myc (5.4-fold) or c-MycΔ (threefold). The response of the CAD, CDK4, and HSP60 genes was similar to that in IMR90 cells, with approximately three- and twofold induction by wild-type c-Myc and by c-MycMBIIΔ, respectively. Thus, TERT, CAD, CDK4 and HSP60 expression is sustained in immortalized cell lines in the absence of c-Myc, and the modest Myc-dependent transactivation of these genes does not depend on the MBII domain.
L-Myc is a weak activator of TERT but not of other Myc targets in IMR90 cells.
We were next interested in whether or not other Myc family proteins had a similar capacity to activate endogenous promoters. L-Myc has been described as having both weak oncogenic activity and poor transactivation in transient assays (4, 6). We transduced a FLAG-tagged L-Myc protein into IMR90 cells, which was expressed at the same level as c-Myc as determined by Western blotting (Fig. 1B). L-Myc reproducibly activated the TERT gene to approximately 20% of the level induced by c-Myc (Fig. 1B). On the other hand, other Myc targets (CAD, CDK4, and HSP60) were up-regulated by L-Myc to the same extent as by c-Myc. In contrast to its response in transient assays, the activity of L-Myc in reconstituted c-myc-null fibroblasts paralleled that of c-Myc, where all four Myc target genes were induced two- to threefold. Thus, L-Myc exhibits transactivation activity similar to that of c-Myc, with the noteworthy exception of activation of the TERT gene in primary human fibroblasts.
TERT activation correlates with TRRAP binding.
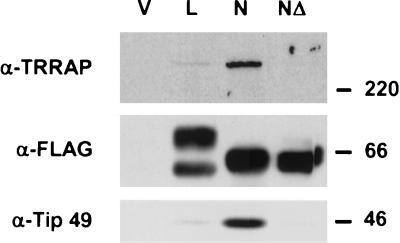
The data above establish a strong requirement for the MBII of c-Myc for TERT activation in primary cells. Previously it was reported that the MBII domain in c- and N-Myc was involved in an interaction with several nuclear cofactors, including TRRAP, TIP48/49, and BAF53 (35, 39, 50). We were interested in establishing a direct link between recruitment of individual cofactors by the Myc family proteins and the activation of Myc-responsive genes. We first tested if L-Myc bound to the same cofactors to which other Myc family proteins did. To this end, FLAG-tagged L-Myc and N-Myc proteins were transiently expressed in HEK 293 cells and were tested for binding to endogenous nuclear cofactors. L-Myc binding to TRRAP and TIP49 was substantially reduced compared to that of N-Myc (Fig. 2). However, binding of nuclear cofactors to L-Myc was still detectable in comparison to the situation for N-MycMBIIΔ, which showed no binding to the nuclear cofactors tested. This weak binding of L-Myc to TRRAP correlates well with its impaired ability to activate the silent TERT gene in IMR90 cells, but as with c-MycMBIIΔ, it does not correlate with the ability of L-Myc to activate other Myc target genes.
FIG. 2.
Recruitment of nuclear cofactors by L-Myc and N-Myc. HEK 293 cells were transiently transfected with FLAG-tagged constructs encoding proteins shown on the top. Normalized cell lysates were prepared and immunoprecipitated with anti-FLAG antibodies. Immunoprecipitated complexes were resolved on a gradient SDS- polyacrylamide gel and were probed by Western blotting with several antibodies indicated on the left. Protein molecular masses are shown on the right in kilodaltons.
E1A-N-MycΔ chimeras selectively bind to TRRAP.
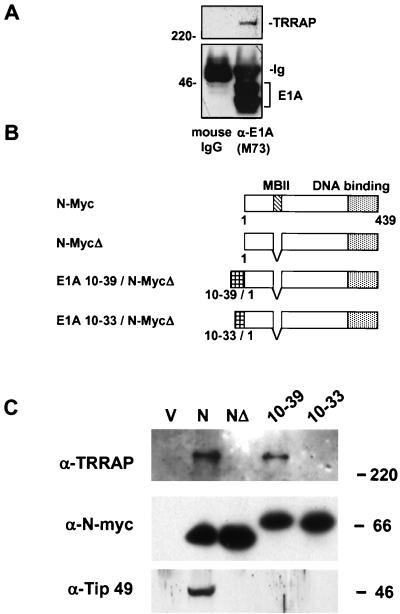
Further support for a link between TRRAP recruitment and TERT activation came from an unexpected direction through studies of the adenoviral oncoprotein E1A. E1A shares many of the biological activities of c-Myc, including oncogenic transformation of primary cells in cooperation with the H-rasG12V oncogene, blocking of differentiation, and promotion of apoptosis (20). E1A is thought to predominantly act through the E2F family of transcription factors (42). It was previously shown that TRRAP bound to the E2F transactivation domain (35) and that TRRAP recruitment was important for E2F activity (33). Since E1A and E2F bind to many of the same cellular cofactors, we tested if E1A bound to TRRAP. As shown in Fig. 3A, immunoprecipitates of E1A from HEK 293 cells indeed contained TRRAP when assayed by Western blotting. The biological consequences of E1A binding to TRRAP are presently unknown, but E1A may modify SAGA function in adenovirus-infected cells or bind to other TRRAP-containing complexes (23).
FIG. 3.
E1A-N-MycΔ chimeras selectively precipitate TRRAP from HEK 293 cells. (A) Normalized whole-cell lysates were prepared from HEK 293 cells that constitutively express the adenovirus E1A protein. Immunoprecipitates were resolved on an SDS-polyacrylamide gel and were probed for the presence of TRRAP in Western blotting using homemade rabbit anti-TRRAP antisera. Results of these studies demonstrate that TRRAP specifically associates with the E1A protein in vivo. 220, 220 kDa. (B) Schematic representation of the E1A-N-MycΔ fusion proteins. E1A portion of the chimera, MBII domain, and DNA binding domain are shown by striped, hatched, and dotted boxes, respectively. (C) Recruitment of nuclear cofactors by N-Myc-derived proteins. HEK 293 cells were transiently transfected with FLAG-tagged constructs encoding proteins shown in panel B. Normalized cell lysates were prepared and immunoprecipitated with anti-FLAG antibodies. Immunoprecipitated complexes were resolved on a gradient SDS-polyacrylamide gel and probed by Western blotting with several antibodies, indicated on the left. Protein molecular masses are shown on the right in kilodaltons.
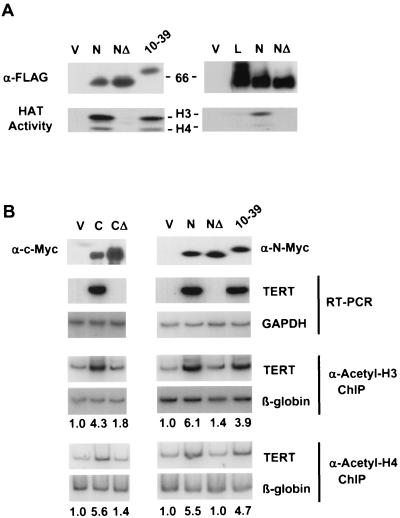
It was previously shown that chimeric proteins with the E1A N terminus fused to the c-Myc DNA binding domain were potent oncogenes (41). The binding of E1A to TRRAP prompted us to test if this interaction could explain the oncogenic activity of E1A-Myc chimeras and be used to molecularly complement the defect in TRRAP binding of the MBII mutations in Myc. We used a FLAG-tagged N-Myc expression vector, since N-Myc binds avidly to TRRAP, and we also derived a deletion mutant in the N-Myc MBII domain that is defective in both TRRAP binding and oncogenic activity (Fig. 2 and see below). We then constructed a series of chimeras in which various segments of the E1A N terminus were fused to the N-MycMBIIΔ protein (Fig. 3B).
We first tested if a specific E1A domain could restore TRRAP binding to N-MycMBIIΔ. Proteins were transiently expressed in HEK 293 cells, immunoprecipitated through a FLAG epitope tag, and assayed for TRRAP binding by Western blotting. Preliminary mapping demonstrated that E1A amino acids 1 to 86 could restore TRRAP binding (data not shown), which was subsequently narrowed to E1A amino acids 10 to 39 (Fig. 3C), although binding was consistently weaker than to wild-type N-Myc. Fusion of E1A amino acids 10 to 33 to N-MycMBIIΔ failed to restore binding (Fig. 3C), implicating E1A amino acids 33 to 39 as critical for TRRAP interaction. We also tested the E1A-N-Myc chimeras for binding to another Myc cofactor, TIP49. There was no enhancement of TIP49 binding to the E1A-N-MycΔ chimeras compared to that to N-MycMBIIΔ (Fig. 3C). Thus, a small E1A domain selectively reconstitutes binding of TRRAP to an N-Myc mutant with a deletion of the MBII region but not binding to another Myc-associated complex(es).
E1A-MycΔ chimeras activate the silent TERT gene.
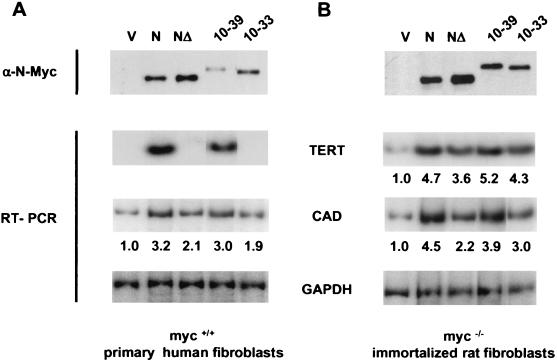
The E1A-N-MycΔ chimeras, wild-type N-Myc, and N-MycΔ were each introduced into IMR90 and c-myc-null fibroblasts to assay their ability to activate target gene expression. Remarkably, the E1A(10-39)-N-MycMBIIΔ fusion protein that bound to TRRAP could also activate the silent TERT gene in IMR90, similar to wild-type N-Myc (Fig. 4A). In contrast, the N-MycMBIIΔ mutant and the E1A(10-33)-N-MycMBIIΔ fusion proteins were completely defective in TERT activation. The response of the endogenous CAD gene to N-Myc and the E1A-N-Myc chimeras was similar to that for c-Myc in IMR90 cells. A representative experiment (Fig. 4A) demonstrated that N-Myc provided a 3.2-fold activation of CAD, whereas N-MycMBIIΔ induced CAD 2.1-fold. Each of the E1A-N-MycMBIIΔ chimeras activated CAD to a similar extent, including the E1A(10-33)-N-MycMBIIΔ fusion that failed to activate TERT in IMR90. All proteins were expressed at similar levels, although N-MycΔ was reproducibly expressed at a higher level than was wild-type N-Myc and although the E1A chimeras were expressed at a lower level (Fig. 4A, top gel). These data provide a further link between recruitment of TRRAP by Myc and its ability to activate transcription of the silent TERT locus.
FIG. 4.
E1A-N-MycΔ chimeras restore the transactivation potential of N-MycΔ. IMR90 (A) and HO15.9 (B) cells were infected with the retroviral vectors carrying cDNAs for proteins shown in Fig. 3B. To determine protein expression, normalized lysates from infected cells were immunoprecipitated with anti-FLAG antibodies and were resolved on an SDS-8% polyacrylamide gel followed by Western blotting with N-Myc-specific antibodies. Quantitative PCR was performed on cDNA synthesized on RNA isolated from cells expressing the constructs indicated on the top. PCR was done using radiolabeled oligonucleotides specific for cDNA of human (A) or rat (B) genes shown in the center. Quantitation of PCR was done using the Molecular Dynamics Phosphor Imaging System. Numbers below a panel indicate an induction (n-fold) determined by dividing a TERT- or CAD-specific signal by a corresponding GAPDH-specific signal and by the ratio of these signals in the vector (V) lane. A representative experiment is shown.
In c-myc-null fibroblasts, the response of the CAD and TERT genes to the N-Myc and E1A-N-MycMBIIΔ proteins was slightly higher than that in primary human fibroblasts. As shown in Fig. 4B, expression of CAD and TERT was induced 4.5- and 4.7-fold by N-Myc, and 2.2- and 3.6-fold, respectively, by N-MycMBIIΔ. The E1A(10-39)- and E1A(10-33)-N-MycMBIIΔ proteins enhanced CAD and TERT expression to a level similar to that induced by wild-type N-Myc. There was no correlation between TERT activation and CAD activation in c-myc-null fibroblasts by fusion proteins that can activate TERT, i.e., E1A(10-39)-N-MycMBIIΔ, and cannot activate TERT, i.e., E1A(10-33)-N-MycMBIIΔ, in IMR90 cells (compare Fig. 4A and B).
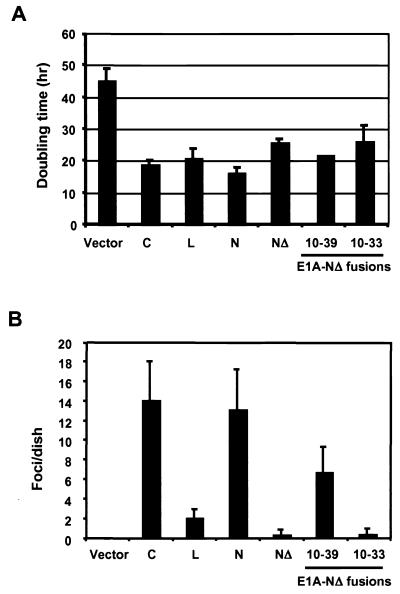
It is noteworthy that the ability of N-MycMBIIΔ to induce transcription of basally expressed genes in c-myc-null fibroblasts (exemplified by the CAD and TERT genes) correlates well with its ability to complement the growth defect of these cells. As shown in Fig. 5A, N-Myc completely rescues the slow-growth phenotype of c-myc-null cells, decreasing their doubling time from 45 to 16 h. Expression of N-MycMBIIΔ also resulted in a decrease of the doubling time to approximately 26 h. The E1A-N-MycMBIIΔ chimeras both restored the growth rate of c-myc-null cells to levels that were intermediate between those of N-Myc and N-MycMBIIΔ. Expression of E1A(10-39)-N-MycΔ provided a doubling time of 21 ± 0.3 h versus 26 ± 5 h for E1A(10-33)-N-MycΔ. Interestingly, L-Myc, despite poor binding to TRRAP, restored the growth rate of c-myc-null cells to a level nearly identical (20 h) to that resulting from the expression of c-Myc or N-Myc (Fig. 5A).
FIG. 5.
Distinct phenotypes of c-Myc, L-Myc, N-Myc, and E1A-N-MycΔ chimeras in cell growth and primary cell transformation. (A) Doubling times of myc-null cells reconstituted with the indicated Myc-derived proteins. (B) Oncogenic activity of the Myc family proteins and E1A-N-MycΔ chimeras in primary rat embryo cells. c-myc, L-myc, N-myc, N-mycΔ, or E1A-N-mycΔ chimeras were cotransfected with H-rasG12V into early passage rat embryo fibroblasts, and foci were scored after 18 days.
N-Myc, c-Myc, and E1A-N-MycMBIIΔ proteins recruit HAT activity to the TERT promoter in IMR90 cells.
The data above strongly suggest that recruitment of a TRRAP-containing complex(es) by Myc is required for activation of the TERT gene in primary human fibroblasts. Several recent studies have demonstrated that TRRAP may exist in a complex with GCN5, a histone H3-specific acetyltransferase (36, 38); TIP60, a histone H4-specific HAT (29); or a novel undefined H4 HAT complex (39). Moreover, recruitment of H4 (and to a lesser extent H3) HAT activity correlated with the activation of several Myc-responsive genes in starved cells after serum stimulation (8, 21). We were interested in whether Myc was able to recruit a specific HAT activity to the nucleosomes on the TERT promoter in IMR90 cells. We first tested if interactions with TRRAP were crucial for recruiting HAT activity by Myc. To this end, complexes containing FLAG-tagged N-Myc, N-MycMBIIΔ, and E1A(10-39)-N-MycMBIIΔ proteins were transiently expressed in HEK 293 cells, immunoprecipitated using FLAG-specific antibodies, eluted from the beads, and assayed for HAT activity and the presence of TRRAP. As shown in Fig. 6A, N-Myc coimmunoprecipitates TRRAP and both H3- and H4-specific acetyltransferase activity. In contrast, the N-MycΔ protein fails to recruit TRRAP and any HAT activity, whereas E1A(10-39)-N-MycΔ recruits both TRRAP (Fig. 2) and H3- and H4-HATs. The HAT activity recruited by E1A(10-39)-N-MycΔ was not as robust as that for wild-type N-Myc, consistent with the smaller amount of TRRAP recruited by this fusion. L-Myc also recruited a reduced level of HAT activity compared to that recruited by N-Myc, but the level was higher than that for N-MycMBIIΔ (Fig. 6A).
FIG. 6.
Recruitment of histone acetyltransferase activity by Myc family proteins. (A) Complexes containing the transiently expressed FLAG-tagged proteins designated on the top were immunoprecipitated from lysed HEK 293 cells using anti-FLAG antibodies. FLAG-tagged proteins were eluted from the beads and were assayed for HAT activity on equal amounts of core histones isolated from HeLa cells. Reaction mixtures were resolved on gradient SDS-4 to 15% polyacrylamide gels and were transferred to a nitrocellulose membrane. The upper section of the membrane was probed by Western blotting with the antibodies designated on the left. Protein molecular weights are shown on the right. The bottom portion of the membrane was subjected to fluorography for HAT activity. Positions of histones H3 and H4 are shown on the right. 66, 66 kDa. (B) IMR90 cells were infected with retroviral vectors carrying the cDNAs indicated on the top. To determine protein expression, normalized lysates from infected cells were immunoprecipitated with anti-FLAG antibodies, resolved on SDS-8% polyacrylamide gel, followed by Western blotting with anti-N-Myc antibodies. cDNA was synthesized on RNA isolated from cells infected with the constructs indicated above and was used as a template in PCR with radiolabeled oligonucleotides specific for human TERT or GAPDH cDNA. Cells infected with the constructs indicated above were cross-linked and lysed, and chromatin was immunoprecipitated (ChIP) with either anti-acetylated H3 or anti-acetylated H4 histone antibodies followed by the reversion of the cross-linking and DNA isolation. Isolated DNA was used in PCR with radiolabeled oligonucleotides flanking Myc binding sites in the promoter of the human TERT gene or a DNA region of similar size in the third intron of a silent β-globin gene. Quantitation of PCR products was performed using the Molecular Dynamics Phosphor Imaging System. Numbers below a panel indicate a difference (n-fold) in a signal intensity determined by dividing a TERT-specific signal by a corresponding globin-specific signal and by the ratio of these signals in the vector lane. RT-PCR, reverse transcriptase PCR.
We were next interested in whether N-Myc and E1A(10-39)-N-MycMBIIΔ recruited histone H3- and/or H4-specific acetyltransferase activity to the TERT promoter. Antibodies specific to acetylated histone H3 or H4 were used to immunoprecipitate cross-linked chromatin from IMR90 cells expressing N-Myc, N-MycMBIIΔ, E1A(10-39)-N-MycMBIIΔ, c-Myc, c-MycMBIIΔ, or vector. After reversal of the cross-linking, DNA was purified and subjected to quantitative PCR with oligonucleotides corresponding to the region of the TERT promoter encompassing two Myc binding sites or to a region of similar size in the third intron of a silent β-globin gene for a control purposes. As shown in Fig. 6B, there was a significant enrichment of the TERT promoter sequences (3.9- to 6.1-fold) in the samples immunoprecipitated with anti-acetylated H3 and anti-acetylated H4 antibodies from all cells in which the TERT gene was activated [c-Myc, N-Myc, or E1A(10-39)-N-MycMBIIΔ] compared to the empty vector, using β-globin as a normalization control. There was very little or no enrichment in samples obtained from cells expressing c-MycΔ or N-MycMBIIΔ, in which the TERT gene is silent. We assume that the signal from the β-globin gene represents a measure of nonspecific DNA binding to the beads during the immunoprecipitation step, since this gene should be silent in primary fibroblasts. However, we cannot exclude the possibility that there is a low level of histone acetylation even on silent genes, including the silent TERT locus. Taken together our data demonstrate that Myc-dependent activation of the silent TERT gene in IMR90 cells requires recruitment of TRRAP-containing complexes to the TERT promoter and that this recruitment is accompanied by increased acetylation of histones H3 and H4 in the promoter area.
TRRAP binding correlates strongly with Myc family oncogenic activity.
One of the most revealing assays for Myc oncogenic function is the transformation of primary rodent cells in cooperation with the H-rasG12V oncogene (31). This assay was originally used to demonstrate the potent transforming activity of E1A-Myc chimeras (41), so we were interested in determining if the E1A-MycMBIIΔ proteins described above would also transform cells and how this activity related to that of L-Myc. Each expression construct was cotransfected with H-rasG12V into early passage rat embryo fibroblasts (31), and foci of transformed cells were scored after 18 to 21 days. As shown in Fig. 5B, c-Myc and N-Myc are highly oncogenic, whereas deletion of the N-Myc MBII abolishes transforming activity. L-Myc has a significantly weaker transforming activity, consistent with earlier reports (6). Fusion of E1A sequences 10 to 39 to N-MycMBIIΔ partially rescued N-Myc transforming activity. In contrast, the fusion of E1A(10-33) to N-MycMBIIΔ was as defective as N-MycMBIIΔ (Fig. 5B). Thus, mapping of E1A segments that rescue the oncogenic activity of Myc in primary cells identifies the same region (10-39) as that required for activation of the silent TERT gene and the defective E1A(10-33) fusions define amino acids 33 to 39 as a critical boundary for both activities.
The fusion of E1A sequences to N-MycMBIIΔ mutants restored binding to TRRAP, but we wanted to demonstrate a more direct involvement of TRRAP in the biological activity of the chimeras. We previously described a segment of TRRAP (amino acids 1261 to 1579) that is a dominant inhibitor of Myc oncogenic activity when ectopically expressed in conjunction with the H-rasG12V oncogene. Cotransfection of plasmids expressing TRRAP(1261-1579) and E1A(10-39)-N-MycMBIIΔ proteins strongly inhibited focus formation in rat embryo fibroblasts (90%) compared to cotransfection of E1A(10-39)-N-MycMBIIΔ with the empty vector (data not shown). These results support the coimmunoprecipitation experiments and argue for a direct involvement of TRRAP in the oncogenic activity of E1A-MycMBIIΔ chimeras.
DISCUSSION
Recruitment of TRRAP by Myc family proteins.
It has long been known that Myc-mediated cellular phenotypes largely depend on the integrity of the Myc transactivation domain, particularly MBII. Several recent studies defined nuclear factors capable of interaction with MBII, thus providing molecular insight into mechanisms of Myc action (35, 50). TRRAP, an integral component of human SAGA and TIP60 complexes (29, 48), is the first polypeptide shown to interact with MBII (35). Human SAGA-related complexes contain GCN5/PCAF and specifically acetylate histone H3, whereas the TIP60 complex acetylates predominantly histone H4. Recently, another TRRAP-containing complex with H4 HAT activity that does not contain TIP60 was reported (39). Results obtained from histone acetylation in vitro and chromatin immunoprecipitation in cultured cells strongly suggest that Myc interacts with at least two distinct TRRAP-containing complexes containing HATs with different histone specificity. A third TRRAP-containing complex that lacks HAT activity has also been described recently (23). Other Myc-interacting proteins include TIP48, TIP49, and BAF53 (39, 50), which can be involved in other TRRAP-containing (29) or non-TRRAP complexes (LeRoy and Wang, personal communication). In our study, it was important to determine the individual involvement of these proteins in Myc-dependent pathways. We found that L-Myc interacts with TRRAP/TIP49 polypeptides and recruits HAT activity much more weakly than c- or N-Myc, which correlates well with the weak oncogenic activity of L-Myc in the H-ras cooperation assay.
We used a small fragment of adenoviral E1A protein (amino acids 10 to 39) to selectively restore an interaction with TRRAP in the context of a MBII deletion without detectable concomitant binding to TIP49 (Fig. 3C). This portion of E1A does not overlap the conserved CR1, CR2, and CR3 regions implicated in other protein-protein interactions and E1A functions (3). A previous study of E1A-interacting proteins identified a 400-kDa band in immunoprecipitates from adenovirus-infected cells that was dependent on amino acids 4 to 49 (2), overlapping the 33-39 boundary that defines TRRAP recruitment to the E1A-N-MycMBIIΔ fusion proteins. An E1A mutant with a deletion of amino acids 26 to 35 was defective for p400 binding but retained the ability to transform baby rat kidney cells in combination with H-rasG12V (2). However, more detailed analysis of the E1AΔ26-35 mutant reveals a substantial deficiency in transforming activity and TRRAP binding compared to what is found in wild-type E1A (15). It is not yet clear if p400 is TRRAP or is a recently described Swi/Snf-related protein of similar migration (23). It is tempting to speculate that E1A binding to TRRAP might alter its activity or the activity of TRRAP-containing, chromatin-modifying complexes. E1A itself does not activate TERT expression (49), presumably because it lacks a DNA binding domain that would target the protein to the TERT locus.
An array of complexes have been found to contain TRRAP and/or bind to Myc proteins (reviewed in the introduction). At face value, the observation that the E1A(10-39)-N-MycMBIIΔ protein binds to TRRAP but not to TIP49 argues that the chimera does not bind to either the TIP60 or p400 complex, both of which contain TIP49 (23, 29). It was recently shown that a TRRAP-associated H4 HAT complex can be distinguished biochemically from the TIP60 complex, although the enzyme itself has not been identified (39). It is tempting to speculate that this latter complex is recruited by the E1A-N-MycMBIIΔ chimera.
TRRAP-dependent activation of Myc-responsive genes.
Telomerase is a two-component ribonucleoprotein DNA polymerase composed of telomerase RNA and TERT (reviewed in reference 25). Telomerase synthesizes short DNA repeats at chromosomal ends and is required for maintaining genomic stability (reviewed in reference 1). Although telomerase is active in many fetal tissues during embryonic development, its activity is gradually repressed in most somatic cells shortly after birth. However, the TERT gene becomes active again during cell immortalization and in the vast majority of human cancers (25). Despite an extensive search for TERT transcriptional activators, Myc is the only factor at present that has been shown to activate expression of a silent TERT gene by binding to its promoter (49). However, the molecular mechanism of this activation by Myc was not elucidated. We showed that overexpression of c-Myc, N-Myc, and, to a much lesser extent, L-Myc activates expression of the silent TERT gene. The use of Myc mutants and E1A-Myc fusion proteins provides compelling evidence that TERT activation and nucleosome modification directly result from the recruitment of TRRAP and its associated histone H3 and H4 HAT activities by the Myc transactivation domain.
An important finding from these studies is the striking difference between primary and immortalized exponentially growing cells in their requirement for TRRAP-containing complexes for activation of the TERT gene. In primary cells, the TERT gene is strongly repressed through unknown mechanisms, whereas in immortalized cells TERT is basally expressed even in the absence of Myc. Presumably, other transcription factors such as Sp1 are responsible for basal promoter activity in c-myc-null cells, but these factors are excluded from activation of TERT in primary cells, even though factors such as Sp1 are ubiquitously expressed. Repression of TERT may involve active histone deacetylation, since trichostatin A can activate TERT expression in different cell types and since the Mad/Max complex can bind to the E box in the TERT promoter (52). However, trichostatin A might also activate other genes that directly or indirectly derepress TERT, including Myc itself.
In contrast to the silent TERT gene, induction of basally expressed genes CAD, CDK4, and HSP60 in exponentially growing IMR90 cells and of CAD, CDK4, HSP60, and TERT in exponentially growing c-myc-null cells did not depend strictly on the MBII domain that is required for recruitment of TRRAP and HAT activity by Myc. This observation is consistent with a previous study of Myc target genes in which Myc binding to the promoter of the active TERT gene in an exponentially growing neuroblastoma cell line occurred without a concomitant change in histone acetylation (16). Moreover, recent studies demonstrated the ability of Myc to activate transcription of the CAD gene through mechanisms independent from histone acetylation (17). We found that Myc and the E1A-N-MycMBIIΔ proteins were sometimes more potent than MycMBIIΔ in the induction of the CAD, CDK4, HSP60, and TERT genes (in HO15.19 cells), suggesting that TRRAP complexes may play a limited role in transcriptional activation of these genes. Furthermore, recent reports show that TRRAP and its associated H4 (but not H3) HAT activities may have a more significant role in the induction of Myc target genes during the serum stimulation of starved cells (21), as opposed to the exponentially growing cells studied here.
The strict dependence of TERT activation on the MBII domain and TRRAP/HAT recruitment in primary fibroblasts correlates well with the similar dependence on the MBII domain in the oncogenic transformation of primary cells in the Myc/H-ras cooperation assay. This link is further supported by the weak binding of L-Myc to TRRAP and its limited recruitment of HAT activity, which correlates with its relatively weak oncogenic activity in primary fibroblasts. An H-ras oncogene alone can transform immortalized cells but requires a cooperating myc oncogene to transform primary cells. Since regulation of the CAD, CDK4, and HSP60 promoters by Myc shows only a minimal dependence on MBII, it is tempting to suggest that genes in this class may not be the rate-limiting targets of Myc in primary cells. On the other hand, it was shown that L-Myc and MycMBIIΔ were capable of a complete or partial growth enhancement of c-myc-null fibroblasts (11, 32; this study), and this enhancement correlated with the induction of the CAD, CDK4, HSP60, and TERT genes (Fig. 3B and 6A). This observation supports the idea that Myc promotes cell growth and causes cellular transformation through the activation of different sets of genes. One attractive model is that the transformation of primary cells may require the activation of strongly repressed TERT-like genes, which can be achieved only through HAT-mediated chromatin modifications. However, at the present time we do not know any other Myc-responsive genes that are silent in normal fibroblasts and activated in transformed or immortalized cells. On the other hand, TERT itself is unlikely to be the rate-limiting Myc target, because TERT overexpression is not able to replace Myc in the transformation of rat fibroblasts (24) and because cells derived from mice lacking telomerase RNA can still be transformed by cooperating myc and H-ras oncogenes (7). A screen for other genes that are strongly repressed in primary cells and activated by Myc overexpression may identify critical Myc target genes involved in oncogenesis.
Acknowledgments
M.A.N., S.C., and J.P. made equal contributions to this study.
We are grateful to Heather Van Buskirk for critical reading of the manuscript. We thank H. Land and B. Amati for communicating experiments prior to publication.
This work was supported by a grant from the New Jersey Commission on Cancer Research to S.C. and by grants from the National Institutes of Health to M.D.C.
REFERENCES
- 1.Artandi, S. E., and R. A. DePinho. 2000. A critical role for telomeres in suppressing and facilitating carcinogenesis. Curr. Opin. Genet. Dev. 10:39-46. [DOI] [PubMed] [Google Scholar]
- 2.Barbeau, D., R. Charbonneau, S. G. Whalen, S. T. Bayley, and P. E. Branton. 1994. Functional interactions within adenovirus E1A protein complexes. Oncogene 9:359-373. [PubMed] [Google Scholar]
- 3.Barbeau, D., R. C. Marcellus, S. Bacchetti, S. T. Bayley, and P. E. Branton. 1992. Quantitative analysis of regions of adenovirus E1A products involved in interactions with cellular proteins. Biochem. Cell Biol. 70:1123-1134. [DOI] [PubMed] [Google Scholar]
- 4.Barrett, J., M. J. Birrer, G. J. Kato, H. Dosaka-Akita, and C. V. Dang. 1994. Activation domains of L-Myc and c-Myc determine their transforming potencies in rat embryo cells. Mol. Cell. Biol. 12:3130-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bello-Fernandez, C., G. Packham, and J. L. Cleveland. 1993. The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc. Natl. Acad. Sci. USA 90:7804-7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Birrer, M. J., S. Segal, J. S. DeGreve, F. Kaye, E. A. Sausville, and J. D. Minna. 1988. L-myc cooperates with ras to transform primary rat embryo fibroblasts. Mol. Cell. Biol. 8:2668-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blasco, M. A., H. W. Lee, M. P. Hande, E. Samper, P. M. Lansdorp, R. A. DePinho, and C. W. Greider. 1997. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 91:25-34. [DOI] [PubMed] [Google Scholar]
- 8.Bouchard, C., O. Dittrich, A. Kiemaier, K. Dohmann, A. Menkel, M. Eilers, and B. Luscher. 2001. Regulation of cyclin D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP recruitment and histone acetylation at the cyclin D2 promoter. Genes Dev. 15:2042-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyd, K. E., and P. J. Farnham. 1997. Myc versus USF: discrimination at the cad gene is determined by core promoter elements. Mol. Cell. Biol. 17:2529-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brough, D. E., T. J. Hofmann, K. B. Ellwood, R. A. Townley, and M. D. Cole. 1995. An essential domain of the c-Myc protein interacts with a nuclear factor that is also required for E1A-mediated transformation. Mol. Cell. Biol. 15:1536-1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bush, A., M. Mateyak, K. Dugan, A. Obaya, S. Adachi, J. Sedivy, and M. D. Cole. 1998. c-myc null cells misregulate cad and gadd45 but not other proposed c-myc targets. Genes Dev. 12:3797-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cole, M. D., and S. B. McMahon. 1999. The Myc oncoprotein: a critical evaluation of transactivation and target gene regulation. Oncogene 18:2916-2924. [DOI] [PubMed] [Google Scholar]
- 13.Dang, C. V. 1999. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol. Cell. Biol. 19:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis, A. C., M. Wims, G. D. Spotts, S. R. Hann, and A. Bradley. 1993. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes Dev. 7:671-682. [DOI] [PubMed] [Google Scholar]
- 15.Deleu, L., S. Shellard, K. Alevizopoulos, B. Amati, and H. Land. 2001. Recruitment of TRRAP required for oncogenic transformation by E1A. Oncogene 20:8270-8275. [DOI] [PubMed] [Google Scholar]
- 16.Eberhardy, S., C. D'Cunha, and P. Farnham. 2000. Direct examination of histone acetylation on Myc target genes using chromatin immunoprecipitation. J. Biol. Chem. 275:33798-33805. [DOI] [PubMed] [Google Scholar]
- 17.Eberhardy, S. R., and P. J. Farnham. 2001. c-Myc mediates activation of the cad promoter via a post-RNA polymerase II recruitment mechanism. J. Biol. Chem. 276:48562-48571. [DOI] [PubMed] [Google Scholar]
- 18.Evan, G. I., A. H. Wyllie, C. S. Gilbert, T. D. Littlewood, H. Land, M. Brooks, C. M. Waters, L. Z. Penn, and D. C. Hancock. 1992. Induction of apoptosis in fibroblasts by c-myc protein. Cell 69:119-128. [DOI] [PubMed] [Google Scholar]
- 19.Facchini, L. M., and L. Z. Penn. 1998. The molecular role of Myc in growth and transformation: recent discoveries lead to new insights. FASEB J. 12:633-651. [PubMed] [Google Scholar]
- 20.Flint, J., and T. Shenk. 1997. Viral transactivating proteins. Annu. Rev. Genet. 31:177-212. [DOI] [PubMed] [Google Scholar]
- 21.Frank, S. R., M. Schroeder, P. Fernandez, S. Taubert, and B. Amati. 2001. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 15:2069-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freytag, S. O., C. V. Dang, and W. M. F. Lee. 1990. Definition of the activities and properties of c-myc required to inhibit cell differentiation. Cell Growth Differ. 1:339-343. [PubMed] [Google Scholar]
- 23.Fuchs, M., J. Gerber, R. Drapkin, S. Sif, T. Ikura, V. Ogryzko, W. S. Lane, Y. Nakatani, and D. M. Livingston. 2001. The p400 complex is an essential E1A transformation target. Cell 106:297-307. [DOI] [PubMed] [Google Scholar]
- 24.Greenberg, R., R. O'Hagan, H. Deng, Q. Xiao, S. Hann, R. Adams, S. Lichtsteiner, L. Chin, G. Morin, and R. DePinho. 1999. Telomerase reverse transcriptase gene is a direct target of c-Myc but is not functionally equivalent in cellular transformation. Oncogene 18:1219-1226. [DOI] [PubMed] [Google Scholar]
- 25.Greider, C. 1999. Telomerase activation. One step on the road to cancer? Trends Genet. 15:109-112. [DOI] [PubMed] [Google Scholar]
- 26.Hatton, K. S., K. Mahon, L. Chin, F.-C. Chiu, H.-W. Lee, D. Peng, S. D. Morgenbesser, J. Horner, and R. A. DePinho. 1996. Expression and activity of L-Myc in normal mouse development. Mol. Cell. Biol. 16:1794-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henriksson, M., and B. Luscher. 1996. Proteins of the Myc network: essential regulators of cell growth and differentiation. Adv. Cancer Res. 68:109-182. [DOI] [PubMed] [Google Scholar]
- 28.Hermeking, H., C. Rago, M. Schuhmacher, Q. Li, J. F. Barett, A. J. Obaya, B. C. O'Connell, M. K. Mateyak, W. Tam, F. Kohlhuber, C. V. Dang, J. M. Sedivy, D. Eick, B. Vogelstein, and K. W. Kinzler. 2000. Identification of CDK4 as a target of c-MYC. Proc. Natl. Acad. Sci. USA 97:2229-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ikura, T., V. V. Ogryzko, M. Grigoriev, R. Groisman, J. Wang, M. Horikoshi, R. Scully, J. Qin, and Y. Nakatani. 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102:463-473. [DOI] [PubMed] [Google Scholar]
- 30.Kuttler, F., P. Ame, H. Clark, C. Haughey, C. Mougin, J. Y. Cahn, C. V. Dang, M. Raffeld, and T. Fest. 2001. c-myc box II mutations in Burkitt's lymphoma-derived alleles reduce cell-transformation activity and lower response to broad apoptotic stimuli. Oncogene 20:6084-6094. [DOI] [PubMed] [Google Scholar]
- 31.Land, H., L. F. Parada, and R. A. Weinberg. 1983. Tumorigenic conversion of primary embryo fibroblasts requires at least 2 cooperating oncogenes. Nature 304:596-602. [DOI] [PubMed] [Google Scholar]
- 32.Landay, M., S. K. Oster, F. Khosravi, L. E. Grove, X. Yin, J. Sedivy, L. Z. Penn, and E. V. Prochownik. 2000. Promotion of growth and apoptosis in c-myc nullizygous fibroblasts by other members of the myc oncoprotein family. Cell Death Differ. 7:697-705. [DOI] [PubMed] [Google Scholar]
- 33.Lang, S. E., S. B. McMahon, M. D. Cole, and P. Hearing. 2001. E2F transcriptional activation requires TRRAP and GCN5 cofactors. J. Biol. Chem. 276:32627-32634. [DOI] [PubMed] [Google Scholar]
- 34.Mateyak, M. K., A. J. Obaya, S. Adachi, and J. M. Sedivy. 1997. Phenotypes of Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 8:1039-1048. [PubMed] [Google Scholar]
- 35.McMahon, S. B., H. A. Van Buskirk, K. A. Dugan, T. D. Copeland, and M. D. Cole. 1998. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 94:363-374. [DOI] [PubMed] [Google Scholar]
- 36.McMahon, S. B., M. A. Wood, and M. D. Cole. 2000. The essential cofactor TRRAP recruits the histone acetylase hGCN5 to c-Myc. Mol. Cell. Biol. 20:556-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogryzko, V. V., T. Kotani, X. Zhang, R. L. Schiltz, T. Howard, J. Quin, and Y. Nakatani. 1998. Histone-like TAFs within the PCAF histone acetylase complex. Cell 94:35-44. [DOI] [PubMed] [Google Scholar]
- 38.Park, J., S. Kunjibettu, S. B. McMahon, and M. D. Cole. 2001. The ATM-related domain of TRRAP is required for histone acetyltransferase recruitment and Myc-dependent oncogenesis. Genes Dev. 15:1619-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park, J., M. A. Wood, and M. D. Cole. 2002. BAF53 forms distinct nuclear complexes and functions as a critical c-Myc-interacting nuclear cofactor for oncogenic transformation. Mol. Cell. Biol. 22:1307-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ralston, R. 1991. Complementation of transforming domains of E1a/myc chimaeras. Nature 353:866-868. [DOI] [PubMed] [Google Scholar]
- 42.Reichel, R., I. Kovesdi, and J. R. Nevins. 1987. Developmental control of a promoter-specific factor that is also regulated by the E1A gene product. Cell 13:501-506. [DOI] [PubMed] [Google Scholar]
- 43.Sawai, S. A., K. Shimono, Y. Yakamatsu, C. Palmes, K. Hanaoka, and H. Kondoh. 1993. Defects of embryonic organogenesis resulting from targeted disruption of the N-myc gene in the mouse. Development 117:1445-1455. [DOI] [PubMed] [Google Scholar]
- 44.Sommer, A., K. Bousset, E. Kremmer, M. Austen, and B. Luscher. 1998. Identification and characterization of specific DNA-binding complexes containing members of the Myc/Max/Mad network of transcriptional regulators. J. Biol. Chem. 273:6632-6642. [DOI] [PubMed] [Google Scholar]
- 45.Stanton, B. R., A. S. Perkins, L. Tessarollo, D. A. Sassoon, and L. F. Parada. 1992. Loss of N-myc function results in embryonic lethality and failure of the epithelial component of the embryo to develop. Genes Dev. 6:2235-2247. [DOI] [PubMed] [Google Scholar]
- 46.Stone, J., T. de Lange, G. Ramsay, E. Jakobovits, J. M. Bishop, H. Varmus, and W. Lee. 1987. Definition of regions in human c-myc that are involved in transformation and nuclear localization. Mol. Cell. Biol. 7:1697-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 48.Vassilev, A., J. Yamauchi, T. Kotani, C. Prives, M. L. Avantaggiati, J. Qin, and Y. Nakatani. 1998. The 400 kDa subunit of the PCAF histone acetylase complex belongs to the ATM superfamily. Mol. Cell 2:869-875. [DOI] [PubMed] [Google Scholar]
- 49.Wang, J., L. Xie, S. Allan, D. Beach, and G. Hannon. 1998. Myc activates telomerase. Genes Dev. 12:1769-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wood, M. A., S. B. McMahon, and M. D. Cole. 2000. An ATPase/helicase complex is an essential cofactor for oncogenic transformation by c-Myc. Mol. Cell 5:321-330. [DOI] [PubMed] [Google Scholar]
- 51.Xiao, Q., G. Claassen, J. Shi, S. Adachi, J. Sedivy, and S. R. Hann. 1998. Transactivation-defective c-MycS retains the ability to regulate proliferation and apoptosis. Genes Dev. 12:3803-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu, D., N. Popov, M. Hou, Q. Wang, M. Bjorkholm, A. Gruber, A. R. Menkel, and M. Henriksson. 2001. Switch from Myc/Max to Mad1/Max binding and decrease in histone acetylation at the telomerase reverse transcriptase promoter during differentiation of HL60 cells. Proc. Natl. Acad. Sci. USA 98:3826-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]