Abstract
The tetraspanins are a family of integral membrane proteins with four transmembrane domains. These molecules form multimolecular networks on the surfaces of many different cell types. Gene-targeting studies have revealed a role for tetraspanins in B- and T-lymphocyte function. We have isolated and deleted a novel tetraspanin, Tssc6, which is expressed exclusively in hematopoietic and lymphoid organs. Using a gene-trapping strategy, we generated an embryonic stem (ES) cell line with an insertion in the Tssc6 locus. Mice were derived from these ES cells and, using RNase protection and reverse transcription-PCR, we demonstrated that the insertion resulted in a null mutation of the Tssc6 allele. Mice homozygous for the gene trap insertion (Tssc6gt/gt mice) were viable and fertile, with normal steady-state hematopoiesis. Furthermore, responses to hemolysis and granulocyte colony-stimulating factor-induced granulopoiesis were equivalent to those of wild-type mice. Lymphoid development was normal in Tssc6gt/gt mice. Whereas Tssc6gt/gt B cells responded normally to lipopolysaccharide, anti-CD40, and anti-immunoglobulin M stimulation, Tssc6gt/gt T cells showed enhanced responses to concanavalin A, anti-CD3, and anti-CD28. This increased proliferation by Tssc6-deleted T lymphocytes was due to increased interleukin 2 production following T-cell receptor stimulation. These results demonstrate that Tssc6 is not required for normal development of the hematopoietic system but may play a role in the negative regulation of peripheral T-lymphocyte proliferation.
The tetraspanins (transmembrane 4 superfamily) are a large family of integral membrane proteins conserved through evolution (39). Tetraspanin proteins contain four highly conserved hydrophobic transmembrane domains. There are two extracellular loops of unequal size and short, intracytoplasmic amino and carboxy termini. The small extracellular loop lies between transmembrane domains 1 and 2, and the large extracellular loop, which confers much of the functional specificity, lies between transmembrane domains 3 and 4. In contrast to the transmembrane domains, the extracellular domains of the family show considerable divergence. There are, however, three motifs—CCG, PXSC, and EGC—containing four highly conserved cysteine residues in the major extracellular domain. The tetraspanins are conceptualized to form a multiprotein network, or “web,” in the cell membrane, interacting with tetraspanin family members, other integral membrane proteins, and intracytoplasmic signaling molecules (7, 17, 39). Functional predictions derived from a structural analysis of the large extracellular loop suggest that two low-polarity regions in the loop may provide the binding sites for multiple protein partners (8).
Tetraspanin associations were initially investigated by immunoprecipitating complexes in cell membrane lysates treated with mild detergents. Such complexes were invariably large and contained multiple proteins, including other tetraspanins. More recently, associations within the tetraspanin network have been dissected according to the ability of tetraspanin-containing complexes to withstand disruption by detergents of graded hydrophobicity (3). Tetraspanin complexes held to be direct and highly specific include those between integrins α3β1 and α6β1 and tetraspanin CD151, α4β1 and CD81 (3), and CD9 and CD81 and the novel immunoglobulin superfamily proteins EWI-2 and EWI-F (the prostaglandin F2 alpha receptor regulatory protein) (3, 31).
Tetraspanins have functional roles in cell motility, membrane fusion, proliferation, and adaptive immunity (7, 14, 17, 39). In some cases, nontetraspanin molecules incorporated into the tetraspanin web may be responsible for the functional effect; the direct role, if any, of the tetraspanin is not yet clear. Many adhesion molecules and, in particular, the β1 integrins, form molecular associations with tetraspanin molecules (3, 7). It is this partnership that likely underpins the role of tetraspanins in cell motility and cancer metastasis (38). Tetraspanins are also proposed to link integrins to cytoplasmic signaling molecules, thereby diversifying integrin function (7, 38). An essential role for CD9 in sperm-egg fusion was revealed by the infertility of CD9 knockout mice. CD9 is also suggested to participate in megakaryocyte membrane fusion (4) and (with CD81) in myotubule formation (33).
Tetraspanin associations have been observed with many lymphocyte cell surface proteins, usually under mild detergent conditions. Coimmunoprecipitating molecules include CD2, CD4 and CD8, and major histocompatibility complex class II (MHC-II). The tetraspanin CD81 is a member of the B-cell receptor complex comprising CD19, CD21, and Leu13. In this context, tetraspanins have been implicated in the control of lymphocyte activation and proliferation (14, 15, 17). Studies on CD81 and CD37 knockout mice have revealed a role for tetraspanins in the immune response. CD81-null B lymphocytes have variably altered proliferation when stimulated in vitro, while T lymphocytes are hyperproliferative to a range of mitogens (20). When immunized with a T-cell-dependent antigen, CD81-null mice were unable to mount an effective immune response (15, 16, 20). CD81 was determined to be important for effective B- and T-cell interaction and interleukin 4 (IL-4) production by T cells (15). Deletion of CD37 also had a negative effect on B-cell-T-cell collaboration and B-cell function in response to immunization with T-cell-independent antigens (9).
Several groups, using different approaches, have identified the human TSSC6 gene, also known as PHEMX. The locus was present on a fragment of 11p15 chromosome that suppressed the growth of a rhabdomyosarcoma cell line (hence, tumor-suppressing subchromosomal transferable fragment cDNA 6) (22). The murine Tssc6 gene was mapped to a syntenic region of distal chromosome 7 within a 1-Mb imprinted domain, although Tssc6 itself was not imprinted (22). Tssc6, designated Phemx, was also identified as an expressed sequence tag in cDNA prepared from the extraembryonic tissue of a 7.5-day-old mouse embryo (10). The Tssc6 protein was confirmed to be a member of the tetraspanin superfamily by alignment of its four transmembrane domains with those of other family members, by the identification of conserved polar amino acids in the hydrophobic transmembrane domains, and by virtue of its four highly conserved cysteines in the large extracellular loop (26). The Tssc6 gene is adjacent to the CD81 locus and is most homologous to this gene, suggesting that one of the loci may have arisen by gene duplication (13, 22, 26).
Northern analysis revealed that Tssc6 transcripts are confined to the hematopoietic compartment (21, 26). Tssc6 expression was identified in multiple hematopoietic cell lines (26) and in flow-sorted erythroid cells, granulocytes, bone marrow B and T lymphocytes, and macrophages cultured from bone marrow (21). Transcripts were also detected in pluripotential Lin− c-Kit+ Sca-1+ cells (21). In the developing murine embryo, Tssc6 mRNA was identified in blood cells in yolk sac blood islands and fetal liver (21, 26). Further analysis of the distribution of Tssc6 in lymphoid subsets awaits the development of monoclonal antibodies.
In this study, we report the analysis of mice deficient in Tssc6. These mice were generated from an embryonic stem (ES) cell line bearing a gene trap insertion in the Tssc6 locus.
MATERIALS AND METHODS
Derivation of the gene trap ES cell line 26F8.
W9.5 ES cells, derived from a 129/Sv.C3-+c+p inbred strain (32), were electroporated with 30 μg of linearized gene trap vector pMS-1 (generously provided by Marjo Salminen and Peter Gruss, Max Planck Institute of Biophysical Chemistry, Göttingen, Germany). Cells were selected in 150 μg of G418 (GibcoBRL, Grand Island, N.Y.)/ml, and surviving clones were picked and expanded. Each clone was differentiated and examined for expression of the gene trap vector in hematopoietic cells by staining for β-galactosidase activity essentially as described previously (30). Briefly, undifferentiated ES cells were dissociated with trypsin (Commonwealth Serum Laboratories, Victoria, Australia) depleted of feeder cells and cultured in suspension culture at 15,000 cells/ml in differentiation medium (1× Iscove's modified Dulbecco's medium with 15% fetal calf serum [selected batch; GibcoBRL], 5% protein-free hybridoma medium II [GibcoBRL], 2 mM glutamine [GibcoBRL], 0.05 mg of ascorbic acid [Sigma Chemical Co., St. Louis, Mo.]/ml, and 0.3% α-monothioglycerol) in ultra-low cluster plates (Corning Costar Corp, Cambridge, Mass.) at 37°C in 8% CO2 in air. After 4 days, 5 to 10 embryoid bodies were seeded in each well of a 24-well tissue culture dish (Corning) in differentiation medium. After a further 5 days, the differentiation cultures were fixed and stained for β-galactosidase activity as described previously (30). From this screen, the gene trap ES cell line 26F8 was selected for further study because lacZ staining was restricted to hematopoietic cells in differentiation cultures and in chimeric embryos.
Molecular characterization of the gene trap insertion.
Mice heterozygous for the gene trap insertion 26F8 (Tssc6gt/+ mice) were obtained by mating chimeras generated by injecting 26F8 ES cells into BALB/c blastocysts. Southern blots of tail DNA digested with HindIII were probed with a 1.1-kb SacI-ClaI β-galactosidase cDNA fragment. This demonstrated that a single integration of the gene trap vector had occurred. The molecular cloning of the gene trap fusion transcript by 5′ rapid amplification of cDNA ends with RNA prepared from the fetal livers of Tssc6gt/+ mice and its identification as the recently discovered gene Tssc6 (also known as Phemx) have been previously described (26). Analysis of the full-length Tssc6 protein demonstrated that it is a member of the tetraspanin superfamily (26). To map the insertion of the gene trap vector into the Tssc6 genomic locus, DNA prepared from mice bearing the 26F8 insertion and their littermates was digested with multiple restriction enzymes, transferred to a nylon membrane, and hybridized with multiple probes obtained from the pMS-1 vector and from Tssc6 cDNA. The results were compared with the restriction enzyme map of the Tssc6 genomic locus (22) (GenBank accession no. AJ251788). Mice heterozygous and homozygous for the gene trap insertion (Tssc6gt/+ and Tssc6gt/gt, respectively) were distinguished by Southern blotting of SacI-digested tail DNA probed with a 276-bp cDNA fragment of Tssc6 (nucleotides [nt] 73 through 349) (see Fig. 1A).
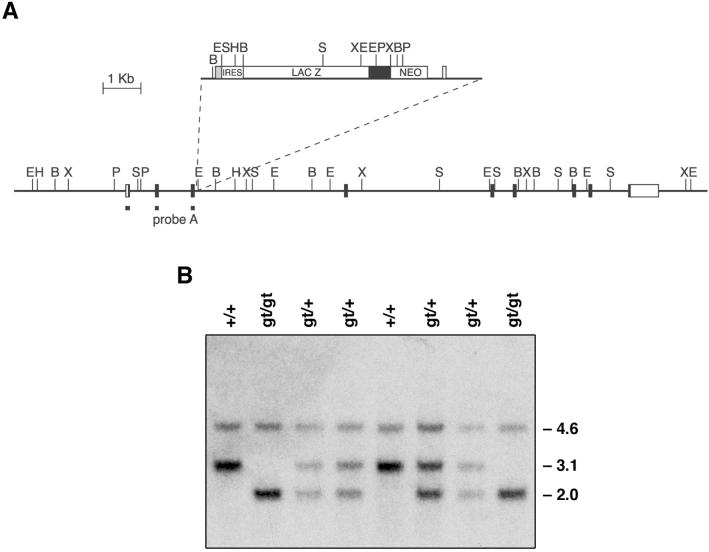
FIG. 1.
Mapping of the disruption of the Tssc6 locus by insertion of the gene trap vector pMS-1 and genotyping of Tssc6 mutant mice. (A) The pMS-1 vector, with the vector backbone shown as a line. The site of insertion of the gene trap vector into intron 3 of the Tssc6 genomic locus is indicated. Extensive mapping of the Tssc6 locus upstream and downstream of the insertion site did not reveal any additional rearrangements. Exons are shown as boxes, with coding regions filled. Restriction enzyme sites are indicated as follows: E, EcoRI; H, HindIII; B, BamHI; X, XbaI; P, PstI; S, SacI. (B) Genotyping of Tssc6 mutant mice. A Southern blot of tail DNA digested with SacI was probed with the fragment shown in panel A. The sizes of the endogenous Tssc6 bands (4.6 and 3.1 kb) and the altered Tssc6 allele (2.0 kb) are indicated. Lanes: +/+, wild-type; gt/+, heterozygous for the Tssc6 gene trap insertion; gt/gt, homozygous for the Tssc6 gene trap insertion.
Analysis of Tssc6 expression.
Poly(A)+ RNA was prepared from the fetal liver, kidney, liver, spleen, thymus, and lymph nodes of wild-type (Tssc6+/+), Tssc6gt/+, and Tssc6gt/gt mice, and Northern analysis was performed as previously described (23). Identical blots were prepared and hybridized to 5′ and 3′ Tssc6 cDNA fragments as well as to a β-galactosidase probe. The 5′ probe was as shown in Fig. 1A, and the 3′ probe was a 333-bp NcoI fragment from a Tssc6 cDNA clone which lacked exon 7 (26). Blots were then stripped and reprobed with a 1.1-kb fragment of chicken glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA. RNase protection analysis was performed with the RPAIII kit (Ambion, Austin, Tex.) as described previously (24). Ten micrograms of RNA was used in each hybridization reaction. The RNase protection probes, illustrated diagrammatically in Fig. 2A, were a 315-bp Tssc6 cDNA fragment (nt 650 through 965) and a 270-bp Tssc6 cDNA fragment (nt 197 through 467), both cloned into pBluescript II SK− (Stratagene, La Jolla, Calif.). A 245-bp fragment of mouse β-actin cDNA, labeled to a 50-fold-lower specific activity, was used to estimate RNA loading. Reverse transcription (RT)-PCR was performed as described previously (26). The oligonucleotide primers used to amplify Tssc6 were used as described previously (26). To detect the gene trap fusion transcript, oligonucleotide primers 5′-ATGGCCACCATGGTGGTTGTC-3′ and 5′-TTGTCGACCTGTTGGTCTGAAACTCAGCCT-3′ (which lies in the gene trap vector) were used, with PCR conditions as described above, except that the annealing temperature was 61°C. The reaction products were electrophoresed through 2% agarose gels, transferred to a nylon membrane, and probed with a radiolabeled internal oligonucleotide, 5′-CTGTGATTGGCCATGCATCGTTGGAGA-3′. Amplification of hypoxanthine phosphoribosyltransferase transcripts was used to estimate the amount of cDNA present in each sample, with oligonucleotides and PCR conditions as described previously (25), except that 24 cycles of PCR were carried out.
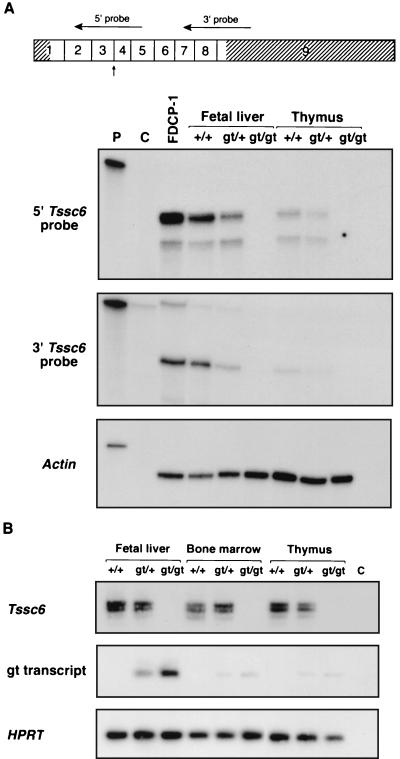
FIG. 2.
The gene trap insertion in Tssc6 results in a null allele. (A) RNase protection analysis of RNA prepared from the fetal liver and thymus of Tssc6gt/gt mice and littermates. At the top of the panel, the Tssc6 cDNA is shown, with the nine exons indicated by boxes. The 5′ and 3′ untranslated regions are hatched. Horizontal arrows indicate the position of the 5′ and 3′ riboprobes. The vertical arrow shows the point of fusion with the gene trap vector. With both 5′ and 3′ riboprobes, protected bands in RNA prepared from the fetal liver and thymus of Tssc6+/+ and Tssc6gt/+ mice are shown, but in RNA from Tssc6gt/gt mice, bands are absent. Lanes: P, undigested, full-length probe; C, probe after RNase digestion; FDCP-1, RNA from FDCP-1 cells, which are known to express Tssc6; +/+, wild-type; gt/+, heterozygous for the Tssc6 gene trap insertion; gt/gt, homozygous for the Tssc6 gene trap insertion. (B) RT-PCR to amplify Tssc6 transcripts and gene trap fusion transcripts from the fetal liver, bone marrow, and thymus of Tssc6gt/gt mice and their littermates. The multiple bands amplified with Tssc6 primers were due to alternate splice variants (26). Hypoxanthine phosphoribosyltransferase transcripts were amplified to estimate the amount of cDNA present in each reaction. Lane C, no cDNA control.
Mice and histologic analysis.
Mice used for all analyses were between 6 and 12 weeks of age unless otherwise stated and were either BALB/c × 129Sv or backcrossed onto C57BL/6. In each experiment, littermates were used as controls. Mice were bred and maintained in standard animal facilities. Sections of brain, salivary glands, thymus, lymph nodes, lung, heart, liver, spleen, kidney, intestinal tract, pancreas, genitourinary tract, muscle, sternum, and femur from Tssc6gt/gt mice and littermates were stained with hematoxylin and eosin and examined with a light microscope.
Flow cytometry.
Single-cell suspensions were prepared by passing solid organs through a fine wire mesh. Spleen, bone marrow, and peripheral blood cell suspensions were depleted of red blood cells by hypotonic lysis with 156 mM ammonium chloride, resuspended in buffer solution (balanced salt solution containing Na-K-Ca-Mg at a molar ratio of 121:3:2:1, buffered to pH 7.2 with HEPES and potassium phosphate, containing 3% bovine calf serum [BCS; HyClone, Logan, Utah]), and then stained with a saturating concentration of the appropriate antibody on ice for 30 min. The monoclonal antibodies used were either biotin or fluorescein isothiocyanate conjugates specific for the following murine markers: B220 (RA3-6B2), immunoglobulin M (IgM) (ly 5.1), IgD (11-26c.2a), CD19 (1D3), CD21 (7G6), CD40 (FGK45), MHC-II (M5/14), CD23 (B3B4), CD3ɛ (145-C211), CD8 (53.6.7), CD4 (YTS-197), CD5 (53-7.3), Thy 1 (T3.24.1), ΤCRαβ (H57-697.1), TCRγδ (GL3), CD2 (RM2-5), CD44 (1 M7.81), CD25 (PC61), CD62L (MEL-14), CD69 (H1.2F3), GR-1 (RB6-8C5), Mac-1 (M1/70), and Ter 119 (all generously provided by A. Strasser, The Walter and Eliza Hall Institute, Victoria, Australia). Biotinylated antibodies were revealed with streptavidin-phycoerythrin (Caltag, Burlingame, Calif.). Dead cells were excluded by propidium iodide (Sigma) uptake, and lymphocyte subpopulations were determined after gating on typical lymphocyte forward and side scatter properties. Cells were analyzed on a FACScan flow cytometer (Becton Dickinson, San Jose, Calif.).
Hematologic analysis.
Mice were bled from the retro-orbital sinus, and peripheral cell counts were performed with an ADVIA 120 automatic cell analyzer (Bayer, Tarrytown, N.Y.). Differential cell counts and morphological analyses were performed manually on May-Grünwald-Giemsa-stained peripheral blood smears or cytocentrifuge preparations of spleen and bone marrow cell suspensions. For progenitor cell assays, adult bone marrow cells at a final concentration of 25,000 cells/ml were added to triplicate 36-mm-diameter tissue culture dishes containing 1 ml of semisolid medium (Dulbecco's modified Eagle's medium with 20% selected BCS and 0.3% agar) and final concentrations of one of the following growth factors: 100 ng of murine stem cell factor (SCF; produced in Pichia pastoris)/ml, 10 ng of recombinant murine macrophage colony-stimulating factor (M-CSF; Cetus, Emeryville, Calif.)/ml, 10 ng of recombinant murine granulocyte-macrophage colony-stimulating factor (GM-CSF; Schering, Bloomfield, N.J.)/ml, 10 ng of human granulocyte colony-stimulating factor (G-CSF; Amgen, Thousand Oaks, Calif.)/ml, 10 ng of murine IL-3 (PeproTech, Rocky Hill, N.J.)/ml, or 2 IU of murine thrombopoietin (TPO; produced in CHO cells)/ml. Plates were incubated at 37°C in 10% CO2 in air for 7 days before being fixed, floated onto glass slides, dried, and then stained for acetylcholinesterase activity and then with Luxol fast blue and hematoxylin. Total and differential colony counts were performed by microscopic examination of the stained culture slides. Bone marrow transplantation experiments conducted to determine the activity of CFU in the spleen (CFU-S) utilized cells from the femurs of one to three Tssc6gt/gt and littermate donor mice. Lethally irradiated C57BL/6 mice (two 5.50-Gy doses, 4 h apart) received between 25 × 103 and 200 × 103 cells via retro-orbital injection. Twelve days postinjection, spleens were fixed in either 70% ethanol-glacial acetic acid-10% neutral buffered formalin (100:5:12 [vol/vol/vol]) or Bouin's reagent and macroscopic colonies were counted.
PHZ-induced anemia.
Fifteen Tssc6gt/gt mice and littermate controls were injected intraperitoneally (i.p.) with 60 mg of phenylhydrazine (PHZ; Sigma)/kg on days 0 and 1. At specified time points, cohorts of three mice of each genotype were bled via the retro-orbital plexus for hematocrit determination (microcapillary tube centrifugation) and then killed, and their spleens were weighed.
Response to G-CSF injection.
Four sex- and weight-matched Tssc6gt/gt and Tssc6+/+ mice were injected subcutaneously with 2.5 μg of recombinant human G-CSF (Filgrastim; Amgen) twice a day for 5 days. G-CSF was diluted in 0.2 ml of normal saline with 5% BCS. Control animals received normal saline with 5% BCS alone. On the sixth day, animals were killed and bled. Total and differential cell counts on blood and spleen were performed. Blood (25 μl) and spleen (25,000 nucleated cells) were cultured and analyzed for progenitor cell content as described above. Triplicate cultures were stimulated with a combination of 10 ng of recombinant murine IL-3/ml, 10 ng of M-CSF/ml, and 2 IU of human erythropoietin (Amgen)/ml.
Acute inflammatory response to casein.
Casein C5890 (Sigma), containing Bacillus sp. bacteria (19), was prepared as a 0.2% solution in 0.2 ml of mouse tonicity phosphate-buffered saline (MT-PBS) and injected i.p. into four mice of each genotype. Mice were killed 3 h postinjection, and their abdomens were lavaged with 3 ml of MT-PBS (19). Total and differential counts on the harvested abdominal fluid were performed by manual methods.
Proliferation assays.
Cell suspensions were prepared from the pooled spleens or lymph nodes of three Tssc6+/+ or Tssc6gt/gt mice. Splenic B and T cells were purified by selecting cells which failed to bind fluoroscein isothiocyanate-labeled GR-1, Mac-1, and Ter 119 antibodies and either Thy-1 or B220 antibodies by using flow cytometric sorting (MoFlo; Cytomation, Fort Collins, Colo.). The purities of sorted B and T lymphocytes were both ≥94% when immunophenotyped with antibodies specific for the enriched population. T-cell subsets were separated by negative selection using magnetic beads (Dynabeads M-450; Dynal, Oslo, Norway). The purity of the enriched cell population was above 95%. Triplicate wells in a flat-bottomed 96-well plate (Becton Dickinson, Paramus, N.J.) received 105 cells and the specified mitogen in 200 μl of media (Kelso Dulbecco's modified Eagle's medium with 10% BCS and 50 μM 2-mercaptoethanol). Mitogens were anti-CD40 (FGK45), anti-IgM [F(ab′)2; Jackson ImmunoResearch Laboratories, West Grove, Pa.], lipopolysaccharide (LPS), concanavalin A, phorbol myristate acetate (PMA), ionomycin (all from Sigma), IL-2 (PeproTech), anti-CD3 (145 C211), and anti-CD28 (37.51; BD Pharmingen, San Diego, Calif.). The latter two mitogens were adsorbed to the plate by overnight incubation at 4°C in a humidified chamber. Cells were stimulated for 48 to 72 h at 37°C and pulsed for the last 6 h with 1 μCi of [3H]thymidine (Amersham, Little Chalfont, Buckinghamshire, United Kingdom). Treated cells were harvested (cell harvester; Inotech, Dottikon, Switzerland), and [3H]thymidine incorporation was measured with a scintillation counter (Top Count; Packard, Meriden, Conn.).
IL-2 measurement.
Lymph node-derived T lymphocytes were enriched using magnetic beads, and 2 × 105 cells/well were stimulated as described above with 10 μg of anti-CD3/ml. Supernatants from duplicate wells were determined using a Quantikine M IL-2 enzyme-linked immunosorbent assay (ELISA) (R&D Systems, Minneapolis, Minn.) following the manufacturer's instructions.
Analysis of protein tyrosine phosphorylation.
Enriched T-lymphocyte populations were suspended in MT-PBS, and 2 × 106 cells/well were stimulated at 37°C in 12-well plates coated with 10 μg of anti-CD3/ml for various times. Cells were lysed in situ with 2× lysis buffer (1× lysis buffer contains 0.1% Triton X-100, 0.5% sodium dodecyl sulfate, 50 mM dithiothreitol, 20 mM Tris HCl [pH 7.5], 150 mM NaCl, 50 mM sodium fluoride, 2 mM sodium orthovanadate, and protease inhibitors [Complete Cocktail tablets; Roche]), clarified, and then concentrated using filter units (Millipore, Bedford, Mass.). Protein was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride membranes (Osmonics, Westborough, Mass.), and blotted with anti-phosphotyrosine clone 4G10 (Upstate Biotechnology, Lake Placid, N.Y.), followed by anti-mouse horseradish peroxidase (HRP) conjugate (Silenus, Melbourne, Australia).
Immunization experiments.
Seven mice of each genotype were injected i.p. with 100, 10, or 1 μg of NP ([4-hydroxy-3-nitrophenyl]acetyl) coupled to keyhole limpet hemocyanin (KLH) (NP18-KLH [NP18/KLH conjugation ratio, 18:1]) precipitated in alum. In one experiment, mice were immunized with 20 μg of NP18-KLH in MT-PBS. Mice were bled at days 0, 14, and 42 postimmunization and then given an i.p. booster on day 42 with 20 μg of NP18-KLH in MT-PBS (mice initially immunized with 100 μg of NP18-KLH in alum were given an i.p. booster with 100 μg of NP18-KLH) and bled on day 49. One mouse of each genotype was sacrificed at day 14, and the spleen and an inguinal lymph node were processed for histology. T-cell-independent responses were induced by immunizing six mice of each genotype with an i.p. injection of 50 μg of NP-LPS in 200 μl of MT-PBS. Three mice of each genotype were bled and killed at day 7 and day 14 postimmunization.
Immunoglobulin detection.
NP-specific antibodies were measured by ELISA on 96-well plates (Dynex, Chantilly, Va.) coated with solutions of 20 μg/ml of either NP17-bovine serum albumin or NP2-bovine serum albumin to detect total and high-affinity anti-NP antibodies, respectively. Serially diluted serum samples were applied to the washed plates and incubated for at least 4 h. HRP-labeled anti-mouse IgG1 and anti-IgM (Southern Biotechnology Associates, Birmingham, Ala.) at a concentration of 1 μg/ml were applied for 4 h before washing and signal detection by 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) substrate (Sigma). NP-specific monoclonal IgG1 was used to generate a standard curve, from which relative units were derived. Basal serum immunoglobulin levels were assayed similarly, with the following modifications. Plates were coated with anti-mouse total immunoglobulin (Silenus) at 2 μg/ml in 0.2 M carbonate buffer. The isotype-specific anti-IgG2a (Southern Biotechnology) and anti-IgA (Silenus) secondary antibodies were biotinylated. For these assays, a 1-h incubation of streptavidin-labeled HRP (Southern Biotechnology) was performed prior to substrate addition and color development. Standards were generated from the standard curves of purified, polyclonal anti-isotype antibody (Sigma).
Statistical analysis.
Significant differences between wild-type and gene-trapped mice were determined by Student's t test for paired samples. A P value of <0.05 was considered to be statistically significant.
RESULTS
Generation of Tssc6-null mice.
In a screen to identify genes expressed during early blood development, we isolated an ES cell line in which the gene trap vector pMS-1 (28) had integrated into the mouse Tssc6 locus. Analysis of the integration event by the 5′ rapid amplification of cDNA ends method yielded a 351-nt sequence which contained the previously unidentified first exon and the second and third exons of Tssc6 (26). The insertion site was mapped by Southern blot analysis using vector probes, Tssc6 cDNA probes, and the genomic sequence of the locus (22). This revealed that a single copy of the gene trap vector had integrated into intron 3 (Fig. 1A).
Tssc6gt/+ mice were mated, generating viable, fertile offspring which were genotyped as shown in Fig. 1B. Tssc6gt/gt mice were born in normal Mendelian numbers. In order to determine the effect of the gene trap insertion on Tssc6 transcription, Northern blots, prepared by using poly(A)+ RNA from the tissues of Tssc6gt/gt mice and their littermates, were probed with Tssc6 cDNA probes. In contrast to Tssc6+/+ and Tssc6gt/+ mice, no Tssc6 transcripts were detected in the fetal liver, spleen, thymus, or kidney from Tssc6gt/gt mice (data not shown). RNase protection was performed in order to detect low-level Tssc6 transcription. In keeping with the Northern blotting results, a riboprobe 3′ of the insertion site did not protect a transcript in the Tssc6gt/gt fetal liver or thymus. A 5′ riboprobe, which crossed the insertion site, was predicted to protect a truncated transcript in the Tssc6gt/+ and Tssc6gt/gt mice, but no such transcript was seen, suggesting that the insertion destabilized the Tssc6 fusion transcript (Fig. 2A). The absence of a full-length Tssc6 transcript in tissues from the Tssc6gt/gt mice was further confirmed by RT-PCR. Using primers designed to amplify the fusion transcript, we detected a faint band in cDNA prepared from Tssc6gt/+ and Tssc6gt/gt tissues (Fig. 2B). These results indicate that the gene trap insertion into the Tssc6 locus has acted as a mutagen, preventing the production of full-length Tssc6 transcripts and resulting in a null allele.
Hematopoiesis in the steady state or during challenge is normal in Tssc6gt/gt mice.
Having established that the gene trap insertion resulted in a null mutation of the Tssc6 locus, Tssc6gt/gt mice were examined for defects in the hematopoietic and immune systems. The mutant mice were of normal appearance and were healthy and fertile. Gross pathology and histological examination of embryonic and adult tissues did not reveal any abnormalities. Total cell counts of peripheral blood, bone marrow, and spleen were similar to those for Tssc6gt/gt mice and littermates, and differential counts did not reveal differences in hematopoietic cell types (Table 1). Similar results were obtained when the analysis was repeated on 6-month-old mice (data not shown). Bone marrow progenitor cells were assayed in clonal cultures stimulated with SCF, IL-3, GM-CSF, G-CSF, M-CSF, or TPO (Table 2). The numbers and types of colonies present did not differ between Tssc6gt/gt mice and littermates. To examine a more primitive progenitor, we performed a CFU-S assay. The number of CFU-S was not significantly different between the marrow cells of the Tssc6gt/gt mice and littermate control donors (Tssc6+/+, 8.8 ± 1.6 CFU-Sd12, n = 5; Tssc6gt/gt, 7.2 ± 1.2 CFU-Sd12, n = 6).
TABLE 1.
Hematological profile of Tssc6-null micea
| Hematological parameter | Value for the following genotype:
|
|
|---|---|---|
| Tssc6+/+ | Tssc6gt/gt | |
| Peripheral blood | ||
| Hematocrit (%) | 46 ± 57 | 47 ± 2 |
| Red blood cell reticulocytes (%) | 4 ± 2 | 3 ± 1 |
| White blood cell count (106/ml) | 6 ± 2 | 6 ± 2 |
| Neutrophils | 1 ± 0.4 | 1 ± 0.3 |
| Lymphocytes | 5 ± 2 | 5 ± 2 |
| Monocytes | 0.2 ± 0.2 | 0.1 ± 0.1 |
| Eosinophils | 0.1 ± 0.2 | 0.1 ± 0.1 |
| Platelets (106/ml) | 958 ± 212 | 1,014 ± 227 |
| Bone marrow | ||
| Cellularity (106 cells/femur) | 31 ± 14 | 33 ± 13 |
| Blasts (%) | 2 ± 1 | 2 ± 1 |
| Promyelocytes/myelocytes (%) | 4 ± 3 | 4 ± 2 |
| Metamyelocytes/neutrophils (%) | 46 ± 5 | 41 ± 8 |
| Lymphocytes (%) | 28 ± 5 | 28 ± 7 |
| Monocytes (%) | 6 ± 2 | 6 ± 2 |
| Eosinophils (%) | 3 ± 2 | 4 ± 1 |
| Nucleated erythroid cells (%) | 11 ± 3 | 15 ± 7 |
| Spleen | ||
| Wt (mg) | 79 ± 18 | 67 ± 16 |
| Blasts (%) | 1 ± 1 | 1 ± 1 |
| Promyelocytes/myelocytes (%) | 0.5 ± 1 | 1 ± 1 |
| Metamyelocytes/neutrophils (%) | 4 ± 2 | 3 ± 2 |
| Lymphocytes (%) | 88 ± 6 | 88 ± 3 |
| Monocytes (%) | 2 ± 1 | 2 ± 2 |
| Eosinophils (%) | 0.5 ± 1 | 1 ± 1 |
| Nucleated erythroid cells (%) | 4 ± 3 | 4 ± 3 |
Data given are means ± standard deviations of the specified hematological parameters from 6 to 12 mice of each genotype.
TABLE 2.
Culture of bone marrow cells
| Stimulus | Genotype | No. of coloniesa
|
|||||
|---|---|---|---|---|---|---|---|
| Blast | G | GM | M | Eo | Meg | ||
| GM-CSF | Tssc6+/+ | 0 | 18 | 9 | 17 | 3 | 0 |
| Tssc6gt/gt | 0 | 19 | 6 | 21 | 0 | 0 | |
| G-CSF | Tssc6+/+ | 0 | 10 | 0 | 0 | 0 | 0 |
| Tssc6gt/gt | 0 | 12 | 1 | 0 | 0 | 0 | |
| M-CSF | Tssc6+/+ | 0 | 3 | 5 | 35 | 0 | 0 |
| Tssc6gt/gt | 0 | 1 | 1 | 34 | 0 | 0 | |
| IL-3 | Tssc6+/+ | 8 | 17 | 14 | 19 | 4 | 8 |
| Tssc6gt/gt | 3 | 26 | 12 | 18 | 6 | 3 | |
| SCF | Tssc6+/+ | 12 | 27 | 1 | 0 | 0 | 0 |
| Tssc6gt/gt | 5 | 18 | 0 | 1 | 0 | 0 | |
| TPO | Tssc6+/+ | 0 | 0 | 0 | 0 | 0 | 5 |
| Tssc6gt/gt | 0 | 0 | 0 | 0 | 0 | 3 | |
Total numbers of colonies present in stained cultures of 25,000 cells. Representative data from one of three comparable experiments are given. G, granulocyte; GM, granulocyte-macrophage; M, macrophage; Eo, eosinophil; Meg, megakaryocyte.
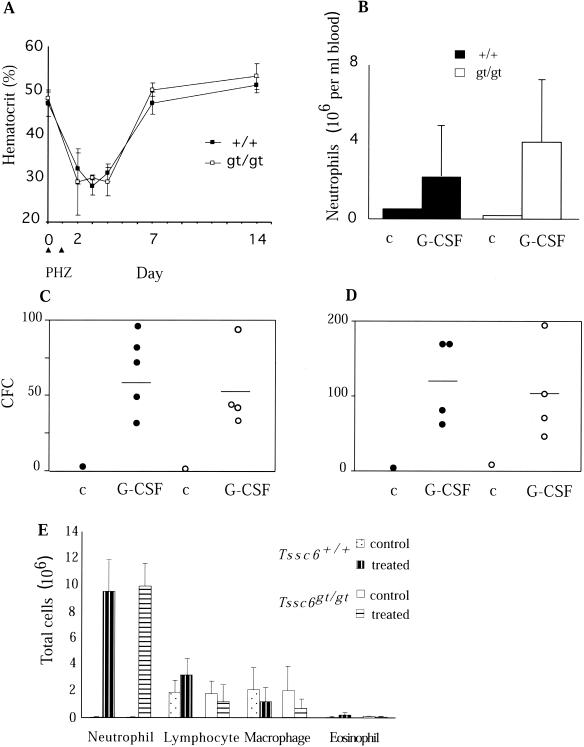
The response of Tssc6gt/gt mice to challenge with PHZ, which causes hemolytic anemia, was tested. Administration of PHZ resulted in a 40% reduction in the hematocrit levels of control and mutant mice. The rate and extent of erythroid recovery in Tssc6+/+ and Tssc6gt/gt mice were similar (Fig. 3A). The increase in spleen size in PHZ-treated mice, reflecting extramedullary hematopoiesis, was also similar in Tssc6gt/gt mice and controls (data not shown). G-CSF was injected over a period of 5 days to induce mobilization of progenitor cells and mature neutrophils from the marrow and spleen into the circulation (27). The induced neutrophilic leukocytosis in Tssc6gt/gt mice and littermates was similar (Fig. 3B). Similarly, the increase in progenitor cell numbers present in peripheral blood and spleen was not altered by Tssc6 deletion (Fig. 3C and D).
FIG. 3.
Responses to diverse stimuli resulting in perturbations of steady-state hematopoiesis are unaltered in Tssc6gt/gt mice. (A) Response to PHZ-induced hemolytic anemia in Tssc6gt/gt mice. PHZ was administered i.p. to Tssc6+/+ (+/+) and Tssc6gt/gt (gt/gt) mice on days 1 and 2, and recovery in hematocrit was assessed at the indicated time points. Values represent the means and standard deviations of the cohorts of three mice analyzed at each time point. (B) Mobilization of mature neutrophils and progenitor cells in the peripheral blood and spleens of Tssc6gt/gt mice. Mice were injected subcutaneously with 2.5 μg of G-CSF twice daily for five consecutive days and analyzed on the sixth day. Peripheral blood neutrophil numbers in Tssc6gt/gt and Tssc6+/+ mice treated with G-CSF were compared to those for a sex-, weight-, and genotype-matched, untreated control mouse (c). Means and standard deviations of four treated mice of each genotype are given. (C and D) Spleen and peripheral blood colony-forming cell (CFC) numbers, respectively. Open (Tssc6gt/gt mice) and filled (Tssc6+/+ mice) circles represent the mean CFC number in triplicate cultures of spleen (25,000 cells) and peripheral blood (25 μl) in agar cultures stimulated by SCF, IL-3, and erythropoietin. For treated mice, the bar depicts the mean number of CFCs from four mice. (E) Mobilization of neutrophils to the peritoneal cavity. Tssc6gt/gt and Tssc6+/+ mice were injected i.p. with casein. Three hours later, the mice were killed and the peritoneal exudate was collected and examined. Data represent the means and standard deviations of absolute cell numbers in the exudate from four treated mice and six untreated controls.
The ability of the mutant mice to mount an acute inflammatory response was tested by i.p. injection of casein. The casein solution was nonsterile and delivered a low-grade bacterial infection, as well as the foreign protein, to generate inflammation (19). Depletion of mature neutrophils from the bone marrow storage pool was comparable between genotypes (data not shown), and the cellular profile of the peritoneal exudate indicated a normal inflammatory response in Tssc6gt/gt mice (Fig. 3E). The phagocytic ability of granulocytes appeared normal, as indicated by the percentage of neutrophils that had phagocytosed bacteria (data not shown).
Lymphocyte development is normal in Tssc6 mutant mice.
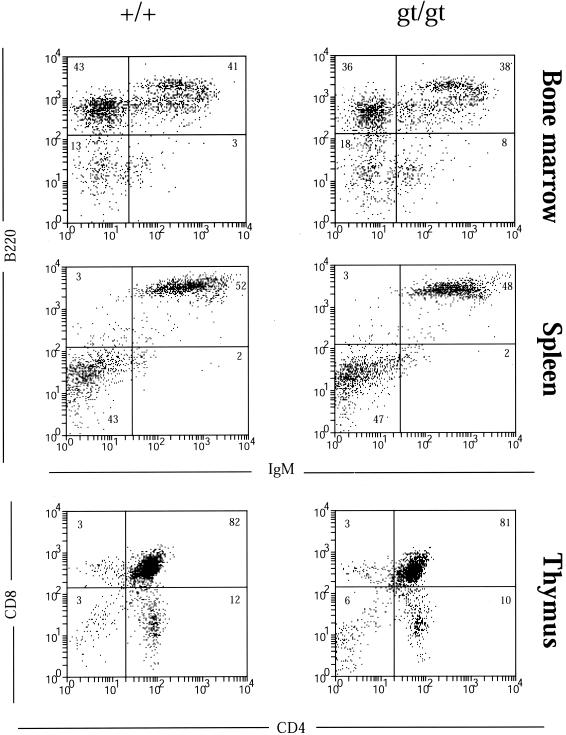
Northern analysis has shown Tssc6 expression in lymphoid organs, in T- and B-cell blasts, and in most lymphoid cell lines (26). The thymi, spleens, and lymph nodes of Tssc6gt/gt mice were of normal size and architecture. Examination of the lymphoid populations in bone marrow, thymus, spleen, and peripheral lymph nodes was performed using 6 to 12 mice of each genotype (Table 3). Representative profiles for each organ are presented in Fig. 4. The percentage of lymphoid cells expressing the B-cell surface antigens B220, IgM, CD19, CD21, CD40, MHC-II, and CD23 in bone marrow and secondary lymphoid tissues did not differ between Tssc6gt/gt mice and Tssc6+/+ littermates (Table 3, Fig. 4, and data not shown). Determination of pro-B-, pre-B- and immature B-cell subsets in the marrow revealed normal B-lymphocyte development in Tssc6gt/gt mice. Peritoneal B lymphocytes, comprising conventional B2 cells (IgM+ B220high) and B1 cells (IgM+ CD5+ IgD− B220low), were also normal (Table 3). T-cell development in the thymus and mature populations in lymph nodes, analyzed by CD4, CD8, CD3, TCRαβ, and TCRγδ expression, were comparable between Tssc6gt/gt mice and Tssc6+/+ littermates (Table 3 and data not shown). The expression of the activation markers CD44, MEL-14, CD69, and CD25 on lymph node-derived T lymphocytes revealed no difference in the number of effector or memory cells between Tssc6gt/gt and littermate mice (data not shown).
TABLE 3.
Lymphocyte populations in Tssc6-deficient micea
| Tissue | Lymphocyte subset | Genotype
|
|
|---|---|---|---|
| Tssc6+/+ | Tssc6gt/gt | ||
| Bone marrow | B220+ IgM− | 46 ± 9 | 45 ± 9 |
| B220low IgM+ | 23 ± 6 | 23 ± 5 | |
| B220high IgM+ | 16 ± 5 | 12 ± 4 | |
| Blood | B220+ IgM+ | 36 ± 12 | 39 ± 11 |
| CD4+ | 34 ± 11 | 35 ± 8 | |
| CD8+ | 15 ± 3 | 15 ± 4 | |
| Spleen | B220+ IgM+ | 50 ± 5 | 47 ± 5 |
| CD4+ | 17 ± 2 | 19 ± 4 | |
| CD8+ | 15 ± 2 | 16 ± 3 | |
| Lymph node | B220+ IgM+ | 24 ± 2 | 20 ± 2 |
| CD4+ | 49 ± 2 | 52 ± 2 | |
| CD8+ | 21 ± 3 | 23 ± 2 | |
| Peritoneal cavity | B220low CD5+ | 43 ± 18 | 37 ± 18 |
| B220high IgM+ | 26 ± 21 | 22 ± 14 | |
Values (means ± standard deviations) represent the percentage of lymphocytes (based on forward and side scatter properties) that expressed the indicated cell surface marker. Values are derived from five animals of each genotype.
FIG. 4.
Flow cytometric analysis of lymphocyte populations in Tssc6+/+ (+/+) and Tssc6gt/gt (gt/gt) mice. Lymphocytes were gated by using forward and side scatter properties. Shown are typical profiles obtained from one of five experiments. Percentages of cells positive for each marker are indicated.
T-cell proliferative responses are exaggerated in vitro.
In vitro assays of lymphocyte proliferation allow examination of B- and T-cell responses to cell surface receptor stimulation by mitogenic agents. Negatively-selected B- and T-cell populations from the spleen or lymph node were used for these assays. Levels of B-lymphocyte proliferation in response to anti-IgM, LPS, and anti-CD40 were equivalent between Tssc6gt/gt and Tssc6+/+ cells (data not shown).
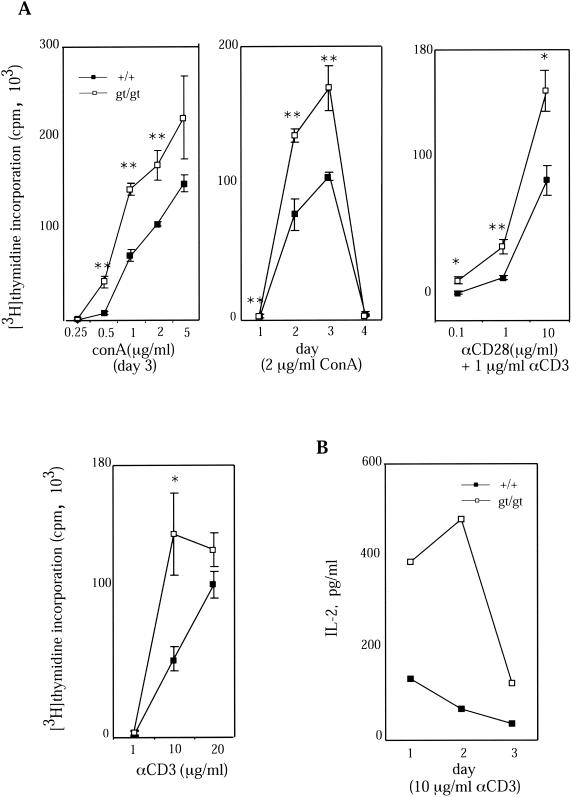
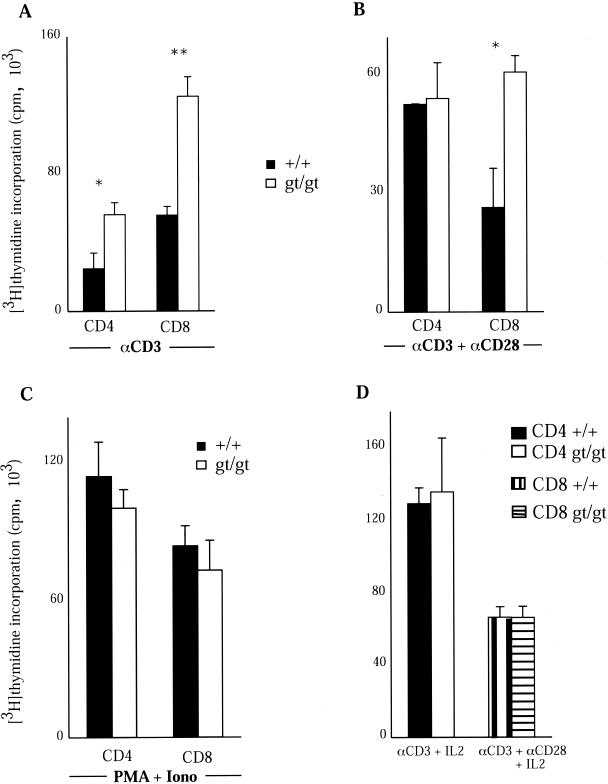
Increased proliferation of Tssc6gt/gt T cells was evident when splenocytes were cultured with different concentrations of concanavalin A. This difference was seen at concentrations of mitogen above 0.25 μg/ml and was evident on days 1 through 3 of the assay (Fig. 5A). There was also an ∼1.5- to 2-fold increase in proliferation by Tssc6gt/gt T lymphocytes in response to T-cell receptor (TCR) ligation with anti-CD3 that was maintained after ligation of the costimulatory molecule CD28 (Fig. 5A). IL-2 in the supernatant of Tssc6gt/gt and control T-cell cultures was measured at multiple time points after the addition of anti-CD3. At all time points studied, IL-2 present in the stimulated Tssc6gt/gt T-cell cultures was significantly higher than in control cultures (Fig. 5B). To further characterize the abnormal proliferative response of Tssc6gt/gt T cells, the proliferation of CD4 and CD8 T cells from wild-type and mutant mice was analyzed. Both CD4 and CD8 T cells from Tssc6gt/gt mice were hyperproliferative in response to TCR ligation. However, activating the cells by cross-linking the TCR and CD28 eliminated the hyperproliferation of Tssc6gt/gt CD4 but not CD8 T cells (Fig. 6). In keeping with the observation that IL-2 production is increased in anti-CD3-stimulated Tssc6gt/gt T cells, the addition of a saturating concentration of IL-2 to CD4 and CD8 T-cell cultures abolished the proliferative advantage displayed by these cells (Fig. 6). Interestingly, when the TCR was bypassed by stimulation with PMA and ionomycin, there was no significant difference between the proliferation rates of Tssc6+/+ and Tssc6gt/gt cells of CD4 and CD8 lineage (Fig. 6).
FIG. 5.
Proliferation of purified T-lymphocyte populations from Tssc6gt/gt mice. (A) T lymphocytes were negatively enriched from the pooled spleens of three mice of each genotype, and 105 cells/well were cultured with surface-adsorbed anti-CD28 and/or anti-CD3. Unseparated splenocytes (105 cells/well) were used for concanavalin A (ConA) stimulation. Proliferative responses of the Tssc6gt/gt (gt/gt) and Tssc6+/+ (+/+) cells for one of at least three comparable experiments are shown. Tritiated thymidine incorporation by unstimulated cells was below 1,000 cpm. Data points symbolize the means for tritiated thymidine incorporation of triplicate wells, and bars represent the standard deviations. ∗, P < 0.05; ∗∗, P < 0.005. (B) Supernatants from T-cell cultures were measured by ELISA for IL-2 secretion following stimulation with 10 μg of anti-CD3/ml. One of two experiments is depicted.
FIG. 6.
Proliferation responses of CD4 and CD8 T-cell subsets. T-cell subsets were cultured at a concentration of 105 cells/well and stimulated with either 10 μg of anti-CD3/ml (A), 1 μg of anti-CD3/ml and 10 μg of anti-CD28/ml (B), or 10 ng of PMA/ml and 100 ng of ionomycin (Iono)/ml (C) for 4 days, with the maximal tritiated thymidine incorporation shown. ∗, P < 0.05; ∗∗, P < 0.005. (D) Proliferative response when IL-2 at a concentration of 100 IU/ml was added to CD8 cells stimulated with anti-CD3 and anti-CD28 or CD4 cells stimulated with anti-CD3. Each value represents the mean and standard deviation obtained from triplicate cultures.
Total intracellular protein tyrosine phosphorylation was compared in wild-type and Tssc6-deleted T lymphocytes following ligation of the CD3/TCR complex. The pattern of tyrosine phosphorylation in resting cells and at 2, 5, and 15 min poststimulation was comparable in Tssc6+/+ and Tssc6gt/gt cells (data not shown).
Normal serum immunoglobulin levels and antibody response to T-cell-dependent and -independent antigens in Tssc6gt/gt mice.
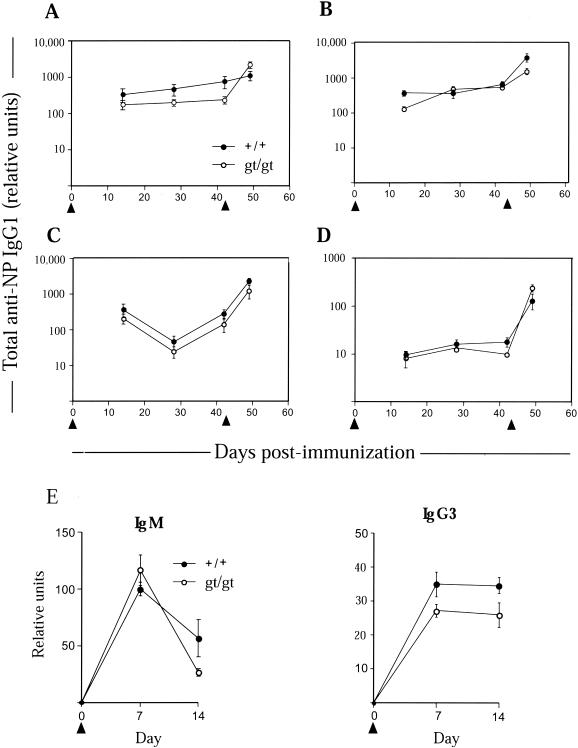
There was no significant difference between the basal level of serum immunoglobulins in Tssc6gt/gt mice and littermate control mice at both 6 to 12 weeks and 6 months of age (data not shown). Since these titers represent an accumulation of immunoglobulin over time and are regulated at many levels, we also determined immunoglobulin production in response to defined stimuli. Tssc6gt/gt mice and littermate controls were immunized with the T-cell-dependent antigen NP18-KLH precipitated in alum. To explore subtle differences in B- and T-cell interaction, we varied the dose of antigen and also examined the effect of suboptimal costimulation (i.e., immunization without adjuvant). Primary and secondary immune responses to a titrated dose of NP18-KLH both with and without adjuvant in Tssc6gt/gt mice and controls were similar (Fig. 7A through D). Moreover, affinity maturation, as determined by the temporal production of high-affinity, NP-specific IgG1, was unaltered in the mutant mice (data not shown). Comparison of follicular hyperplasia and germinal center development in histological sections of an inguinal lymph node and spleen obtained during the late primary response did not demonstrate differences between Tssc6gt/gt mice and controls (data not shown). The immune response to the T-cell-independent antigen NP-LPS was also normal in Tssc6gt/gt mice (Fig. 7E).
FIG. 7.
Humoral response of Tssc6gt/gt mice to immunization with T-cell-dependent and T-cell-independent antigens. The T-cell-dependent antigen NP18-KLH was administered i.p. (arrowheads) at the following doses for the primary response. (A) One hundred micrograms of NP18-KLH precipitated in alum. (B) Ten micrograms of NP18-KLH precipitated in alum. (C) One microgram of NP18-KLH precipitated in alum. (D) Twenty micrograms of NP18-KLH without adjuvant. Boosting occurred where indicated by the second arrow. (E) Fifty micrograms of NP-LPS injected i.p. on day 0 into each mouse to generate a T-cell-independent immune response. Anti-NP IgM and IgG3 were measured by ELISA at the indicated time points. Data represent the means and standard errors of the means for six mice (panels A through D) or cohorts of three mice (panel E) of each genotype.
DISCUSSION
Tssc6 is a recently identified member of the tetraspanin superfamily expressed exclusively in the hematopoietic system. Mice with a mutation of Tssc6 were generated from a gene trap ES cell line, and analysis of the consequences of the gene trap insertion demonstrated that it had resulted in a null mutation of the Tssc6 allele. We therefore examined the Tssc6gt/gt mice to reveal the role of this molecule in hematopoiesis. In comparison with littermates, no alterations in steady-state hematopoiesis or in the hematopoietic response to challenge with PHZ, G-CSF, or peritoneal inflammation were observed. Examination of the lymphoid populations in the peripheral blood, thymus, spleen, and peritoneum of adult Tssc6gt/gt mice did not reveal any abnormalities.
While in vitro responses by Tssc6gt/gt B cells to proliferative stimuli were comparable with those from littermates, T cells from Tssc6gt/gt showed a hyperproliferative response to concanavalin A and cross-linking of CD3 and CD28. The increased proliferation of Tssc6-deleted cells was due to a higher level of IL-2 production. The T-cell hyperproliferative response was not due to a greater percentage of activated and/or memory cells in the Tssc6gt/gt mice, as expression of CD44 and CD62L was normal. A normal response of Tssc6gt/gt T lymphocytes to stimulation with PMA and ionomycin established that the hyperproliferative response required TCR stimulation. When the Tssc6gt/gt T-lymphocyte response was resolved into CD4 and CD8 subsets, the proliferative advantage of Tssc6gt/gt CD4, but not CD8, T cells could be counteracted by optimal costimulation. This may be because, in comparison with CD8 cells, CD4 T cells are known to be more sensitive to CD28 activation (1, 2, 5, 29) and produce higher quantities of IL-2 upon TCR and CD28 stimulation (2, 5, 29). In keeping with this, addition of exogenous IL-2 to T-cell cultures abrogated the difference in proliferation between Tssc6gt/gt and Tssc6+/+ CD4 and CD8 T cells in response to CD3 and CD3 plus CD28, respectively. Despite the in vitro data, Tssc6gt/gt mice showed a normal humoral response when immunized with the T-cell-dependent antigen NP18-KLH.
Analysis of three tetraspanin knockouts now points to a common role for tetraspanins in the control of lymphocyte proliferation. A T-cell hyperproliferative response to TCR cross-linking has been observed in T cells derived from mice with null mutations in CD81 (20), CD37 (M. Wright, unpublished data), and Tssc6. By contrast, the proliferative responses of B cells in the latter two models were entirely normal (9; Wright, unpublished). Conversely, CD81-deficient B cells were hypoproliferative, presumably because of the poor expression of the B-cell costimulatory molecule CD19. CD81 is known to be a key member of a B-cell signal transduction complex that includes CD19 and Leu13 (6, 18). The data from targeted mice suggests that this role of CD81 in B-cell biology is specific to this particular tetraspanin and not common to all members of the family.
The notion that tetraspanins influence T-cell activation and proliferation was previously suggested by the response of T cells to the cross-linking of cell surface tetraspanins. Monoclonal antibodies against CD9, CD53, CD81, and CD82 are capable of costimulating T cells, and one anti-CD53 antibody was shown to be fully mitogenic for rat T cells (11, 12, 34, 35, 37). Tetraspanin-mediated costimulation occurs via a mechanism different from that by which costimulation through the classical CD28 pathway occurs. As distinct from that of CD28, CD81 signaling is inhibited by cyclosporine and generates a different cytokine repertoire upon costimulatory engagement (37). Moreover, cross-linking of CD9 and CD28 can synergize in providing costimulatory signals (40). Hence, the data from monoclonal antibody cross-linking studies suggest that the role of tetraspanins in T-cell activation is stimulatory whereas data from tetraspanin-deficient mice are most consistent with a regulatory function.
The answer to this paradox may lie in the ability of tetraspanins to form multimolecular complexes with other proteins. Of particular interest are recent reports that tetraspanins can associate with cholesterol, sphingolipid, and kinase-rich microdomains known as rafts. Specifically, the tetraspanins CD9, CD63, and CD81 can associate with rafts in an epidermoid carcinoma cell line (3) and in T cells CD9 has been shown to be present in rafts (40). A recent paper by Viola and colleagues (36) showed that T-cell costimulation through CD28 is mediated by the reorganization of membrane lipid rafts. Clustering of kinase-rich lipid rafts around the TCR complex occurs following CD28 engagement and results in an increased stability of tyrosine phosphorylation. Cross-linking of raft-associated tetraspanins could also lead to reorganization and clustering of lipid rafts and explain the costimulatory effect of tetraspanin antibodies. It has been argued that the primary function of tetraspanins is that of a molecular facilitator which organizes the microdomains of membrane proteins (17). Therefore, if a key role of tetraspanins in T cells is to organize lipid raft microdomains, the absence of these molecular organizers in a tetraspanin knockout may lead to the disruption of raft compartmentalization and an enhanced proliferative response.
To date, 18 tetraspanins have been shown to be expressed on lymphocytes (26). The mild phenotype observed in Tssc6gt/gt mice may therefore be a consequence of functional redundancy between members of the tetraspanin superfamily. Analysis of mouse strains with deletions of multiple tetraspanin molecules will further define the role of these proteins in immune regulation and may provide insights into additional functions of this large and evolutionarily conserved protein family.
Acknowledgments
We acknowledge the animal husbandry skills of Angie Porter and Annette Feeney. Marilyn Ibrahim, Sandra Mifsud, Ladina DiRago, and Amanda Light provided expert technical assistance. We are appreciative of the advice of Andreas Strasser and Raffi Gugasyan.
This work was supported by the National Health and Medical Research Council of Australia and the Sylvia and Charles Viertel Charitable Foundation.
REFERENCES
- 1.Abe, R., P. Vandenberghe, N. Craighead, D. S. Smoot, K. P. Lee, and C. H. June. 1995. Distinct signal transduction in mouse CD4+ and CD8+ splenic T cells after CD28 receptor ligation. J. Immunol. 154:985-997. [PubMed] [Google Scholar]
- 2.Cannons, J. L., P. Lau, B. Ghumman, M. A. DeBenedette, H. Yagita, K. Okumura, and T. H. Watts. 2001. 4-1Bb ligand induces cell division, sustains survival, and enhances effector function of CD4 and CD8 T cells with similar efficacy. J. Immunol. 167:1313-1324. [DOI] [PubMed] [Google Scholar]
- 3.Claas, C., C. S. Stipp, and M. E. Hemler. 2001. Evaluation of prototype TM4SF protein complexes and their relation to lipid rafts. J. Biol. Chem. 276:7974-7984. [DOI] [PubMed] [Google Scholar]
- 4.Clay, D., E. Rubinstein, Z. Mishal, A. Anjo, M. Prenant, C. Jasmin, C. Boucheix, and M. C. Le Bousse-Kerdiles. 2001. CD9 and megakaryocyte differentiation. Blood 97:1982-1989. [DOI] [PubMed] [Google Scholar]
- 5.Deeths, M. J., and M. F. Mescher. 1997. B7-1-dependent co-stimulation results in qualitatively and quantitatively different responses by CD4+ and CD8+ T cells. Eur. J. Immunol. 27:598-608. [DOI] [PubMed] [Google Scholar]
- 6.Fearon, D. T. 1993. The CD19-CR2-TAPA-1 complex, CD45 and signaling by the antigen receptor of B lymphocytes. Curr. Opin. Immunol. 5:341-348. [DOI] [PubMed] [Google Scholar]
- 7.Hemler, M. E. 1998. Integrin associated proteins. Curr. Opin. Cell Biol. 10:578-585. [DOI] [PubMed] [Google Scholar]
- 8.Kitadokoro, K., D. Bordo, G. Galli, R. Petracca, F. Falugi, S. Abrignani, G. Grandi, and M. Bolognesi. 2001. CD81 extracellular domain 3D structure: insight into the tetraspanin superfamily structural motifs. EMBO J. 20:12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knobeloch, K. P., M. D. Wright, A. F. Ochsenbein, O. Liesenfeld, J. Lohler, R. M. Zinkernagel, I. Horak, and Z. Orinska. 2000. Targeted inactivation of the tetraspanin CD37 impairs T-cell-dependent B-cell response under suboptimal costimulatory conditions. Mol. Cell. Biol. 20:5363-5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ko, M. S., T. A. Threat, X. Wang, J. H. Horton, Y. Cui, X. Wang, E. Pryor, J. Paris, J. Wells-Smith, J. R. Kitchen, L. B. Rowe, J. Eppig, T. Satoh, L. Brant, H. Fujiwara, S. Yotsumoto, and H. Nakashima. 1998. Genome-wide mapping of unselected transcripts from extraembryonic tissue of 7.5-day mouse embryos reveals enrichment in the t-complex and under-representation on the X chromosome. Hum. Mol. Genet. 7:1967-1978. [DOI] [PubMed] [Google Scholar]
- 11.Lagaudriere-Gesbert, C., F. Le Naour, S. Lebel-Binay, M. Billard, E. Lemichez, P. Boquet, C. Boucheix, H. Conjeaud, and E. Rubinstein. 1997. Functional analysis of four tetraspans, CD9, CD53, CD81, and CD82, suggests a common role in costimulation, cell adhesion, and migration: only CD9 upregulates HB-EGF activity. Cell. Immunol. 182:105-112. [DOI] [PubMed] [Google Scholar]
- 12.Lebel-Binay, S., C. Lagaudriere, D. Fradelizi, and H. Conjeaud. 1995. CD82, member of the tetra-span-transmembrane protein family, is a costimulatory protein for T cell activation. J. Immunol. 155:101-110. [PubMed] [Google Scholar]
- 13.Lee, M. P., S. Brandenburg, G. M. Landes, M. Adams, G. Miller, and A. P. Feinberg. 1999. Two novel genes in the center of the 11p15 imprinted domain escape genomic imprinting. Hum. Mol. Genet. 8:683-690. [DOI] [PubMed] [Google Scholar]
- 14.Levy, S., S. C. Todd, and H. T. Maecker. 1998. CD81 (TAPA-1): a molecule involved in signal transduction and cell adhesion in the immune system. Annu. Rev. Immunol. 16:89-109. [DOI] [PubMed] [Google Scholar]
- 15.Maecker, H. T., M. S. Do, and S. Levy. 1998. CD81 on B cells promotes interleukin 4 secretion and antibody production during T helper type 2 immune responses. Proc. Natl. Acad. Sci. USA 95:2458-2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maecker, H. T., and S. Levy. 1997. Normal lymphocyte development but delayed humoral immune response in CD81-null mice. J. Exp. Med. 185:1505-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maecker, H. T., S. C. Todd, and S. Levy. 1997. The tetraspanin superfamily: molecular facilitators. FASEB J. 11:428-442. [PubMed] [Google Scholar]
- 18.Matsumoto, A. K., D. R. Martin, R. H. Carter, L. B. Klickstein, J. M. Ahearn, and D. T. Fearon. 1993. Functional dissection of the CD21/CD19/TAPA-1/Leu-13 complex of B lymphocytes. J. Exp. Med. 178:1407-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Metcalf, D., L. Robb, A. R. Dunn, S. Mifsud, and L. Di Rago. 1996. Role of granulocyte-macrophage colony-stimulating factor and granulocyte colony-stimulating factor in the development of an acute neutrophil inflammatory response in mice. Blood 88:3755-3764. [PubMed] [Google Scholar]
- 20.Miyazaki, T., U. Muller, and K. S. Campbell. 1997. Normal development but differentially altered proliferative responses of lymphocytes in mice lacking CD81. EMBO J. 16:4217-4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicholson, R. H., S. Pantano, J. F. Eliason, A. Galy, S. Weiler, J. Kaplan, M. R. Hughes, and M. S. Ko. 2000. Phemx, a novel mouse gene expressed in hematopoietic cells maps to the imprinted cluster on distal chromosome 7. Genomics 68:13-21. [DOI] [PubMed] [Google Scholar]
- 22.Paulsen, M., O. El-Maarri, S. Engemann, M. Strödicke, O. Franck, K. Davies, R. Reinhardt, W. Reik, and J. Walter. 2000. Sequence conservation and variability of imprinting in the Beckwith-Wiedemann syndrome gene cluster in human and mouse. Hum. Mol. Genet. 9:1829-1841. [DOI] [PubMed] [Google Scholar]
- 23.Robb, L., D. J. Hilton, T. A. Willson, and C. G. Begley. 1996. Structural analysis of the gene encoding the murine interleukin-11 receptor α-chain and a related locus. J. Biol. Chem. 271:13754-13761. [DOI] [PubMed] [Google Scholar]
- 24.Robb, L., R. Li, L. Hartley, H. N. Nandurkar, F. Koentgen, and C. G. Begley. 1998. Infertility in female mice lacking the receptor for interleukin 11 is due to a defective uterine response to implantation. Nat. Med. 4:303-308. [DOI] [PubMed] [Google Scholar]
- 25.Robb, L., I. Lyons, R. Li, L. Hartley, R. P. Harvey, F. Kontgen, and C. G. Begley. 1995. Absence of yolk sac hematopoiesis from mice with a targeted disruption of the scl gene. Proc. Natl. Acad. Sci. USA 92:7075-7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robb, L., J. M. Tarrant, J. Groom, M. Ibrahim, R. Li, B. Borobokas, and M. D. Wright. 2001. Molecular characterisation of mouse and human TSSC6: evidence that TSSC6 is a genuine member of the tetraspanin superfamily and is expressed specifically in haematopoietic organs. Biochim. Biophys. Acta 1522:31-41. [DOI] [PubMed] [Google Scholar]
- 27.Roberts, A. W., and D. Metcalf. 1994. Granulocyte colony-stimulating factor induces selective elevations of progenitor cells in the peripheral blood of mice. Exp. Hematol. (New York) 22:1156-1163. [PubMed] [Google Scholar]
- 28.Salminen, M., B. I. Meyer, and P. Gruss. 1998. Efficient poly A trap approach allows the capture of genes specifically active in differentiated embryonic stem cells and in mouse embryos. Dev. Dyn. 212:326-333. [DOI] [PubMed] [Google Scholar]
- 29.Shuford, W. W., K. Klussman, D. D. Tritchler, D. T. Loo, J. Chalupny, A. W. Siadak, T. J. Brown, J. Emswiler, H. Raecho, C. P. Larsen, T. C. Pearson, J. A. Ledbetter, A. Aruffo, and R. S. Mittler. 1997. 4-1BB costimulatory signals preferentially induce CD8+ T cell proliferation and lead to the amplification in vivo of cytotoxic T cell responses. J. Exp. Med. 186:47-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stanford, W. L., G. Caruana, K. A. Vallis, M. Inamdar, M. Hidaka, V. L. Bautch, and A. Bernstein. 1998. Expression trapping: identification of novel genes expressed in hematopoietic and endothelial lineages by gene trapping in ES cells. Blood 92:4622-4631. [PubMed] [Google Scholar]
- 31.Stipp, C. S., T. V. Kolesnikova, and M. E. Hemler. 2001. EWI-2 is a major CD9 and CD81 partner and member of a novel Ig protein subfamily. J. Biol. Chem. 276:40545-40554. [DOI] [PubMed] [Google Scholar]
- 32.Szabo, P., and J. R. Mann. 1994. Expression and methylation of imprinted genes during in vitro differentiation of mouse parthenogenetic and androgenetic embryonic stem cell lines. Development 120:1651-1660. [DOI] [PubMed] [Google Scholar]
- 33.Tachibana, I., and M. E. Hemler. 1999. Role of transmembrane 4 superfamily (TM4SF) proteins CD9 and CD81 in muscle cell fusion and myotube maintenance. J. Cell Biol. 146:893-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tai, X. G., K. Toyooka, Y. Yashiro, R. Abe, C. S. Park, T. Hamaoka, M. Kobayashi, S. Neben, and H. Fujiwara. 1997. CD9-mediated costimulation of TCR-triggered naive T cells leads to activation followed by apoptosis. J. Immunol. 159:3799-3807. [PubMed] [Google Scholar]
- 35.Tai, X. G., Y. Yashiro, R. Abe, K. Toyooka, C. R. Wood, J. Morris, A. Long, S. Ono, M. Kobayashi, T. Hamaoka, S. Neben, and H. Fujiwara. 1996. A role for CD9 molecules in T cell activation. J. Exp. Med. 184:753-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Viola, A., S. Schroeder, Y. Sakakibara, and A. Lanzavecchia. 1999. T lymphocyte costimulation mediated by reorganization of membrane microdomains. Science 283:649-650. [DOI] [PubMed] [Google Scholar]
- 37.Witherden, D. A., R. Boismenu, and W. L. Havran. 2000. CD81 and CD28 costimulate T cells through distinct pathways. J. Immunol. 165:1902-1909. [DOI] [PubMed] [Google Scholar]
- 38.Woods, A., and J. R. Couchman. 2000. Integrin modulation by lateral association. J. Biol. Chem. 275:24233-24236. [DOI] [PubMed] [Google Scholar]
- 39.Wright, M. D., and M. G. Tomlison. 1994. The ins and outs of the transmembrane 4 superfamily. Immunol. Today 15:588-594. [DOI] [PubMed] [Google Scholar]
- 40.Yashiro-Ohtani, Y., X. Y. Zhou, K. Toyo-Oka, X. G. Tai, C. S. Park, T. Hamaoka, R. Abe, K. Miyake, and H. Fujiwara. 2000. Non-CD28 costimulatory molecules present in T cell rafts induce T cell costimulation by enhancing the association of TCR with rafts. J. Immunol. 164:1251-1259. [DOI] [PubMed] [Google Scholar]