Abstract
The Dorsal morphogen directs formation of the Drosophila dorsoventral axis by both activating and repressing transcription. It contains an N-terminal Rel homology domain (RHD), which is responsible for DNA binding and regulated nuclear import, and a C-terminal domain (CTD) that contains activation and repression motifs. To determine if the RHD has a direct role in transcriptional control, we analyzed a series of RHD mutations in S2 cells and embryos. Two classes of mutations (termed class I and class II mutations) that alter activation without affecting DNA binding or nuclear import were identified. The two classes appear to define distinct protein interaction surfaces on opposite faces of the RHD. Class I mutations enhance an apparently inhibitory interaction between the RHD and the CTD and eliminate both activation and repression by Dorsal. In contrast, class II mutations result in increased activation in S2 cells but severely decreased activation in embryos and have little effect on repression. Analysis of the cuticles of class II mutant embryos suggests that, in the absence of Dorsal-mediated activation, Dorsal-mediated repression is not sufficient to pattern the embryo. These results provide some of the first evidence that the RHD plays an active role in transcriptional regulation in intact multicellular organisms.
Dorsal is a maternal morphogen that is crucial for the establishment of the dorsoventral axis during Drosophila embryogenesis (12, 16, 35). In the blastoderm embryo, Dorsal forms a dorsoventral nuclear concentration gradient, with the highest concentrations on the ventral side of the embryo. Dorsal directs pattern formation by directly regulating the transcription of a number of zygotically active genes, which in turn direct the differentiation of the germ layers. For example, Dorsal activates twist (twi) and snail (sna) in ventral nuclei, thus determining the mesoderm. At the same time, this factor represses zerknüllt (zen) and decapentaplegic (dpp) in ventral nuclei. This restricts their expression to dorsal nuclei, thereby establishing the spatial limits of the dorsal ectoderm and amnioserosa.
Dorsal is a member of the Rel family of transcription factors (22). In addition to Dorsal, members of this family include the vertebrate protein NF-κB and the Drosophila proteins Dif and Relish. Rel family proteins are characterized by an N-terminal ≈300-amino-acid Rel homology domain (RHD), which is responsible for protein dimerization, DNA binding, and regulated nuclear import. Rel family proteins can function as either homodimers or heterodimers. For example, Dorsal functions in the embryo as a homodimer (23, 26), while NF-κB, a vertebrate Rel family factor, is a heterodimer of p50 and p65 (44). Not only do Dorsal and NF-κB exhibit sequence homology, they are also regulated by a conserved pathway. Both are initially retained in the cytoplasm due to an interaction with a cytoplasmic inhibitor. The regulated phosphorylation and consequent ubiquitin- and proteasome-dependent destruction of the inhibitor then allows nuclear uptake of the Rel family protein.
While the RHD mediates dimerization, DNA binding, and regulated nuclear import, the transcriptional regulatory functions of Rel family factors are generally thought to reside outside the RHD. For example, the NF-κB subunit p65 contains multiple activation domains in the region C terminal to the RHD (5, 19). Likewise, a number of studies indicate that the region of Dorsal C terminal to the RHD (termed the C-terminal domain [CTD]) is required for transcriptional activation. For example, deletion of the extreme C-terminal end of Dorsal results in decreased levels of activation in yeast cells (3), cultured Drosophila cells (40), and Drosophila embryos (27). In addition, fusion proteins consisting of the Gal4 DNA-binding domain fused to the Dorsal CTD mediate activation of Gal4 binding site-containing reporters in Drosophila embryos (17). Furthermore, the CTD has been found to interact with the TAFII110 and TAFII60 subunits of the TFIID complex, and this interaction appears to be required for Dorsal-mediated activation in vitro (38, 46).
While studies of activation by Rel family proteins have generally focused on the activation domains outside the RHD, a number of studies suggest that the RHD does more than passively tether activation domains to the template. For example, while the Dorsal RHD is not sufficient for simple activation of a Dorsal binding site-containing reporter, it interacts with Twist synergistically to activate a reporter containing adjacent Dorsal and Twist binding sites (40). Furthermore, the RHDs in both Dorsal (1) and p65 (45) have the inherent ability to interact with Drosophila CREB binding protein (dCBP), a transcriptional coactivator protein, and these interactions appear to play roles in transcriptional activation.
To dissect the roles of the RHD in activation from its roles in dimerization, DNA binding, and nuclear import, we have carried out an alanine scan mutagenesis of this domain. Analysis of the mutant proteins in cultured cells and in Drosophila embryos resulted in the identification of two classes of RHD mutations that resolve activation from DNA binding and nuclear import. The two classes of mutations appear to define two separate surfaces on the RHD. One of the classes of mutations may block activation, at least in part, by strengthening an inhibitory interaction between the RHD and the CTD. This analysis adds to a growing body of evidence suggesting that eukaryotic transcription factors are not strictly modular entities with independent DNA-binding and regulatory domains. Rather, it appears that many DNA-binding domains may actively participate in transcriptional control, at least in part by modulating the activity of linked regulatory domains.
MATERIALS AND METHODS
Site-directed mutagenesis.
Mutations in the Dorsal RHD were generated with the QuickChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocol with pAR-Dorsal(1-379) (40) as the template. The mutations and the integrity of the entire coding region for each mutant were confirmed by DNA sequencing.
Cotransfection assays.
Plasmid pPac-Dorsal (40) was used for the expression of Dorsal in S2 cells. Mutants were introduced into this plasmid with a BstXI fragment containing the mutation to replace the wild-type counterpart. pPac-Twist (40) and pPac-Cactus (3) were used for the expression of Twist and Cactus, respectively, in S2 cells. The DE5 reporter, G5 reporter, control Renilla luciferase reporter, and pPac-Gal4(1-147) have been described previously (9). pPac-Gal4-CTD was constructed by inserting a PCR fragment encoding residues 330 to 678 of Dorsal between the BamHI and KpnI sites of pPac-Gal4(1-147). Plasmids were introduced into S2 cells by using calcium phosphate as described previously (11).
For reporter assays, 5 μg of luciferase reporter was used for each transfection, and 0.1 μg of the control Renilla luciferase reporter was also included as an internal control to normalize transcription efficiency. The amounts of pPac-Dorsal, pPac-Twist, and pPac-Cactus are indicated in the figure legends. Total plasmid DNA was brought to 20 μg with pBluescript carrier DNA. The dual luciferase reporter assay (Promega) was performed following the manufacturer's protocol. All firefly luciferase activities were normalized to the control Renilla luciferase activities, and the basal activity was set to 1. All cotransfection experiments were done in duplicate, with the standard deviation indicated.
Integrated reporter assays.
The DE5 reporter was integrated into the genome of S2 cells with the Drosophila expression system (Invitrogen) following the manufacturer's instructions, and 300 μg of hygromycin per ml was used for selection. Dorsal and mutants were cloned into the SmaI site of pRM-Ha-3 (3) and integrated into the genome together with the reporter. A total of 107 cells were treated with 0, 100, or 500 μM CuSO4, and luciferase activity was measured 2 days postinduction.
Protein-protein interaction assays.
pGEX-dCBP(781-1159) (1), pGEX-Cactus (41), pGEX-Twist (25), and pGEX-CTD (17) were used to express glutathione S-transferase (GST) fusion proteins in Escherichia coli. Purification of fusion proteins was performed as described previously (40). Baculovirus expression and purification of Flag-Groucho was performed as described previously (8). Dorsal380 and mutants were labeled with [35S]methionine with the TNT T7-coupled reticulocyte lysate system (Promega) according to the manufacturer's protocols. pAR-Dorsal(1-379) (40) or mutants were used as the templates.
Binding assays were performed essentially as described previously (40); 2 μg of a fusion protein was immobilized on glutathione-agarose beads (GST-dCBP, GST-Cactus, GST-Twist, and GST-CTD) or Flag-agarose beads (Flag-Groucho) in 600 μl of HEMNK buffer (40 mM HEPES [pH 7.5], 0.2 mM EDTA, 5 mM MgCl2, 0.2 mM EDTA, 0.5% NP-40, 100 mM KCl, 1 mM dithiothreitol) and incubated with 10 μl of in vitro translation product for 1 h at 4°C. The beads were then washed five times with 1 ml of HEMNK, eluted with 40 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, and resolved by SDS-10% PAGE. The gel was then dried and exposed to a phosphorimaging screen. The scanned image was visualized, and the intensity of the bands was calculated with Image Quant software (Molecular Dynamics).
DNase I footprinting assays.
Flag-tagged Dorsal380 or Dorsal380 mutants were made by inserting the Dorsal380 coding sequences between the NotI and XbaI sites of the transfer vector pVL1392 (PharMingen). Recombinant baculovirus was obtained with the baculovirus expression vector system (PharMingen) according to the manufacturer's protocol. Protein expression and purification of Flag-Dorsal380 or mutants with anti-Flag affinity beads were performed as previously described (8). The concentration of purified proteins was determined by comparing the intensity of proteins bands in Coomassie-stained gels to a bovine serum albumin standard.
The DNase I footprinting assays were performed as described previously (37). A ≈500-bp NheI/NcoI fragment from the DE5 reporter and a ≈500-bp XhoI/SacI fragment from pBS-zenVRR(2 × 180) (generously provided by M. Levine) were used as probes.
Generation of transgenic flies.
The P-element expression vector was constructed as follows: 4.5 kb of genomic DNA from the dorsal (dl) 5′-flanking region (13) was fused to sequences encoding wild-type or mutant forms of Dorsal followed by the nt1 epitope (17). These fragments of DNA were then inserted between the KpnI and BamHI sites of pHWZ128 (32), leaving the hsp70 poly(A) signal intact.
Fly stocks of dl1/SM6; P[Dorsal/nt1]/TM3 were generated by P-element transformation of w1118 flies, followed by crossing with dl1/SM6 flies as described previously (17). dl1/dl1 females were selected to collect embryos that were devoid of endogenous Dorsal. dl1 is a null allele.
Antibody staining and in situ hybridization of whole-mount embryos.
Embryos (0 to 3 h) from multiple independent transformation lines of each construct were collected and fixed. Antisense RNA probes were labeled with digoxigenin, and in situ hybridization was carried out as described previously (42). Whole-mount antibody staining was carried out with the Vectastain ABC kit (Vector Laboratories, Inc.) with anti-nt1 monoclonal antibody (7).
RESULTS
Mutagenesis of Dorsal RHD.
To explore the role of the Dorsal RHD in activation, we mutagenized this domain. Since the ≈300-amino-acid RHD folds into a compact globular structure stabilized by long-range interactions (10, 21, 36), we decided against deletion analysis for fear that deletions would compromise the integrity of the RHD. Instead, we generated multiple point mutations targeting residues on the surface of the domain. The three-dimensional structure of Dorsal has not been determined. However, the structures of the closely related p50 and p65 RHDs are available (10, 21, 36) and were used for planning the mutagenesis of the Dorsal RHD. In general, residues with side chains that are more than 50% surface exposed were selected for mutagenesis. To reduce the number of mutants that needed to be analyzed, amino acids that cluster together were mutated in groups. In most cases, no more than three amino acids were mutated at a time, although in a few cases mutants containing either four or five altered amino acids were generated.
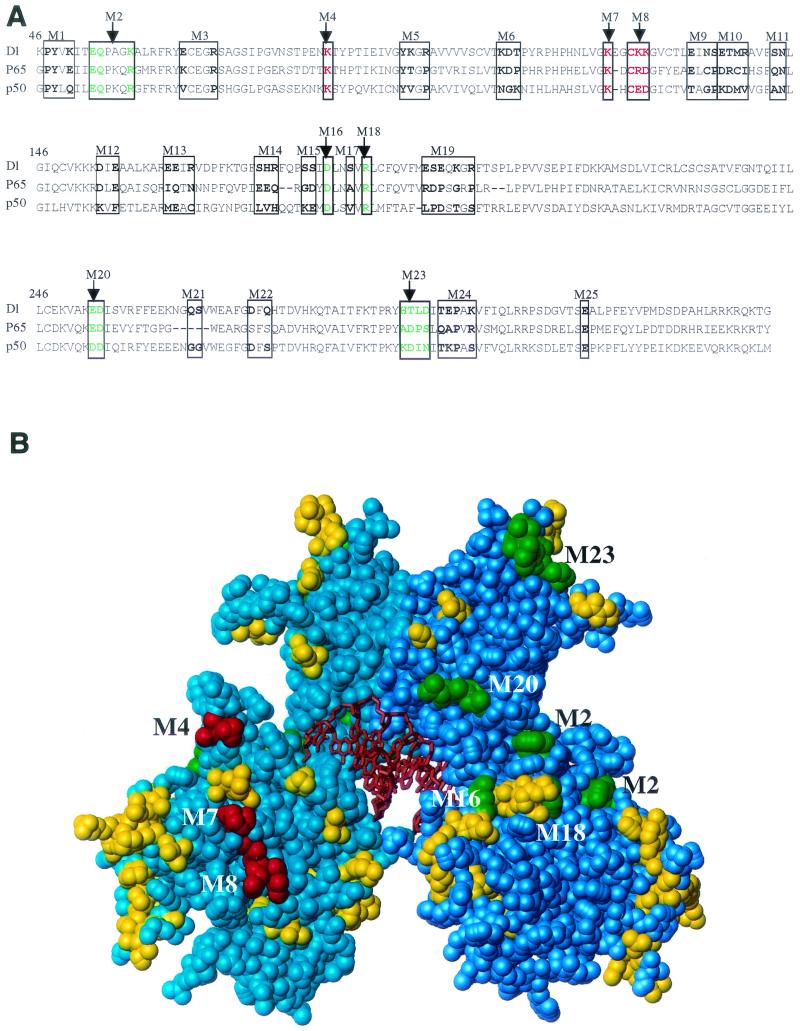
Altogether, we generated 27 mutants, targeting a total of 61 amino acid residues covering a large fraction of the surface of the RHD (Table 1 and Fig. 1). Most of the mutations converted wild-type residues to alanines, although in four cases (M4′, M7′, M16, and M18) charged residues were converted to residues of the opposite charge. Amino acid residues close to the DNA-binding and dimerization interfaces of the RHD were avoided in the mutagenesis.
TABLE 1.
Mutagenesis of the Dorsal RHD
| Mutant | Amino acid(s) mutated | Exposed surfacea | Phenotypic classb |
|---|---|---|---|
| M1 | P47A, W48A, K50A | 0.65 | Neutral |
| M2 | E53A, Q54A, K58A | 0.60 | I |
| M3 | E65A, R69A | 0.78 | Neutral |
| M4 | K84A | 0.83 | II |
| M4′ | K84E | 0.83 | II |
| M5 | W94A, K95A, R97A | 0.56 | Neutral |
| M6 | K107A, D108A, T109A | 0.81 | Neutral |
| M7 | K121A | 0.85 | II |
| M7′ | K121E | 0.85 | II |
| M8 | C124A, K125A, K126A | 0.87 | II |
| M9 | E132A, N134A, S135A | 0.60 | Neutral |
| M10 | E136A, T137A, M138A, R139A | 0.74 | Neutral |
| M11 | S143A, N144A | 0.54 | Neutral |
| M12 | D154A, E156A | 0.73 | Neutral |
| M13 | E163A, E164A, R166A | 0.81 | Neutral |
| M14 | S175A, H176A, R177A | 0.54 | Neutral |
| M15 | S181A, S182A | 0.88 | Neutral |
| M16 | D184K | 0.81 | I |
| M17 | S187A | 0.43 | Neutral |
| M18 | R189E | 0.63 | Ic |
| M19 | E197A, S198A, E199A, K201, R203A | 0.81 | Neutral |
| M20 | E253A, D254A | 0.64 | I |
| M21 | Q266A, S267A | 0.98 | Neutral |
| M22 | D274A, Q276A | 0.81 | Neutral |
| M23 | H294A, T295A, L296A, D297A | 0.71 | I |
| M24 | T299A, E300A, P301A, K303A | 0.94 | Neutral |
| M25 | E318A | 0.91 | Neutral |
Fraction of the total surface area of the mutated residues accessible to water. The calculation (6) was carried out by using the structure of the p50 homodimer (21).
Phenotypic class assignments are based on the data in Fig. 2B. Mutants that were at least twofold more active than the wild type are defined as class II. Mutants that were at least twofold less active than the wild type are defined as class I. The remaining mutants are defined as neutral.
This mutant showed severely reduced DNA-binding activity.
FIG. 1.
Mutagenesis of the Dorsal RHD. (A) Sequence alignment of the Dorsal RHD with those of p65 and p50. The amino acids that were mutated are indicated in bold and boxed. Amino acids changed in class I and class II mutants are colored green and red, respectively. (B) Space-filling model of the RHD. The RHD homodimer structure is based on the coordinates determined for p50 (Protein Data Bank identification no. 1NF-K). The two polypeptide chains are shown in cyan and blue, and the DNA is shown in orange. The amino acids that were altered in the mutants are shaded green (class I mutations), red (class II mutations), and yellow (phenotypically silent mutations).
Cotransfection assays reveal two classes of mutants with altered ability to activate transcription.
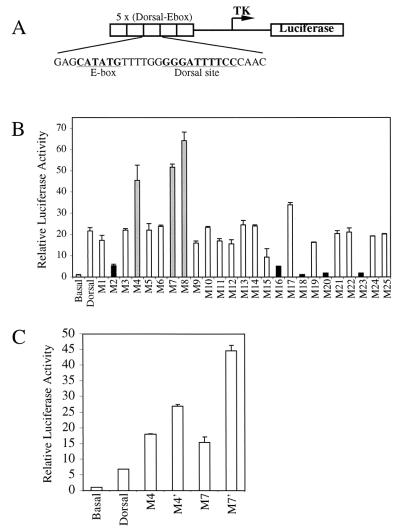
The Dorsal mutants were examined for their ability to activate transcription in a transient-transfection assay with the DE5 reporter (Fig. 2A) (3, 40). This reporter contains both Twist and Dorsal binding sites. It is weakly activated by Dorsal alone but is strongly activated by the combination of Dorsal and Twist. The reporter was cotransfected into S2 cells along with vectors encoding Twist and wild-type Dorsal or one of the Dorsal mutants (Fig. 2B). The majority of the mutants gave approximately wild-type levels of stimulation. However, five mutants (M2, M16, M18, M20, and M23) (Fig. 2B) were compromised in their ability to activate transcription in this assay. An examination of a molecular model of the RHD (Fig. 1B) shows that the altered amino acids in these mutants, hereafter referred to as class I mutants, are all on the same face of the RHD, suggesting that they define a positively acting interaction surface.
FIG. 2.
Cotransfection assays reveal two classes of mutants. (A) Structure of the DE5 reporter. (B) Activation of the DE5 reporter by Dorsal mutants. S2 cells were transfected with 60 ng of a plasmid encoding the wild-type or indicated mutant forms of Dorsal together with 20 ng of a plasmid encoding Twist, 5 μg of the DE5 reporter, and 0.1 μg of a plasmid containing the Renilla luciferase gene under the control of the herpes simplex virus thymidine kinase (TK) promoter. Cell extracts were prepared and assayed for luciferase activity 2 days posttransfection. Firefly luciferase activities were first divided by the Renilla luciferase activities to normalize for variations in transfection efficiency. The normalized values were then divided by the basal value (no activators) to obtain the relative luciferase activity. Results shown are the averages of duplicate assays, and error bars show the standard deviation. Three mutants (class II mutants M4, M7, and M8; gray bars) were found to give significantly better activation than wild-type Dorsal, while five mutants (class I mutants M2, M16, M18, M20, and M23; black bars) yielded significantly reduced Dorsal-mediated activation. (C) Effect of replacing the alanine substitutions in the class II mutants with glutamic acid substitutions. The mutants were analyzed as described for panel B.
The cotransfection assays also revealed a second class of mutants with altered ability to activate transcription. Three of the mutants (M4, M7, and M8) (Fig. 2B) consistently yielded two- to threefold greater activation than that achieved with wild-type Dorsal. Examination of the RHD model (Fig. 1B) reveals that the amino acids affected in these superactive mutants cluster together on the opposite face of the domain from the amino acids altered in the class I mutants. Thus, these mutants, hereafter referred to as class II mutants, may define a second interface on the RHD. All three class II mutations eliminated lysine side chains, suggesting that electrostatic interactions are important for stabilizing the interaction with the hypothetical target of this interacting surface. This conclusion is supported by the finding that replacement of two of these lysines, K84 and K121, by glutamate, an amino acid of the opposite charge, resulted in an even stronger superactive phenotype than did the corresponding alanine substitutions (Fig. 2C).
Class I and II mutations do not generally inactivate the RHD.
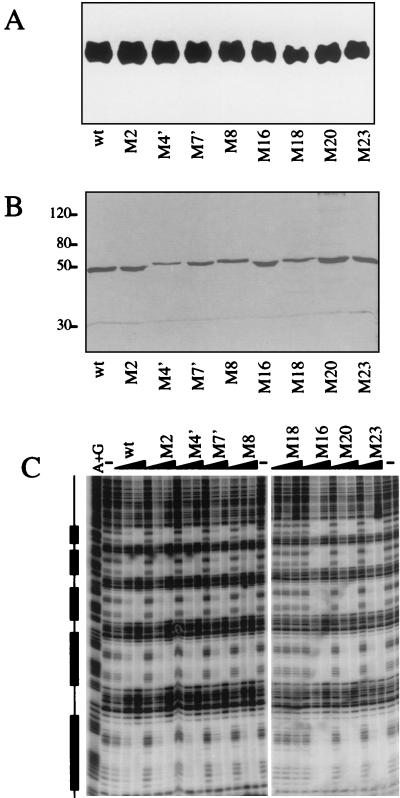
Although we exercised caution in designing our mutants by trying to select surface amino acids that would not play critical roles in stabilizing the RHD fold, it was still possible that some of our mutations destabilized the RHD or perturbed its folding. To address these possibilities, we examined the expression, nuclear localization, and DNA-binding activity of the mutants. When nuclear extracts of S2 cells transfected with equal amounts of DNA encoding Dorsal RHD mutants were analyzed by anti-Dorsal immunoblotting (Fig. 3A), the wild-type and mutant proteins were all found to accumulate in the nucleus to similar levels.
FIG. 3.
Effects of the mutations on protein stability and DNA-binding activity. (A) Mutations do not affect levels of nuclear Dorsal protein. S2 cells (50 ml) were transiently transfected with 2 μg of wild-type (wt) Dorsal380 or the indicated mutant forms of Dorsal380. Cells were harvested 2 days posttransfection, and nuclear extracts were prepared. The proteins were resolved by SDS-10% PAGE, transferred to a polyvinylidene difluoride membrane, and probed with anti-Dorsal antibody. (B) Purification of recombinant Dorsal mutants. Flag-tagged Dorsal380 or the indicated Dorsal380 mutants were immunopurified to homogeneity from Sf9 cells infected with recombinant baculoviruses. Proteins were resolved by SDS-10% PAGE and silver stained. (C) With one exception, the mutations do not alter DNA-binding activity. The purified proteins shown in panel B were assayed for binding to the DE5 enhancer by a DNase I footprinting assay. The boxes to the left indicate the Dorsal binding sites. Each protein was assayed at 2, 10, and 50 ng. Lane A+G, A+G chemical sequencing ladder; lane −, no-protein control.
To assess the effects of the mutations on DNA-binding activity, we carried out DNase I footprinting assays. Recombinant mutant proteins were purified to homogeneity (Fig. 3B). With either the DE5 enhancer (Fig. 3C) or the zen ventral repression region (data not shown) as a probe, we observed nearly wild-type DNA-binding activity for all the class II mutants and four of the five class I mutants. Only mutation M18 showed severely reduced DNA binding. Thus, the expression, localization, and DNA-binding data indicate that, with the possible exception of M18, the mutants do not have reduced stability or structural integrity.
Mutations do not interfere with binding to known Dorsal-interacting proteins.
As discussed above, the mutations in the RHD may define surfaces for interaction with other regulatory proteins. We therefore examined the effects of these mutations on proteins thought to interact with the Dorsal RHD and positively or negatively regulate Dorsal function.
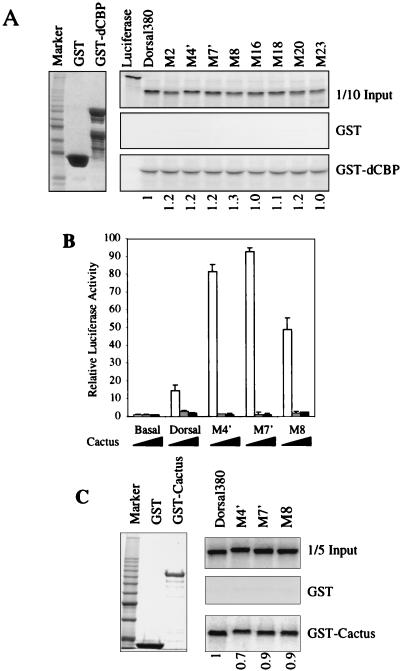
The RHD binds to the coactivator protein dCBP and may stimulate Dorsal-mediated activation in S2 cells and in the embryo (1). It is possible that the class I mutations impair Dorsal-mediated activation by interfering with the interaction between Dorsal and dCBP. We therefore examined the ability of the RHD mutants to bind dCBP in vitro. In agreement with previous findings, we detected a specific interaction between the wild-type Dorsal RHD (Dorsal380) and a GST fusion protein containing a fragment of dCBP (amino acids 781 to 1159). However, none of the mutations that altered activation in S2 cells had a significant effect on this interaction (Fig. 4A). Therefore, it is unlikely that our mutations altered activation by altering the affinity of Dorsal for dCBP.
FIG. 4.
Mutations do not affect interactions of Dorsal with known partner proteins. (A) Mutations do not affect binding of Dorsal to the coactivator dCBP. Left, purified GST and GST-dCBP(781-1159) were resolved by SDS-PAGE and visualized by staining with Coomassie blue. Right, in vitro-translated and 35S-labeled luciferase, Dorsal380 (a C-terminal truncation of Dorsal containing the intact RHD), and the indicated Dorsal380 mutants were incubated with equal amounts of GST or GST-dCBP immobilized on glutathione beads and washed extensively. The eluted proteins were resolved by SDS-PAGE and imaged by autoradiography. One-tenth of the input protein amount is shown for comparison. (B) Class II mutants are still responsive to Cactus in vivo. S2 cells was cotransfected with 60 ng of a plasmid encoding full-length Dorsal or the indicated Dorsal mutants together with 20 ng of a plasmid encoding Twist, 5 μg of the DE5 reporter, and 0 ng (white bars), 100 ng (gray bars), or 500 ng (black bars) of a plasmid encoding Cactus. Luciferase activity was determined as described in the legend to Fig. 1A. (C) Class II mutants bind Cactus in vitro. Left, GST and GST-Cactus purified from E. coli and stained with Coomassie blue. Right, In vitro-translated and 35S-labeled Dorsal380 or the indicated Dorsal380 mutants were incubated with either GST or GST-Cactus immobilized on glutathione beads and washed extensively. The eluted proteins were resolved by SDS-PAGE and imaged by autoradiography. One-fifth of the input protein amount is shown for comparison. Quantification of the interaction is shown below the lanes, with wild-type binding arbitrarily set to 1.
The superactivity of the class II mutants suggests that they are impaired in their ability to interact with a negative regulator of Dorsal activity. One well-characterized negative regulator of Dorsal is Cactus, which binds the RHD and inhibits Dorsal nuclear uptake (20, 30). In cotransfection assays, activation by Dorsal was blocked by simultaneous overexpression of Cactus (Fig. 4B). However, the class II mutants responded to Cactus as well as wild-type Dorsal. Furthermore, GST pulldown assays confirmed that the wild-type RHD and the class II mutants bound to Cactus with comparable affinity (Fig. 4C). Thus, these mutations do not impair the interaction with Cactus.
We also examined the interaction between the RHD mutants and three other factors known to interact with the RHD, Ubc9 (3), Twist (40), and Groucho (14). We did not detect any effects of the mutations on these interactions (data not shown). In conclusion, our results strongly suggest that the two surfaces defined by the two classes of mutations define interaction surfaces for novel regulatory interactions.
Noncovalent interaction between the RHD and CTD of Dorsal.
Eukaryotic transcriptional activators are often viewed as modular entities, with independent DNA-binding and activation domains (29, 43). It was thus surprising to find that class I mutations, which leave the known functions of the RHD intact (e.g., DNA binding and nuclear import), do not activate transcription despite the presence of a presumably functional activation domain in the CTD. Previous studies of this activation domain have shown that it functions independently of the RHD in embryos (17), but similar studies have not been carried out with S2 cells.
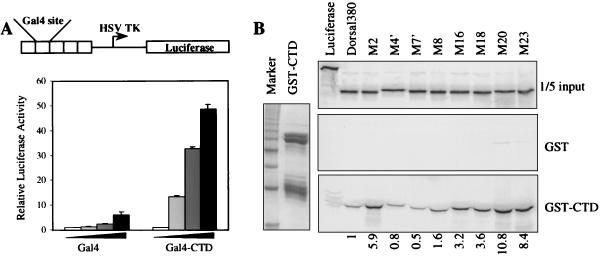
To demonstrate that the CTD functions as an activation domain in S2 cells, we generated a chimeric factor consisting of the Gal4 DNA-binding domain fused to the Dorsal CTD. The addition of the Dorsal CTD to the Gal4 DNA-binding domain was found to result in a potent activator (Fig. 5A). These findings imply that the class I mutant RHD serves to restrain activation by the CTD. To explore this possibility further, we looked for a noncovalent interaction between the RHD and the CTD. We found that the RHD bound to a GST fusion protein containing the Dorsal CTD (Fig. 5B). Moreover, the class I mutations (M2, M16, M18, M20, and M23) resulted in a 3- to 10-fold increase in CTD binding. In contrast, the class II mutations (M4′, M7′, and M8) each had less than a twofold effect on CTD binding. These findings suggest that binding of the CTD to the RHD negatively regulates activation by the CTD and that the surface defined by the class I mutations negatively modulates this interaction.
FIG. 5.
Interaction between RHD and CTD. (A) Dorsal CTD can activate transcription. Top, schematic diagram of the G5 luciferase reporter. The firefly luciferase gene is under the control of five copies of a module containing Gal4 binding sites. These are inserted immediately upstream of the herpes simplex virus thymidine kinase (HSV TK) core promoter. Bottom, S2 cells were transfected with 5 μg of the G5 luciferase reporter and 0.1 μg of a plasmid containing the Renilla luciferase gene under the control of the herpes simplex virus thymidine kinase promoter, together with 0 ng, 150 ng, 500 ng, or 2 μg of pPac-Gal4 or pPac-Gal4-CTD. (B) The RHD binds the CTD. Left, purified GST-CTD. Right, in vitro-translated and 35S-labeled Dorsal380 or the indicated Dorsal380 mutants were incubated with either GST or GST-Dorsal-CTD immobilized on glutathione beads and washed extensively. The eluted proteins were resolved by SDS-PAGE and imaged by autoradiography. One-fifth of the input protein amount is shown for comparison. Quantification of the interaction is shown below the lanes, with wild-type binding arbitrarily set to 1.
Class I mutations prevent both activation and repression in the embryo.
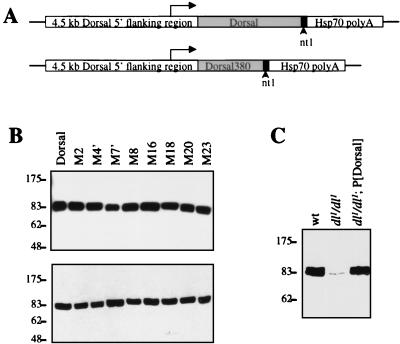
To determine if the regulatory regions in the RHD that we identified with transient-transfection assays are also functional during Drosophila embryogenesis, transgenes encoding Dorsal variants were introduced into the germ line by P-element-mediated transformation. Maternal expression of cDNAs encoding the Dorsal variants was directed by a 4.5-kb segment of 5′-flanking DNA from the dl locus. This region appears to contain all the cis-regulatory elements required for normal maternal expression of dl (13). The encoded proteins contain a 19-amino-acid epitope (nt1) at the C-terminal end, against which we have a monoclonal antibody, thereby providing a way to monitor levels of expression (Fig. 6A).
FIG. 6.
Transformation of germ line with P-elements encoding Dorsal mutants. (A) Structure of expression vectors. A 4.5-kb region from the endogenous dl locus was used to direct the expression of full-length Dorsal, full-length Dorsal mutants, and Dorsal380. The 19-amino-acid nt1 epitope was appended to the C terminus of the encoded proteins. The constructs also contained the hsp70 polyadenylation signal. (B) Analysis of the expression level of dl transgenes. Forty embryos from mothers bearing transgenes encoding the indicated nt1-tagged forms of Dorsal were homogenized in SDS-PAGE loading buffer, resolved by SDS-PAGE, transferred to a polyvinylidene difluoride membrane, and probed with anti-nt1 antibody. (C) Comparison of the expression levels of endogenous and transgenic Dorsal proteins. Embryos with the indicated maternal genotypes were analyzed as in panel B except with anti-Dorsal antibody.
Flies containing transgenes often show considerable differences in expression levels due to position effects. To ensure that we were comparing transformant lines with equal levels of expression, embryos from numerous lines were collected and expression levels were compared by immunoblotting with anti-nt1 antibody. We identified a set of lines that displayed similar levels of expression (Fig. 6B). The set contained two independent lines for each class I and each class II mutant. In the analysis described below, the two transformant lines of each Dorsal mutant were found to give very similar results.
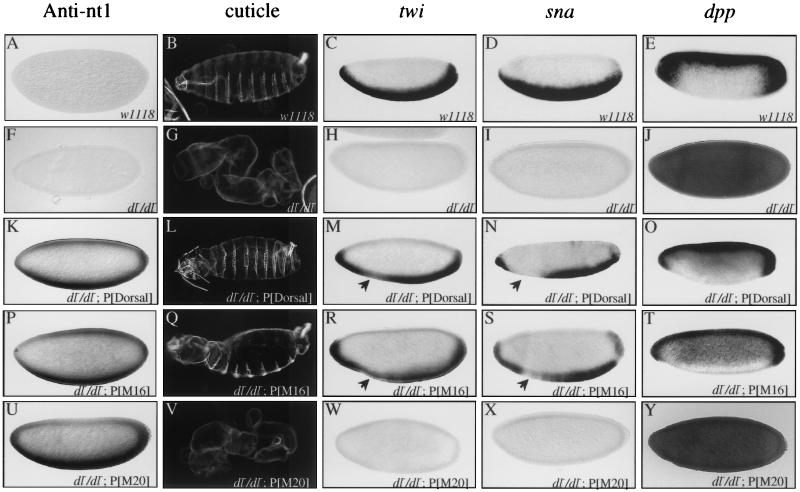
First, we examined rescue of the null dl1 mutant phenotype by the wild-type dl transgene. The cuticle phenotypes due to loss-of-function alleles of dl are often described with a scale ranging from D0 (completely dorsalized) to D3 (weakly dorsalized) (2). The cuticles of embryos laid by dl1 females lack ventral denticle belts, Filzkörper, and a head skeleton and instead consist only of a tube of dorsal epidermis (the D0 phenotype) (Fig. 7, compare panels B and G). The wild-type transgene was found to rescue the ventral denticle belts and Filzkörper and partially restore the head skeleton (yielding a D2 phenotype) (Fig.7L). However, the head was abnormal and hatching was not observed. twi and sna, two genes that are normally activated on the ventral side of the embryo by Dorsal, are not expressed in embryos lacking maternal Dorsal protein (compare panels C and D with H and I). In situ hybridization experiments indicate that the wild-type transgene largely rescued twi and sna expression (Fig. 7M and N). However, reduced levels of expression were observed around the region of the prospective cephalic furrow. Examination of the expression patterns of genes that are repressed by Dorsal, such as dpp (compare panels E and O) and zen (data not shown), showed that the recombinant Dorsal was indistinguishable from endogenous Dorsal in its ability to repress transcription in the blastoderm embryo.
FIG. 7.
Class I mutations abolish Dorsal function in the embryo. (A, F, K, P, and U) Whole-mount anti-nt1 antibody staining of embryos laid by mothers with the indicated genotypes. (B, G, L, Q, and V) Cuticle preparations of embryos laid by mothers with the indicated genotypes. (C, H, M, R, and W) Whole-mount in situ hybridization with digoxigenin-labeled antisense riboprobe against the twi mRNA to embryos laid by mothers with the indicated genotypes. (D, I, N, S, and X) Whole-mount in situ hybridization with digoxigenin-labeled antisense riboprobe against the sna mRNA to embryos laid by mothers with the indicated genotypes. (E, J, O, T, and Y) Whole-mount in situ hybridization with digoxigenin-labeled antisense riboprobe against the dpp mRNA to embryos laid by mothers with the indicated genotypes. Embryos are oriented with the anterior to the left and the dorsal side up. Arrows in panels M, N, R, and S indicate the positions in the embryos where twi and sna expression is reduced.
The partial rescue observed here indicates that the dl transgene is not providing full activity. Comparison of the expression level of the transgene with the expression level of the endogenous dl gene by anti-Dorsal immunoblotting indicates that the recombinant and endogenous proteins were expressed at very similar levels (Fig. 6C). This is consistent with the observation that the recombinant protein is just as active as the endogenous protein in transcriptional repression. It thus appears that the C-terminal epitope tag reduces the ability of Dorsal to activate transcription. This is consistent with previously published results indicating that Dorsal contains an activation domain at the extreme C-terminal end of the protein (40).
We then proceeded to examine the class I mutants. Staining of embryos with anti-nt1 antibody (Fig. 7, panels P and U, and data not shown) showed that the mutant proteins were specifically localized to the ventral nuclei in a pattern very similar to that observed for the wild-type recombinant protein (panel K). This finding that the mutant proteins are localized to the nucleus in a properly regulated manner provides further evidence that the mutant proteins are folded properly.
As shown above (Fig. 2), the class I mutations reduced transcriptional activation in S2 cells. Similarly, in situ hybridization experiments showed that four of the five class I mutants (M2, M18, M20, and M23) were completely unable to activate twi and sna transcription in the embryo. Since these four mutations yielded indistinguishable phenotypes, we show data for only one (M20) (Fig. 7W and X). Although these mutants were selected solely on the basis of their inability to activate transcription, we found that they were also unable to repress the transcription of dpp (Fig. 7Y) and zen (data not shown). In accord with this inability to activate or repress transcription, these mutants also had no ability to rescue the D0 cuticle phenotype (Fig. 7V). The fifth class I mutant (M16) had more modest effects in the cotransfection assays than did the other four mutations in this class (Fig. 2B). The M16 mutation also had a relatively weak effect on binding to the CTD (Fig. 5B). This mutant rescued pattern formation as well as the wild-type recombinant protein (Fig. 7Q to T).
Mutants that show superactivation in S2 cells exhibit reduced ability to activate transcription in embryos.
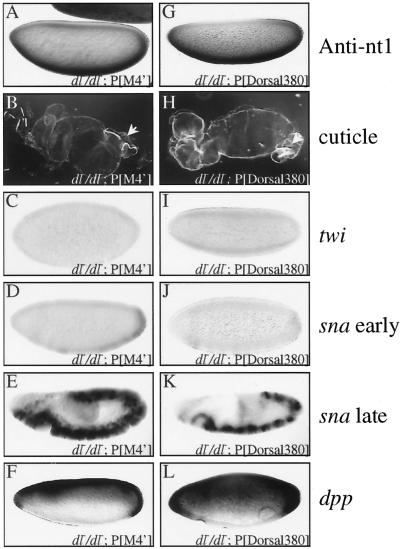
Examination of the embryonic phenotypes of the class II mutants (M4′, M7′, and M8), which exhibit superactivity in S2 cells, yielded unexpected results. Since all three class II mutants gave very similar phenotypes, we show data for only one (M4′). Even though the class II mutants were localized to ventral nuclei similarly to the product of the wild-type transgene (Fig. 8A), they all rescued dorsoventral patterning to a significantly lesser extent than wild-type Dorsal. In situ hybridization revealed very weak (nearly undetectable) expression of twi and sna in the blastoderm embryo (Fig. 8C and D). But unlike the embryos containing the class I mutants, these embryos exhibited detectable mesoderm-specific sna expression during germ band elongation (Fig. 8E). Also unlike the class I mutants, the class II mutants were able to repress transcription of genes such as dpp (Fig. 8F) and zen (data not shown) on the ventral side of the embryo. In accord with the greatly reduced ability of the class II mutants to activate transcription relative to the wild-type transgene, the cuticles were only weakly rescued (Fig. 8B). They displayed a D1 phenotype, exhibiting Filzkörper (arrow) but lacking ventral denticle bands.
FIG. 8.
Class II mutants and Dorsal380 show reduced activation in vivo. (A and G) Whole-mount anti-nt1 antibody staining of embryos laid by mothers with the indicated genotypes. (B and H) Cuticle preparations of embryos laid by mothers with the indicated genotypes. Filzkörper are indicated by arrows. (C and I) Whole-mount in situ hybridization with digoxigenin-labeled antisense riboprobe against the twi mRNA to embryos laid by mothers with the indicated genotypes. (D, E, J, and K) Whole-mount in situ hybridization with digoxigenin-labeled antisense riboprobe against the sna mRNA to embryos laid by mothers with the indicated genotypes. (F and L) Whole-mount in situ hybridization with digoxigenin-labeled antisense riboprobe against the dpp mRNA to embryos laid by mothers with the indicated genotypes. All in situ hybridizations show blastoderm stage embryos except panels D and J, which show germ band-elongated embryos.
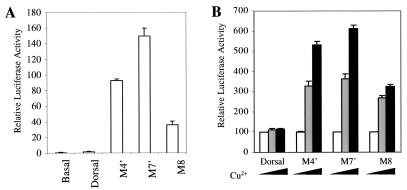
The different activities of the class II mutants in S2 cells versus embryos may be due to differences in template structure in the two systems. Transiently transfected reporter genes in S2 cells may not adopt the same chromatin structure as endogenous Dorsal target genes in embryos. We therefore further tested the ability of the class II mutants to activate transcription in S2 cells with a stably integrated luciferase reporter containing the DE5 enhancer. The cells also contained integrated expression constructs encoding wild-type Dorsal or class II Dorsal mutants under the control of the Cu2+-inducible metallothionein promoter. The results obtained with the integrated reporter (Fig. 9B) were in close accord with results obtained in transient-transfection assays (Fig. 9A). In both cases, there was little activation by wild-type Dorsal. This is due, most likely, to the absence of Twist, since previous studies have indicated that Dorsal activates the DE5 reporter very poorly in the absence of Twist (42). However, with both the transiently and stably transfected reporters, the class II mutants yielded significant levels of activation. Thus, in S2 cells, the class II mutants were able to superactivate transcription of stably integrated templates. This indicates that the difference in the behavior of these mutants in S2 cells and embryos cannot be ascribed to the difference between unintegrated and integrated templates.
FIG. 9.
Comparison of unintegrated and integrated templates. (A) Class II mutants can activate transcription of unintegrated templates in the absence of Twist. S2 cells were cotransfected with 5 μg of DE5 reporter together with 1 μg of either wild-type full-length Dorsal or class II mutants. (B) Class II mutants can activate transcription of integrated templates. The DE5 reporter was stably integrated into the genome of S2 cells. Full-length Dorsal or the indicated Dorsal mutants under the control of the metallothionein promoter were also integrated into the genome. Luciferase activity was measured 2 days after induction with CuSO4. For each mutant, luciferase activity in the absence of CuSO4 was set to 100. White bars, no CuSO4; gray bars, 100 μM CuSO4; black bars, 500 μM CuSO4.
CTD is required for activation in the embryo.
A number of previous studies as well as data presented above (Fig. 5A) indicate that the Dorsal CTD contains an activation domain. However, our discovery of a set of RHD mutations (the class I mutations) that abolish activation even in the presence of the CTD could be interpreted to suggest that the C-terminal activation domain does not contribute to activation by Dorsal in embryos. To determine the contribution of the RHD and CTD to activation in the embryo, we generated transgenic fly lines expressing nt1-tagged Dorsal380, which contains the RHD but lacks the CTD (Fig. 6A). Lines expressing levels of Dorsal380 similar to the level of recombinant Dorsal in our other transgenic lines were identified by immunoblotting (data not shown). Staining of embryos from these lines with the anti-nt1 antibody revealed specific localization to the ventral nuclei (Fig. 8G). In situ hybridization revealed very weak (nearly undetectable) expression of twi and sna in the blastoderm embryo (Fig. 8I and J) but detectable mesoderm-specific sna expression during germ band elongation (Fig. 8K). Thus, while the RHD is sufficient for very weak activation, the CTD greatly potentiates Dorsal-mediated activation.
Dorsal380 also exhibited a reduced ability to repress the transcription of genes such as dpp (Fig. 8L) and zen (data not shown). While these genes are clearly repressed ventrally, the domains of expression are broader than they are in embryos expressing full-length Dorsal. This finding is in agreement with previous results suggesting that the CTD of Dorsal contains a region that mediates an interaction with the corepressor Groucho (17).
In agreement with the reduced ability of Dorsal380 to activate and repress transcription, this truncated form of Dorsal also had a reduced ability to rescue dorsoventral patterning of the cuticle (Fig. 8H). Like the class II mutant embryos, the cuticles of the Dorsal380 embryos displayed a D1 phenotype, exhibiting Filzkörper but lacking ventral denticle bands.
DISCUSSION
Dorsal RHD mutants that modulate activation potential.
We have identified two classes of Dorsal RHD mutations affecting Dorsal-mediated activation. Class I mutations result in a loss of activation in both S2 cells and embryos, while class II mutations result in superactivation in S2 cells and reduced activation in embryos. The mutants appear to have wild-type stability, are localized to the nucleus in a regulated manner, and, with a single exception, have wild-type DNA-binding activity. Thus, our findings indicate that, in addition to its well-characterized roles in dimerization, DNA binding, and regulated nuclear localization, the RHD must have an active role in transcriptional control. This is the first time such a role has been demonstrated in an intact multicellular organism.
The two classes of mutations appear to define surfaces for two distinct regulatory interactions. This conclusion is suggested by the observation that the class I and class II mutations reside on opposite faces of the RHD. However, the mutations do not alter binding affinity to any previously known RHD-interacting proteins, including the positively acting factors dCBP and Twist. In principle, our screen should have yielded mutants with compromised ability to bind these two proteins, since previous studies suggest that both of these Dorsal-interacting proteins have roles in Dorsal-mediated activation (1, 40). It is possible that the Dorsal-Twist and Dorsal-dCBP interactions involve numerous weak interactions, so that mutations in one or a few closely spaced interacting residues do not have measurable effects on binding affinity.
Communication between DNA-binding domains and activation domains.
Our class I mutations block activation in the context of full-length Dorsal protein despite the presence of an activation domain in the CTD. As we have demonstrated here, this activation domain can function independently of the RHD. These findings are inconsistent with a simple model for transcriptional control in which the sole function of a DNA-binding domain is to tether one or more activation domains to the template to allow them to interact either directly or indirectly with the transcriptional machinery or chromatin template. However, our findings are in accord with previous studies suggesting that DNA-binding domains often contain interaction surfaces that send regulatory signals to attached activation domains (31). For example, certain mutations in the glucocorticoid receptor DNA-binding domain prevent activation while not interfering with DNA binding despite the presence of independent activation domains in the factor (39). It has been suggested that the amino acids affected in these glucocorticoid receptor mutants play roles in transmitting an allosteric signal from the DNA to the activation domain, which serves to stimulate activation domain function. Rel homology domains are also thought to undergo conformational changes upon binding to DNA, as shown by changes in both protease sensitivity and circular dichroism spectra, which could modulate the activity of linked activation domains (19, 24, 34).
The analysis of the Dorsal RHD class I mutations presented here suggests a novel mechanism by which a DNA-binding domain could regulate a linked activation domain. In particular, we have found that the RHD binds the CTD and that mutations that increase the affinity of this interaction weaken the ability of Dorsal to function as a transcriptional activator. These findings strongly suggest that noncovalent interactions between the RHD and the CTD downregulate activation by the CTD. In effect, the RHD may be serving as a decoy for the activation domain. When the activation domain interacts with the RHD, it may be unavailable to interact with the general machinery.
While the class I mutant RHD binds the CTD, the wild-type RHD also binds the CTD, albeit with a lower affinity. Thus, the wild-type RHD may also be able to downregulate CTD function, and the phenotype of the class I mutants may represent a heightening of this wild-type function. Given the complexity of eukaryotic genomes and the relatively low DNA-binding specificity of sequence-specific factors such as Dorsal, this downregulation may be necessary to prevent high levels of inappropriate activation by Dorsal when it binds to adventitious sites that occur throughout the genome. When Dorsal binds to bona fide Dorsal-responsive enhancers, however, it may find itself in the context of other transcriptional activators that can cooperate with Dorsal via enhanceosome formation to yield high levels of activation.
In addition to preventing activation, class I mutations also prevent repression. One possible interpretation of this observation is that the same RHD-CTD interaction that prevents activation also prevents repression, possibly by interfering with the repression domain in the CTD (17).
Uncoupling of Dorsal-mediated repression from Dorsal-mediated activation.
In contrast to the class I mutants, which behave similarly in S2 cells and embryos, the class II mutants displayed significantly different behavior in the two settings. While the class II mutants are more potent activators than wild-type Dorsal in S2 cells, they have a severely attenuated ability to activate transcription in embryos. These findings suggest that the surface defined by the class II mutations interacts with a variety of coregulatory proteins, including both positive and negative regulators. In embryos, a positive coregulator may be the dominant interacting protein, and thus perturbing the interaction surface results in reduced levels of activation. In contrast, in S2 cells, a negative coregulator may dominate, resulting in superactivation upon disruption of the interaction surface. These mutations may interfere with multiple interactions by influencing the conformation of the RHD. This possibility is suggested by previous work showing that binding of the RHD to DNA induces a change in the rate of protease cleavage at certain protease-hypersensitive sites in the RHD. These hypersensitive sites map to the region of the RHD defined by our class II mutations (34). Perhaps the class II mutations alter the ability of the RHD to undergo a DNA-induced conformational change, thereby altering the affinity of the factor for multiple coregulators.
Because the class II Dorsal mutants are able to repress but unable to activate transcription in the blastoderm embryo, they allow us to assess the developmental consequences of uncoupling Dorsal-mediated activation from Dorsal-mediated repression. Despite the ability of class II mutants to repress dpp in the ventral ectoderm, these mutants are unable to rescue the ventral ectoderm, as evidenced by the absence of ventral denticle belts. These findings support the idea that Dpp can diffuse ventrally to block ventral ectoderm formation in the ventrolateral region (4, 15). In the wild-type embryo, Dorsal is competent to activate transcription and therefore turns on short gastrulation (sog) and brinker in the ventrolateral region (33). Sog then functions upstream while Brinker functions downstream of Dpp receptors to block Dpp signaling and allow ventral ectoderm formation in the ventolateral region (4, 18, 28). Our finding that a repression-competent but activation-defective form of Dorsal is not sufficient to allow ventral ectoderm formation suggests that Dorsal-mediated activation is of primary importance in the subdivision of the embryo into developmental domains. In contrast, Dorsal-mediated repression may be a relatively recent adaptation that ensures the complete shutdown of Dpp signaling in the ventral and ventrolateral regions. The development of Dorsal alleles that are able to activate but unable to repress transcription would allow further testing of these ideas.
Acknowledgments
We thank Stephen Smale and Uptal Banerjee for critical reading of the manuscript, Ruth Steward for providing us with a genomic clone containing the dl 5′-flanking region, and Larry Zipursky for providing the anti-nt1 antibody. We also thank Benjamin Chen and Jonathan Wojciak for technical assistance.
This work was supported by National Institutes of Health grant GM44522.
REFERENCES
- 1.Akimaru, H., D. X. Hou, and S. Ishii. 1997. Drosophila CBP is required for dorsal-dependent twist gene expression. Nat. Genet. 17:211-214. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, K. V., and C. Nusslein-Volhard. 1986. Dorsal-group genes of Drosophila, p. 177-194. In J. Gall (ed.), Gametogenesis and the early embryo. A. R. Liss, New York, N.Y.
- 3.Bhaskar, V., S. A. Valentine, and A. J. Courey. 2000. A functional interaction between dorsal and components of the Smt3 conjugation machinery. J. Biol. Chem. 275:4033-4040. [DOI] [PubMed] [Google Scholar]
- 4.Biehs, B., V. Francois, and E. Bier. 1996. The Drosophila short gastrulation gene prevents Dpp from autoactivating and suppressing neurogenesis in the neuroectoderm. Genes Dev. 10:2922-2934. [DOI] [PubMed] [Google Scholar]
- 5.Blair, W. S., H. P. Bogerd, S. J. Madore, and B. R. Cullen. 1994. Mutational analysis of the transcription activation domain of RelA: identification of a highly synergistic minimal acidic activation module. Mol. Cell. Biol. 14:7226-7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowie, J. U., R. Luthy, and D. Eisenberg. 1991. A method to identify protein sequences that fold into a known three-dimensional structure. Science 253:164-170. [DOI] [PubMed] [Google Scholar]
- 7.Cagan, R. L., H. Kramer, A. C. Hart, and S. L. Zipursky. 1992. The bride of sevenless and sevenless interaction: internalization of a transmembrane ligand. Cell 69:393-399. [DOI] [PubMed] [Google Scholar]
- 8.Chen, G., and A. J. Courey. 1999. Baculovirus-transfer vector for eukaryotic expression and immunoaffinity purification of Gal4-fusion proteins. BioTechniques 26:808-810, 812, 814. [DOI] [PubMed] [Google Scholar]
- 9.Chen, G., P. H. Nguyen, and A. J. Courey. 1998. A role for Groucho tetramerization in transcriptional repression. Mol. Cell. Biol. 18:7259-7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, Y. Q., S. Ghosh, and G. Ghosh. 1998. A novel DNA recognition mode by the NF-κB p65 homodimer. Nat. Struct. Biol. 5:67-73. [DOI] [PubMed] [Google Scholar]
- 11.Courey, A. J., D. A. Holtzman, S. P. Jackson, and R. Tjian. 1989. Synergistic activation by the glutamine-rich domains of human transcription factor Sp1. Cell 59:827-836. [DOI] [PubMed] [Google Scholar]
- 12.Courey, A. J., and J. D. Huang. 1995. The establishment and interpretation of transcription factor gradients in the Drosophila embryo. Biochim. Biophys. Acta 1261:1-18. [DOI] [PubMed] [Google Scholar]
- 13.Drier, E. A., L. H. Huang, and R. Steward. 1999. Nuclear import of the Drosophila Rel protein Dorsal is regulated by phosphorylation. Genes Dev. 13:556-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubnicoff, T., S. A. Valentine, G. Chen, T. Shi, J. A. Lengyel, Z. Paroush, and A. J. Courey. 1997. Conversion of dorsal from an activator to a repressor by the global corepressor Groucho. Genes Dev. 11:2952-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferguson, E. L., and K. V. Anderson. 1992. Decapentaplegic acts as a morphogen to organize dorsal-ventral pattern in the Drosophila embryo. Cell 71:451-461. [DOI] [PubMed] [Google Scholar]
- 16.Flores-Saaib, R. D., and A. J. Courey. 2000. Regulation of dorso/ventral patterning in the Drosophila embryo by multiple dorsal-interacting proteins. Cell Biochem. Biophys. 33:1-17. [DOI] [PubMed] [Google Scholar]
- 17.Flores-Saaib, R. D., S. Jia, and A. J. Courey. 2001. Activation and repression by the C-terminal domain of Dorsal. Development 128:1869-1879. [DOI] [PubMed] [Google Scholar]
- 18.Francois, V., M. Solloway, J. W. O'Neill, J. Emery, and E. Bier. 1994. Dorsal-ventral patterning of the Drosophila embryo depends on a putative negative growth factor encoded by the short gastrulation gene. Genes Dev. 8:2602-2616. [DOI] [PubMed] [Google Scholar]
- 19.Fujita, T., G. P. Nolan, S. Ghosh, and D. Baltimore. 1992. Independent modes of transcriptional activation by the p50 and p65 subunits of NF-κB. Genes Dev. 6:775-787. [DOI] [PubMed] [Google Scholar]
- 20.Geisler, R., A. Bergmann, Y. Hiromi, and C. Nusslein-Volhard. 1992. cactus, a gene involved in dorsoventral pattern formation of Drosophila, is related to the IκB gene family of vertebrates. Cell 71:613-621. [DOI] [PubMed] [Google Scholar]
- 21.Ghosh, G., G. van Duyne, S. Ghosh, and P. B. Sigler. 1995. Structure of NF-κB p50 homodimer bound to a κB site. Nature 373:303-310. [DOI] [PubMed] [Google Scholar]
- 22.Ghosh, S., M. J. May, and E. B. Kopp. 1998. NF-κB and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 16:225-260. [DOI] [PubMed] [Google Scholar]
- 23.Govind, S., A. M. Whalen, and R. Steward. 1992. In vivo self-association of the Drosophila rel-protein Dorsal. Proc. Natl. Acad. Sci. USA 89:7861-7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hay, R. T., and J. Nicholson. 1993. DNA-binding alters the protease susceptibility of the p50 subunit of NF-κB. Nucleic Acids Res. 21:4592-4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ip, Y. T., R. E. Park, D. Kosman, E. Bier, and M. Levine. 1992. The dorsal gradient morphogen regulates stripes of rhomboid expression in the presumptive neuroectoderm of the Drosophila embryo. Genes Dev. 6:1728-1739. [DOI] [PubMed] [Google Scholar]
- 26.Isoda, K., and C. Nusslein-Volhard. 1994. Disulfide cross-linking in crude embryonic lysates reveals three complexes of the Drosophila morphogen dorsal and its inhibitor cactus. Proc. Natl. Acad. Sci. USA 91:5350-5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Isoda, K., S. Roth, and C. Nusslein-Volhard. 1992. The functional domains of the Drosophila morphogen dorsal: evidence from the analysis of mutants. Genes Dev. 6:619-630. [DOI] [PubMed] [Google Scholar]
- 28.Jazwinska, A., C. Rushlow, and S. Roth. 1999. The role of brinker in mediating the graded response to Dpp in early Drosophila embryos. Development 126:3323-3334. [DOI] [PubMed] [Google Scholar]
- 29.Johnson, P. F., and S. L. McKnight. 1989. Eukaryotic transcriptional regulatory proteins. Annu. Rev. Biochem. 58:799-839. [DOI] [PubMed] [Google Scholar]
- 30.Kidd, S. 1992. Characterization of the Drosophila cactus locus and analysis of interactions between cactus and dorsal proteins. Cell 71:623-635. [DOI] [PubMed] [Google Scholar]
- 31.Lefstin, J. A., and K. R. Yamamoto. 1998. Allosteric effects of DNA on transcriptional regulators. Nature 392:885-888. [DOI] [PubMed] [Google Scholar]
- 32.Liaw, G. J., K. M. Rudolph, J. D. Huang, T. Dubnicoff, A. J. Courey, and J. A. Lengyel. 1995. The torso response element binds GAGA and NTF-1/Elf-1, and regulates tailless by relief of repression. Genes Dev. 9:3163-3176. [DOI] [PubMed] [Google Scholar]
- 33.Markstein, M., P. Markstein, V. Markstein, and M. S. Levine. 2002. Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc. Natl. Acad. Sci. USA 99:763-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matthews, J. R., J. Nicholson, E. Jaffray, S. M. Kelly, N. C. Price, and R. T. Hay. 1995. Conformational changes induced by DNA binding of NF-κB. Nucleic Acids Res. 23:3393-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morisato, D., and K. V. Anderson. 1995. Signaling pathways that establish the dorsal-ventral pattern of the Drosophila embryo. Annu. Rev. Genet. 29:371-399. [DOI] [PubMed] [Google Scholar]
- 36.Muller, C. W., F. A. Rey, M. Sodeoka, G. L. Verdine, and S. C. Harrison. 1995. Structure of the NF-κB p50 homodimer bound to DNA. Nature 373:311-317. [DOI] [PubMed] [Google Scholar]
- 37.Pan, D. J., J. D. Huang, and A. J. Courey. 1991. Functional analysis of the Drosophila twist promoter reveals a dorsal-binding ventral activator region. Genes Dev. 5:1892-1901. [DOI] [PubMed] [Google Scholar]
- 38.Pham, A. D., S. Muller, and F. Sauer. 1999. Mesoderm-determining transcription in Drosophila is alleviated by mutations in TAF(II)60 and TAF(II)110. Mech. Dev. 84:3-16. [DOI] [PubMed] [Google Scholar]
- 39.Schena, M., L. P. Freedman, and K. R. Yamamoto. 1989. Mutations in the glucocorticoid receptor zinc finger region that distinguish interdigitated DNA-binding and transcriptional enhancement activities. Genes Dev. 3:1590-1601. [DOI] [PubMed] [Google Scholar]
- 40.Shirokawa, J. M., and A. J. Courey. 1997. A direct contact between the Dorsal Rel homology domain and Twist may mediate transcriptional synergy. Mol. Cell. Biol. 17:3345-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tatei, K., and M. Levine. 1995. Specificity of Rel-inhibitor interactions in Drosophila embryos. Mol. Cell. Biol. 15:3627-3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tautz, D., and C. Pfeifle. 1989. A nonradioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma 98:81-85. [DOI] [PubMed] [Google Scholar]
- 43.Triezenberg, S. J. 1995. Structure and function of transcriptional activation domains. Curr. Opin. Genet. Dev. 5:190-196. [DOI] [PubMed] [Google Scholar]
- 44.Urban, M. B., R. Schreck, and P. A. Baeuerle. 1991. NF-κB contacts DNA by a heterodimer of the p50 and p65 subunit. EMBO J. 10:1817-1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhong, H., R. E. Voll, and S. Ghosh. 1998. Phosphorylation of NF-κB p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol. Cell 1:661-671. [DOI] [PubMed] [Google Scholar]
- 46.Zhou, J., J. Zwicker, P. Szymanski, M. Levine, and R. Tjian. 1998. TAFII mutations disrupt Dorsal activation in the Drosophila embryo. Proc. Natl. Acad. Sci. USA 95:13483-13488. [DOI] [PMC free article] [PubMed] [Google Scholar]