Abstract
E47 and Pip are proteins crucial for proper B-cell development. E47 and Pip cooperatively bind to adjacent sites in the immunoglobulin kappa chain 3′ enhancer and generate a potent transcriptional synergy. We generated protein-DNA computer models to visualize E47 and Pip bound to DNA. These models predict precise interactions between the two proteins. We tested predictions deduced from these models by mutagenesis studies and found evidence for novel direct interactions between the E47 helix-loop-helix domain (Arg 357 or Asp 358) and the Pip N terminus (Leu 24). We also found that precise spatial alignment of the binding sites was necessary for transcriptional synergy and cooperative DNA binding. A Pip dominant negative mutant that cannot synergize with E47 inhibited enhancer activity in plasmacytoma cells and could not activate transcription in pre-B cells. Using electrophoretic mobility shift assays, we found that Pip can bind to the heavy-chain intron enhancer region. In addition, we found that in fibroblasts Pip greatly increased E47 induction of germ line Iμ transcripts associated with somatic rearrangement and isotype class switching. However, a Pip dominant negative mutant inhibited germ line Iμ transcripts. The importance of these results for late B-cell functions is discussed.
During B-cell development, cells progress through an ordered series of steps, including pro-B-, pre-B-, B-, and plasma-cell stages. These stages can be defined by expression of specific cell surface markers and ordered rearrangement of immunoglobulin (Ig) heavy-chain and light-chain genes (28). The heavy-chain genes usually rearrange first, early in B-cell development during the change from the pro-B- to the pre-B-cell stage. Ig light-chain genes (kappa and lambda) are unrearranged and transcriptionally silent at the pro-B-cell stage but undergo somatic rearrangement during the pre-B- to B-cell transition to produce a productive light-chain gene. B cells subsequently undergo class switch recombination to produce antibodies with different effector functions.
A variety of studies indicate that enhancers at the Ig heavy-chain and light-chain loci are very important for proper B-cell development (23, 59, 63). These enhancers play crucial roles not only in Ig transcription but also in somatic rearrangement, isotype class switch recombination, somatic mutation, and control of chromatin structure (5, 35, 37, 43, 48). A variety of transcription factors [E2A, EBF, PU.1, BSAP(Pax-5), Pip, and IKAROS] are known to control development of the B-cell lineage (18, 33, 42), and many of these factors bind to the multiple Ig gene enhancers and regulate their activities.
The E2A gene product binds to heavy-chain and light-chain gene enhancers and is responsible for controlling early B-cell development (1, 70, 71). E2A belongs to the helix-loop-helix (HLH) class of transcription factors, which are necessary for numerous developmental processes, including myogenesis, hematopoiesis, neurogenesis, and sex determination (2, 9, 39, 44, 67). The E2A gene encodes three gene products (E12, E47, and E2-5), which differ either at their N-terminal regions or within the basic HLH (bHLH) region by differential RNA processing. E2A proteins can form either homodimers or heterodimers with other HLH proteins. However, in B cells E47 primarily forms homodimers (60).
Although the E2A proteins are ubiquitously expressed, E2A knockouts primarily affect B-cell development and arrest B-cell differentiation at an early stage (1, 70, 71). The E2A proteins are crucial for proper somatic rearrangement of Ig and T-cell receptor genes (3, 8, 56, 58), and ectopic expression of E2A in non-B cells can induce sterile Iμ transcripts associated with somatic rearrangement of Ig heavy-chain genes (8, 58). Late in B-cell development, E2A is also implicated in Ig class switch recombination (54). Therefore, E2A is crucial for both early and late functions in B-cell development.
Another protein needed for B-cell development, Pip, is an interferon regulatory factor (IRF)-related protein expressed primarily in B-lymphoid cells and variously called NF-EM5, LSIRF, IRF4, or ICSAT (12, 40, 42, 51, 68). Pip binds to Ig light-chain enhancers via a winged HTH domain and is expressed at lower levels in pre-B cells than in plasma cells, when Pip expression increases dramatically (6, 12). Mutation of the Pip gene by homologous recombination yields mice with normal numbers of B and T cells, but these mice show greatly reduced serum Ig concentrations (42). B- and T-cell function is compromised, and knockout mice fail to mount detectable antibody responses (42). These results indicate that Pip is very important for late B-cell functions.
Pip binds to DNA very poorly on its own but is efficiently recruited to DNA by interaction with the Ets-related factor PU.1 (13, 51, 52). Interestingly, Pip also cooperatively binds to DNA in association with the E2A protein E47 (45). This cooperative interaction leads to a 100-fold transcriptional synergy between Pip and E47. Since E47 is required for both early and late B-cell functions but Pip is needed only for late B-cell development, the functional synergy between E47 and Pip is likely to be important for some late B-cell function. The identity of this function, however, is unknown. Understanding the mechanism of interaction between E47-Pip is likely to be important not only for understanding enhancer function but also for deciphering the late B-cell functions mediated by both proteins during hematopoietic development.
We prepared computer-generated molecular models to approximate the structures of the DNA binding domains of E47 and Pip bound to adjacent sites on the Igκ 3′ enhancer. These models predicted close association of the Pip N terminus and the E47 HLH regions. They also suggested that nucleotide insertions between the E47 and Pip sites would disrupt E47-Pip interaction. We show here that these predictions are accurate. We found that a single-base-pair insertion between the E47 and Pip sites severely compromised cooperative DNA binding and transcriptional synergy. We found that specific amino acid residues within the Pip amino-terminal region are needed for transcriptional synergy and efficient cooperative DNA binding with E47. We also identified residues within the E47 HLH region that are needed for synergy with Pip. These E47 and Pip contacts represent a novel mechanism of interaction between HLH and IRF domain proteins. As expected, Pip mutants that cannot synergize with E47 acted as dominant negative mutants in plasma cells and failed to stimulate enhancer activity in pre-B cells. Interestingly, a previously unidentified Pip site within the Ig heavy-chain enhancer region apparently mediates E47-Pip synergy which induces germ line Iμ transcripts associated with somatic rearrangement and isotype class switching. The implications of our results for late B-cell functions is discussed.
MATERIALS AND METHODS
DNA constructs.
Reporter plasmid (E47-Pip)4LBKCAT is identical to reporter plasmid 7 reported previously (50). Preparation of the CoreTKCAT plasmid was previously described (49). Oligonucleotide sequences in the Ins1, Ins2, Ins5, and Ins10 reporter constructs (see Fig. 2) were inserted into the LBKCAT reporter as BamHI-BglII fragments and multimerized into four copies as described previously (50). Mutant E47 and Pip DNA constructs were prepared by PCR with appropriate oligonucleotide primers or by overlap extension PCR (29) with primers containing the appropriate mutant sequences. Specific details of mutant construction are available upon request.
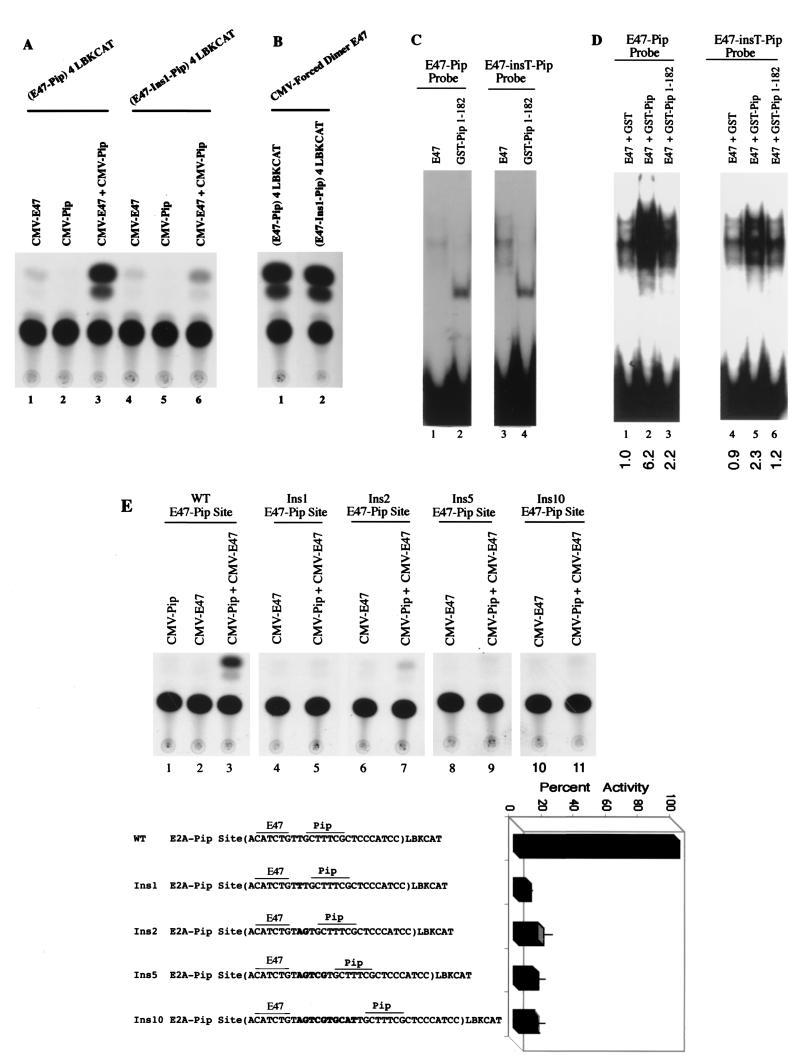
FIG. 2.
A single-base-pair insertion destroys E47-Pip synergy. (A) NIH 3T3 cells were transfected with reporter plasmids containing either a multimerized (four copies) E47-Pip motif (lanes 1 to 3) or a multimerized motif with a single-base-pair insertion between the E47 and Pip sites (lanes 4 to 6). Reporter plasmids were cotransfected with E47 or Pip alone or with E47 plus Pip. CAT activities show a dramatic loss in transcriptional synergy by E47 and Pip with the single-base-pair insertion reporter. (B) The single-base-pair insertion did not compromise the E47 binding site. The wild-type and 1-bp insertion reporters were cotransfected with a forced dimer E47 expression vector. Each reporter was equally activated, indicating that the single-base-pair insertion did not reduce the ability of E47 alone to bind to DNA. (C) The single-base-pair insertion does not disrupt separate DNA binding by E47 and Pip. EMSA was performed with wild-type or 1-bp insertion probes with either recombinant E47 or GST-Pip 1-182. Identities of proteins and probes are indicated above the lanes. (D) The single-base-pair insertion reduces E47-Pip cooperative DNA binding. EMSA was performed with either the wild-type E47-Pip DNA binding probe (lanes 1 to 3) or the 1-bp insert probe (lanes 4 to 6). Above each lane are indicated the proteins included in each assay and the probe used. Relative binding values determined by phosphorimager quantification are shown below each lane. (E) All insertions between the E47 and Pip sites in the Igκ 3′ enhancer abolish transcriptional synergy between E47 and Pip. NIH 3T3 cells were transfected with reporter plasmids containing either wild-type (WT) E47-Pip sites or insertions of 1, 2, 5, or 10 bp between the sites. CAT activities are shown in the top panel. Above each lane are shown the identities of cotransfected expression plasmids. The bottom panel shows the E47 and Pip sequences in the reporter plasmids (inserted nucleotides are in boldface), and the histogram shows percent activity of each reporter, with the wild-type reporter activity defined as 100%. Error bars represent standard deviations of the means.
Cell culture and transfections.
NIH 3T3 cells were grown in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum and transfected by the calcium phosphate coprecipitation method (24). Cells were harvested at 48 h posttransfection. Each transfection mixture contained 1 μg of a β-galactosidase-expressing plasmid to normalize transfection efficiencies, 3 to 5 μg of reporter plasmid, and 3 to 5 μg of either empty expression vector (cytomegalovirus [CMV]) or CMV effectors. Transfection efficiencies were normalized by using β-galactosidase activity, and chloramphenicol acetyltransferase (CAT) assays followed by thin-layer chromatography were performed as described by Gorman et al. (22). Percent CAT activity was calculated by scintillation counting of the acetylated product and substrate spots. In each case transfections were performed three to five times. S194 and 3-1 cells were grown and transfected as previously described (50).
Molecular models.
Initial molecular models of the DNA binding domains of E47 and Pip on the wild-type and single-insertion mutant Igκ enhancer sites were generated as follows: (i) ideal double-stranded B-form DNA helices corresponding to the sequence 5′ACATCTGTT(T)GCTTTCGCTCCCATCC3′, where (T) indicates the insertion, were generated with the program Namot2 (http://www.t10.lanl.gov/namot); (ii) using the graphical display program Setor (16), the coordinates for residues 335 to 392 of the homodimeric bHLH DNA binding domain of E47 were manually docked onto the E2A recognition site CATCTG of the enhancer, matching contacts observed to the corresponding sequence CACCTG in the crystallographic structure of the E47-DNA complex (14); and (iii) the coordinates for residues 7 to 111 of the winged HTH DNA binding domain of IRF-1 (used as a homology model for residues 23 to 127 of Pip; see reference 15 for a sequence alignment) were docked onto the Pip recognition site GCTTTCG of the enhancer, matching contacts observed to the corresponding sequence TTTC in the crystallographic structure of the IRF-1-DNA complex (pdb file 1IF1) (15). To include the effects of nucleic acid bending, the coordinates of the two DNA-protein complexes were merged such that the 3′ T (bold) of the E47-DNA complex (AACACCTGGCT) was superimposed upon the 5′ A (bold) of the IRF-1-DNA complex (ACTTTCACTTCTC). Underlined sequences are the core E47 and IRF-1 binding sites, respectively. This served to orient the two complexes, with their crystallographically observed DNA bending, into the juxtaposition of the E74 and Pip recognition sites within the Igκ enhancer.
EMSA.
Electrophoretic mobility shift assays (EMSA) were performed with about 0.1 ng (15,000 cpm) of either the Igκ E47-Pip DNA probe (ACATCTGTTGCTTTCGCTCCCATCC), the μE4 DNA probe (CCAGGTGGTGTTTTGCTCAGCCTGG), or a 220-bp HinfI-HinfI DNA fragment containing the heavy-chain intron enhancer core. Reaction mixtures consisted of 20 μl containing 2 μg of poly(dI-dC), 10 mM Tris-HCl (pH 7.5), 50 mM NaCl, 1 mM dithiothreitol, 1 mM EDTA, 5% glycerol, and various proteins. Glutathione S-transferase (GST) fusion proteins were prepared as described by Kaelin et al. (32), and EMSA reaction mixtures contained 0.1 to 2 μg of each protein. Samples were fractionated on 4% polyacrylamide gels in 0.25× Tris-borate-EDTA buffer.
Reverse transcription-PCR (RT-PCR).
Transfected NIH 3T3 cells were harvested 2 days after transfection, and RNA was prepared by the Trizol method according to the specifications of the manufacturer (GibcoBRL). Twenty-five micrograms of total RNA was treated with DNase I and then copied into cDNA by using Superscript II (Stratagene) according to the manufacturer's recommended procedures. After digestion of RNA with RNase H, the cDNA was diluted 10-fold. One microliter and two successive fourfold dilutions were subjected to PCR with outer Iμ primers (forward, ACCTGGGAATGTATGGTTGTGGCTT; reverse, ATGCAGATCTCTGTTTTTGCCTCC) for 20 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s. Five microliters of the first PCR mixture was subjected to PCR with the inner Iμ primers (forward, GGTGGCTTTGAAGGAACAATTCCAC; reverse, TCTGAACCTTCAAGGATGCTCTTG) and trace amounts of [α-32P]dCTP, using the same cycle parameters. PCR with the actin primers (forward, GCCGCACTCGTCATTGACAATGG; reverse, CCCGTTCAGTCAGGATCTTCATGAG) was performed only through the first PCR step. PCR products were separated on 8% polyacrylamide gels, dried, exposed to X-ray film, and quantitated with a Bio-Rad phosphorimager.
RESULTS
Models of E47 and Pip on DNA.
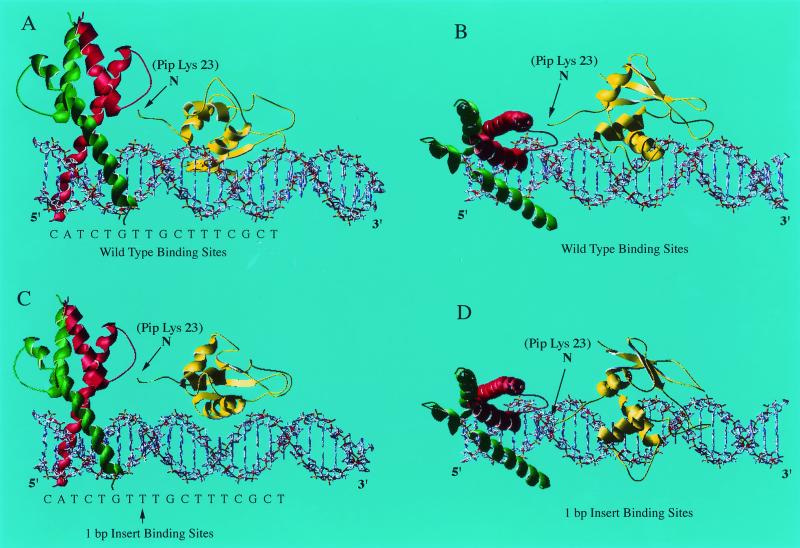
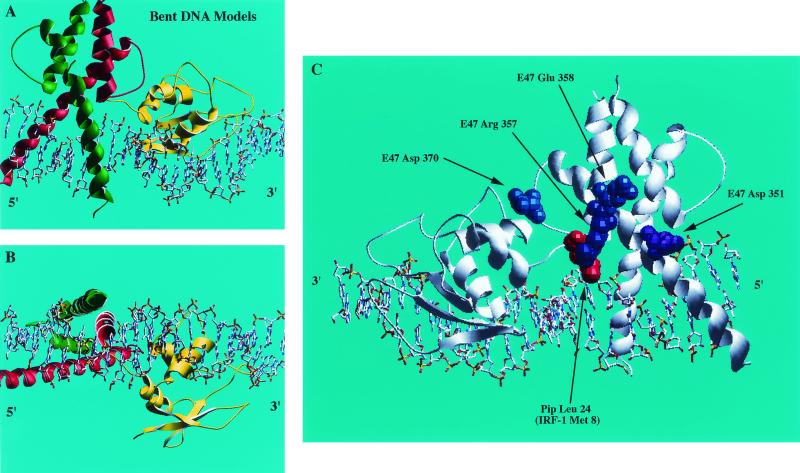
Our previous work showed that E47 and Pip bind to adjacent DNA sequences within the Igκ 3′ enhancer (45). The close proximity of the E47 and Pip binding sites, their cooperative DNA binding, and their transcriptional synergy suggested an intimate relationship between the proteins on DNA. To visualize how these proteins might interact when bound to the Igκ 3′ enhancer, we prepared models based upon the existing crystal structure of E47 (14) in association with the DNA binding domain of the Pip relative IRF-1 (15). We reasoned that the high degree of homology between the Pip and IRF-1 DNA binding domains (12) would make IRF-1 a suitable model for Pip in our studies. The first six residues of IRF-1 in the published structure appear to be disordered in the crystal complex, and thus our model corresponds to Pip residues 23 to 127 (15).
The E47-Pip (IRF-1)-DNA model generated suggested close associations between the proteins on DNA (Fig. 1A and B). In particular, the model suggested that the Pip (IRF-1) N-terminal tail makes direct contacts with the HLH domain of E47. If E47-Pip synergy depended upon such protein-protein contacts, nucleotide insertions between the E47 and Pip binding sites would be expected to compromise these interactions. Therefore, we also modeled E47 and IRF-1 on the Igκ 3′ enhancer sequence with a 1-bp insertion between the E47 and Pip binding sites (Fig. 1C and D). In this model, the Pip N terminus is rotated by about 36° and translated away by about 3.4 Å to a distinct position in relation to E47 (compare Fig. 1B and D). If the synergy between E47 and Pip requires interaction between the E47 HLH region and the Pip N-terminal sequence, this mutation would be expected to disrupt those interactions.
FIG. 1.
Models of E47 and Pip on the Igκ 3′ enhancer binding sites. (A and B) An E47 bHLH homodimer (red and green) derived from the known E47 crystal structure (14) was positioned on the ideal B-form DNA of the wild-type E-box motif within the Igκ 3′ enhancer. The winged HTH DNA binding domain from IRF-1 was used as a substitute for the Pip DNA binding domain. The known IRF-1 crystal structure (15) was used to model residues 23 to 117 of the Pip DNA binding domain (yellow). Views from the side (A) and bottom (B) of the structure are shown. The arrow shows the N terminus of the IRF structure, which correlates to Pip Lys 23. The sequence of the E47 and Pip motif from the Igκ 3′ enhancer is shown below the model. (C and D) The same E47 and IRF-1 (Pip) DNA binding domains were modeled on the Igκ 3′ enhancer binding sites with a single nucleotide inserted between the E47 and Pip sites. Views from the side (C) and bottom (D) of the structure are shown. The positions of the Pip N terminus and the 1-bp insertion between the E47 and Pip sites are indicated by arrows.
Nucleotide insertions between the E47 and Pip sites destroy transcriptional synergy and reduce cooperative DNA binding.
We prepared reporter constructs containing either multimers of the wild-type E47-Pip binding site or multimers with a 1-bp insertion between their binding sites and tested their function by transient-expression assays in NIH 3T3 cells. Similar to previous results (45), the wild-type E47-Pip site reporter showed little activity with either E47 or Pip cDNA alone but yielded a potent (50-fold) transcriptional synergy when the two were transfected together (Fig. 2A lanes 1 to 3). Interestingly, the 1-bp insertion construct nearly completely abolished the E47-Pip transcriptional synergy (lanes 4 to 6). This loss in synergy was not due to an inability of E47 to bind DNA alone, because both constructs were equally activated by a forced dimer E47 construct (head-to-tail copies of E47 connected by a flexible linker sequence) (61) which shows high activity on the Igκ 3′ enhancer E47 sequence (Fig. 2B). In addition, EMSA showed comparable binding of E47 to both the wild-type and 1-bp insertion probes (Fig. 2C, lanes 1 and 3). Full-length Pip does not bind to DNA efficiently, but a truncated Pip protein binds to DNA well (6, 7, 12, 45). We found that a truncated Pip protein (Pip 1-182) bound equally to wild-type and 1-bp insertion probes (Fig. 2C, lanes 2 and 4). Therefore, the 1-bp insertion did not disrupt the E47 or Pip binding sites. Our previous work showed a 10- to 15-fold cooperative binding effect between E47 and Pip on the Igκ 3′ enhancer sequence (45). When E47 and Pip were assayed together, phosphorimager quantification showed an approximate threefold reduction in cooperative DNA binding with the 1-bp insertion sequence probe compared to the wild-type probe (Fig. 2D [see quantitation below each lane]). Therefore, we conclude that the single-base-pair insertion between the E47 and Pip sites does not disrupt either binding site but instead causes a reduction in cooperative DNA binding between E47 and Pip. This loss in cooperative DNA binding is paralleled by a dramatic loss in transcriptional synergy.
To be certain that the loss in synergy was not due to some peculiar feature of the 1-bp insertion construct, we prepared additional insertion constructs containing either 2-, 5-, or 10-bp insertions between the E47 and Pip sites. Like the 1-bp insertion construct, each of these constructs resulted in a dramatic loss of transcriptional synergy between E47 and Pip (Fig. 2E).
The Pip N terminus is required for transcriptional synergy and maximal cooperative DNA binding.
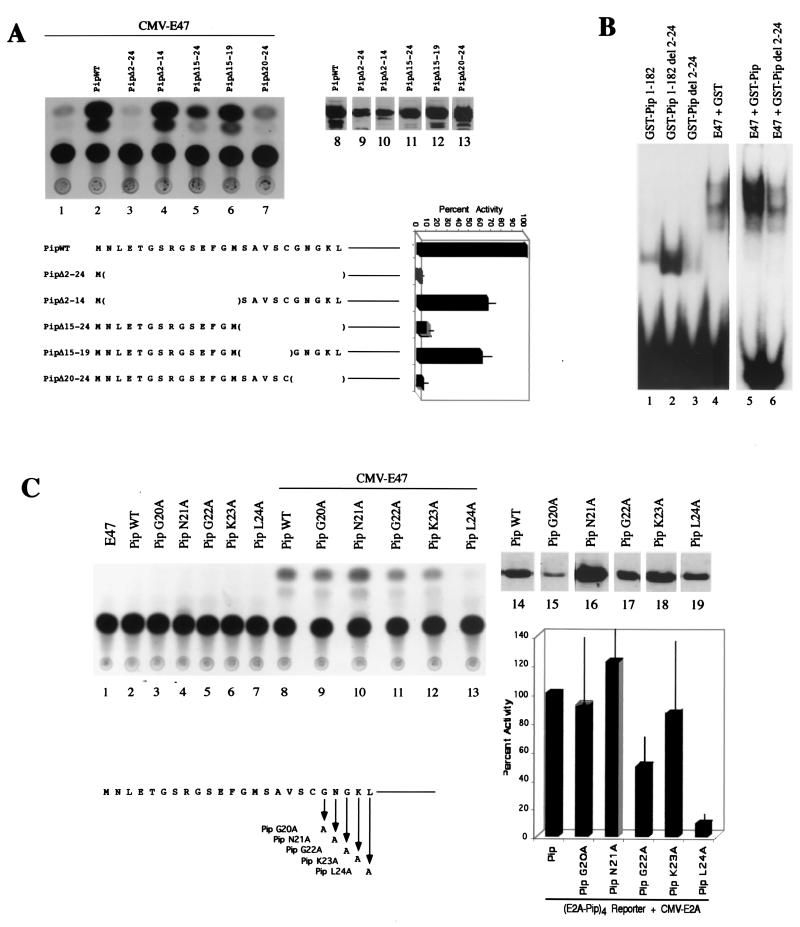
Our E47-Pip (IRF-1)-DNA model shown in Fig. 1 suggested that the Pip N-terminal region might be involved in direct protein-protein interactions with E47. To test this idea, we prepared several constructs with various deletions within the Pip N-terminal 24 amino acids, and tested their ability to activate transcription in association with E47. We found that deletion of Pip residues 2 to 24 resulted in complete loss of transcriptional synergy compared to that with wild-type Pip (Fig. 3A, lanes 2 and 3). Deletion of residues 2 to 14 had a modest effect on synergy, but a great loss was observed upon deletion of residues 15 to 24 (lanes 4 and 5). Deletion of Pip residues 15 to 19 had a small effect on synergy, but synergy was lost upon deletion of residues 20 to 24 (lanes 6 and 7). Western blots of extracts from transfected cells with Pip antisera indicated appropriate expression of each mutant construct (lanes 8 to 13). In summary, our results indicate that Pip residues 20 to 24 are crucial for transcriptional synergy with E47.
FIG. 3.
Pip N-terminal sequences are necessary for E47-Pip synergy. (A) Various Pip N-terminal deletion mutants, shown in the bottom panel, were transfected into NIH 3T3 cells along with an E47 expression plasmid and the wild-type (WT) E47-Pip reporter plasmid. CAT activities shown in the top left panel (lanes 1 to 7) indicate that all constructs that lack Pip sequences between residues 20 and 24 show low transcriptional activity. Lanes 8 to 13 show Western blot data for cell extracts of transfected cells probed with anti-Pip antisera, confirming protein expression. The identity of each transfected Pip mutant is shown above each lane. The bottom right panel shows percent activity of each Pip mutant, with wild-type Pip defined as 100%. Error bars represent standard deviations of the means. (B) Deletion of Pip residues 2 to 24 does not affect Pip DNA binding directly but greatly reduces E47-Pip cooperative DNA binding. EMSA with the wild-type E47-Pip DNA probe and various Pip mutant proteins alone or with E47 is shown. The identity of each protein is shown above the lanes. (C) Pip residue Leu 24 is necessary for E47-Pip transcriptional synergy. NIH 3T3 cells were transfected with the E47-Pip reporter plasmid and various Pip mutants either alone (lanes 2 to 7) or with the E47 expression plasmid (lanes 8 to 13). A representative CAT assay is shown in the top panel, and a histogram summarizing the data is shown in the lower right panel. Error bars represent standard deviations of the means. The Pip N-terminal sequence is shown in the bottom left panel. Lanes 14 to 19 show Western blot data for extracts isolated from transfected cells probed with anti-Pip antisera. The identity of each Pip protein is shown above the lanes.
By comparison to the IRF-1 structure, the Pip winged HTH DNA binding domain resides between residues 26 and 127 (12, 15). This raised the possibility that deletion of sequences adjacent to the DNA binding domain might reduce the ability of Pip to bind to DNA. However, EMSA studies showed that the Pip 1-182Δ2-24 protein bound to DNA even more efficiently than the Pip 1-182 protein (Fig. 3B, lanes 1 and 2). In contrast, cooperative DNA binding in association with E47 was greatly compromised by deletion of Pip residues 2 to 24 (lanes 5 and 6). Therefore, our results strongly indicate that Pip residues 20 to 24 are necessary for Pip transcriptional synergy with E47 and that the Pip N-terminal sequences are also necessary for maximal cooperative DNA binding with E47.
To more precisely identify the Pip residues needed for transcriptional synergy with E47, we prepared individual alanine point mutations between Pip residues 20 and 24. These mutant proteins were assayed by transfection into NIH 3T3 cells. Mutation of Pip Leu 24 resulted in a dramatic loss in transcriptional synergy (Fig. 3C, lane 13), while a twofold reduction was observed when Gly 22 was mutated (lane 11). Mutation of Gly 20, Asp 21, or Lys 23 had minimal impact on transcriptional synergy with E47 (lanes 9, 10, and 12). Thus, the most important residue within this region is Pip Leu 24. Again, Western blots of extracts isolated from transfected cells indicated appropriate expression of each Pip mutant protein (lanes 14 to 19).
Identification of E47 residues important for synergy with Pip.
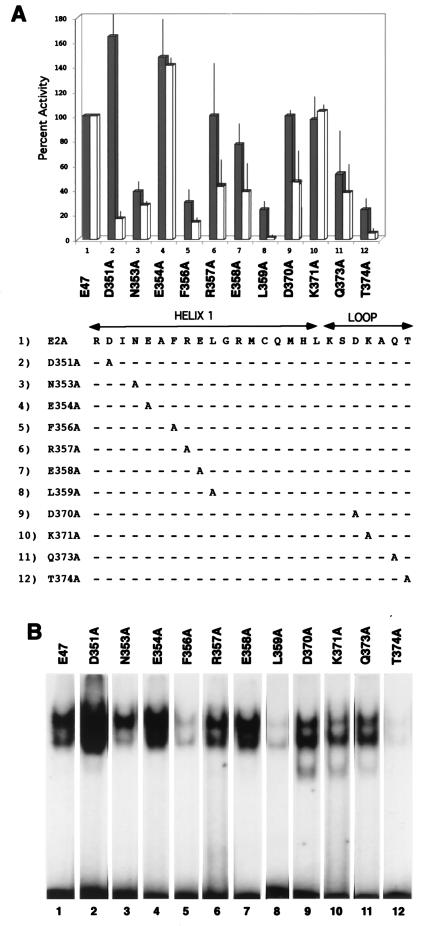
Our E47-Pip-DNA model in Fig. 1 suggested that the Pip N-terminal region might contact sequences within the E47 HLH region. The closest contacts appeared to lie within either E47 helix 1 or the loop sequence. Therefore, we prepared individual alanine point mutations within E47 helix 1 residues 351, 353, 356, 357, 358, and 359 and loop residues 370 to 374 (Fig. 4A). Since these mutations could directly affect E47 homodimerization properties or E47 DNA binding alone, it was important to explore the activity of each E47 mutant in the absence of Pip. We transfected the E47-Pip reporter construct into NIH 3T3 cells with each E47 mutant in the absence and presence of Pip. The activity of each mutant was compared to that of wild-type E47 alone or to that of wild-type E47 plus Pip (Fig. 4A). Our goal was to identify E47 mutants that activated comparably to wild-type E47 alone but substantially lost synergy in the presence of Pip.
FIG. 4.
Identification of E47 residues involved in synergy with Pip. (A) Individual E47 constructs were transfected into NIH 3T3 cells with the E47-Pip-responsive reporter either alone (dark bars) or with the Pip expression plasmid (white bars). CAT activity is normalized to either wild-type E47 alone (dark bars) (defined as 100%) or wild-type E47 plus Pip (white bars) (defined as 100%). The bottom panel shows the E47 helix 1 and loop sequences and the specific amino acids mutated in each construct. Dashes in the bottom panel indicate amino acid identity. (B) Each E47 mutant was prepared in bacteria as a GST fusion protein and used in EMSA with the kappa E47-Pip site as a probe. Identities of proteins added are shown above the lanes.
Several mutants with mutations within the helix 1 region lost synergy with Pip. These mutants include D351A, N353A, F356A, R357A, E358A, and L359A. Many of these mutants lost E47 activity alone, indicating an effect on either E47 dimerization or DNA binding. For instance, Phe 356 and Leu 359 mutations are known to disrupt dimerization and DNA binding (65). Leu 359 faces the hydrophobic interior of the helix, and Phe 356 stacks across the dimer interface and interacts with Lys 375, which along with Asn 353 contacts the phosphodiester backbone of DNA (14). As expected, EMSA studies showed greatly reduced DNA binding with E47 mutants F356A, L359A, and T374A (Fig. 4B, lanes 5, 8, and 12) and moderate reductions with mutant N353A (lane 3). Therefore, the loss of activity of these mutants is unlikely to result from disruptions in Pip interaction. Several of the helix 1 mutants, however, did not affect E47 activity alone, indicating a potentially specific effect on E47-Pip synergy. These mutants are D351A, R357A, and E358A. These residues lie within stripes of either acidic (Asp 351 and Glu 358) or basic (Arg 357) residues on the surface of helix 1 (14). EMSA studies showed that these mutants bound to DNA efficiently (Fig. 4B, lanes 2, 6, and 7), In fact, D351A bound to DNA even better than wild-type E47, consistent with its stronger transcriptional activity (Fig. 4A, lane 2).
Three of the E47 loop mutants (D370A, Q373A, and T374A) showed reduced synergy with Pip. Gln 373 participates in hydrogen bonds with Gly 360, Gln 364, and Gln 381 (14) to stabilize the loop and orientation of helices 1 and 2. Therefore, loss of activity with the Q373A mutation may not relate to E47-Pip synergy. This mutant bound to DNA with slightly reduced efficiency (Fig. 4B, lane 11). Thr 374 hydrogen bonds with Leu 377 and showed a greatly reduced ability to bind to DNA (lane 12). The most conclusive results were obtained with D370A, because this mutation did not affect E47 activity alone (Fig. 4A) and bound to DNA efficiently (Fig. 4B, lane 9).
In summary, our DNA binding and transfection data indicate that E47 residues Asp 351, Arg 357, Glu 358, and Asp 370 are involved in synergy with Pip.
Refinement of the E47-Pip-DNA model.
Our initial protein-DNA models were prepared using linear DNA molecules without calculating bends introduced by the transcription factors. To better visualize the binding to E47 and Pip (IRF-1) on DNA, we prepared models which included bends in the DNA predicted from the published crystal structures (14, 15). Although difficult to build due to the small overlap between the DNA sequences in the two crystal structures relative to that of the Igκ 3′ enhancer, combined with possible end effects, this model suggests that the enhancer may become slightly bowed around the two bound proteins (Fig. 5A and B). This would result in the Pip N-terminal sequences coming in even closer proximity with E47 helix 1 and loop sequences than suggested with idealized DNA. Interestingly, the position of Pip Leu 24 (IRF-1 Met 8) in this model is very near E47 residue Arg 357 and possibly Glu 358 (Fig. 5C). In addition, E47 Asp 370 lies near sequences at the C terminus of helix α2 and the following loop L2 in the Pip DNA binding domain (15). E47 Asp 351, which loses synergy with Pip when mutated, does not directly interact with IRF-1 sequences in our model. However, Pip extends an additional 22 amino acids amino terminal to the comparable IRF-1 residue (Met 8), making it possible that these sequences contact E47 Asp 351. In general, there is a good correspondence between the proximity of E47 and Pip sequences in our models and their impact on transcriptional synergy when mutated. Therefore, we believe that our models are good approximations of the structures of E47 and Pip bound to adjacent sites within the Igκ 3′ enhancer.
FIG. 5.
Refinement of the E47-Pip (IRF-1)-DNA model. (A and B) The possible effects of DNA bending were modeled by docking the structures of E47 and IRF-1 complexes such that their respective DNA structures were aligned to correspond with that of the Igκ 3′ enhancer. Overlap of the DNA helices can be seen between the two proteins. Shown are side (A) and bottom (B) views of the models. (C) Locations of E47 and Pip residues that disrupt synergy when mutated. The model is viewed from the opposite side as in panels A and B and Fig. 1 to best visualize E47 and Pip residues involved in E47-Pip transcriptional synergy. E47 mutations that affect synergy with Pip are labeled and shown in blue. The position of Pip Leu 24 (IRF-1 Met 8), which is necessary for synergy with E47, is labeled and shown in red.
Function within the B-cell lineage.
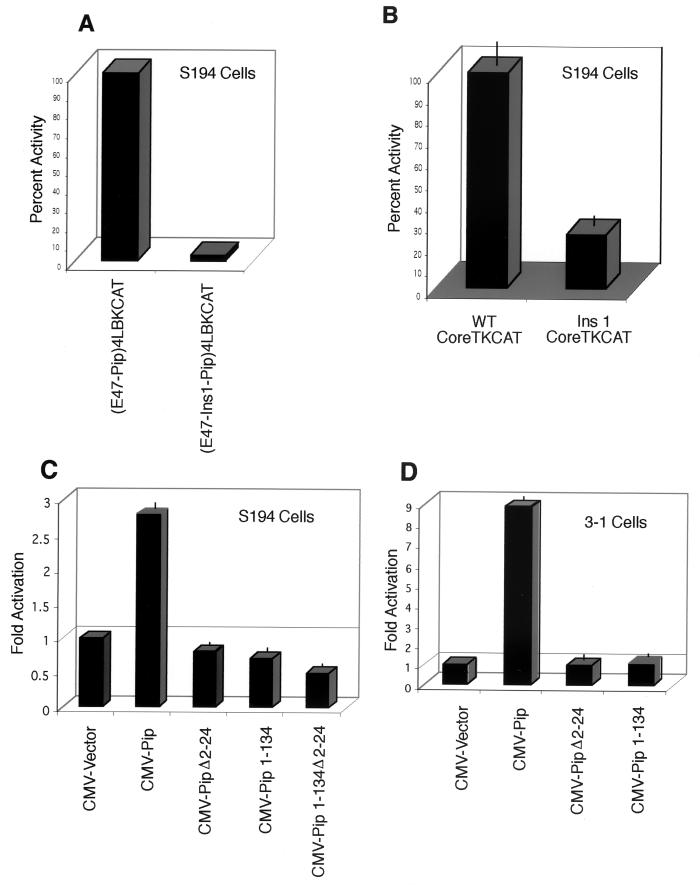
All of our transfection studies described above were performed with NIH 3T3 cells. To test whether similar activities are present within the B-cell lineage, we performed experiments with cells representative of either the plasma cell stage (S194 cells) or the pre-B-cell stage (3-1 cells). We previously observed (45, 50) that a reporter containing multimerized E47-Pip binding sites shows high transcriptional activity in S194 plasmacytoma cells. These cells express E47 and Pip proteins. As expected, transfection of the (E47-Pip)4LBKCAT reporter construct into S194 plasmacytoma cells resulted in high levels of transcriptional activity (Fig. 6A). However, insertion of a single base pair between the E47 and Pip binding sites [construct (E47-Ins1-Pip)4LBKCAT] abolished transcriptional activity (Fig. 6A), similar to our results with NIH 3T3 cells (Fig. 2). Previously we showed that mutation of either the E47 or the Pip site in the context of the intact enhancer core resulted in a 75% reduction in enhancer activity in plasmacytoma cells (45). We tested the effect of the single nucleotide insertion between the E47 and Pip sites in the context of the enhancer core and found a similar 75% decrease in enhancer function (Fig. 6B). Therefore, E47-Pip synergy plays an important role in overall enhancer function. Since S194 cells apparently support E47-Pip synergy, one would predict that Pip proteins that cannot synergize with E47 would function as dominant negative mutants in these cells. Indeed, while transfection of wild-type Pip led to a nearly threefold further increase in transcription, Pip mutants lacking residues 2 to 24, or mutants containing only the Pip DNA binding domain (and thus lacking the activation domain), failed to activate transcription and instead led to a reduction in transcriptional activity (Fig. 6C). 3-1 pre-B cells contain low levels of Pip protein and do not support high levels of E47-Pip synergy (50). Transfection of wild-type Pip into these cells resulted in a ninefold transcriptional activation through the E47-Pip binding sites (Fig. 6D). On the other hand, a Pip mutant lacking the N-terminal 24 amino acids and a mutant containing only the Pip DNA binding domain both failed to activate transcription in 3-1 cells (Fig. 6D).
FIG. 6.
E47-Pip transcriptional synergy in B-cell lines. (A) A single-base-pair insertion between the E47 and Pip sites abolishes endogenous E47-Pip synergy in plasma cells. S194 cells were transfected with either the wild-type E47-Pip-dependent reporter plasmid or the reporter plasmid with a 1-bp insertion between the sites. Activity with the wild-type reporter is defined as 100%. (B) E47-Pip synergy is important for 3′ enhancer activity. Plasmids containing either the entire 3′ enhancer core (WTCoreTKCAT), or with a 1-bp insertion between the E47 and Pip sites (Ins1CoreTKCAT) were transfected into S194 cells. Activity with CoreTKCAT is defined as 100%. (C) Pip mutants that cannot synergize with E47 act as dominant negative mutants in plasmacytoma cells. S194 cells were transfected with the wild-type E47-Pip dependent reporter plasmid and either wild-type Pip expression plasmid or mutant Pip expression plasmids. The identity of each Pip expression plasmid is shown below the lanes. (D) Pip mutants that cannot synergize with E47 fail to activate transcription in pre-B cells. 3-1 pre-B cells were transfected with the wild-type E47-Pip-dependent reporter plasmid and various Pip expression constructs. The identity of each Pip expression plasmid is shown below the lanes.
Pip can bind to the Ig heavy-chain intron enhancer.
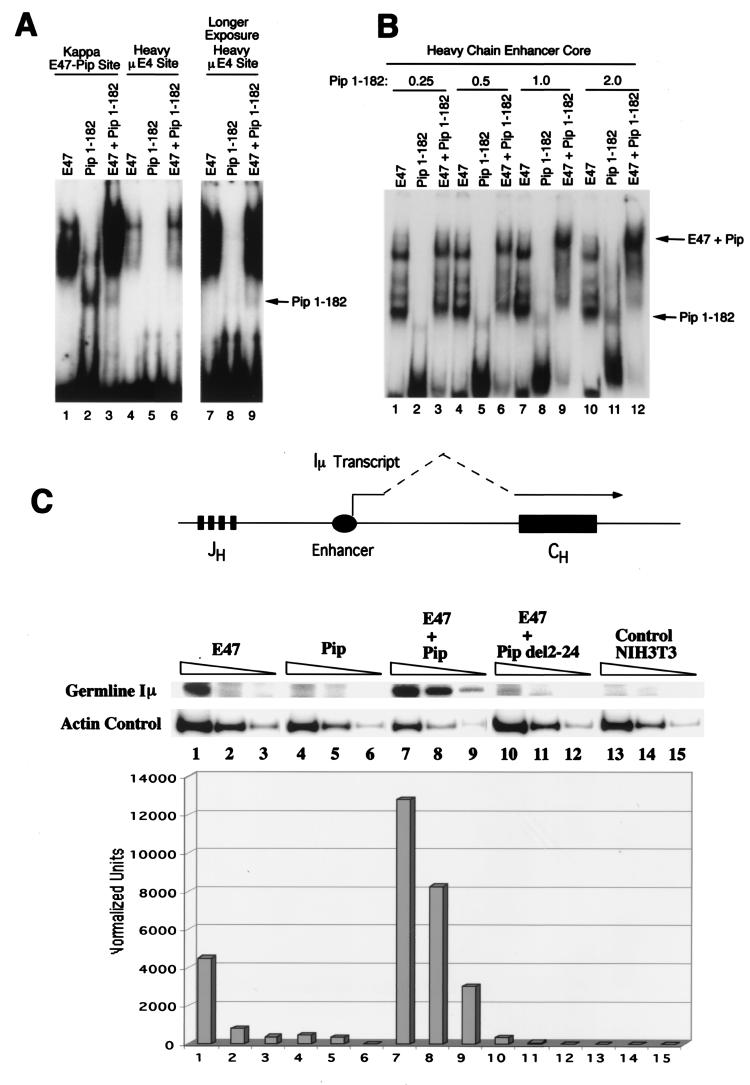
Pip has not previously been shown to bind to the Ig heavy-chain enhancer. However, preliminary chromatin immunoprecipitation studies provided evidence for Pip binding sites within the heavy-chain enhancer region (data not shown). Interestingly, the heavy-chain enhancer μE4 site shows similarity to the E47-Pip site from the kappa 3′ enhancer. Therefore, we performed EMSA with the μE4 heavy-chain site as probe, using the kappa E47-Pip site as a positive control (Fig. 7A, lanes 1 to 6). E47 efficiently bound to the μE4 probe, but Pip 1-182 bound weakly (lanes 4 to 9). Coincubation of both proteins resulted in a modest increase in DNA binding on the μE4 probe (lanes 4 and 6). This sequence may therefore represent a weak E47-Pip motif. We searched for the existence of other potential Pip sites by using a 220-bp HinfI DNA fragment that encompasses the entire heavy-chain enhancer core region. This probe resulted in efficient binding by both E47 and Pip (Fig. 7B, lanes 1 and 2). Interestingly, increasing amounts of Pip protein in the presence of E47 resulted in progressive decreases in complex mobility, suggesting the presence of multiple Pip sites within the heavy-chain enhancer fragment (Fig. 4B, lanes 3, 6, 9, and 12). The potential significance of these Pip sites within the heavy-chain enhancer was explored in experiments described below.
FIG. 7.
E47-Pip synergy at the Ig heavy-chain locus. (A) The heavy-chain μE4 site is a weak E47-Pip motif. EMSA was performed with either the kappa E47-Pip DNA probe or the μE4 site probe. Proteins added are indicated above the lanes. The arrow points to a weak DNA-Pip complex with the μE4 probe. (B) Pip binds to the heavy-chain intron enhancer region. EMSA was performed with a 220-bp HinfI DNA fragment containing the entire heavy-chain intron enhancer core. Proteins added are indicated above the lanes. The mass in micrograms of Pip 1-182 added is shown at the top. Arrows point to either the Pip 1-182-DNA complex or the E47-Pip-DNA complex. (C) E47-Pip synergy stimulates endogenous sterile Iμ transcripts. NIH 3T3 cells were transfected with plasmids expressing E47, Pip, E47 plus Pip, or E47 plus PipΔ2-24. RNA was isolated from transfected cells and subjected to RT-PCR with primers specific for sterile Iμ or actin transcripts. The top panel shows a map of the heavy-chain locus and diagrams Iμ transcripts. The middle panel shows results of RT-PCR assays using fourfold-increasing amounts of cDNA with either Iμ-specific or actin-specific primers. The data in the middle panel were quantitated by phosphorimager analysis, normalized for transfection efficiency by β-galactosidase activity, and then normalized to the actin control. The histogram in the bottom panel shows the normalized level of Iμ transcripts in each lane. The identities of the constructs used in each transfection are shown above the lanes.
Pip can stimulate E47-dependent induction of Iμ sterile transcripts at the heavy-chain locus.
Previously it was shown that E47 transfection into either pre-T cells or fibroblasts induces expression of germ line Ig heavy-chain transcripts from the Iμ promoter adjacent to the heavy-chain intron enhancer (Fig. 7C) (8, 58). In light of the above EMSA data showing Pip binding to the Ig heavy-chain enhancer, we sought to determine whether E47-Pip synergy might augment expression of E47-induced Iμ transcripts. NIH 3T3 cells were transfected with plasmids expressing either E47, Pip, E47 plus Pip, or E47 plus PipΔ2-24 (which cannot synergize with E47 [Fig. 3A]). RNA was isolated from transfected cells and subjected to RT-PCR with primers specific for heavy-chain sterile Iμ transcripts or with actin as a control. As previously reported (8, 58), E47 induced expression of endogenous Iμ transcripts (Fig. 7C, compare lanes 1 to 3 with lanes 13 to 15). Transfection of Pip alone did not induce these transcripts (lanes 4 to 6). However, the combination of E47 plus Pip resulted in a dramatic 5- to 10-fold induction of endogenous Iμ transcripts (lanes 7 to 9). If this induction is due to E47-Pip synergy rather than each protein individually stimulating expression, the PipΔ2-24 mutant should function as a dominant negative mutant. Importantly, PipΔ2-24 cotransfected with E47 resulted in reduced levels of Iμ transcripts compared to E47 alone (Fig. 7C, lanes 10 to 12). The lack of induction by Pip alone and the dominant negative effect of PipΔ2-24 indicate that the increased Iμ expression is due to E47-Pip synergy rather than to each protein functioning independently. We conclude that E47-Pip synergy can stimulate expression of endogenous germ line Iμ transcripts involved in somatic rearrangement and isotype class switch recombination of Ig genes.
DISCUSSION
Mechanism of E47-Pip interactions.
Our results define a novel mechanism of interaction between HLH and IRF domain proteins. E47 residues important for E47-Pip synergy include Asp 351, Arg 357, Glu 358, and Asp 370. Three of these residues (Asp 351, Arg 357, and Glu 358) lie on the outer surface of E47 helix 1 (14). These residues apparently make important direct contacts with Pip N-terminal sequences. Indeed, our molecular model in Fig. 5 suggests close contacts between Pip residue Leu 24 and E47 residue Arg 357 (and possibly Glu 358). Pip Leu 24 and E47 Arg 357 may contact one another through hydrophobic interactions between their long aliphatic side chains. Although E47 Asp 351 is more distant from Pip in the model in Fig. 5, this aspartate may contact residues in the N terminus of Pip that are not present in the corresponding IRF-1 structure. Alternatively, substitution of Asp 351 may indirectly perturb interactions of E47 and Pip, perhaps by perturbing the HLH structure. Interestingly, this substitution also led to an enhancement of DNA binding by E47 (Fig. 4B).
Our data also indicate a role of E47 Asp 370 in synergy with Pip. This E47 loop residue is predicted by our model (Fig. 5) to make contacts within the Pip DNA binding domain in the vicinity of Pip residues 76 to 82. These residues (LFKGKFG) form a positively charged cluster at the C terminus of helix α2 and within loop L2 of Pip, with the latter being followed by the recognition helix α3 (15). Further mutagenesis studies will be required to establish the mechanism of E47 Asp 370 interaction with regions of Pip beyond its N terminus.
The 10- to 15-fold increase in E47 DNA binding in the presence of Pip (45) could be the primary mechanism responsible for E47-Pip transcriptional synergy. However, it is clear that synergy requires both the E47 and Pip activation domains (45), suggesting that a simple increase in DNA binding by E47 and Pip is not sufficient for synergy. Other functions mediated by the E47 and Pip activation domains are required. We cannot exclude the possibility that contacts between E47 and Pip on DNA generate a novel interaction surface for a coactivator protein responsible for potent transactivation function. Such interactions could recruit coactivators to the complex or could stabilize interactions with the SAGA complex, which is known to interact with E47 (38).
Our results also indicate that the Pip N-terminal 24 amino acids inhibit Pip DNA binding when Pip is assayed alone (Fig. 3B). Deletion of these residues enabled Pip to bind to DNA better than unmutated Pip. Similar results were observed by Yee et al. (69), who showed that deletion of Pip residues 1 to 19 increased DNA binding two- to fivefold. In contrast, while deletion of Pip residues 2 to 24 increased Pip binding alone, this deletion greatly reduced cooperative DNA binding in association with E47. Interestingly, nuclear magnetic resonance measurements by Yee and coworkers (69) indicated that residues 1 to 19 of Pip may be unstructured in the isolated protein. Similarly, the N-terminal six residues in the IRF-1 crystal structure (comparable to Pip residues 1 to 22) used in our computer models appear to be flexible as evident by a lack of observable electron density (15). A possible explanation for these Pip DNA binding properties is that the unstructured N-terminal tail plays an inhibitory role in protein-DNA interaction, perhaps through electrostatic effects (69). However, in association with E47, at least some of these sequences may take on a more ordered structure and increase Pip-E47-DNA interactions. This is consistent with our observation that even a single-base-pair insertion between the E47 and Pip sites in the Igκ 3′ enhancer eliminates synergy. Thus, a precisely defined interface appears to be required for the cooperative interaction between the two proteins.
Interaction with other proteins.
E2A and Pip are both known to interact with a variety of proteins. E2A proteins can be positively or negatively regulated by association with Id, Erg-3, Ets-1, Fli-1, Lmx1.1, Lmx1.2, Pitx proteins, and Pip (4, 31, 45, 46, 53, 55). Similarly, Pip can be positively regulated by interaction with PU.1 (12, 51, 52) or negatively regulated by interaction with BCL6 or Blimp (26, 27).
Pip interaction with PU.1 results in cooperative DNA binding and transcriptional synergy (12, 13, 51, 52) similar to those observed here with E47. However, the transcriptional synergy between E47 and Pip is much stronger (50- to 100-fold) than the PU.1-Pip synergy (5-fold) (52), and different segments of Pip are important. Cooperative DNA binding between PU.1 and Pip requires Pip C-terminal sequences, particularly residues 398 and 399 (6, 7), while we find here that Pip N-terminal sequences are needed for maximal binding with E47. The distinct mechanisms of PU.1-Pip and E47-Pip DNA binding probably relate to the opposite orientations of the Pip site relative to the PU.1 and E47 sites, respectively (45, 50, 51). In addition to the N-terminal and C-terminal regions of Pip involved in PU.1 and E47 interaction, Pip also interacts with other IRF proteins via residues 245 to 412, defined as the IAD region (41, 64). Thus, Pip functions can be modified by interaction with a variety of distinct proteins.
The E47-Pip interaction that we observe here defines a novel mechanism that may be utilized by other members of the HLH and IRF families. Helix 1 is highly conserved between E47, E12, HEB, and daughterless (14). All of these proteins contain an aspartate residue at the position comparable to E47 Asp 351. Similarly, all contain a basic residue at the position comparable to E47 Arg 357 and acidic residues at positions comparable to E47 Glu 358 and Asp 370. Similarly, Pip Leu 24 is conserved at the comparable position in IRF family members ICSBP and ISGF3γ (12). These homologies suggest that other HLH-IRF family member synergies might exist. Indeed, we previously showed that ICSBP can synergize with E47 in transcription (45). This synergy could potentially take place in macrophages and lymphoid cell types, where both ICSBP and E2A proteins are coexpressed (36). IRF-1, which contains a methionine at the position of Pip Leu 24, does not synergize with E47 (unpublished data). It will be very interesting to determine whether ISGF3γ can synergize with E47, because like E2A, ISGF3γ is expressed in most cell types.
E47-Pip synergy and B-cell functions.
Both E47 and Pip levels increase at late stages of B-cell development or after activation of primary B cells (12, 19, 25, 27, 40, 54). We found that E47 and Pip can both bind to the Igκ 3′ and the heavy-chain intron enhancers. What is the functional significance of the E47-Pip synergy that we observed at the kappa and heavy-chain loci? E47 and Pip are clearly important for enhancer activity and transcription of Ig genes. However, increased expression upon B-cell activation may be important for other crucial B-cell functions. E47 function on somatic rearrangement of heavy-chain and light-chain genes is well documented (1, 3, 20, 56). E47 can induce sterile Iμ transcripts at the heavy-chain locus as well as some germ line V gene transcripts (8, 20, 58). The fact that Pip can greatly augment the ability of E47 to induce endogenous sterile Iμ transcripts (Fig. 7C) suggests that E47-Pip synergy might contribute to Ig gene somatic rearrangement. However, Pip knockouts do not inhibit Ig rearrangements (42), indicating that E47-Pip synergy is not essential for this function. Instead, a later B-cell function may require E47-Pip synergy.
Late in B-cell development, E47 is needed for the heavy-chain class switch recombination necessary for conversion of IgM molecules formed in the primary immune response into effector classes needed for secondary immune responses (IgG, IgA, and IgM) (21, 54). E2A proteins are highly expressed in the dark zone of germinal centers where Ig class switch recombination occurs (21, 57). It is intriguing that activation of B cells through anti-CD40 and interleukin-4 treatment stimulates both E47 and Pip protein levels and that these treatments also stimulate class switch recombination in primary B cells (25, 27, 54). The parallels between increased E47 and Pip protein levels and increased switch recombination suggest that E47-Pip synergy may be involved in class switch recombination. Our observed augmentation of sterile Iμ transcript expression by E47-Pip synergy (Fig. 7C) is consistent with this idea. Iμ transcripts might thus be regulated by E47-Pip synergy, while sterile germ line transcripts at other constant regions might be controlled by other factors. For instance, sterile transcripts at other constant regions appear to be controlled by factors such as C/EBP, BCL-6, STAT6, BSAP, NF-κB, and PU.1 (10, 11, 17, 30, 34, 47, 62, 66). Mutation of these factors might affect a subset of germ line transcripts and limit switch recombination to a subset of constant regions. Since Iμ transcripts are required for all initial switch recombinations, mutations in either E47 or Pip would be expected to have a more universal effect. Such a scenario could be tested by determining whether dominant negative Pip mutants that cannot synergize with E47 inhibit heavy-chain class switch recombination. This result would be consistent with the inability of Pip knockout mutant animals to mount secondary immune responses (42). The availability of Pip dominant negative mutant Δ2-24, which cannot synergize with E47, will enable us in the future to test the in vivo functional consequences of E47-Pip synergy.
Acknowledgments
We thank T. Kadesch for information on E47-Pip synergy and Iμ transcripts and R. Grosschedl for the forced dimer E47 construct. We thank T. Ellenberger for providing coordinates of the E47-DNA crystal complex. We also thank R. Perry, N. G. Avadhani, and J. Pehrson for critically reading the manuscript.
This work was supported by NIH grant GM42415 to M.L.A. and a CIMR Scientist Award to L.P.M.
REFERENCES
- 1.Bain, G., E. C. R. Maandag, D. J. Izon, D. Amsen, A. M. Kruisbeek, B. C. Weintraub, I. Krop, M. S. Schlissel, A. J. Feeney, M. van Roon, M. van der Valk, H. P. J. te Riele, A. Berns, and C. Murre. 1994. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell 79:885-892. [DOI] [PubMed] [Google Scholar]
- 2.Bain, G., and C. Murre. 1998. The role of E-proteins in B- and T-lymphocyte development. Semin. Immunol. 10:143-153. [DOI] [PubMed] [Google Scholar]
- 3.Bain, G., W. J. Romanow, K. Albers, W. L. Havran, and C. Murre. 1999. Positive and negative regulation of V(D)J recombination by the E2A proteins. J. Exp. Med. 189:289-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benezra, R., R. L. Davis, D. Lockshon, D. L. Turner, and H. Weintraub. 1990. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell 61:49-59. [DOI] [PubMed] [Google Scholar]
- 5.Betz, A. G., C. Milstein, A. Gonzalel-Fernandez, R. Pannell, T. Larson, and M. S. Neuberger. 1994. Elements regulating somatic hypermutation of an Igκ gene: critical role of the intron enhancer/matrix attachment region. Cell 77:239-248. [DOI] [PubMed] [Google Scholar]
- 6.Brass, A. L., E. Kehrli, C. F. Eisenbeis, U. Storb, and H. Singh. 1996. Pip, a lymphoid-restricted IRF, contains a regulatory domain that is important for autoinhibition and ternary complex formation with the Ets factor PU.1. Genes Dev. 10:2335-2347. [DOI] [PubMed] [Google Scholar]
- 7.Brass, A. L., A. Q. Zhu, and H. Singh. 1999. Assembly requirements of PU.1-Pip (IRF-4) activator complexes: inhibiting function in vivo using fused dimers. EMBO J. 18:977-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi, J., C.-P. Shen, H. Radomska, L. Eckhardt, and T. Kadesch. 1996. E47 activates the Ig-heavy chain and TdT loci in non-B cells. EMBO J. 15:5014-5021. [PMC free article] [PubMed] [Google Scholar]
- 9.Cline, T. W. 1989. The affairs of daughterless and the promiscuity of developmental regulators. Cell 59:231-234. [DOI] [PubMed] [Google Scholar]
- 10.Cunningham, K., H. Ackerly, F. Alt, and W. Dunnick. 1998. Potential regulatory elements for germline transcription in or near murine Sγ1. Int. Immunol. 10:527-536. [DOI] [PubMed] [Google Scholar]
- 11.Delphin, S., and J. Stavnezer. 1995. Characterization of an interleukin 4 (IL-4) responsive region in the immunoglobulin heavy chain germline ɛ promoter: regulation by NF-IL-4, a C/EBP family member and NF-κB/p50. J. Exp. Med. 181:181-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisenbeis, C. F., H. Singh, and U. Storb. 1995. Pip, a novel IRF family member, is a lymphoid-specific PU.1-dependent transcriptional activator. Genes Dev. 9:1377-1387. [DOI] [PubMed] [Google Scholar]
- 13.Eisenbeis, C. F., H. Singh, and U. Storb. 1993. PU.1 is a component of a multiprotein complex which binds an essential site in the murine immunoglobulin λ2-4 enhancer. Mol. Cell. Biol. 13:6452-6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellenberger, T., D. Fass, M. Arnaud, and S. C. Harrison. 1994. Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer. Genes Dev. 8:970-980. [DOI] [PubMed] [Google Scholar]
- 15.Escalante, C. R., J. Yie, D. Thanos, and A. K. Aggarwal. 1998. Structure of IFR-1 with bound DNA reveals determinants of interferon regulation. Nature 391:103-106. [DOI] [PubMed] [Google Scholar]
- 16.Evans, V. 1993. SETOR: hardware lighted three-dimensional solid model representations of macromolecules. J. Mol. Graphics 11:134-138. [DOI] [PubMed] [Google Scholar]
- 17.Ford, G. S., C. H. Yin, B. Barnhart, K. Sztam, and L. R. Covey. 1998. CD40 ligand exerts differential effects on the expression of Iγ transcripts in subclones of an IgM+ human B cell lymphoma line. J. Immunol. 160:595-605. [PubMed] [Google Scholar]
- 18.Glimcher, L. H., and H. Singh. 1999. Transcription factors in lymphocyte development—T and B cells get together. Cell 96:13-23. [DOI] [PubMed] [Google Scholar]
- 19.Glynne, R., S. Akkaraju, J. I. Healy, J. Rayner, C. C. Goodnow, and D. H. Mack. 2000. How self-tolerance and the immunosuppresive drug FK506 prevent B-cell mitogenesis. Nature 403:672-676. [DOI] [PubMed] [Google Scholar]
- 20.Goebel, P., N. Janney, J. R. Valenzuela, W. J. R. Romanow, C. Murre, and A. J. Feeney. 2001. Localized gene-specific induction of accessibility to V(D)J recombination induced by E2A and early B cell factor in nonlymphoid cells. J. Exp. Med. 194:645-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldfarb, A. N., J. P. Flores, and K. Lewandowska. 1996. Involvement of the E2A basic helix-loop-helix protein in immunoglobulin heavy chain class switching. Mol. Immunol. 33:947-956. [DOI] [PubMed] [Google Scholar]
- 22.Gorman, C. M., L. F. Moffat, and B. H. Howard. 1982. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol. Cell. Biol. 2:1044-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorman, J. R., N. van der Stoep, R. Monroe, M. Cogne, L. Davidson, and F. W. Alt. 1996. The Igκ 3′ enhancer influences the ratio of Igκ versus Igλ B lymphocytes. Immunity 5:241-252. [DOI] [PubMed] [Google Scholar]
- 24.Graham, F. L., and A. J. Van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 25.Grumont, R. J., and S. Gerondakis. 2000. Rel induces interferon regulatory factor 4 (IRF-4) expression in lymphocytes: modulation of interferon-regulated gene expression by rel/nuclear factor kappa B. J. Exp. Med. 191:1281-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta, S., A. Anthony, and A. B. Pernis. 2001. Stage-specific modulation of IFN-regulatory factor 4 function by Kruppel-type zinc finger proteins. J. Immunol. 166:6104-6111. [DOI] [PubMed] [Google Scholar]
- 27.Gupta, S., M. Jiang, A. Anthony, and A. Pernis. 1999. Lineage-specific modulation of interleukin 4 signaling by interferon regulatory factor 4. J. Exp. Med. 190:1837-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hardy, R. R., C. E. Carmack, S. A. Shinton, J. D. Kemp, and K. Hayakawa. 1991. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J. Exp. Med. 173:1213-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 30.Iciek, L. A., S. A. Delphin, and J. Stavnezer. 1997. CD40 cross-linking induces Igɛ germline transcripts in B cells via activation of NF-κB. J. Immunol. 158:4769-4779. [PubMed] [Google Scholar]
- 31.Johnson, J. D., W. Zhang, A. Rudnick, W. J. Rutter, and M. S. German. 1997. Transcriptional synergy between LIM-homeodomain proteins and basic helix-loop-helix proteins: the LIMZ domain determines specificity. Mol. Cell. Biol. 17:3488-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaelin, W. G. J., D. C. Pallas, J. A. DeCaprio, F. J. Kaye, and D. M. Livingston. 1991. Identification of cellular proteins that can interact specifically with the T/E1A-binding region of the retinoblastoma gene product. Cell 64:521-532. [DOI] [PubMed] [Google Scholar]
- 33.Kee, B. L., and C. Murre. 2001. Transcription factor regulation of B lineage commitment. Curr. Opin. Immunol. 13:180-185. [DOI] [PubMed] [Google Scholar]
- 34.Lin, S., and J. Stavnezer. 1996. Activation of NF-κB/Rel by CD40 engagement induces the mouse germline immunoglobulin Cγ1 promoter. Mol. Cell. Biol. 16:4591-4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maizels, N. 1995. Somatic hypermutation: how many mechanisms diversify V region sequences? Cell 83:9-12. [DOI] [PubMed] [Google Scholar]
- 36.Mamane, Y., C. Heylbroeck, P. Genin, M. Algarte, M. J. Servant, C. LePage, C. DeLuca, H. Kwon, R. Lin, and J. Hiscott. 1999. Interferon regulatory factors: the next generation. Gene 237:1-14. [DOI] [PubMed] [Google Scholar]
- 37.Manis, J. P., N. van der Stoep, M. Tian, R. Ferrini, L. Davidson, A. Bottaro, and F. W. Alt. 1998. Class switching in B cells lacking the 3′ immunoglobulin heavy chain enhancers. J. Exp. Med. 188:1421-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Massari, M. E., P. A. Grant, M. G. Pray-Grant, S. L. Berger, J. L. Workman, and C. Murre. 1999. A conserved motif present in a class of helix-loop-helix proteins activates transcription by direct recruitment of the SAGA complex. Mol. Cell 4:63-73. [DOI] [PubMed] [Google Scholar]
- 39.Massari, M. E., and C. Murre. 2000. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 20:429-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsuyama, T., A. Grossman, H.-W. Mittrucker, D. P. Siderovski, F. Kiefer, T. Kawakami, C. D. Richardson, T. Taniguchi, S. K. Yoshinaga, and T. W. Mak. 1995. Molecular cloning of LSIRF, a lymphoid-specific member of the interferon regulatory factor family that binds the interferon-stimulated response element (ISRE). Nucleic Acids Res. 23:2127-2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Merero, D., S. Hashmueli, B. Koren, A. Azriel, A. Oumard, S. Kirchhoff, H. Hauser, S. Nagulapalli, M. L. Atchison, and B.-Z. Levi. 1999. Protein-protein and DNA-protein interactions affect the activity of lymphoid specific IFN regulatory factors. J. Immunol. 163:6468-6478. [PubMed] [Google Scholar]
- 42.Mittrucker, H.-W., T. Matsuyama, A. Grossman, T. M. Kundig, J. Potter, A. Shahinian, A. Wakeham, B. Patterson, P. S. Ohashi, and T. W. Mak. 1997. Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science 275:540-543. [DOI] [PubMed] [Google Scholar]
- 43.Mostoslavsky, R., N. Singh, A. Kirillov, R. Pelanda, H. Cedar, A. Chess, and Y. Bergman. 1998. κ chain monoallelic demethylation and the establishment of allelic exclusion. Genes Dev. 12:1801-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murre, C., G. Bain, M. A. V. Dijk, I. Engel, B. A. Furnari, M. E. Massari, J. R. Matthews, M. W. Quong, R. R. Rivera, and M. H. Stuiver. 1994. Structure and function of helix-loop-helix proteins. Biochim. Biophys. Acta 1218:129-135. [DOI] [PubMed] [Google Scholar]
- 45.Nagulapalli, S., and M. L. Atchison. 1998. Transcription factor Pip can enhance DNA binding by E47, leading to transcriptional synergy involving multiple protein domains. Mol. Cell. Biol. 18:4639-4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nelson, B., G. Tian, B. Erman, J. Gregoire, R. Maki, B. Graves, and R. Sen. 1993. Regulation of lymphoid-specific immunoglobulin mu heavy chain enhancer by ETS-domain proteins. Science 261:82-86. [DOI] [PubMed] [Google Scholar]
- 47.Oettgen, H. C. 2000. Regulation of the IgE isotype switch: new insights on cytokine signals and the functions of ɛ germline transcripts. Curr. Opin. Immunol. 12:618-623. [DOI] [PubMed] [Google Scholar]
- 48.Pan, Q., C. Petit-Frere, J. Stavnezer, and L. Hammarstrom. 2000. Regulation of the promoter for human immunoglobulin γ3 germ-line transcription and its interaction with the 3′α enhancer. Eur. J. Immmunol. 30:1019-1029. [DOI] [PubMed] [Google Scholar]
- 49.Pongubala, J. M. R., and M. L. Atchison. 1995. Activating transcription factor 1 and cyclic AMP response element modulator can modulate the activity of the immunoglobulin κ 3′ enhancer. J. Biol. Chem. 270:10304-10313. [DOI] [PubMed] [Google Scholar]
- 50.Pongubala, J. M. R., and M. L. Atchison. 1991. Functional characterization of the developmentally controlled immunoglobulin kappa 3′ enhancer: regulation by Id, a repressor of helix-loop-helix transcription factors. Mol. Cell. Biol. 11:1040-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pongubala, J. M. R., S. Nagulapalli, M. J. Klemsz, S. R. McKercher, R. A. Maki, and M. L. Atchison. 1992. PU.1 recruits a second nuclear factor to a site important for immunoglobulin κ 3′ enhancer activity. Mol. Cell. Biol. 12:368-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pongubala, J. M. R., C. Van Beveren, S. Nagulapalli, M. J. Klemsz, S. R. McKercher, R. A. Maki, and M. L. Atchison. 1993. Effect of PU.1 phosphorylation on interaction with NF-EM5 and transcriptional activation. Science 259:1622-1625. [DOI] [PubMed] [Google Scholar]
- 53.Poulin, G., M. Lebel, M. Chamberland, F. W. Paradis, and J. Drouin. 2000. Specific protein-protein interaction between basic helix-loop-helix transcription factors and homeoproteins of the Pitx family. Mol. Cell. Biol. 20:4826-4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Quong, M. W., D. P. Harris, S. L. Swain, and C. Murre. 1999. E2A activity is induced during B-cell activation to promote immunoglobulin class switch recombination. EMBO J. 18:6307-6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rivera, R. R., M. H. Stuiver, R. Steenbergen, and C. Murre. 1993. Ets proteins: new factors that regulate immunoglobulin heavy-chain gene expression. Mol. Cell. Biol. 13:7163-7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Romanow, W. J., A. W. Langerak, P. Goebel, I. L. M. Wolvers-Tettero, J. M. van Dongen, A. J. Feeney, and C. Murre. 2000. E2A and EBF act in synergy with the V(D)J recombinase to generate a diverse immunoglobulin repertoire in nonlymphoid cells. Mol. Cell 5:343-353. [DOI] [PubMed] [Google Scholar]
- 57.Rutherford, M. N., and D. P. LeBrun. 1998. Restricted expression of E2A protein in primary human tissues correlates with proliferation and differentiation. Am. J. Pathol. 153:165-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schlissel, M., A. Voronova, and D. Baltimore. 1991. Helix loop helix transcription factor-E47 activates germ-line immunoglobulin heavy-chain gene transcription and rearrangement in a pre-T cell line. Genes Dev. 5:1367-1376. [DOI] [PubMed] [Google Scholar]
- 59.Serwe, M., and F. Sablitzky. 1993. V(D)J recombination in B cells is impaired but not blocked by targeted deletion of the immunoglobulin heavy chain enhancer. EMBO J. 12:2321-2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shen, C.-P., and T. Kadesch. 1995. B-cell-specific DNA binding by an E47 homodimer. Mol. Cell. Biol. 15:4518-4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sigvardsson, M., M. O'Riordan, and R. Grosschedl. 1997. EBF and E47 collaborate to induce expression of the endogenous immunoglobulin surrogate light chain genes. Immunity 7:25-36. [DOI] [PubMed] [Google Scholar]
- 62.Stutz, A. M. W., M. 1999. Functional synergism of STAT6 with either NF-κB or PU.1 to mediate IL-4-induced activation of germline gene transcription. J. Immunol. 163:4383-4391. [PubMed] [Google Scholar]
- 63.Takeda, S., Y. R. Zou, H. Bluethmann, D. Kitamura, U. Muller, and K. Rajewsky. 1993. Deletion of the immunoglobulin κ chain intron enhancer abolishes κ chain gene rearrangement in cis but not λ chain gene rearrangement in trans. EMBO J. 12:2329-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Veals, S. A., T. Santa Maria, and D. E. Levy. 1993. Two domains of ISGF3γ that mediate protein-DNA and protein-protein interactions during transcription factor assembly contribute to DNA-binding specificity. Mol. Cell. Biol. 13:196-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Voronova, A., and D. Baltimore. 1990. Mutations that disrupt DNA binding and dimer formation in the E47 helix-loop-helix protein map to distinct domains. Proc. Natl. Acad. Sci. USA 87:4722-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Warren, W. D., K. L. Roberts, L. A. Linehan, and M. T. Berton. 1999. Regulation of the germline Cγ1 promoter by CD40 ligand and IL-4: dual role for tandem NF-κB binding sites. Mol. Immunol. 36:31-44. [DOI] [PubMed] [Google Scholar]
- 67.Weintraub, H. 1993. The myoD family and myogenesis: redundancy, networks, and thresholds. Cell 75:1241-1244. [DOI] [PubMed] [Google Scholar]
- 68.Yamagata, T., J. Nishida, T. Tanaka, R. Sakai, K. Mitani, M. Yoshida, T. Taniguchi, Y. Yazaki, and H. Hirai. 1996. A novel interferon regulatory factor family transcription factor, ICSAT/Pip/LSIRF, that negatively regulates the activity of interferon-regulated genes. Mol. Cell. Biol. 16:1283-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yee, A. A., P. Yin, D. P. Siderovski, T. W. Mak, D. W. Litchfield, and C. H. Arrowsmith. 1998. Cooperative interaction between the DNA-binding domains of PU.1 and IRF4. J. Mol. Biol. 279:1075-1083. [DOI] [PubMed] [Google Scholar]
- 70.Zhuang, Y., P. Cheng, and H. Weintraub. 1996. B-lymphocyte development is regulated by the combined dosage of three basic helix-loop-helix genes, E2A, E2-2, and HEB. Mol. Cell. Biol. 16:2898-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhuang, Y., P. Soriano, and H. Weintraub. 1994. The helix-loop-helix gene E2A is required for B cell formation. Cell 79:875-884. [DOI] [PubMed] [Google Scholar]