Abstract
Steroidogenic factor 1 (SF-1) is an orphan nuclear receptor with no known ligand. We showed previously that phosphorylation at serine 203 located N′-terminal to the ligand binding domain (LBD) enhanced cofactor recruitment, analogous to the ligand-mediated recruitment in ligand-dependent receptors. In this study, results of biochemical analyses and an LBD helix assembly assay suggest that the SF-1 LBD adopts an active conformation, with helices 1 and 12 packed against the predicted alpha-helical bundle, in the apparent absence of ligand. Fine mapping of the previously defined proximal activation function in SF-1 showed that the activation function mapped fully to helix 1 of the LBD. Limited proteolyses demonstrate that phosphorylation of S203 in the hinge region mimics the stabilizing effects of ligand on the LBD. Moreover, similar effects were observed in an SF-1/thyroid hormone LBD chimera receptor, illustrating that the S203 phosphorylation effects are transferable to a heterologous ligand-dependent receptor. Our collective data suggest that the hinge together with helix 1 is an individualized specific motif, which is tightly associated with its cognate LBD. For SF-1, we find that this intramolecular association and hence receptor activity are further enhanced by mitogen-activated protein kinase phosphorylation, thus mimicking many of the ligand-induced changes observed for ligand-dependent receptors.
To date, just under 50 members of the nuclear receptor gene superfamily have been identified (27, 42). Cognate ligands have been identified for about half of these receptors, leaving the remaining members of this family as orphans. Whether all of these orphan receptors are activated by bona fide in vivo ligands is still unclear. One such orphan, steroidogenic factor 1 (SF-1; officially referred as NR5A1), is known to regulate a wide variety of genes in multiple endocrine cell types and is also required for early endocrine organogenesis (reviewed in references 10 and 33). In contrast to ligand-dependent receptors, whose activity is controlled by ligand binding, SF-1 is constitutively active in the apparent absence of ligand in many cellular contexts.
While no regulatory ligand has been identified for SF-1, indirect evidence suggests that extracellular signaling events may modulate SF-1 activity. This suggestion stems from the fact that many SF-1 target genes are stimulated by peptide hormone signaling via the second messenger cyclic AMP (cAMP). More importantly, cAMP-induced up-regulation of these target genes is mediated primarily through SF-1 (13, 15, 24). However SF-1 does not appear to be phosphorylated directly by the cAMP/protein kinase A pathway but, rather, is phosphorylated by the mitogen-activated protein kinase (MAPK) at a single serine residue (S203) located in the large central hinge region between the DNA binding domain (DBD) and the ligand binding domain (LBD) (11, 26). Activation of the MAPK pathway modulates SF-1 transcription activity by increasing cofactor recruitment and thus may provide a ligand-independent method of regulating SF-1 activity (11). In the last several years, evidence has directly linked cAMP/PKA signaling to the Ras-Raf-MEK-ERK/MAPK pathway via activation of Ras-related small GTPases (23, 41), thereby supporting our hypothesis that peptide hormone receptor signaling may modulate SF-1 activity via an Erk2-like phosphorylation. Indeed, at least two peptide hormone signaling events upstream of SF-1 are mediated through MAPK signaling, including the gonadotropin-releasing hormone and the adrenocorticotropic hormone pathways (20, 38).
In addition to S203 phosphorylation, deletion analyses of SF-1 have mapped an activation function domain in the vicinity of the hinge and N′-terminal region of the LBD (residues 185 to 260), which overlaps with the MAPK phosphorylation site (6, 11). This activation domain (referred to as AF1 or proximal activation region), together with the classical activation function 2 (AF2), located at the C′ terminus of the LBD, are required for coactivator enhancement of SF-1 activity (6, 15, 16, 22). In certain contexts, the hinge region of SF-1 also exhibits an independent repressor domain (31), which resembles closely a motif found in the glucocorticoid receptor (GR) that mediates repression at dimeric sites (14). The close proximity between an activation domain and a MAPK site identified in SF-1 can also be found in other ligand-dependent nuclear receptors where the AF1 and MAPK site(s) are located in the N′ terminus or A/B domain. For these nuclear receptors, phosphorylation is implicated in modulating both ligand-dependent and ligand-independent activity (18, 46, 56).
The hinge domain of nuclear receptors has been loosely defined as the region between the DBD and LBD. Prior to structural information revealing that the LBD is an alpha-helical bundle, the C′-terminal boundary of the “hinge” region was assigned to the beginning of helix 3 of the LBD; in SF-1 this corresponds to residue 260 (see Fig 1A). Subsequently, high-resolution crystal structures of the thyroid hormone (TRβ) and retinoic acid nuclear receptors incorporated the proximal portion of the hinge region into the LBD structure as the first and second helices (40, 52, 53). The so-called hinge region was reassigned to residues adjacent to helix 1 and has been viewed as a flexible unstructured region. More recently, Pissios et al. showed that the highly divergent helix 1 is specific to each receptor and assembles in trans with the remainder of the LBD only after addition of either hormone or corepressor NcoR peptides (34). Since ligand does not interact directly with helix 1, assembly of helix 1 with the remainder of the LBD in trans was proposed to stabilize the overall structure of the LBD. Taken together, these results suggest that the hinge is more than a simple flexible connector between the LBD and DBD and, instead, may participate in conjunction with helix 1 to promote an active conformation.
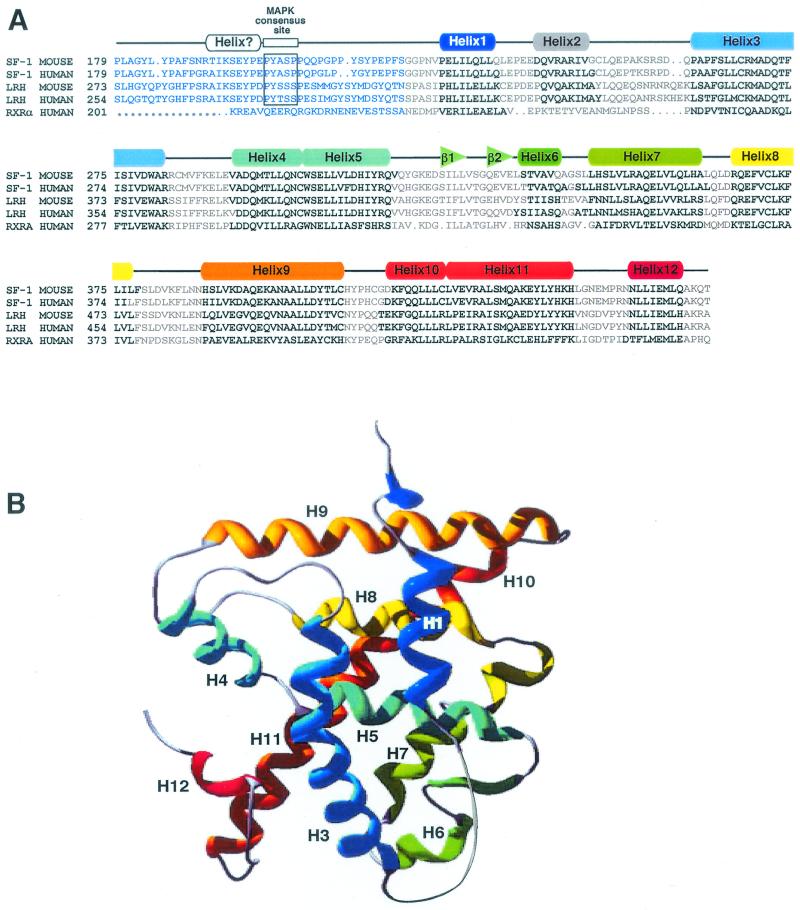
FIG. 1.
Predicted secondary structure of the hinge-LBD region of SF-1. (A) Sequence alignment of SF-1 hinge-LBD is shown for two mammalian species and the other member of subclass V family of nuclear receptors, LRH1. Included in this alignment are human and mouse sequences of SF-1 and LRH1 and the human sequence of RXRα. The sequence numbering is indicated at the beginning of every new line. Predicted alpha-helices and beta-turns are indicated in bold and above in rainbow-colored boxes (rectangles and triangles, respectively) ranging from 1 to 12 for the alpha-helices. (B) Modeling of SF-1 predicts a helical bundle with a ligand binding polar pocket. A three-dimensional model of the SF-1 LBD was obtained using the WHATIF program (50). Helices are indicated in rainbow colors and numbered corresponding to the coloring shown in panel A, with blue lettering used to depict the hinge region.
Our previous work established that phosphorylation of the hinge region modulates SF-1 activity by indirectly influencing the LBD (11). In this study, we investigated the role of the hinge-helix 1 region and asked how S203 phosphorylation might affect the structural integrity of SF-1. To do this, we used biochemical and cellular studies to indirectly assess the protein stability of the LBD before and after S203 phosphorylation. Similar analyses were performed with a SF-1/TRβ chimera to determine if modification of the SF-1 hinge region would influence a heterologous LBD. Our findings suggest that the hinge-helix 1 region is specific for each receptor and that modification of this hinge region by phosphorylation enhances the overall stability and transcriptional activity of SF-1.
MATERIALS AND METHODS
Plasmids.
Glutathione S-transferase GST- and Gal4-SF-1 fusions plasmids were constructed by subcloning of the PCR-amplified EcoRI-XbaI fragments into pGEX-4T1 and pM vectors (Clontech). Following amplification of the SF-1/TRβ chimera construct by PCR, fragments were subcloned into pM with EcoRI-SalI and in pGEX-4T1 with EcoRI-XhoI. VP16-SF-1 and VP16-SF-1/TRβ constructs were obtained by subcloning PCR products into pVP16 (Clontech) with EcoR1-XhoI. For all pcDNA3 constructs, PCR fragments containing an in-frame ATG for either SF-1-LBD or TRβ-LBD were subcloned into pcDNA3 (Invitrogen) with EcoRI-XhoI. Mutations of SF-1 helix 1 were obtained with the Quickchange site-directed mutagenesis kit as specified by the manufacturer (Stratagene). The SF-1 PCR fragments were subcloned into the bacterial expression plasmid pBH4 (donated by W. Lim, University of California San Francisco) which contains an in-frame N′-terminal six-histidine tag. The E202 TRβ expression plasmid was a generous gift from J. Baxter, University of California San Francisco.
Cell culture, RT-PCR, and Western blot analyses.
NIH 3T3 δRaf-1:ER and Y1 mouse adrenal cortical cells were grown in Dulbecco modified Eagle medium supplemented with 10% bovine calf serum. For transfections, δRaf-1:ER or Y1 cells were plated the day before into 24-well dishes at 50% density. Transfections were performed by Fugene (Roche) treatment for 24 hs. Typically 200 ng of Gal4-luciferase reporter construct (54), 100 ng of β-galactosidase expression vector, and 100 ng of expression vectors were used. To activate the MAPK pathway, cells were treated with 10 μM β-estradiol for 5 h before being harvested and assayed for luciferase (PharMingen International) and β-galactosidase activity to monitor for transfection efficiency. In experiments using the wild-type or the S203A mutant SF-1/TRβ constructs, 1 μM T3 hormone dissolved in dimethyl sulfoxide was added before harvesting the cells. All experiments were performed in triplicates and repeated at least three times.
For reverse transcription-PCR (RT-PCR), total RNA from NIH 3T3 δRaf-1:ER cells transfected with the Gal4-SF-1 or Gal4-SF-1/TRβ fusion constructs were isolated using the RNeasy protocol (Qiagen). PCRs were conducted with Gal4 primers or actin-specific primers. RT-PCR of actin transcripts served as a reference to normalize the Gal4 PCR reactions, as well as controlling for genomic DNA contamination. Western blot assays of Y1 cells transfected with MKP-1 or not transfected were performed using a S203 phosphospecific SF-1 antibody (Phospho-SF-1) at a 1:1,000 dilution on whole cellular extracts. Total SF-1 was detected with SF-1 antibody (provided by K. Morohashi).
Protein expression and purification.
All GST fusion proteins were expressed from the parent pGEX-4T1 vector, containing mSF-1, hTRβ, SF-1/TRβ, SF-1 helix 1, or the SF-1-hinge/TRβ-helix 1, in BL21DE3 (pLysS) at 30°C and induced for 4 h with 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at an optical density at 600 nm of 0.6. Bacterial pellets were resuspended in 10 mM Tris-HCl (pH 8.0)-150 mM NaCl-1 mM EDTA-1 mM dithiothreitol-1 mM phenylmethylsulfonyl fluoride protease inhibitors (Roche), frozen-thawed, incubated with 0.1 mg of lysozyme per ml, and lysed by sonication. N-Sarcosyl was added to lysates (final concentration, 1.5%), and debris was removed by centrifugation. Samples were incubated with 100 μl of a 50% slurry of glutathione-agarose 4B beads (Pharmacia) for 1 h at 4°C, after which the beads were washed extensively and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12% polyacrylamide). His-tagged recombinant SF-1 (residues 179 to 462) proteins were expressed in BL21DE3(pLysS) and induced with IPTG. Harvested bacteria were frozen-thawed in 50 mM Tris-HCl (pH 8.0)-100 mM NaCl-5% glycerol-protease inhibitors, and cleared lysates were subjected to Talon (Clontech) chromatography. His6-tagged proteins were eluted with imidazole and subsequently cleaved with recombinant tobacco etch virus protease (TEV) (2 h at 4°C). Purified SF-1 protein was further resolved on a MonoQ column. The purity and homogeneity of expressed SF-1-LBD proteins were assessed using both SDS-PAGE and native PAGE, and determined to be >99% pure.
GST pulldown and helix assembly assays.
For in vitro kinasing of GST fusion proteins, bound proteins were incubated with recombinant Erk2 kinase, under conditions recommended by the manufacturer (New England Biolabs). Bound proteins were washed extensively in buffer containing 20 mM Tris-HCl (pH 8.0), 0.1 M NaCl, 0.01% NP-40, 10% glycerol, 1 mM EDTA, 1 mM dithiothreitol, 0.1 mM phenylmethylsulfonyl fluoride, and a protease inhibitor cocktail before being incubated with in vitro-transcribed and -translated GRIP for 2 h at 4°C. For all GST pulldown assays, GST-protein fusions were incubated with in vitro-transcribed and translated products for 16 h at 4°C in the buffer described above except that it contained 20% glycerol and 2 mg of bovine serum albumin per ml without NP-40. Bound proteins were washed four times with their respective buffers supplemented with 0.01% NP-40 in the helix assembly assay. The proteins were eluted and analyzed by SDS-PAGE and autoradiography.
In vitro transcription and translation, limited proteolysis.
In vitro-translated proteins were generated from in vitro-transcribed pSG5-GRIP, pcDNA3-SF-1-helix 2-12, and pcDNA3-SF-1/TRβ-helix 2-12, using the TNT T7-coupled reticulocyte lysate system in the presence of [35S]methionine as specified by the manufacturer (Pharmacia). For limited proteolysis experiments of SF-1 hinge-helix 1, purified His6-tagged SF-1 protein spanning residues 179 to 462 was incubated in the absence or presence of Erk2 and then with Talon beads and extensively washed to eliminate residual Erk2 prior to digestion with chymotrypsin (0.125 μg) for 5 to 15 min, all reactions were stopped, and the products were analyzed by SDS-PAGE (15% polyacrylamide), blotted onto a polyrinylidene difluoride Immobilon filter, and stained with Ponceau S. Edman degradation sequencing was used to identify the N′-terminal sequence of all visibly stained peptides. In vitro-transcribed and -translated [35S]methionine-labeled HA epitope-tagged full-length SF-1 and SF-1/TRβ proteins were immunoprecipitated using an HA antibody (Babco) and then incubated with protein A-Sepharose (Sigma). Bound proteins were washed and incubated in control buffer with either hormone or recombinant Erk2. Following treatments, bound proteins were washed and equilibrated in chymotrypsin buffer (80 mM Tris-HCl [pH 7.8], 10 mM CaCl2) and then incubated with increasing amounts of chymotrypsin for 1 h at 30°C. Reactions were stopped, and the products were analyzed by SDS-PAGE and autoradiography.
Multiple sequence alignment and modeling.
All sequences were obtained from the nuclear receptor database NucleaRDB (12) and selected based on their amino acid similarity to the mouse SF-1 sequence. The mouse SF-1 sequence was corrected at amino acid 256 (R for G) according to the original published sequence. Multiple sequence alignment was performed with an iterative profile-based alignment procedure using the WHAT IF sequence alignment module (50). The profile was created based on an optimal alignment of the sequence of mouse SF-1, with high gap penalties assigned in the region spanning all 12 putative helices. The Meta server of PredictProtein (43) was used to run several programs for prediction of secondary structure, PHDSec (44), PROF (32), SAM-T99 (17), and Sspro (35). The three-dimensional model of the mSF-1 LBD was constructed with the WHAT IF program (50) based on the structure of the hRXRα receptor: sequences of mSF-1 were aligned against the sequence of the PDB entry of the crystal structure of the liganded LBD, chain A (7). Models were built using the previously described protocol (51).
RESULTS
Position of helix 1 or the activation function domain in a three-dimensional model of the SF-1 LBD.
To assess how the SF-1-proximal activation function relates to the LBD and helix 12 (AF2), we used multiple algorithms to model the secondary structure of the hinge and N′-terminal regions of the LBD region, as well as the LBD. Secondary-structure predictions indicate that the proximal activation function region and the MAPK site reside in an unstructured hinge region, which extends into the first of 12 putative alpha-helices within the LBD of SF-1 (residues 220 to 462, Fig. 1A). The high sequence similarity to RXRα also allowed us to generate a three-dimensional model of SF-1 based on the crystal structure of RXRα bound by 9-cis-retinoic acid (7). Although this model lacks predictive information about the hinge region, it supports the assumption that helices 1 through 12 are in the active configuration similar to a ligand-dependent receptor when bound by an agonist and pack tightly against the LBD helical bundle, as would be observed for a liganded receptor (Fig. 1A). Moreover, a modest-size hydrophobic cavity (340 Å) is predicted to be formed by residues located on the second beta-turn and helices 3, 5, 7, and 11 with three polar side chains projecting into this cavity (S304 and R314 from helix 5 and Y439 from helix 11) (Fig. 1). We also note that a moderate degree of conservation (36%) exists between side chains lining the predicted ligand binding pocket of SF-1 and those found in RXRα, including a conserved arginine 314 in helix 5 (R316 in RXRα) positioned opposite to helix 12. Similar to previous findings that helix 1 is tightly associated with the LBD when bound by ligand or corepressor peptides (34), our best-fit model of SF-1 predicts that the proximal activation function domain, including helix 1, is tightly associated with the LBD.
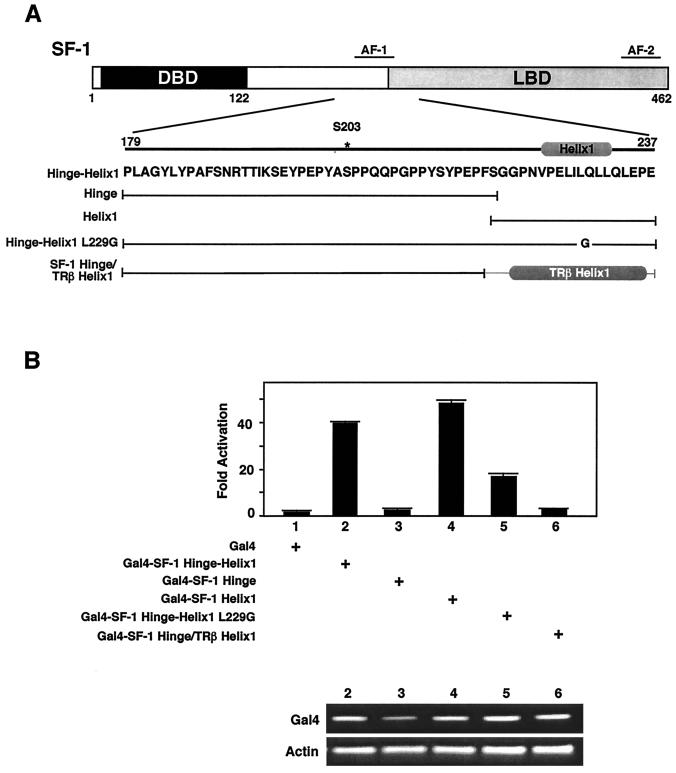
Finer mapping of the previously defined proximal activation function (residues 185 to 260) (11, 31) using a standard Gal4 DBD system showed that the activation function mapped fully to helix 1 of the LBD (Fig. 2). In contrast, the hinge region (residues 179 to 219) of SF-1 was inactive. As predicted, disruption of helix 1 attenuated the activation function, as shown by the presence of L229G and L227G mutants (Fig. 2B and data not shown). Replacing helix 1 of SF-1 with that of TRβ fails to recapitulate these results, suggesting that an alpha-helical motif is not sufficient to confer activity. The observed differences in the intrinsic activity of SF-1 helix 1 and TRβ appear not to stem from differential expression levels, since all Gal4 constructs transcripts were expressed equivalently (Fig. 2B). Therefore, in SF-1, the predicted helix 1 contains an activation function domain, and we have now modified our original nomenclature and refer to this activation function domain as activation function helix 1. (AFH1).
FIG. 2.
Helix 1 of SF-1 contains an activation function AFH1 domain. (A) A schematic diagram of the major domains of the SF-1 protein is shown, with the amino acid sequence of the hinge-helix 1 region presented below. (B) Activities of constructs diagrammed in panel A are shown after cotransfection into NIH 3T3 cells with the luciferase reporter containing four tandem Gal4 binding sites, pGAL-RE-TK (54). Luciferase activity is expressed as fold activation, with the empty Gal4-DBD expression vector taken to be 1. To monitor for gross differences in expression levels of each Gal4 construct used, RT-PCR was performed using primers for both the Gal4 DBD present in all constructs and for actin; the results are shown in the lower panel for all corresponding constructs transfected (upper panel).
Helix 1 of SF-1 assembles with helices 2 to 12 in the apparent absence of ligand.
Our three-dimensional model assumes that SF-1 adopts an active conformation that is similar to a liganded LBD. To experimentally address this assumption, we asked if helix 1 of SF-1 would assemble with helices 2 to 12, as shown previously for ligand-dependent receptors (34). Indeed, GST pulldown experiments showed that the hinge and helix 1 of SF-1 (residues 179 to 240) specifically recruited helices 2 to 12 (238 to 462) of SF-1 but not helices 2 to 12 of TRβ (Fig. 3A and data not shown). These in vitro results were confirmed in cellular transfection experiments in NIH 3T3 cells, using a similar two-component system consisting of Gal4 DBD fused to the hinge-helix 1 region of SF-1 and helices 2 to 12 of SF-1 fused to the activation domain of VP16 (Fig. 3B). Similar to results obtained in GST pulldown assays, we observed an association of hinge-helix 1 and helices 2 to 12 as judged by increased reporter activity. No association was observed between helix 1 of SF-1 with either unrelated helices 2 to 12 (TRβ; Fig. 3B) or helices 1 to 12 of SF-1 (data not shown). Two mutations predicted to disrupt helix 1, L227G and L229G, failed to exhibit appreciable assembly in trans (Fig. 3B and data not shown). For SF-1, the ability of helix 1 to assemble in trans with the remainder of the LBD in the apparent absence of ligand supports our assumption that SF-1 adopts an active conformation and illustrates that, similar to other receptors, helix 1 of SF-1 is specific for its cognate LBD.
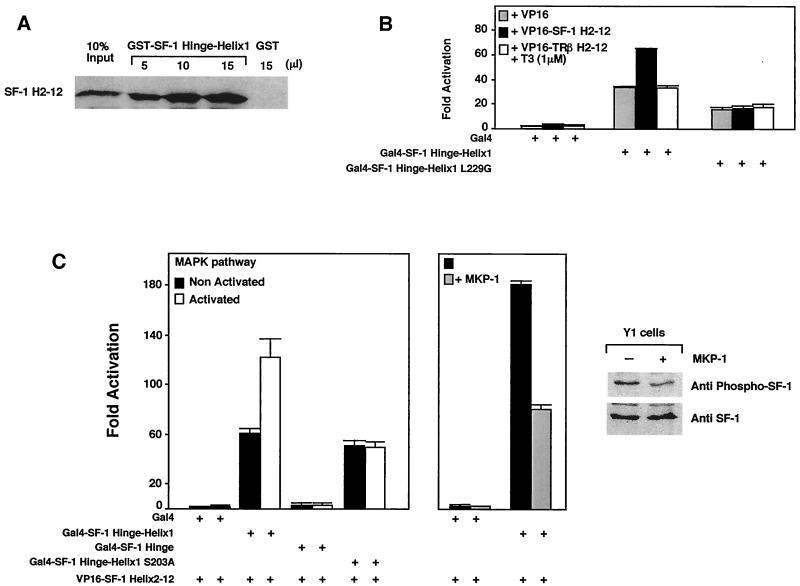
FIG. 3.
Phosphorylation of SF-1 enhances recruitment of the LBD. (A) In vitro assembly of increasing amounts of purified recombinant GST-SF-1 hinge-helix 1 protein with the in vitro-transcribed and translated [35S]methionine-labeled remainder of the LBD protein (SF-1 H2-12) were assessed by a standard GST pulldown assay. Ten percent of the labeled input is shown as indicated (10% input), along with a single pulldown with control GST protein (GST). (B) Association of SF-1 hinge-helix 1 with the remainder of the LBD was tested in a mammalian two-hybrid experiment using NIH 3T3 δRaf-1:ER cells. The activation domain VP16 fused to SF-1 helices 2 to 12 (VP16-SF-1 H2-12) and the SF-1 hinge-helix 1 fused to the Gal4 binding domain were cotransfected with the Gal4-responsive luciferase reporter pGAL-RE-TK in δRaf-1:ER cells for 24 h. VP-16 fused to helices 2 to 12 of TRβ was also used in these experiments in the presence of T3 (1 μM). Similar results were obtained with the mouse Y1 adrenocortical cell line, and assembly was shown to be dependent on VP16 (data not shown). (C) MAPK pathway activation increases helix assembly of SF-1 in vivo. The luciferase activity of the pGAL-RE-TK (Gal4 reporter) is shown for a mammalian two-hybrid expression system containing VP16-SF-1 H2-12 with different deletions, and mutants of SF-1 hinge-helix 1 fused to Gal4 DBD were cotransfected in the δRaf-1:ER or in Y1 cells for 24 h. Activity was also measured in an S203A mutant harboring a nonphosphorylatable alanine residue. To activate the MAPK pathway, δRaf-1:ER cells were treated with 10−5 M β-estradiol for 5 h. To inhibit the MAPK pathway, the MAPK inhibitor MAPK phosphatase (MPK-1) expression vector was cotransfected in Y1 cells grown in standard growth media. The degree of SF-1 phosphorylation was evaluated by using an S203-phosphospecific SF-1 antibody compared with signals obtained with a SF-1 antibody, with (+) our without (−) transfection of the MPK-1. A representative Western blot is shown in the far-right panel.
Phosphorylation of the hinge region modulates activity of the SF-1 LBD.
In addition to the activation function associated with the hinge-helix 1 region, our previous work showed that phosphorylation of S203 in the hinge region modulates SF-1 activity by increasing cofactor recruitment (11). We hypothesized that S203 phosphorylation would enhance the helix assembly of SF-1, similar to the stabilizing effects of ligand for other receptors. To induce the MAPK cascade and increase S203 phosphorylation in SF-1, we used an inducible Raf1 NIH 3T3 cell line (δRaf-1:ER) (45). We first showed that activation of the MAPK cascade does not increase the activity of the AFH1 alone (data not shown). However, assembly of hinge-helix 1 and helices 2 to 12 of SF-1 increased significantly after activation of the MAPK cascade. This MAPK-induced increase was not observed when using a nonphosphorylatable S203A mutant, thus confirming that SF-1 S203 is the major residue targeted by the MAPK pathway (Fig. 3C). We also tested the effect of a MAPK-specific phosphatase, MKP-1, in Y1 adrenocortical cells. Helix assembly of SF-1 was consistently higher in these cells than in δRaf-1:ER cells, and cotransfection of the MAPK-specific inhibitor, MKP-1, markedly reduced helix assembly (Fig. 3C). Using a phosphospecific SF-1 S203 antibody (M. Desclozeaux and H. A. Ingraham, unpublished data), Western blotting demonstrated that the level of phospho-SF-1 decreased when MKP-1 was transfected in the Y1 cells (Fig. 3C, right panel). Conversely, the degree of SF-1 phosphorylation increased when the MAPK pathway was activated in the δRaf:ER cells (data not shown). We noted that phosphorylation of S203 by Erk2 did not increase the recruitment of helices 2 to 12 by hinge-helix 1 SF-1 in GST pulldown assays (data not shown). This result may reflect the inability to exceed the already high levels of in vitro helix assembly observed for SF-1 or, alternatively, may suggest that the S203 phosphorylation increases the activity of the assembled receptor complex. Nonetheless, our collective data suggest that phosphorylation of the hinge domain (S203) enhances the assembled LBD activity.
MAPK phosphorylation of S203 modulates a heterologous LBD.
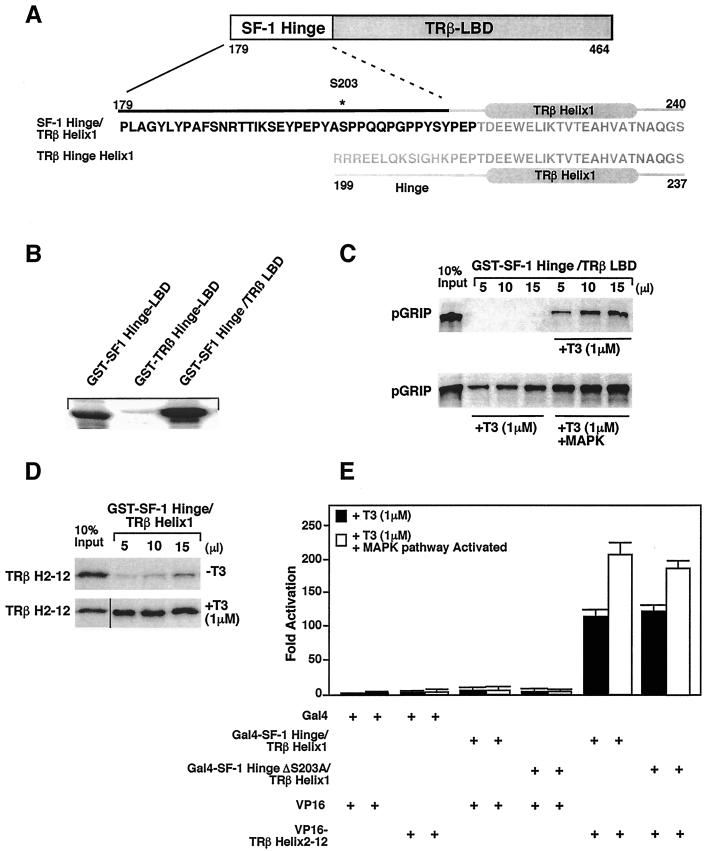
To test whether the effects of S203 phosphorylation observed for SF-1 could be transferred to a heterologous LBD, we created a chimera that included the SF-1 hinge region and the TRβ LBD. This SF-1-hinge/TRβ-LBD chimera (referred to as SF-1/TRβ) was fused at conserved residues PEP, which are located N′-terminal to their respective helices 1 (Fig. 4A). In vitro phosphorylation of the SF-1/TRβ chimera using recombinant Erk2 kinase was observed only when the SF-1 hinge containing S203 was present (Fig. 4B). GST pulldown experiments showed that this SF-1/TRβ chimera was capable of recruiting the nuclear receptor coactivator GRIP-1 in a ligand-dependent manner (Fig. 4C). Moreover, phosphorylation of the SF-1/TRβ chimera increased cofactor recruitment, as reported for SF-1 (Fig. 4C, lower panel) (11). GST pulldown experiments showed that helix assembly of this SF-1/TRβ chimera was ligand dependent, in contrast to the ligand-independent assembly of SF-1 (Fig. 4D). These results are similar to those obtained previously with just the TRβ hinge-helix 1 and TRβ helices 2 to 12 (34). Furthermore, assembly of the SF-1/TRβ chimera is specific for the LBD of TRβ; no assembly was observed with helices 2 to 12 of SF-1 (data not shown). In contrast to the inherent transcriptional activity of the helix 1 region of SF-1, the chimera SF-1 hinge/TRβ helix 1 displays no basal transcription activity (Fig. 4E). Assays performed with the δRaf-1:ER cells confirmed that assembly in trans is absolutely dependent on T3 (Fig. 4E). As with SF-1, activation of the MAPK pathway increased the helix assembly of the SF-1/TRβ chimera to a similar extent to that observed for SF-1 (Fig. 4E). However, slightly less assembly was observed with the corresponding S203A mutant. This result is markedly different from the dramatic reduction in assembly observed for the SF-1 S203A mutant (Fig. 3C) and suggests that secondary effects such as cofactor phosphorylation participate in the assembly of the SF-1/TRβ chimera in the cellular context. We conclude that phosphorylation of the SF-1 hinge region modulates cofactor recruitment of a heterologous LBD.
FIG. 4.
MAPK phosphorylation modulates SF-1/TRβ chimera. (A). A schematic representation of different domains of the SF-1/TRβ chimera protein is shown, with detailed sequence and secondary structure of the hinge-helix 1 region indicated below the diagram. (B) Erk2 phosphorylates the SF-1/TRβ chimera in vitro. GST fusion proteins containing the SF-1 hinge-LBD, the SF-1 hinge/TRβ LBD, or the TRβ LBD were phosphorylated in vitro by Erk2, as described in Materials and Methods. (C) Phosphorylation increases recruitment of the nuclear receptor coactivator pGRIP by SF-1/TRβ only in the presence of the T3 ligand (+T3). Associations were assessed by GST pulldown assays using the GST-SF-1 hinge/TRβ LBD recombinant protein and in vitro-transcribed and translated [35S]methionine-labeled GRIP1 in the absence or presence of 1 μM T3 (top panel). The bottom panel shows the same experiment done with in vitro-phosphorylated GST-SF-1 hinge/TRβ LBD protein. (D) Effects of SF-1 hinge region on the helix association of SF-1/TRβ in vitro. In vitro assembly assays were performed with purified recombinant GST-SF-1 hinge/TRβ helix 1 protein and the in vitro-transcribed and -translated [35S]methionine-labeled remainder of the TRβ LBD protein (TRβ H2-12). (E) In vivo helix assembly of the SF-1/TRβ chimera using mammalian two-hybrid expression constructs. In the δRaf-1:ER cells, Gal4-SF-1 hinge/TRβ helix 1 or S203A mutant and VP16-TRβ helix 2-12 were cotransfected with pGAL-RE-TK for 24 h in the presence of 1 μM T3 or vehicle alone. The MAPK pathway was activated using 10−5 M β-estradiol for 5 h, and the luciferase activity was measured.
Hinge phosphorylation stabilizes both SF-1 and a SF-1/TRβ chimera.
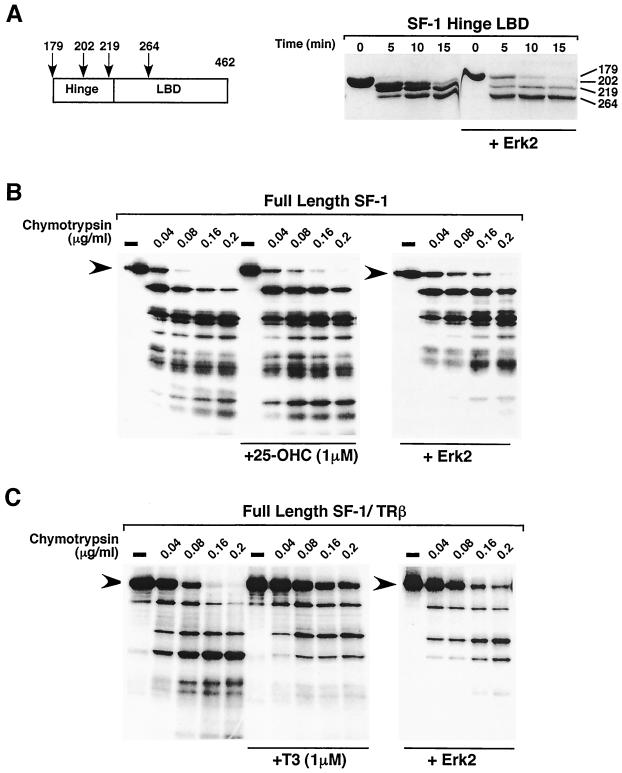
To determine if phosphorylation of the SF-1 hinge domain promotes a conformational change in the LBD, as inferred from the increased activity of assembled helices and increased cofactor recruitment, we performed limited proteolysis on the SF-1 hinge-LBD before and after S203 phosphorylation. We hypothesized that phosphorylation would stabilize SF-1, making it less accessible to proteases akin to the ligand-induced conformational changes observed for ligand-dependent receptors (1, 9, 21, 48). Limited proteolysis was carried out on purified phosphorylated hinge-LBD SF-1 protein (residues 179 to 462) (Fig. 5A). For nonphosphorylated SF-1 protein, we observed four major fragments after limited chymotrypsin digestion. Identical peptide fragments were obtained as judged by Edman sequencing following Erk2 phosphorylation; however, both the dynamics and ratios of the fragments changed dramatically. Specifically, we noted that the full-length hinge-LBD SF-1 fragment persisted much longer whereas cleavage in the hinge region was altered significantly, as evidenced by the reduced proteolytic products beginning at residues 202 or 219, just N′-terminal of helix 1 (Fig. 5A, right panel). Interestingly, while more potential chymotrypsin sites can be identified in the SF-1 hinge region (at residues 184, 186, 198, 213, and 215), these sites are not cleaved in either the unphosphorylated or phosphorylated SF-1 protein. Taken together, these data imply that the hinge is not completely exposed to solvent and that the protease sensitivity of the hinge is reduced further after S203 phosphorylation.
FIG. 5.
Phosphorylation of SF-1 or an SF-1/TRβ chimera reduces protease sensitivity. (A) Purified recombinant SF-1 hinge-helix 1 protein spanning residues 179 to 462 (12.5 μg per point) was phosphorylated with recombinant Erk2 and then incubated with chymotrypsin for 5, 10, or 15 minutes. The four major proteolytic fragments identified by protein sequencing, as described in Materials and Methods, are shown relative to the hinge and LBD regions in SF-1 (left panel). In the right panel, results of chymotrypsin cleavage are shown, with the N′-terminal residue corresponding to each peptide indicated. (B) In vitro-transcribed and -translated [35S]methionine-labeled full-length SF-1 protein was incubated with increasing amounts of chymotrypsin protease in the presence or absence of 25-OH hydroxycholesterol (+25-OHC) (left and middle panels), or after phosphorylation with Erk2 (+Erk2) (right panel). Full-length protein is indicated (black arrowheads). (C) Limited proteolysis with chymotrypsin was carried out on a chimeric protein containing the SF-1 DBD-hinge region fused to the TRβ LBD (full-length SF-1/TRβ) in the absence (left panel) or presence (+T3, middle panel) of T3 and after Erk2 phosphorylation with no T3 (+Erk2, right panel). Full-length protein is indicated (black arrowheads).
Limited proteolysis was also used to determine the effects of phosphorylation on full-length SF-1. A dramatic decrease in protease sensitivity was observed with in vitro-phosphorylated SF-1, as evidenced by the persistence of slower-migrating peptide fragments (Fig. 5B, right panel). By contrast, addition of the candidate SF-1 ligand, 25-hydroxycholesterol, had minimal effect on the proteolytic pattern (Fig. 5B) (5). The lack of an SF-1 ligand precludes a direct comparison of proteolytic patterns generated from phosphorylated and liganded receptors. To circumvent this problem, we used a full-length SF-1/TRβ chimera, which can be phosphorylated at S203 and can also bind the T3 ligand. As predicted from previous analyses of TRβ (34), addition of T3 resulted in marked reduction of the protease sensitivity (Fig 5C). More importantly, in the absence of T3, Erk2 phosphorylation of the SF-1/TRβ chimera exhibited reduced protease sensitivity with an almost identical pattern to that observed for the liganded chimera. The decreased protease sensitivity observed for Erk2-treated SF-1 and SF-1/TRβ implies that S203 phosphorylation induces a conformational change in the LBD, similar to the stabilizing effects of ligand for ligand-dependent receptors. Melting-temperature profiles of purified hinge-LBD SF-1 protein showed that the hinge-LBD of SF-1 unfolds as a single entity with a higher melting temperature (48°C) than that observed for unliganded TRβ (45.8°C) (data not shown). Melting-temperature profiles obtained for three independent preparations of highly enriched S203 phosphorylated protein showed a modest but reproducible increase in the thermal transition temperature (49.8°C), supporting the hypothesis that S203 phosphorylation increases the overall stability of the LBD.
DISCUSSION
In contrast to the well-defined molecular events that trigger ligand-dependent receptor activation, it is not well understood how constitutively active orphan receptors, such as SF-1, are regulated. In this study we found that the LBD of SF-1 is compact and assembles in trans without addition of ligand, suggesting that this orphan receptor normally assumes an active conformation, with helices 1 and 12 packed against the LBD helical bundle. These studies extend our previous work on SF-1 phosphorylation (11) and show that S203 phosphorylation in the hinge contributes to the stabilization of the LBD, analogous to ligand-induced stabilization for ligand-dependent receptors. These stabilizing effects can be transferred to a ligand-dependent LBD, as illustrated in experiments conducted with the SF-1-hinge/TRβ-LBD chimera. Our collective data suggest that the hinge together with helix 1 is an individualized specific motif, which is tightly associated with its cognate LBD. For SF-1, we found that this intramolecular association and hence receptor activity are further enhanced by MAPK phosphorylation, thus mimicking many of the ligand-induced changes observed for ligand-dependent receptors.
Activation of SF-1 versus ligand-dependent receptors.
The structural consequences of ligand binding to the LBD are now well understood for many receptors (39). Ligand acts by stabilizing and remodeling the helical structure of the LBD, with the predominant change being seen in the position of helix 12. Despite some obvious differences between SF-1 and other members of the nuclear receptor gene family, secondary-structure prediction of the SF-1 hinge-LBD region shows it to be remarkably similar to other receptors from helices 3 to 12. Our choice to use liganded RXRα as a template to model SF-1 is partially validated by the fact that assembly of helix 1 with helices 2 to 12 takes place in the absence of ligand and that our proteolysis experiments show helix 12 to be protected. These data agree with our model of SF-1, in which helices 1 and 12 are tightly associated with the LBD helical bundle rather than being loosely associated, as shown for unliganded receptors (34). Our findings cannot exclude the possibility that a small molecule occupies the ligand binding pocket in our experiments, as has now been shown by solved LBD structures of other orphan nuclear receptors (3, 4, 47). However, we conclude that SF-1 exhibits many features of ligand-dependent receptors in the apparent absence of a ligand.
Hinge-helix 1 in nuclear receptor function.
For many nuclear receptors, the N′-terminal AF1 acts synergistically with the AF2 domain in achieving full transcription activity, and together they simultaneously interact with either one coactivator or a coactivator complex (2, 8, 19, 49, 55). However, genetic dissection of these two activation functions show that AF1 and AF2 act independently of one another in vivo, as shown by the distinct development effects following their isolated disruption (28). This finding is consistent with the fact that some nuclear receptor cofactors exhibit specificity for either AF1 or AF2 (reviewed in reference 36). In SF-1, an AF2-independent activation domain is found in helix 1 of the LBD (see above) (11, 31). In this respect, the AFH1 domain in SF-1 is reminiscent of other non-AF-2 activation function domains found in several steroid receptors. For the estrogen receptor alpha (ERα) and GR, this unconventional activation function is referred to as AF2a or tau (τ)2, respectively (29, 30). While helix 1 of SF-1 has very little identity to this τ2 region, it does contain an LXXLL cofactor interaction motif that may facilitate its interaction with the p160 cofactor family, other nuclear receptors, and unidentified cofactors. It will be of interest to determine if there are AFH1-specific coregulators, as shown for the τ2 of GR (58).
Phosphorylation and stabilization of the LBD.
Whether the integration of extracellular signal and nuclear receptor activity plays a more prominent role in regulating SF-1 than for ligand-dependent receptors is not clear. In work described in this paper, we have established that SF-1 is phosphorylated in the Y1 cells using a phospho-S203 SF-1 antibody. The close proximity of phosphorylated S203 to the putative helix 1 suggests that these two regions might participate in stabilization of the LBD. This arrangement is conserved between SF-1 and its closest homologue, the liver-related homologue (LRH1) receptor, with a potential MAPK phosphorylation site located at PYASPP in the hinge region (Fig. 1A). Interestingly, two other orphan receptors, Nurr1 and Nor1, have a potential MAPK site in the hinge region just adjacent to helix 1. Our hypothesis that phosphorylation stabilizes the LBD is supported by the fact that phosphorylation changes the dynamics of proteolysis, increases the activity of an assembled SF-1 LBD, and reproducibly increases the melting transition. Thus, the effects of hinge phosphorylation in SF-1 appear to render the LBD more stable and compact, similar to ligand-dependent receptor activation.
Experiments with the SF-1/TRβ chimera protein demonstrate that phosphorylation of the SF-1 hinge domain further enhances ligand-induced cofactor recruitment and helix assembly. Decreased protease sensitivity was observed after phosphorylation of the SF-1/TRβ chimera in the absence of ligand. We noted that in our helix assembly assays, the SF-1/TRβ chimera receptor requires larger quantities of T3 than does TRβ. Unexpectedly then the loss of the small TRβ hinge region precludes maximal ligand potency (M. Desclozeaux and I. N. Krylova, unpublished data). Although further experiments are needed to examine this effect, these preliminary data suggest that both helix 1 and the very distal hinge region are essential for maximal receptor function. This is consistent with recent results showing that enhanced assembly of TRβ by the coactivators GRIP1 and SRC1 is dependent on helix 1 (57). Because MAPK-induced helix assembly remained intact in the S203A SF-1/TRβ mutant, we speculate that coactivator phosphorylation may indirectly enhance the assembly of the SF-1/TRβ mutant in a cellular context (25). By contrast, the MAPK-induced assembly was absent in the S203A SF-1 mutant, suggesting that MAPK-induced assembly in SF-1 is due primarily to SF-1 phosphorylation.
While the precise structural impact of phosphorylation on the LBD has yet to be determined, it is tempting to speculate that S203 phosphorylation induces conformational changes in the adjacent unstructured hinge region similar to that found when phosphorylated CREB binds to p300 (37). For instance, in SF-1, S203 phosphorylation could nucleate one or two proximal helices in the hinge, allowing the hinge to simultaneously bind and stabilize the LBD. In addition, S203 phosphorylation could induce a change in the AFH1 and thereby modulate specific cofactor recruitment, possibly in an AF2-independent manner, as shown previously for estrogen receptor on MAPK phosphorylation (49). In the present study, we found that the AFH1 activity, by itself, is not enhanced by activation of the MAPK pathway but instead requires the full LBD, suggesting a codependency of AFH1 and AF2 in mediating full SF-1 activity. Further experiments are needed to address both the structural and physiological consequences of SF-1 S203 phosphorylation.
Acknowledgments
We thank H. R. Huber for conceptual assistance with the circular dichroism analysis, W. Lim (UCSF) for providing pBH4- and TEV-expressing vectors, M. McMahon (UCSF) for discussions and for providing the δRaf-1:ER cells, C. W. Turck for performing Edman degradation sequencing of SF-1 hinge-LBD proteolytic peptides, G. Vriend (CMBI, University of Nijmegen, Nijmegen The Netherlands) for providing assistance with the three-dimensional modeling of SF-1, P. Webb and B. West (UCSF) for providing us with all TR and Gal4-luciferase reporter constructs, and H. Der Ou and M. J. Budny for technical help with both HPLC and circular dichroism instrumentation. We are also especially grateful to F. Schaufele for critical reading of the manuscript.
This work was supported by an American Heart Association fellowship to M.D, by American Heart Association and NICHD-RO1 and NICHD-KO2 grants to H.A.I, and by a NIDDK PO1 grant to H.A.I. and R.J.F.
REFERENCES
- 1.Allan, G. F., X. H. Leng, S. Y. Tsai, N. L. Weigel, D. P. Edwards, M. J. Tsai, and B. W. Omalley. 1992. Hormone and antihormone induce distinct conformational changes which are central to steroid receptor activation. J. Biol. Chem. 267:19513-19520. [PubMed] [Google Scholar]
- 2.Benecke, A., P. Chambon, and H. Gronemeyer. 2000. Synergy between estrogen receptor alpha activation functions AF1 and AF2 mediated by transcription intermediary factor TIF2. EMBO Rep. 1:151-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Billas, I. M., L. Moulinier, N. Rochel, and D. Moras. 2001. Crystal structure of the ligand-binding domain of the ultraspiracle protein USP, the ortholog of retinoid X receptors in insects. J. Biol. Chem. 276:7465-7474. [DOI] [PubMed] [Google Scholar]
- 4.Bourguet, W., V. Vivat, J. M. Wurtz, P. Chambon, H. Gronemeyer, and D. Moras. 2000. Crystal structure of a heterodimeric complex of RAR and RXR ligand-binding domains. Mol. Cell 5:289-298. [DOI] [PubMed] [Google Scholar]
- 5.Christenson, L. K., J. M. McAllister, K. O. Martin, N. B. Javitt, T. F. Osborne, and J. F. Strauss III. 1998. Oxysterol regulation of steroidogenic acute regulatory protein gene expression. Structural specificity and transcriptional and posttranscriptional actions. J. Biol. Chem. 273:30729-30735. [DOI] [PubMed] [Google Scholar]
- 6.Crawford, P. A., J. A. Polish, G. Ganpule, and Y. Sadovsky. 1997. The activation function-2 hexamer of steroidogenic factor-1 is required, but not sufficient for potentiation by SRC-1. Mol. Endocrinol. 11:1626-1635. [DOI] [PubMed] [Google Scholar]
- 7.Egea, P. F., A. Mitschler, N. Rochel, M. Ruff, P. Chambon, and D. Moras. 2000. Crystal structure of the human RXRalpha ligand-binding domain bound to its natural ligand: 9-cis-retinoic acid. EMBO J. 19:2592-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glass, C. K., and M. G. Rosenfeld. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14:121-141. [PubMed] [Google Scholar]
- 9.Greenfield, N., V. Vijayanathan, T. J. Thomas, M. A. Gallo, and T. Thomas. 2001. Increase in the stability and helical content of estrogen receptor alpha in the presence of the estrogen response element: analysis by circular dichroism spectroscopy. Biochemistry 40:6646-6652. [DOI] [PubMed] [Google Scholar]
- 10.Hammer, G. D., and H. A. Ingraham. 1999. Steroidogenic factor-1: its role in endocrine organ development and differentiation. Front. Neuroendocrinol. 20:199-223. [DOI] [PubMed] [Google Scholar]
- 11.Hammer, G. D., I. Krylova, Y. X. Zhang, B. D. Darimont, K. Simpson, N. L. Weigel, and H. A. Ingraham 1999. Phosphorylation of the nuclear receptor SF-1 modulates cofactor recruitment: integration of hormone signaling in reproduction and stress. Mol. Cell 3:521-526. [DOI] [PubMed] [Google Scholar]
- 12.Horn, F., G. Vriend, and F. E. Cohen. 2001. Collecting and harvesting biological data: the GPCRDB and NucleaRDB information systems. Nucleic Acids Res. 29:346-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu, M. C., N. C. Hsu, C. I. Pai, C. K. Wang, and B. Chung. 2001. Functions of the upstream and proximal steroidogenic factor 1 (SF-1)-binding sites in the CYP11A1 promoter in basal transcription and hormonal response. Mol. Endocrinol. 15:812-818. [DOI] [PubMed] [Google Scholar]
- 14.Iniguez-Lluhi, J. A., and D. Pearce. 2000. A common motif within the negative regulatory regions of multiple factors inhibits their transcriptional synergy. Mol. Cell. Biol. 20:6040-6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito, M., Y. Park, J. Weck, K. E. Mayo, and J. L. Jameson. 2000. Synergistic activation of the inhibin alpha-promoter by steroidogenic factor-1 and cyclic adenosine 3′, 5′-monophosphate. Mol. Endocrinol. 14:66-81. [DOI] [PubMed] [Google Scholar]
- 16.Jacob, A. L., and J. Lund. 1998. Mutations in the activation function-2 core domain of steroidogenic factor-1 dominantly suppresses PKA-dependent transactivation of the bovine CYP17 gene. J. Biol. Chem. 273:13391-13394. [DOI] [PubMed] [Google Scholar]
- 17.Karplus, K., C. Barrett, and R. Hughey. 1998. Hidden Markov models for detecting remote protein homologies. Bioinformatics 14:846-856. [DOI] [PubMed] [Google Scholar]
- 18.Kumar, R., and E. B. Thompson. 1999. The structure of the nuclear hormone receptors. Steroids 64:310-319. [DOI] [PubMed] [Google Scholar]
- 19.Lanz, R. B., N. J. McKenna, S. A. Onate, U. Albrecht, J. Wong, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1999. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell 97:17-27. [DOI] [PubMed] [Google Scholar]
- 20.Le, T., and B. P. Schimmer. 2001. The regulation of mapks in y1 mouse adrenocortical tumor cells. Endocrinology 142:4282-4287. [DOI] [PubMed] [Google Scholar]
- 21.Leng, X. H., S. Y. Tsai, B. W. Omalley, and M. J. Tsai. 1993. Ligand-dependent conformational changes in thyroid hormone and retinoic acid receptors are potentially enhanced by heterodimerization with retinoic X receptor. J. Steroid Biol. Mol. Biol. 46:643-661. [DOI] [PubMed] [Google Scholar]
- 22.Li, L. A., E. F. Chiang, J. C. Chen, N. C. Hsu, Y. J. Chen, and B. C. Chung. 1999. Function of steroidogenic factor 1 domains in nuclear localization, transactivation, and interaction with transcription factor TFIIB and c-Jun. Mol. Endocrinol. 13:1588-1598. [DOI] [PubMed] [Google Scholar]
- 23.Liebmann, C. 2001. Regulation of MAP kinase activity by peptide receptor signalling pathway: paradigms of multiplicity. Cell Signal. 13:777-785. [DOI] [PubMed] [Google Scholar]
- 24.Liu, Z., and E. R. Simpson. 1997. Steroidogenic factor 1 (SF-1) and SP1 are required for regulation of bovine CYP11A gene expression in bovine luteal cells and adrenal Y1 cells. Mol. Endocrinol. 11:127-137. [DOI] [PubMed] [Google Scholar]
- 25.Lopez, G. N., C. W. Turck, F. Schaufele, M. R. Stallcup, and P. J. Kushner. 2001. Growth factors signal to steroid receptors through mitogen-activated protein kinase regulation of p160 coactivator activity. J. Biol. Chem. 276:22177-22182. [DOI] [PubMed] [Google Scholar]
- 26.Lund, J., M. Bakke, G. Mellgren, K. Morohashi, and S. O. Doskeland. 1997. Transcriptional regulation of the bovine CYP17 gene by cAMP. Steroids 62:43-45. [DOI] [PubMed] [Google Scholar]
- 27.Maglich, J. M., A. Sluder, X. Guan, Y. Shi, D. D. McKee, K. Carrick, K. Kamdar, T. M. Willson, and J. T. Moore. 2001. Comparison of complete nuclear receptor sets from the human, Caenorhabditis elegans and Drosophila genomes. Genome Biol. 2(8):research 0029:1-0029:7. [DOI] [PMC free article] [PubMed]
- 28.Mascrez, B., M. Mark, W. Krezel, V. Dupe, M. LeMeur, N. B. Ghyselinck, and P. Chambon. 2001. Differential contributions of AF-1 and AF-2 activities to the developmental functions of RXR alpha. Development 128:2049-2062. [DOI] [PubMed] [Google Scholar]
- 29.Milhon, J., S. Lee, K. Kohli, D. Chen, H. Hong, and M. R. Stallcup. 1997. Identification of amino acids in the tau 2-region of the mouse glucocorticoid receptor that contribute to hormone binding and transcriptional activation. Mol. Endocrinol. 11:1795-1805. [DOI] [PubMed] [Google Scholar]
- 30.Norris, J. D., D. Fan, S. A. Kerner, and D. P. McDonnell. 1997. Identification of a third autonomous activation domain within the human estrogen receptor. Mol. Endocrinol. 11:747-754. [DOI] [PubMed] [Google Scholar]
- 31.Ou, Q., J. F. Mouillet, X. Yan, C. Dorn, P. A. Crawford, and Y. Sadovsky. 2001. The DEAD box protein DP103 is a regulator of steroidogenic factor-1. Mol. Endocrinol. 15:69-79. [DOI] [PubMed] [Google Scholar]
- 32.Ouali, M., and R. D. King. 2000. Cascaded multiple classifiers for secondary structure prediction. Protein Sci. 9:1162-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parker, K. L., and B. P. Schimmer. 1997. Steroidogenic factor 1: a key determinant of endocrine development and function. Endocr. Rev. 18:361-377. [DOI] [PubMed] [Google Scholar]
- 34.Pissios, P., I. Tzameli, P. Kushner, and D. D. Moore. 2000. Dynamic stabilization of nuclear receptor ligand binding domains by hormone or corepressor binding. Mol. Cell 6:245-253. [DOI] [PubMed] [Google Scholar]
- 35.Pollastri, G., P. Baldi, P. Fariselli, and R. Casadio. 2001. Improved prediction of the number of residue contacts in proteins by recurrent neural networks. Bioinformatics 17(Suppl. 1):S234-S242. [DOI] [PubMed] [Google Scholar]
- 36.Rachez, C., and L. P. Freedman. 2001. Mediator complexes and transcription. Curr. Opin. Cell Biol. 13:274-280. [DOI] [PubMed] [Google Scholar]
- 37.Radhakrishnan, I., G. C. Perez-Alvarado, D. Parker, H. J. Dyson, M. R. Montminy, and P. E. Wright. 1997. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions. Cell 91:741-752. [DOI] [PubMed] [Google Scholar]
- 38.Reiss, N., L. N. Llevi, S. Shacham, D. Harris, R. Seger, and Z. Naor. 1997. Mechanism of mitogen-activated protein kinase activation by gonadotropin-releasing hormone in the pituitary of alphaT3-1 cell line: differential roles of calcium and protein kinase C. Endocrinology. 138:1673-82. [DOI] [PubMed] [Google Scholar]
- 39.Renaud, J. P., and D. Moras. 2000. Structural studies on nuclear receptors. Cell. Mol. Life Sci. 57:1748-1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Renaud, J. P., N. Rochel, M. Ruff, V. Vivat, P. Chambon, H. Gronemeyer, and D. Moras. 1995. Crystal structure of the RAR-gamma ligand-binding domain bound to all-trans retinoic acid. Nature 378:681-689. [DOI] [PubMed] [Google Scholar]
- 41.Richards, J. S. 2001. New signaling pathways for hormones and cyclic adenosine 3′,5′-monophosphate action in endocrine cells. Mol. Endocrinol. 15:209-218. [DOI] [PubMed] [Google Scholar]
- 42.Robinson-Rechavi, M., A. Carpentier, M. Duffraisse, and V. Laudet. 2001. How many nuclear hormone receptors are there in the human genome? Trends Genet. 17:554-556. [DOI] [PubMed] [Google Scholar]
- 43.Rost, B. 1996. PHD: predicting one-dimensional protein structure by profile-based neural networks. Methods Enzymol. 266:525-539. [DOI] [PubMed] [Google Scholar]
- 44.Rost, B., and C. Sander. 1994. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins 19:55-72. [DOI] [PubMed] [Google Scholar]
- 45.Samuels, M. L., M. J. Weber, J. M. Bishop, and M. McMahon. 1993. Conditional transformation of cells and rapid activation of the mitogen-activated protein kinase cascade by an estradiol-dependent human raf-1 protein kinase. Mol. Cell. Biol. 13:6241-6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shao, D. L., and M. A. Lazar. 1999. Modulating nuclear receptor function: may the phos be with you. J. Clin. Investig. 103:1617-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stehlin, C., J. M. Wurtz, A. Steinmetz, E. Greiner, R. Schule, D. Moras, and J. P. Renaud. 2001. X-ray structure of the orphan nuclear receptor RORbeta ligand-binding domain in the active conformation. EMBO J. 20:5822-5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Toney, J. H., L. Wu, A. E. Summerfield, G. Sanyal, B. M. Forman, J. B. Zhu, and H. H. Samuels. 1993. Conformational changes in chicken thyroid hormone receptor-alpha-1 induced by binding to ligand or to DNA. Biochemistry 32:2-6. [DOI] [PubMed] [Google Scholar]
- 49.Tremblay, A., G. B. Tremblay, F. Labrie, and V. Giguere. 1999. Ligand-independent recruitment of SRC-1 to estrogen receptor beta through phosphorylation of activation function AF-1. Mol. Cell 3:513-519. [DOI] [PubMed] [Google Scholar]
- 50.Vriend, G. 1990. WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 8:52-56. [DOI] [PubMed] [Google Scholar]
- 51.Vriend, G., and V. Eijsink. 1993. Prediction and analysis of structure, stability and unfolding of thermolysin-like proteases. J. Comput. Aided Mol. Des. 7:367-396. [DOI] [PubMed] [Google Scholar]
- 52.Wagner, R. L., J. W. Apriletti, M. E. McGrath, B. L. West, J. D. Baxter, and R. J. Fletterick. 1995. A structural role for hormone in the thyroid hormone receptor. Nature. 378:690-697. [DOI] [PubMed] [Google Scholar]
- 53.Wagner, R. L., B. R. Huber, A. K. Shiau, A. Kelly, S. T. Cunha Lima, T. S. Scanlan, J. W. Apriletti, J. D. Baxter, B. L. West, and R. J. Fletterick. 2001. Hormone selectivity in thyroid hormone receptors. Mol. Endocrinol. 15:398-410. [DOI] [PubMed] [Google Scholar]
- 54.Webb, P., C. M. Anderson, C. Valentine, P. Nguyen, A. Marimuthu, B. L. West, J. D. Baxter, and P. J. Kushner. 2000. The nuclear receptor corepressor (N-CoR) contains three isoleucine motifs (I/LXXII) that serve as receptor interaction domains (IDs). Mol. Endocrinol. 14:1976-1985. [DOI] [PubMed] [Google Scholar]
- 55.Webb, P., P. Nguyen, J. Shinsako, C. Anderson, W. Feng, M. P. Nguyen, D. Chen, S. M. Huang, S. Subramanian, E. McKinerney, B. S. Katzenellenbogen, M. R. Stallcup, and P. J. Kushner. 1998. Estrogen receptor activation function 1 works by binding p160 coactivator proteins. Mol. Endocrinol. 12:1605-1618. [DOI] [PubMed] [Google Scholar]
- 56.Weigel, N. L. 1996. Steroid hormone receptors and their regulation by phosphorylation. Biochem J. 319:657-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu, Y., P. Delerive, W. W. Chin, and T. P. Burris. 2002. Requirement of helix 1 and the AF-2 domain of the thyroid hormone receptor for coactivation by PGC-1. J. Biol. Chem. 277:8898-8905. [DOI] [PubMed] [Google Scholar]
- 58.Yang, L., J. Guerrero, H. Hong, D. B. DeFranco, and M. R. Stallcup. 2000. Interaction of the tau2 transcriptional activation domain of glucocorticoid receptor with a novel steroid receptor coactivator, Hic- 5, which localizes to both focal adhesions and the nuclear matrix. Mol. Biol. Cell 11:2007-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]