Abstract
Rsp5p is an ubiquitin-protein ligase of Saccharomyces cerevisiae that has been implicated in numerous processes including transcription, mitochondrial inheritance, and endocytosis. Rsp5p functions at multiple steps of endocytosis, including ubiquitination of substrates and other undefined steps. We propose that one of the roles of Rsp5p in endocytosis involves maintenance and remodeling of the actin cytoskeleton. We report the following. (i) There are genetic interactions between rsp5 and several mutant genes encoding actin cytoskeletal proteins. rsp5 arp2, rsp5 end3, and rsp5 sla2 double mutants all show synthetic growth defects. Overexpressed wild-type RSP5 or mutant rsp5 genes with lesions of some WW domains suppress growth defects of arp2 and end3 cells. The defects in endocytosis, actin cytoskeleton, and morphology of arp2 are also suppressed. (ii) Rsp5p and Sla2p colocalize in abnormal F-actin-containing clumps in arp2 and pan1 mutants. Immunoprecipitation experiments confirmed that Rsp5p and Act1p colocalize in pan1 mutants. (iii) Rsp5p and Sla2p coimmunoprecipitate and partially colocalize to punctate structures in wild-type cells. These studies provide the first evidence for an interaction of an actin cytoskeleton protein with Rsp5p. (iv) rsp5-w1 mutants are resistant to latrunculin A, a drug that sequesters actin monomers and depolymerizes actin filaments, consistent with the fact that Rsp5p is involved in actin cytoskeleton dynamics.
The ubiquitination pathway, by the action of the cascade of the E1 ubiquitin-activating enzyme, the E2 ubiquitin-conjugating enzymes and the E3 ubiquitin ligases, activates and transfers ubiquitin to target proteins. Rsp5p of Saccharomyces cerevisiae belongs to the Nedd4-like hect domain-containing family of E3 proteins (24, 30). In addition to the catalytic hect domain, Rsp5p possesses a C2 domain, responsible for binding Ca2+, lipids and proteins, and three WW domains that mediate protein-protein interactions. Rsp5p interacts with itself in the two-hybrid system and therefore is likely to be a multimeric protein (13). It is localized in uniformly distributed punctate complexes in cells and cofractionates with a nonnuclear, nonmitochondrial organellar fraction (18). Electron microscopy studies indicated that some of these complexes are located at plasma membrane invaginations, likely sites of endocytic vesicle formation, and at the endosome (62).
The best-documented role for Rsp5p is in endocytosis, a process that internalizes solutes, receptors, and plasma membrane transporters via endocytic vesicles and early and late endosomes, thereby delivering the cargo to the vacuole (51). Rsp5p ubiquitinates and down-regulates plasma membrane transporters such as Gap1p, the general amino acid permease (25), Fur4p, the uracil permease (19), and Tat2p, the tryptophan permease (4), and receptors such as Ste2p, the α-factor receptor (14), and Ste3p, the a-factor receptor (50). Ubiquitination of these proteins leads to their internalization and subsequent degradation in the vacuole. Despite this well-defined role in the ubiquitination of substrates resulting in their endocytosis, it seems clear that Rsp5p must play additional roles in later steps of endocytosis. The strongest support for this is the fact that Rsp5p is required for fluid-phase endocytosis, a process not involving cargo ubiquitination (13, 18, 73).
Rsp5p also plays less well characterized roles in other processes including transcription, mitochondrial inheritance, the mitochondrial-cytoplasmic distribution of proteins, minichromosome maintenance, response to anesthetics, regulation of cellular pH, and biosynthesis of unsaturated fatty acids (11, 16, 28, 30, 33, 68, 70, 72). Recent reports also document that Rsp5p-dependent ubiquitination is involved in the vesicular traffic step of sorting of several amino acid permeases at the Golgi apparatus (4, 26, 54). The mechanisms by which Rsp5p affects these various processes remain to be delineated.
Endocytosis requires a properly functioning actin cytoskeleton (21). Specifically, rapid actin filament turnover is necessary (5, 34). Actin cortical patches, major cytoskeletal structures associated with invaginations of the plasma membrane (43), are also essential for normal endocytosis (47). Many cortical-patch components are known, but how they affect endocytosis is not well understood. Mutations of genes encoding actin (ACT1), numerous actin binding proteins, and proteins affecting the actin cytoskeleton, including ARP2, BEE1, VRP1, SLA2, PAN1, END3, and SLA1, cause defects in endocytosis, in addition to causing aberrant cytoskeleton organization.
Arp2p is a component of the Arp2/3 multimeric protein complex crucial for actin dynamics and organization. The Arp2/3 complex from S. cerevisiae consists of seven subunits: two actin-related proteins, Arp2p and Arp3p, and five other proteins, Arc15p, Arc18p, Arc19p, Arc35p, and Arc40p. Of these, only Arc40p has an essential function (67). The Arp2/3 complex functions in actin binding, nucleation of actin polymerization, pointed-end capping, branching of actin filaments, and actin patch movement (44). The nucleating activity of the Arp2/3 complex is activated by Bee1p (66), Myo3p, Myo5p (15, 20, 35), and Abp1p (22). Very recently, Pan1p also was shown to bind and activate the Arp2/3 complex, indicating that Pan1p might recruit the Arp2/3 complex to the endocytic machinery (12). Pan1p interacts with multiple proteins involved in endocytosis, including Sla1p, End3p (59), and several clathrin assembly proteins (63, 64). Some of these interactions are regulated by phosphorylation by Prk1p kinase (71). Sla2p binds to F-actin (40) and is also involved in actin polymerization (37). Sla2p binds Ark1p kinase, which is implicated in signaling to the actin cytoskeleton (10). The combined data have led to the hypothesis that the actin cytoskeleton might function in endocytosis via endocytic vesicle formation, scission, or pinching off the plasma membrane (32, 48). However, the fundamental mechanism underlying actin cytoskeleton function in endocytosis is not fully understood.
Our previous observations indicated that Rsp5p might affect the actin cytoskeleton. RSP5 was identified in the same mutant selection as were VRP1 and PAN1 (73), both encoding actin cytoskeletal proteins. Additionally, rsp5 vrp1 and rsp5 pan1 double mutants showed synthetic growth defects or synthetic lethality (72). Here we provide several lines of evidence supporting the hypothesis that Rsp5p functions in actin cytoskeleton maintenance and dynamics. The data lead us to propose that the function of Rsp5p in the actin cytoskeleton may be important for fluid-phase endocytosis and the steps of endocytosis unrelated to plasma membrane cargo protein ubiquitination. We show the existence of synthetic growth defects of rsp5 with many actin cytoskeleton mutants, suppression of growth defects of some cytoskeletal mutants by an additional copy of the RSP5 gene, and localization of Rsp5p in abnormal actin clumps in these mutants. Furthermore, we also show colocalization and coimmunoprecipitation of Sla2p with Rsp5p in wild-type cells and resistance to LAT-A indicating alteration of actin cytoskeleton dynamics in the rsp5-w1 mutant.
MATERIALS AND METHODS
Strains, media, and growth conditions.
The S. cerevisiae strains used are described in Table 1. YEPD, YEPG, SC, and sporulation media were prepared and genetic manipulations were performed as described by Sherman (53). Yeast strains were transformed by the method of Chen et al. (9). Suppression of the temperature-sensitive phenotype of various mutants by YCpHA-RSP5 was monitored by a drop test. Appropriate transformants were suspended in water, and serial 10-fold dilutions were plated on YEPD. Growth was observed after 2 days of incubation at 34, 37, or 39°C, as indicated. Escherichia coli strain DH5αF′ [F′ supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1] was used for cloning and plasmid propagation.
TABLE 1.
List of S. cerevisiae strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| T8-1D | MATα SUP11 ade2-1 mod5-1 ura3-1 lys2-1 leu2-3,112 his4-519 | 73 |
| TZ23 | MATα SUP11 rsp5-13 ade2-1 mod5-1 ura3-1 lys2-1 leu2-3,112 his4-519 | 72 |
| TZ81 | MATα SUP11 pan1-9 ade2-1 mod5-1 ura3-1 lys2-1 leu2-3,112 his4-519 | 73 |
| BG1 | MATα SUP11 rsp5-Δ ade2-1 mod5-1 ura3-1 lys2-1 leu2-3, 112 his4-519 p[HA-RSP5, CEN, LEU2] | 18 |
| BG2 | MATα SUP11 rsp5-Δ ade2-1 mod5-1 ura3-1 lys2-1 leu2-3, 112 his4-519 p[HA-rsp5-w1, CEN, LEU2] | 18 |
| BG3 | MATα SUP11 rsp5-Δ ade2-1 mod5-1 ura3-1 lys2-1 leu2-3, 112 his4-519 p[HA-rsp5-w2, CEN, LEU2] | 18 |
| BG4 | MATα SUP11 rsp5-Δ ade2-1 mod5-1 ura3-1 lys2-1 leu2-3, 112 his4-519 p[HA-rsp5-w3, CEN, LEU2] | 18 |
| T82-14C | MATa rsp5-13 SUP11 ade2-1 mod5-1 ura3-1 lys2-1 leu1 trp5 | This work |
| SS330 | MATa ade2-101 his3-200 tyr1 ura3-52 | 74 |
| ts817 | MATa ade2-101 his3-200 tyr1 ura3-52 sla2ts | 74 |
| RH144-3D | MATa his4 ura3 leu2 bar1-1 | 49 |
| RH266-1D | MATa his4 ura3 leu2 bar1-1 end3-1 | 49 |
| YPH500 | MATα ade2-101 his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 | 42 |
| YMW82 | MATα ade2-101 his3-Δ200, leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 arp2-1 | 42 |
| MK1 | MATα his3-Δ200 leu2-3, 112 ura3-52 lys2-801 trp1-1 RSP5::HA-RSP5 | M. Kwapisz, unpublished allele replacement |
| MK2 | MATα his3-Δ200 leu2-3, 112 ura3-52 lys2-801 trp1-1 rsp5::HA-rsp5-w1 | M. Kwapisz, unpublished allele replacement |
| MK4 | MATα his3-Δ200 leu2-3, 112 ura3-52 lys2-801 trp1-1 rsp5::HA-rsp5-w3 | M. Kwapisz, unpublished allele replacement |
| MK5 | MATα his3-Δ200 leu2-3, 112 ura3-5 lys2-801 trp1-1 rsp5::HA-rsp5-13 | M. Kwapisz, unpublished allele replacement |
| Y23725 | MAT a/α his3-Δ1/his3-Δ1 leu2-Δ0/leu2-Δ0 lys2-Δ0/LYS2 met15-Δ0/MET15 ura3-Δ0/ura3-Δ0 arp2-Δ(YDL029w::kan/MXY)/ARP2 | Euroscarf |
| Y129912 | MAT α his3-Δ1 leu2-Δ0 lys2-Δ0 ura3-Δ0 end3-Δ (YNL084c::kanMXY) | Euroscarf |
Plasmids and plasmid constructions.
Standard nucleic acid manipulations and plasmid propagations in E. coli were performed as described by Sambrook et al. (52). The plasmids used were YCpHA-RSP5, YCpHA-rsp5-w1, YCpHA-rsp5-w2, YCpHA-rsp5-w3 (18), RB2 (72), and p195gf (61). Plasmid YCpHA-rsp5-13 was constructed by replacing the KpnI-KpnI fragment of YCpHA-RSP5 with a DNA fragment generated by PCR using genomic DNA from the rsp5-13 strain as a template. The DNA sequence of the PCR fragment was confirmed.
Cellular extracts and immunoblot analysis.
Total protein extracts were prepared from T8-1D, BG1, BG2, BG3, and BG4 for immunoprecipitation of HA-Rsp5p and Sla2p and from TZ81 YCpHA-RSP5 for actin immunoprecipitation. Cells were collected by a 5-min centrifugation at 3,000 × g, washed with ice-cold water, and suspended in an equal volume of cold buffer (100 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM EDTA, protease inhibitor mix Complete [Boehringer Mannheim], phenylmethylsulfonyl fluoride). An equal volume of glass beads was added, and the cells were broken by vortexing eight times for 30 s with 30-s intervals at 4°C. To remove cell debris and unbroken cells, the samples were centrifuged for 5 min at 4°C at 3,000 × g. Cellular extracts to study Fur4p degradation were prepared by alkali lysis (61). Immunoblot analysis was performed as previously described (7). The mouse monoclonal antibodies employed were anti-HA, clone 16B12 and clone 12CA5 (BabCo), and anti-actin clone C4 (ICN). Rabbit polyclonal antibodies used were anti-Sla2p from D. Drubin. Primary antibodies were detected with horseradish peroxidase-conjugated goat anti-mouse or anti-rabbit secondary antibodies followed by enhanced chemiluminescence (Amersham).
Endocytosis assay.
Internalization of uracil permease (Fur4p) was observed by monitoring uracil uptake in cells expressing Fur4p from p195gF containing a PGAL10-FUR4 promoter fusion (61). Cells grown at 30°C in galactose-containing medium were shifted to 34°C for 2 h, and samples were taken at various times after cycloheximide addition. A 1-ml volume of yeast culture was incubated with 5 μM [14C]uracil (Amersham) for 20 s at 37°C, filtered through Whatman GF/C filters, and washed twice with ice-cold water, and the radioactivity was measured.
Immunoprecipitation.
A 100-μl volume of cellular extract was incubated with TNET buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM EDTA, 1% Triton X-100), 16B12 anti-HA antibody or anti-myc antibody, and protein A/G agarose beads (Santa Cruz) overnight at 4°C. The precipitated material was pelleted by centrifugation, washed three times with TNET buffer, and solubilized by suspension of the beads in 2× sodium dodecyl sulfate-containing gel-loading buffer (52). The unbound fraction was precipitated with trichloroacetic acid (final concentration, 5%). The trichloroacetic acid pellet was washed with 1 M Tris base and suspended in gel-loading buffer. Total, immunoprecipitated, and unbound proteins were analyzed by immunoblotting.
Immunofluorescence.
Indirect-immunofluorescence microscopy was performed by modification of a previously described procedure (46). Cells were fixed in culture by the addition of formaldehyde to a final concentration of 3.7% for 1 h. Fixed cells were collected by centrifugation, washed, and then converted into spheroplasts. The spheroplasts were washed and applied to microscopic slides coated with poly-l-lysine. For staining of actin cytoskeletal proteins, the slides were additionally treated at −20°C with cold methanol for 6 min and then at −20°C with cold acetone for 30 s. For double immunofluorescence, primary antibodies and the appropriate secondary antibodies were applied together. The antibodies used were anti-HA 16B12 (BabCo), anti-Sla2p-2L and anti-cofilin (both from D. Drubin), and anti-mouse Cy3-conjugated and anti-rabbit fluorescein isothiocyanate-conjugated secondary antibodies from Jackson ImmunoResearch Laboratories, Inc. Cells were viewed by fluorescence microscopy using a Microphot-SA (Nikon). Images were collected using a Photometrix CH350A camera with QED or Lucia G software and processed using Photoshop 6.0 (Adobe) software.
Actin staining.
For actin cytoskeleton staining, we employed a modification of a previously published procedure (1). Cultures (10 ml) were grown overnight in YEPD to an optical density at 600 nm of 0.5. In some experiments, half of the culture was shifted to 37°C for 2 h (see Results) and then shifted to 34°C for 4 h. The cells were fixed for 2 h by addition of formaldehyde in K3PO4 (pH 6.5) to a final concentration of 3.7%. After fixing, the cells were collected by centrifugation, washed three times in 1× phosphate-buffered saline (PBS) (pH 6.5) (52), and then stained with Oregon Green 488-conjugated phalloidin or rhodamine-conjugated phalloidin (Molecular Probes) for 2 h in the dark with gentle agitation. The cells were washed five times with PBS (pH 6.5), stained with 4′,6-diamidino-2-phenylindole (DAPI; 0.5 μg/ml), washed twice with PBS, and resuspended in mounting medium. For combined actin staining and immunofluorescence, the cells were treated as described by Vaduva et al. (60).
LAT-A sensitivity.
Sensitivity to latrunculin A (LAT-A) was assessed by the halo assay (2). Strains MK1, MK2, MK4, and MK5 were grown at 30°C overnight in YEPD. A 10-μl volume of each culture was added to 2 ml of 2× YEPD, which was then mixed with 2 ml of 1% chilled molten agar. The cell suspension was poured onto the surface of a YEPD plate. Four sterile disks were placed on each plate. An equal volume of different dilutions of LAT-A solution was applied to the disks. The plates were incubated for 2 days at 30°C, at which time zones of growth inhibition were observed.
RESULTS
Genetic interactions between RSP5 and genes encoding actin cytoskeletal proteins indicate a role for Rsp5p in the actin cytoskeleton. (i) Synthetic growth defects between rsp5 and mutant genes encoding cytoskeletal proteins.
Synthetic lethality or synthetic growth defects between various mutant genes provide a valuable tool for uncovering protein products that physically interact or function in parallel pathways (23). Our previous studies documented multiple roles for Rsp5p in endocytosis (18) and provided the first genetic indication that Rsp5p might function in the actin cytoskeleton (72). To test the hypothesis that one of the undefined roles for Rsp5p in endocytosis might be to modulate the actin cytoskeleton, we assessed genetic interactions between rsp5 and temperature-sensitive alleles of arp2, end3, or sla2. Arp2p and Sla2p are directly involved in actin-filament nucleation and polymerization (37, 44). End3p interacts physically with Pan1p (58), and Pan1p binds and activates Arp2/3 actin-nucleating complex (12).
Strains containing the rsp5-13 hect domain mutation (72) were mated to strains with the arp2-1, end3-1 or sla2ts817 mutation (42, 49, 74). The resulting diploids were sporulated, tetrads were dissected, and the phenotype of haploid progeny with two mutations was compared with the phenotype of cells containing single mutations and the reference parental strains. rsp5-13 arp2-1 double-mutant cells grew more poorly than cells with the respective single mutations at 34°C on YEPD, indicating a synthetic growth defect for these mutations (Fig. 1A). The rsp5-13 end3-1 mutant was viable, but it grew significantly slower on YEPD at 30°C than did either single mutant (not shown), and it did not grow at 35°C on YEPD (Fig. 1B). Thus, rsp5-13 shows an additive growth defect with end3-1. Likewise, a synthetic growth defect between rsp5-13 and sla2ts817 was observed, since double-mutant progeny could not grow at 30°C (Fig. 1C). Similar synthetic growth defects were observed when arp2, end3, or sla2 was combined with an RSP5 allele with a mutation in the third WW domain (rsp5-w3 [data not shown]). In contrast, we did not observe an additive growth defect of rsp5 and clathrin deficiency in control crosses of rsp5 with a strain harboring the chc1::GAL1-CHC1 promoter fusion (from S. Lemmon) on glucose-containing media (not shown). The genetic interactions between rsp5 and arp2, end3, or sla2 indicate Rsp5p function in the actin cytoskeleton.
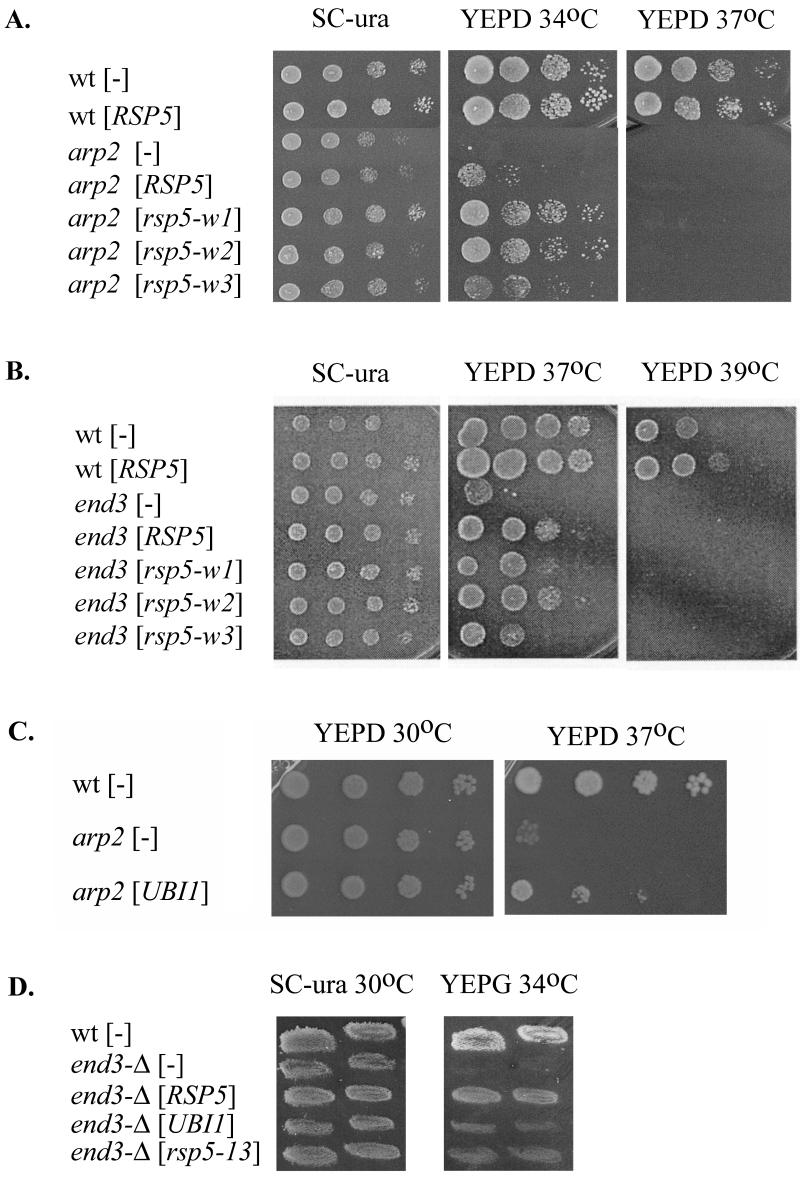
FIG. 1.
Synthetic growth defect caused by a combination of rsp5-13 with arp2, end3, and sla2 mutations. Growth of spore clones from the crosses rsp5-13 × arp2-1 (A), rsp5-13 × end3-1 (B), and rsp5-13 ×sla2ts (C) was tested on YEPD plates incubated at various temperatures. Growth was observed after 2 to 3 days. Note that some spore clones are red, and they display differently from white ones.
(ii) An additional copy of RSP5 suppresses growth defects of arp2 and end3.
Increased expression of some proteins may affect the activity of a second protein of interest by direct physical interaction, by a change in intracellular localization, or by posttranslational modification. For example, a single additional copy of ACT1, encoding actin, was able to suppress vrp1 (73) and led to the verified model that these two proteins physically interact (60). To obtain further information regarding the role of Rsp5p in the actin cytoskeleton and endocytosis, the effect of an additional copy of RSP5 or rsp5 on the temperature-sensitive growth of arp2-1, end3-1, sla2ts817, and pan1-9 mutants (73) was assessed.
The mutants were transformed with YCp plasmids bearing RSP5, rsp5-w or rsp5-13 under the control of the natural RSP5 promoter. rsp5-w alleles possess mutations of individual Rsp5p WW domains and were designed to abolish interactions of Rsp5p WW domains with other proteins (18). The rsp5-13 allele contains a mutation of the Rsp5p hect domain (72). The growth rates of transformants were tested at various temperatures. An additional copy of RSP5 or mutant rsp5 did not influence the growth of cells containing sla2ts817 or pan1-9 (data not shown). However, YCpHA-RSP5 or YCpHA-rsp5-w plasmids suppressed the growth defects at 34°C of the arp2-1 mutant (Fig. 2A). Suppression was most efficient for the rsp5-w1 and rsp5-w2 alleles, and RSP5 and rsp5-w3 were less effective. Western blot analysis showed that mutant Rsp5p-w is stable when cells are cultured at temperatures below 37°C (references 13 and 18 and data not shown). YCpHA-RSP5 and YCpHA-rsp5-w plasmids also suppressed growth defects of end3-1 at 37°C. In this case, end3-1 suppression by rsp5-w3 was less pronounced than suppression by RSP5, rsp5-w1, and rsp5-w2 alleles (Fig. 2B). For both arp2-1 and end3-1, suppression was partial as the transformants were unable to grow at elevated temperatures (37 or 39°C; Fig. 2, right panels) at which wild-type strains grow. YCpHA-rsp5-13 did not suppress arp2-1 (not shown), indicating that the catalytic activity of Rsp5p is necessary for suppression. Further support for this conclusion comes from observation that overexpression of the UBI1 gene, encoding ubiquitin, also suppressed the arp2-1 growth defect with high efficiency at 37°C (Fig. 2C). Taken together, the data indicate that small increases in Rsp5p cellular levels provide functions that are impaired in these mutants that affect the actin cytoskeleton and that ubiquitin-protein ligase activity is necessary for at least arp2 suppression.
FIG. 2.
(A and B) An extra copy of RSP5 can partially suppress the growth defect caused by arp2 (A) and end3 (B). Temperature-sensitive arp2-1 and end3-1 strains were transformed with plasmids carrying RSP5 or rsp5-w genes or vector. (C) The arp2-1 strain was transformed with plasmid bearing UBI1 (RB2) or vector. Serial dilutions (1:10 steps) of transformants were spotted on YEPD and SC-Ura plates and allowed to grow at the indicated temperatures for 3 days. Growth was compared to that of the parental strain. (D) The end3-Δ strain was transformed with YCpHA-RSP5, YCpHA-rsp5-13, a plasmid bearing UBI1 (RB2), or vector. Transformants were replica plated on SC-Ura or YEPG and incubated at 30 or 34°C for 3 days.
To determine whether increased Rsp5p suppresses the various mutations of the actin cytoskeletal components via physical interactions with the mutant proteins or, instead, by bypassing their function by stimulating a parallel pathway, we tested whether arp2-Δ or end3-Δ deletion mutations can be suppressed by a low-copy-number ectopic RSP5. Dissection of tetrads derived from the diploid arp2-Δ/+ (Y23725; Euroscarf) yielded only two viable spores, confirming other reports that the ARP2 gene is essential in some backgrounds (29, 66). The diploid arp2-Δ/+ was transformed with YCpHA-RSP5 and sporulated, and the resulting tetrads were dissected and grown on YEPD at 23°C. Under these conditions, several tetrads yielded four viable spores, two that grew as well as wild-type strains and two that grew very poorly (data not shown). The data indicate that the arp2-Δ deletion is partially suppressed by an additional copy of RSP5. The haploid end3-Δ strain (Y12992; Euroscarf) was used to test suppression by RSP5 since END3 is nonessential (6). Transformants of end3-Δ bearing YCpHA-RSP5 grew better than end3-Δ with vector alone at 34°C on glycerol-containing medium (Fig. 2D). The END3 deletion was also suppressed by the rsp5-13 hect mutant gene, albeit less efficiently. UBI1 also suppressed the end3-Δ growth defect at 34°C but did so weakly compared to suppression by RSP5. Since an additional copy of RSP5 suppresses null arp2 and end3 mutations, suppression is not due to direct interactions of Rsp5p with Arp2p and End3p but, rather, to interactions with other actin cytoskeleton components. Taken together, the results provide support for the hypothesis that Rsp5p functions in actin cytoskeleton organization, modeling, or actin polymerization.
(iii) Low-copy-number vectors with rsp5 suppress the actin cytoskeleton and endocytosis defects of arp2-1.
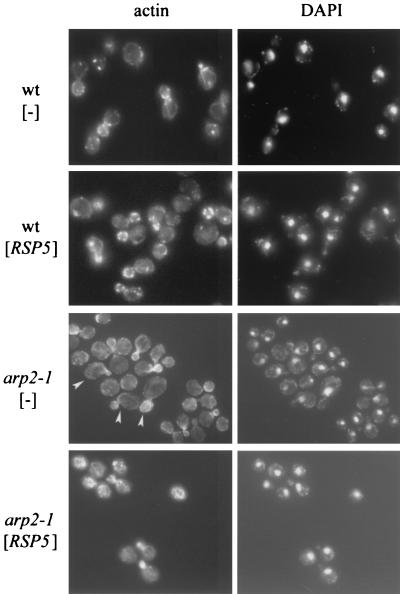
In addition to temperature-sensitive growth, mutant arp2-1 cells have abnormal actin distribution, especially if the cells are incubated at elevated temperatures (42). To determine whether additional Rsp5p corrects the defects in actin organization and morphology, arp2-1 cells containing vector alone or YCpHA-RSP5 were grown at 30°C and shifted for 2 h to 37°C and then for 4 h to 34°C (the temperature at which suppression of growth was observed). Cortical patches were smaller for arp2-1 than for wild-type cells, but cytoplasmic cables were visible. In addition, arp2-1 mother cells often were bigger and mother cells and buds had an abnormal shape (asymmetric cells, beak-shaped buds [Fig. 3]). The abnormal buds contained brightly staining clumps of filamentous actin. In contrast, arp2 cells bearing YCpHA-RSP5 had wild-type morphology and polarized distribution of cortical patches between the mother and the bud (Fig. 3). Therefore, an increase in Rsp5p levels corrects the actin cytoskeleton defects and confers normal shape on arp2-1 cells at elevated temperatures.
FIG. 3.
An additional copy of RSP5 corrects the arp2-1 defects in actin cytoskeleton and morphology. The arp2-1 mutant and parental strain were transformed with vector or YCpHA-RSP5 plasmid. Transformants were grown at 30°C and shifted to 37°C for 2 h and then to 34°C for 4 h. Actin was stained with phalloidin. Cellular DNA was stained with DAPI. Cells that are bigger and have abnormal shape are indicated by arrowheads.
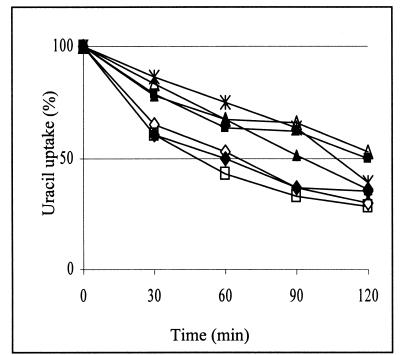
Defects in the actin cytoskeleton often result in abnormal endocytosis (21), and this was observed also in arp2 cells since they are defective in endocytosis of the plasma membrane protein uracil permease, Fur4p (41). To determine whether suppression of the actin cytoskeleton defects in arp2 cells also corrects the defects in endocytosis, we studied the internalization of overexpressed Fur4p in the presence or absence of plasmids encoding wild-type or mutant Rsp5p. The rate of uracil uptake, indicative of uracil permease activity at the plasma membrane, was measured after cycloheximide addition. The arp2 mutant showed retarded decrease of uracil uptake (Fig. 4), documenting defects in Fur4p internalization described previously (41). This delay was also observed in cells containing YCpHA-rsp5-w3, but the phenotype was partially suppressed by YCpHA-RSP5 or YCpHA-rsp5-w1. Moreover, we observed nearly normal internalization of Fur4p in cells with YCpHA-rsp5-w2. In the control experiment we observed that an additional copy of RSP5 did not significantly affect the Fur4p internalization rate for wild-type cells (Fig. 4). These results do not completely parallel the results of suppression of arp2-1 growth defects. For example, rsp5-w1 and rsp5-w2, which suppress the growth defect of arp2-1 at 34°C equally well, affect Fur4p internalization differently. Possibly, the actin cytoskeleton malfunction in arp2-1 affects other processes (mitochondrial inheritance, for example) that are corrected differently by various rsp5-w mutant genes. However, the excellent suppression by rsp5-w2 of Fur4p internalization defects of arp2 is in line with the previous finding that rsp5-w2 does not disrupt endocytic machinery, since it does not cause defects in fluid-phase endocytosis (18). Suppression of the actin cytoskeleton defects of arp2-1 by YCpHA-RSP5 is consistent with its suppression of endocytosis defects.
FIG. 4.
RSP5 and rsp5-w2 suppress endocytosis defects of arp2-1 cells. The wild-type strain bearing vector (◊) or YCpHA-RSP5 (⧫) and the arp2-1 strain bearing vector (▪), YCpHA-RSP5 (▴), YCpHA-rsp5-w1 (*), YCpHA-rsp5-w2 (□), or YCpHA-rsp5-w3 (Δ) and plasmid p195gF were grown at 30°C with galactose as a carbon source and shifted to 34°C for 2 h. Cycloheximide (100 mg/ml) was then added to the media. Uracil uptake at the times indicated after addition of cycloheximide was measured. Representative results of three independent experiments are shown.
Normal subcellular distribution of Rsp5p depends on a functional actin cytoskeleton.
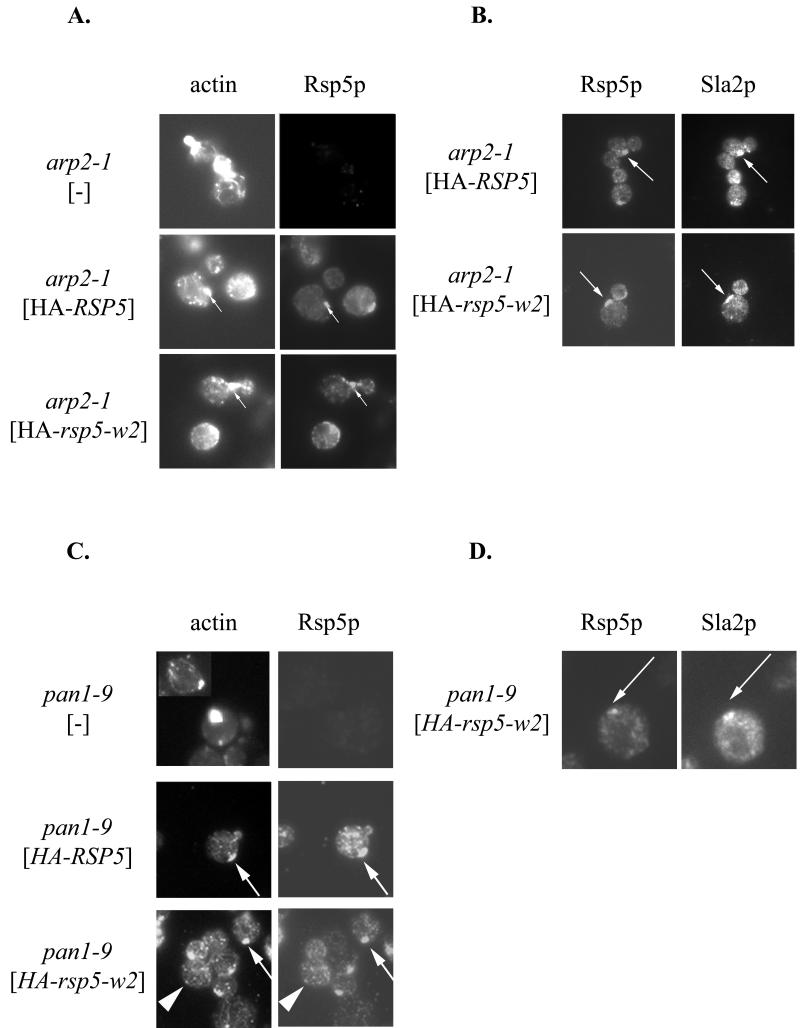
The above genetic studies indicate that there are interactions between Rsp5p and components of the actin cytoskeleton. If the interactions are direct, one would predict that abnormalities in actin cytoskeleton organization would affect Rsp5p localization. We tested whether arp2-1, end3-1, and sla2ts817 mutations affect Rsp5p subcellular distribution. YCpHA-RSP5 and YCpHA-rsp5-w2 were expressed in cells with the wild-type RSP5 chromosomal gene and a mutation of arp2, end3, or sla2. Immunofluorescence examination of HA-Rsp5p localization in arp2-1 grown at 30°C showed that the distribution was abnormal. In addition to small punctate structures, characteristic of Rsp5p localization (18), clumps of Rsp5p, often close to the plasma membrane, were evident in a fraction of cells (Fig. 5A). Abnormal actin clumps in mother cells are characteristic of some arp2 mutants (39), but for arp2-1 mutant cells the clumps have been reported to be in buds (42). Therefore, we used double staining to test if HA-Rsp5p clumps present in arp2-1 cells also contain actin (see Materials and Methods). Clumps of concentrated HA-Rsp5p in arp2 cells partially colocalized with actin clumps (Fig. 5A). Actin clumps in mother cells of arp2-1 mutants observed in this experiment could result from overexpression of RSP5, since they were not observed in arp2-1 cells without YCpHA-RSP5 plasmid. Examination of end3 and sla2 cells expressing HA-RSP5 revealed changes in HA-Rsp5p location. As with arp2 cells, clumps that contained HA-Rsp5p were evident (not shown), but these were not characterized further.
FIG. 5.
Mislocation of Rsp5p in mutants with defective actin cytoskeletons. (A) Rsp5p is localized in clumps that contain F-actin in the arp2-1 mutant. The arp2-1 strain was transformed with YCpHA-RSP5 or YCpHA-rsp5-w2, and the transformants were processed for immunofluorescence using anti-HA antibody and stained with phalloidin. The structures containing HA-Rsp5p and actin are indicated by arrows. (B) Sla2p and Rsp5p colocalize in arp2-1 mutant cells. Transformants were double stained with anti-HA and anti-Sla2p antibody. The structures containing HA-Rsp5p and Sla2p are indicated by arrows. (C) Rsp5p is present in clumps that contain actin in pan1-9 cells. Plasmids bearing HA-RSP5 or HA-rsp5-w were transformed into the pan1-9 mutant. Cells were stained by indirect immunofluorescence using the anti-HA antibody and subsequently stained with phalloidin. The clumps of concentrated HA-Rsp5p colocalizing with actin are indicated by arrows. Some normal actin patches that colocalize with punctate structures of HA-Rsp5p are indicated by arrowheads. Actin clumps are present in almost every cell expressing HA-rsp5-w2 (bottom panel). (D) Sla2p is present in the same clumps as Rsp5p in the pan1-9 mutant. Cells expressing HA-rsp5-w2 were stained by double indirect immunofluorescence using anti-HA and anti-Sla2p antibody. The dots that correspond to HA-Rsp5-w2p and Sla2p are indicated by arrows.
F-actin clumps, similar to those observed in the arp2 mutant cells, were previously reported for end3, sla2, and pan1 mutant cells (6, 27, 57) and also for double prk1-Δ ark1-Δ mutants lacking two protein kinases that regulate the cortical actin cytoskeleton (10). The composition of the actin clumps varies depending on the mutant. They can contain proteins normally found in cortical actin patches, such as cofilin, Sla2p, Sac6p, and Abp1p. To learn if actin clumps in arp2-1 cells, which contain Rsp5p, also contain proteins characteristic of actin patches, we determined the location of Sla2p and cofilin. arp2-1 cells with YCpHA-RSP5 or YCpHA-rsp5-w2 were used for double immunofluorescence with anti-HA antibodies and anti-Sla2p or anticofilin antibodies. Sla2p was present in HA-Rsp5p-containing abnormal structures in arp2 cells (Fig. 5B). In contrast, cofilin was not found in clumps containing actin and Rsp5p but in some cells the signal was in separate cofilin bars (data not shown). Similar cofilin bars were previously described in cells overproducing cofilin (31) and in arp2-Δ mutants (67).
Since some pan1 mutations cause aberrations in the actin cytoskeleton, including formation of actin clumps (57), the pan1-9 mutant strain that was isolated in the same screen as rsp5/mdp1 mutants (73) was studied in more detail. The previously studied pan1-8 mutation did not cause significant changes in the actin cytoskeleton (73). In contrast, phalloidin staining for pan1-9 cells grown at 30°C revealed clumps of polymerized actin (Figure 5C), similar to the situation reported for pan1-4 cells at 37°C (57). The pan1-9 cells containing YCpHA-RSP5 were double stained. The studies showed that actin clumps in pan1-9 cells colocalized with clumps of concentrated HA-Rsp5p (Fig. 5C). The number of cells with clumped actin increased in pan1-9 cells carrying YCpHA-RSP5 or YCpHA-rsp5-w2 (Fig. 5C and data not shown). We also observed some normal-size actin patches corresponding to punctate structures that contain HA-Rsp5p (Fig. 5C). Since the distribution of actin and Rsp5p in pan1-9 cells was similar to the distribution seen in arp2-1 mutants, we checked if the Sla2p and cofilin distribution is abnormal in pan1-9 as in arp2-1 cells. The pan1 cells containing YCpHA-rsp5-w2 were stained with the anti-HA antibody and anti-Sla2p antibody or antibody specific to cofilin. Sla2p colocalized with HA-Rsp5p and actin clumps in pan1-9 cells (Fig. 5D). Cofilin, the major actin-depolymerizing protein, did not colocalize with actin clumps in pan1-9, but instead was observed in separate cofilin bars (data not shown).
Immunoprecipitation studies were performed to confirm that HA-Rsp5p colocalizes with actin in the pan1-9 mutant. HA-Rsp5p was immunoprecipitated from total extracts with anti-HA 16B12 antibody or with a control antibody, anti-myc. Total extracts and immunoprecipitated and unprecipitated proteins were analyzed by immunoblotting using antibodies against HA and actin. HA-Rsp5p was efficiently precipitated from extracts with anti-HA antibody. In contrast, in the control immunoprecipitation with anti-myc antibody, HA-Rsp5p was found exclusively in the unprecipitated fraction (Fig. 6). Immunoblotting with the anti-actin antibody revealed that part of the total actin pool is immunoprecipitated by anti-HA antibody only from extracts that contained HA-Rsp5p. However, the major pool of cellular actin was not immunoprecipitated. The data confirm our immunofluorescence experiments showing partial colocalization of actin and Rsp5p.
FIG. 6.
Rsp5p interacts with actin in the pan1-9 mutant. Total extracts from TZ81 pan1-9 cells carrying HA-RSP5 on a centromeric plasmid or vector were used for immunoprecipitation with 16B12 anti-HA (HA) or with control anti-myc antibody (C). The blot was first incubated with anti-actin and then with anti-HA antibody to detect HA-Rsp5p. T, total extracts; IP, immunoprecipitate; S, supernatant fraction. The immunoglobulin G (IgG) light chain is indicated.
The above results indicate that in numerous mutants, arp2, end3, sla2, and pan1, HA-Rsp5p is abnormally located in clumps. More detailed studies with arp2 and pan1 mutants showed that HA-Rsp5p clumps and actin clumps are the same structures. Moreover, an additional copy of Rsp5p affects the actin cytoskeleton in these mutants by increasing the number of cells with actin clumps. Thus, a polarized actin cytoskeleton is necessary for proper cellular localization of Rsp5p, and Rsp5p affects the actin cytoskeleton.
Rsp5p colocalizes with the actin cytoskeletal component in wild-type cells.
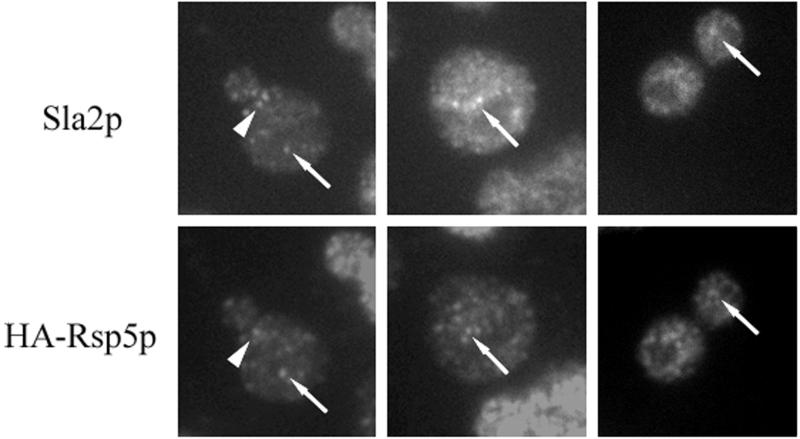
If Sla2p and Rsp5p colocalize in abnormal actin-containing clumps in cells with a disorganized actin cytoskeleton, they may interact in wild-type cells. To test interactions of Sla2p with Rsp5p, we determined the location of these proteins in cells with a single, tagged copy of RSP5 (strain BG1; relevant genotype, rsp5-Δ YCpHA-RSP5). Sla2p, similar to Rsp5p, is located in many punctate complexes in the cell (69). Inspection of the microscopic images revealed that some HA-Rsp5p and Sla2p complexes overlap (Fig. 7). Some of these complexes are polarized in dividing cells (Fig. 7). We also investigated whether the Sla2p location depends on WW domains of Rsp5p and found that in rsp5-w cells (BG2-4) the Sla2p location was not different from that in RSP5 cells (data not shown). The data indicate that Sla2p and Rsp5p partially colocalize in wild-type cells and that some of the Rsp5p pool is located in actin patches.
FIG. 7.
Rsp5p and Sla2p partially colocalize in wild-type cells. Cells were processed for double immunofluorescence with anti-Sla2p and anti-HA antibody. Dots that correspond to both proteins are indicated by arrows. A polarized dot is indicated by an arrowhead.
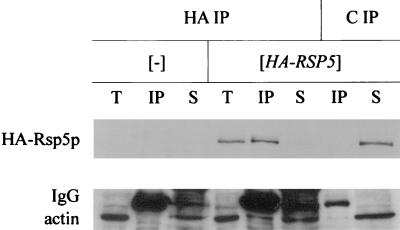
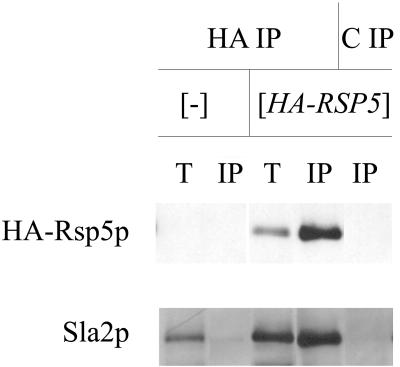
We used immunoprecipitation to verify that Rsp5p and Sla2p interact in wild-type cells. Extracts were prepared from the wild-type strain (T8-1D) and the rsp5-Δ strain containing YCpHA-RSP5 (strain BG1). Proteins from these extracts were immunoprecipitated with anti-HA antibody and control preimmune serum. Total extracts and immunoprecipitated proteins were analyzed by Western blotting. Only in the BG1 strain did the anti-HA antibody react with a protein corresponding to Rsp5p in total extracts and the immunoprecipitated fraction. Reprobing with the polyclonal anti-Sla2p antibody revealed a 116-kDa protein corresponding to Sla2p in total extracts and a pool of Sla2p in the BG1 cell extract immunoprecipitated by anti-HA antibody (Fig. 8). Analysis of immunoprecipitates from rsp5-w1 (BG2), rsp5-w2 (BG3), and rsp5-w3 (BG4) cell extracts in parallel experiments indicated that mutations in individual WW domains of Rsp5p do not prevent Sla2p binding (data not shown). In contrast to studies with pan1 mutant cells, we were unable to coimmunoprecipitate actin and Rsp5p from extracts obtained from wild-type cells.
FIG. 8.
Sla2p forms a complex with HA-Rsp5p. Total extracts from wild-type T8-1D (−) and rsp5-Δ-bearing YCpHA-RSP5 (HA-RSP5) cells were used for immunoprecipitation using anti-HA 16B12 antibody (HA) and control preimmune serum (C). Western blot analyses with anti-Sla2p and with anti-HA antibody were done on the same blot. T, total extracts; IP, immunoprecipitate. One-quarter of the total extract used in immunoprecipitation was loaded onto the gel.
Our data documenting Rsp5p and Sla2p colocalization and coimmunoprecipitation provide the first example of an interaction in the same complex of an actin binding protein with Rsp5p, a ubiquitin ligase. Further investigation is required to determine whether Sla2p interacts directly or indirectly with Rsp5p and whether Sla2p is a substrate for ubiquitination. The data further support the notion that WW domains of Rsp5p are not required for interaction and colocalization of Rsp5p and Sla2p.
LAT-A resistance of rsp5 mutants implicates Rsp5p in actin dynamics.
If Rsp5p functions in actin cytoskeleton dynamics, the actin cytoskeleton organization should be affected by rsp5 mutations. However, this was not evident in previous studies with cells with the mdp1-1/rsp5-17 allele (73). To test the hypothesis further, we studied the effect of different alleles (hect domain and ww mutations) on the actin cytoskeletal organization, but again cells with these mutant rsp5 alleles had a normal actin cytoskeletal organization (data not shown). To assess a possible effect of Rsp5p on the actin cytoskeleton by a different method, we used the drug LAT-A. LAT-A is an actin monomer binding drug, which blocks the polymerization of actin, and the effect of mutations on actin polymerization/depolymerization can be monitored as altered sensitivity or resistance to this drug. For example, cells showing abnormal actin cytoskeleton organization are often hypersensitive to LAT-A (2, 39, 60). Interestingly, sla1 and end3 mutants, each of which is synthetically lethal in combination with pan1 alleles, are resistant to LAT-A (2). The responses to LAT-A of wild-type and rsp5-13, rsp5-w1, and rsp5-w3 isogenic mutant strains (MK1, MK2, MK4, and MK5 strains with genomic RSP5 replaced by mutant rsp5 genes) were tested by using the halo assay. The rsp5-13 cells that have a mutant hect domain had the same sensitivity as parental cells. Also, the rsp5-w3 mutation did not influence LAT-A sensitivity. Interestingly, rsp5-w1 mutations cause cells to be resistant to LAT-A, as determined by measuring the radius of inhibition of growth around the drug (6 mM)-saturated filter disk (2.3 mm for rsp5-w1 versus 3.3 mm for wild-type cells [Fig. 9]). One interpretation of the results is that interaction of the Rsp5p WW1 domain with an unknown protein may be important for actin cytoskeleton dynamics. The conclusion that Rsp5p might be involved in actin cytoskeleton dynamics is also supported by the finding that overexpression of RSP5 causes low levels of resistance of wild-type cells to LAT-A (data not shown).
FIG. 9.
The rsp5-w1 mutant is resistant to LAT-A. Halo assay testing the sensitivity of rsp5-13, rsp5-w1, and rsp5-w3 mutant strains to LAT-A is shown. Concentrations of LAT-A were as indicated. The experiment was performed in triplicate, and representative results are shown.
DISCUSSION
Prior to the present studies, only a few observations linked the ubiquitin system to the actin cytoskeleton. An early study with Drosophila showed that arthrin is a ubiquitin molecule attached to actin (3). It was suggested that the yeast actin cytoskeletal proteins Arc15p and Rvs167p could interact with WW domains similar to those in Rsp5p (8). Recently, genetic interactions between yeast rsp5 and mutations affecting actin cytoskeleton were reported (17, 72). In this paper we provide the first data documenting that the Rsp5p ubiquitin ligase functions in the actin cytoskeleton. This conclusion is supported by genetic results as well as by the demonstration that Rsp5p and components of the actin cytoskeleton colocalize and physically interact in wild-type cells and in cells with actin cytoskeletal mutations. This conclusion is also supported by the finding that Rsp5p affects actin cytoskeleton dynamics, since some rsp5 mutants are resistant to LAT-A, a drug that depolymerizes actin filaments.
Our genetic studies support the model that Rsp5p is involved in actin polymerization. First, we document negative genetic interactions between rsp5 and arp2 or sla2 mutations, directly affecting actin polymerization (37, 44). Moreover, an additional copy of RSP5 suppresses arp2-1 and this suppression depends on ubiquitin-ligase activity. In addition, arp2-1 is efficiently suppressed by the overexpression of ubiquitin. Increased expression of RSP5 suppressed not only the growth defects but also the actin cytoskeleton defects and the endocytosis deficiency of the arp2-1 mutant, also linking these two processes. Mutations in WW1 and WW2 domains that putatively disrupt the interactions of Rsp5p with other proteins enhance the suppression of arp2-1, possibly by releasing Rsp5p from other interactions. On the other hand, WW3 of Rsp5p is necessary for arp2-1 suppression.
How do Rsp5p and the Arp2/3 complex interact to promote actin polymerization? The Arp2/3 complex nucleates actin filament assembly and actin filament branching (44). By itself the Arp2/3 complex nucleates actin polymerization poorly. Its activity is enhanced on binding of activating proteins: Bee1p (66), the type I myosins Myo3p and Myo5p (15, 20, 35), Abp1p (22), and Pan1p (12). Overexpression of BEE1, like overexpression of RSP5, suppresses the arp2-1 mutation (36). Also, as with rsp5, there are synthetic negative interactions of arp2 with bee1 (12). However, as assessed by the two-hybrid assay, Bee1p does not interact with Arp2p directly (39). Rather, Bee1p associates with Arp2p probably through interaction with Arc40p (15) or Arc18p components of Arp2/3 (38). Suppression of arp2-Δ by an additional copy of RSP5 indicates that Rsp5p functions in actin cytoskeleton dynamics independently of Arp2p. By analogy to Bee1p, Rsp5p possibly interacts indirectly with Arp2p via binding to Arc15p or another subunit of the Arp2/3 complex (8). Alternatively, Rsp5p could affect one of the Arp2/3 activators (Bee1p, Pan1p, or others) by altering its stability or function or it may be involved in actin depolymerization or a signaling mechanism regulating the actin cytoskeleton. These hypothetical models for the role of Rsp5p in actin dynamics would be consistent with the formation of filamentous actin clumps in arp2 cells with an extra copy of RSP5 at low temperature and the finding that these clumps contain Rsp5p. Similar overexpression of WASP (a protein homologous to Bee1p) in mammalian cells induces the formation of actin clusters that colocalized with overexpressed protein, indicating a role in actin polymerization (56). Thus, we favor the possibility that Rsp5p influences actin polymerization.
Pan1p, an activator of Arp2/3 complex, may couple actin polymerization with endocytosis (12). The temperature-sensitive pan1-4 mutation, which is nonviable in combination with arp2-1 (12), affects the actin cytoskeleton organization and fluid phase and receptor-mediated endocytosis (58). Pan1p binds to Arp2/3 complex via its C-terminal 532 amino acids (12). Pan1p also interacts with End3p involved in endocytosis (59). Genetic studies support the model that End3p competes with Prk1p to regulate Pan1p function (58, 71). end3-Δ can be suppressed by prk1-Δ via bypassing End3p activity (71). By analogy, our genetic studies show that an additional copy of RSP5 suppresses the end3-Δ mutation, supporting the hypothesis that Rsp5p may prevent Prk1p function. This is not by modifying Pan1p, since ubiquitinated forms of Pan1p were not detected in vivo (13; J. Kamínska, unpublished observation). We were also unable to coimmunoprecipitate Pan1p with Rsp5p (Kamínska, unpublished). Thus, Rsp5p may suppress end3 by interaction with another protein(s) that binds Pan1p.
A potential candidate is Sla2p, which binds Pan1p (J. Cope and D. Drubin, personal communication) in the region adjacent to the acidic activation domain and also serves to couple the actin cytoskeleton and endocytosis (27, 65). We show that Sla2p directly or indirectly interacts with Rsp5p. However, Sla2p, in addition to functioning in the actin cytoskeleton dynamics, may also function in translation (45, 74). Thus, although we favor the model that Rsp5p-Sla2p complexes associate with Pan1p and function in actin polymerization, we cannot rule out the alternative that these complexes function in different processes. Our data that Rsp5p and Sla2p localization depends on proper function of the actin cytoskeleton support the first hypothesis.
Rsp5p probably plays multiple roles in the endocytosis pathway. First, fluid-phase endocytosis, for which there is no protein cargo to serve as a substrate for ubiquitin conjugation, is dependent on Rsp5p (72) and requires Rsp5p ubiquitin ligase catalytic activity and the C2, WW1, and WW3 domains (13, 18). Second, deletion of the C2 Rsp5p domain partially stabilized Fur4p even though Fur4p was ubiquitinated (62). This result is consistent with an earlier report showing that Gap1p internalization is impaired in rsp5-ΔC2 cells even though Gap1p is ubiquitinated by Rsp5p lacking the C2 domain (55). However, deletion of the Rsp5p C2 domain has no effect on α-factor internalization by Ste2p (13) Third, internalization of a chimeric Ste2p-Ub protein requires Rsp5p catalytic activity even though Ste2p-Ub possesses ubiquitin (14). The data indicate that there are at least two distinct roles for Rsp5p ubiquitin ligase activity at the internalization step of endocytosis: (i) regulated ubiquitination of cargo proteins and (ii) ubiquitination of a component of the endocytic machinery. Our present data lead us to suggest that the second ubiquitination-dependent process could involve a protein affecting the Arp2/3 complex and subsequent actin cytoskeleton dynamics required for endocytosis. The target protein that is ubiquitinated remains to be identified.
Our results add new complexity to endocytosis, and now we need to better define the mechanisms by which Rsp5p exerts its functions in endocytosis. However, actin cytoskeleton regulation clearly can be added to the list of processes that are controlled by ubiquitination.
Acknowledgments
This work was supported by State Committee for Scientific Research of Poland grant 6P04B02416 to T.˙Z. and by National Science Foundation and National Institutes of Health grants to A.K.H.
We are grateful to H. Riezman, B. Windsor, R. Li, A. Jacobson, M. Kwapisz, R. Haguenauer-Tsapis, and S. Lemmon for strains and plasmids and to D. Drubin for antibodies. We also thank D. Drubin for communicating unpublished results.
REFERENCES
- 1.Adams, A. E., and J. R. Pringle. 1991. Staining of actin with fluorochrome-conjugated phalloidin. Methods Enzymol. 194:729-731. [DOI] [PubMed] [Google Scholar]
- 2.Ayscough, K. R., J. Stryker, N. Pokala, M. Sanders, P. Crews, and D. G. Drubin. 1997. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J. Cell Biol. 137:399-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ball, E., C. C. Karlik, C. J. Beall, D. L. Saville, J. C. Sparrow, B. Bullard, and E. A. Fyrberg. 1987. Arthrin, a myofibrillar protein of insect flight muscle, is an actin-ubiquitin conjugate. Cell 51:221-228. [DOI] [PubMed] [Google Scholar]
- 4.Beck, T., A. Schmidt, and M. N. Hall. 1999. Starvation induces vacuolar targeting and degradation of the tryptophan permease in yeast. J. Cell Biol. 146:1227-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belmont, L. D., and D. G. Drubin. 1998. The yeast V159N actin mutant reveals roles for actin dynamics in vivo. J. Cell Biol. 142:1289-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bénédetti, H., S. Raths, F. Crausaz, and H. Riezman. 1994. The END3 gene encodes a protein that is required for the internalization step of endocytosis and for actin cytoskeleton organization in yeast. Mol. Biol. Cell 5:1023-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boguta, M., L. A. Hunter, W. C. Shen, E. C. Gillman, N. C. Martin, and A. K. Hopper. 1994. Subcellular locations of MOD5 proteins: mapping of sequences sufficient for targeting to mitochondria and demonstration that mitochondrial and nuclear isoforms commingle in the cytosol. Mol. Cell. Biol. 14:2298-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, A., S. Cheang, X. Espanel, and M. Sudol. 2000. Rsp5 WW domains interact directly with the carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 275:20562-20571. [DOI] [PubMed] [Google Scholar]
- 9.Chen, D. C., B. C. Yang, and T. T. Kuo. 1992. One-step transformation of yeast in stationary phase. Curr. Genet. 21:83-84. [DOI] [PubMed] [Google Scholar]
- 10.Cope, M. J., S. Yang, C. Shang, and D. G. Drubin. 1999. Novel protein kinases Ark1p and Prk1p associate with and regulate the cortical actin cytoskeleton in budding yeast. J. Cell Biol. 144:1203-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de la Fuente, N., A. M. Maldonado, and F. Portillo. 1997. Glucose activation of the yeast plasma membrane H+-ATPase requires the ubiquitin-proteasome proteolytic pathway. FEBS Lett. 411:308-312. [DOI] [PubMed] [Google Scholar]
- 12.Duncan, M. C., M. J. Cope, B. L. Goode, B. Wendland, and D. G. Drubin. 2001. Yeast Eps15-like endocytic protein, Pan1p, activates the Arp2/3 complex. Nat. Cell Biol. 3:687-690. [DOI] [PubMed] [Google Scholar]
- 13.Dunn, R., and L. Hicke. 2001. Domains of the Rsp5 ubiquitin-protein ligase required for receptor-mediated and fluid-phase endocytosis. Mol. Biol. Cell 12:421-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunn, R., and L. Hicke. 2001. Multiple roles for Rsp5p-dependent ubiquitination at the internalization step of endocytosis. J. Biol. Chem. 276:25974-25981. [DOI] [PubMed] [Google Scholar]
- 15.Evangelista, M., B. M. Klebl, A. H. Tong, B. A. Webb, T. Leeuw, E. Leberer, M. Whiteway, D. Y. Thomas, and C. Boone. 2000. A role for myosin-I in actin assembly through interactions with Vrp1p, Bee1p, and the Arp2/3 complex. J. Cell Biol. 148:353-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fisk, H. A., and M. P. Yaffe. 1999. A role for ubiquitination in mitochondrial inheritance in Saccharomyces cerevisiae. J. Cell Biol. 145:1199-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gagny, B., A. Wiederkehr, P. Dumoulin, B. Winsor, H. Riezman, and R. Haguenauer-Tsapis. 2000. A novel EH domain protein of Saccharomyces cerevisiae, Ede1p, involved in endocytosis. J. Cell Sci. 113:3309-3319. [DOI] [PubMed] [Google Scholar]
- 18.Gajewska, B., J. Kamínska, A. Jesionowska, N. C. Martin, A. K. Hopper, and T. ˙Zołądek. 2001. WW domains of Rsp5p define different functions: determination of roles in fluid phase and uracil permease endocytosis in Saccharomyces cerevisiae. Genetics 157:91-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galan, J. M., V. Moreau, B. André, C. Volland, and R. Haguenauer-Tsapis. 1996. Ubiquitination mediated by the Npi1p/Rsp5p ubiquitin-protein ligase is required for endocytosis of the yeast uracil permease. J. Biol. Chem. 271:10946-10952. [DOI] [PubMed] [Google Scholar]
- 20.Geli, M. I., R. Lombardi, B. Schmelzl, and H. Riezman. 2000. An intact SH3 domain is required for myosin I-induced actin polymerization. EMBO J. 19:4281-4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geli, M. I., and H. Riezman. 1998. Endocytic internalization in yeast and animal cells: similar and different. J. Cell Sci. 111:1031-1037. [DOI] [PubMed] [Google Scholar]
- 22.Goode, B. L., A. A. Rodal, G. Barnes, and D. G. Drubin. 2001. Activation of the Arp2/3 complex by the actin filament binding protein Abp1p. J. Cell Biol. 153:627-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guarente, L. 1993. Synthetic enhancement in gene interaction: a genetic tool come of age. Trends Genet. 9:362-366. [DOI] [PubMed] [Google Scholar]
- 24.Harvey, K. F., and S. Kumar. 1999. Nedd4-like proteins: an emerging family of ubiquitin-protein ligases implicated in diverse cellular functions. Trends Cell Biol. 9:166-169. [DOI] [PubMed] [Google Scholar]
- 25.Hein, C., J. Y. Springael, C. Volland, R. Haguenauer-Tsapis, and B. André. 1995. NPl1, an essential yeast gene involved in induced degradation of Gap1 and Fur4 permeases, encodes the Rsp5 ubiquitin-protein ligase. Mol. Microbiol. 18:77-87. [DOI] [PubMed] [Google Scholar]
- 26.Helliwell, S. B., S. Losko, and C. A. Kaiser. 2001. Components of a ubiquitin ligase complex specify polyubiquitination and intracellular trafficking of the general amino acid permease. J. Cell Biol. 153:649-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holtzman, D. A., S. Yang, and D. G. Drubin. 1993. Synthetic-lethal interactions identify two novel genes, SLA1 and SLA2, that control membrane cytoskeleton assembly in Saccharomyces cerevisiae. J. Cell Biol. 122:635-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoppe, T., K. Matuschewski, M. Rape, S. Schlenker, H. D. Ulrich, and S. Jentsch. 2000. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell 102:577-586. [DOI] [PubMed] [Google Scholar]
- 29.Huang, M. E., J. L. Souciet, J. C. Chuat, and F. Galibert. 1996. Identification of ACT4, a novel essential actin-related gene in the yeast Saccharomyces cerevisiae. Yeast 12:839-848. [DOI] [PubMed] [Google Scholar]
- 30.Huibregtse, J. M., M. Scheffner, S. Beaudenon, and P. M. Howley. 1995. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc. Natl. Acad. Sci. USA 92:5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iida, K., and I. Yahara. 1999. Cooperation of two actin-binding proteins, cofilin and Aip1, in Saccharomyces cerevisiae. Genes Cells 4:21-32. [DOI] [PubMed] [Google Scholar]
- 32.Jeng, R. L., and M. D. Welch. 2001. Cytoskeleton: actin and endocytosis—no longer the weakest link. Curr. Biol. 11:R691-R694. [DOI] [PubMed] [Google Scholar]
- 33.Kamínska, J., A. Tobiasz, M. Gniewosz, and T. ˙Zołądek. 2000. The growth of mdp1/rsp5 mutants of Saccharomyces cerevisiae is affected by mutations in the ATP-binding domain of the plasma membrane H+-ATPase. Gene 242:133-140. [DOI] [PubMed] [Google Scholar]
- 34.Lappalainen, P., and D. G. Drubin. 1997. Cofilin promotes rapid actin filament turnover in vivo. Nature 388:78-82. [DOI] [PubMed] [Google Scholar]
- 35.Lechler, T., A. Shevchenko, and R. Li. 2000. Direct involvement of yeast type I myosins in Cdc42-dependent actin polymerization. J. Cell Biol. 148:363-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li, R. 1997. Bee1, a yeast protein with homology to Wiskott-Aldrich syndrome protein, is critical for the assembly of cortical actin cytoskeleton. J. Cell Biol. 136:649-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li, R., Y. Zheng, and D. G. Drubin. 1995. Regulation of cortical actin cytoskeleton assembly during polarized cell growth in budding yeast. J. Cell Biol. 128:599-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Machesky, L. M., and R. H. Insall. 1998. Scar and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr. Biol. 8:1347-1356. [DOI] [PubMed] [Google Scholar]
- 39.Madania, A., P. Dumoulin, S. Grava, H. Kitamoto, C. Scharer-Brodbeck, A. Soulard, V. Moreau, and B. Winsor. 1999. The Saccharomyces cerevisiae homologue of human Wiskott-Aldrich syndrome protein Las17p interacts with the Arp2/3 complex. Mol. Biol. Cell 10:3521-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCann, R. O., and S. W. Craig. 1997. The I/LWEQ module: a conserved sequence that signifies F-actin binding in functionally diverse proteins from yeast to mammals. Proc. Natl. Acad. Sci. USA 94:5679-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moreau, V., J. M. Galan, G. Devilliers, R. Haguenauer-Tsapis, and B. Winsor. 1997. The yeast actin-related protein Arp2p is required for the internalization step of endocytosis. Mol. Biol. Cell 8:1361-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moreau, V., A. Madania, R. P. Martin, and B. Winson. 1996. The Saccharomyces cerevisiae actin-related protein Arp2 is involved in the actin cytoskeleton. J. Cell Biol. 134:117-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mulholland, J., D. Preuss, A. Moon, A. Wong, D. Drubin, and D. Botstein. 1994. Ultrastructure of the yeast actin cytoskeleton and its association with the plasma membrane. J. Cell Biol. 125:381-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mullins, R. D., J. A. Heuser, and T. D. Pollard. 1998. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. USA 95:6181-6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palecek, J., J. Hasek, and H. Ruis. 2001. Rpg1p/Tif32p, a subunit of translation initiation factor 3, interacts with actin-associated protein Sla2p. Biochem. Biophys. Res. Commun. 282:1244-1250. [DOI] [PubMed] [Google Scholar]
- 46.Pringle, J. R., A. E. Adams, D. G. Drubin, and B. K. Haarer. 1991. Immunofluorescence methods for yeast. Methods Enzymol. 194:565-602. [DOI] [PubMed] [Google Scholar]
- 47.Pruyne, D., and A. Bretscher. 2000. Polarization of cell growth in yeast. J. Cell Sci. 113:571-585. [DOI] [PubMed] [Google Scholar]
- 48.Qualmann, B., M. M. Kessels, and R. B. Kelly. 2000. Molecular links between endocytosis and the actin cytoskeleton. J. Cell Biol. 150:F111-F116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raths, S., J. Rohrer, F. Crausaz, and H. Riezman. 1993. end3 and end4: two mutants defective in receptor-mediated and fluid phase endocytosis in Saccharomyces cerevisiae. J. Cell Biol. 120:55-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roth, A. F., and N. G. Davis. 2000. Ubiquitination of the PEST-like endocytosis signal of the yeast a-factor receptor. J. Biol. Chem. 275:8143-8153. [DOI] [PubMed] [Google Scholar]
- 51.Rotin, D., O. Staub, and R. Haguenauer-Tsapis. 2000. Ubiquitination and endocytosis of plasma membrane proteins: role of Nedd4/Rsp5p family of ubiquitin-protein ligases. J. Membr. Biol. 176:1-17. [DOI] [PubMed] [Google Scholar]
- 52.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 53.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 54.Soetens, O., J. O. De Craene, and B. André. 2001. Ubiquitin is required for sorting to the vacuole of the yeast general amino acid permease, Gap1. J. Biol. Chem. 276:43949-43957. [DOI] [PubMed] [Google Scholar]
- 55.Springael, J. Y., J. O. De Craene, and B. André. 1999. The yeast Npi1/Rsp5 ubiquitin ligase lacking its N-terminal C2 domain is competent for ubiquitination but not for subsequent endocytosis of the Gap1 permease. Biochem. Biophys. Res. Commun. 257:561-566. [DOI] [PubMed] [Google Scholar]
- 56.Symons, M., J. M. Derry, B. Karlak, S. Jiang, V. Lemahieu, F. Mccormick, U. Francke, and A. Abo. 1996. Wiskott-Aldrich syndrome protein, a novel effector for the GTPase CDC42Hs, is implicated in actin polymerization. Cell 84:723-734. [DOI] [PubMed] [Google Scholar]
- 57.Tang, H. Y., and M. Cai. 1996. The EH-domain-containing protein Pan1 is required for normal organization of the actin cytoskeleton in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:4897-4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tang, H. Y., A. Munn, and M. Cai. 1997. EH domain proteins Pan1p and End3p are components of a complex that plays a dual role in organization of the cortical actin cytoskeleton and endocytosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:4294-4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tang, H. Y., J. Xu, and M. Cai. 2000. Pan1p, End3p, and S1a1p, three yeast proteins required for normal cortical actin cytoskeleton organization, associate with each other and play essential roles in cell wall morphogenesis. Mol. Cell. Biol. 20:12-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vaduva, G., N. C. Martin, and A. K. Hopper. 1997. Actin-binding verprolin is a polarity development protein required for the morphogenesis and function of the yeast actin cytoskeleton. J. Cell Biol. 139:1821-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Volland, C., D. Urban-Grimal, G. Geraud, and R. Haguenauer-Tsapis. 1994. Endocytosis and degradation of the yeast uracil permease under adverse conditions. J. Biol. Chem. 269:9833-9841. [PubMed] [Google Scholar]
- 62.Wang, G., J. M. McCaffery, B. Wendland, S. Dupre, R. Haguenauer-Tsapis, and J. M. Huibregtse. 2001. Localization of the Rsp5p ubiquitin-protein ligase at multiple sites within the endocytic pathway. Mol. Cell. Biol. 21:3564-3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wendland, B., and S. D. Emr. 1998. Pan1, yeast eps15, functions as a multivalent adaptor that coordinates protein-protein interactions essential for endocytosis. J. Cell Biol. 141:71-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wendland, B., K. E. Steece, and S. D. Emr. 1999. Yeast epsins contain an essential N-terminal ENTH domain, bind clathrin and are required for endocytosis. EMBO J. 18:4383-4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wesp, A., L. Hicke, J. Palecek, R. Lombardi, T. Aust, A. L. Munn, and H. Riezman. 1997. End4p/Sla2p interacts with actin-associated proteins for endocytosis in Saccharomyces cerevisiae. Mol. Biol. Cell 8:2291-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Winter, D., T. Lechler, and R. Li. 1999. Activation of the yeast Arp2/3 complex by Bee1p, a WASP-family protein. Curr. Biol. 9:501-504. [DOI] [PubMed] [Google Scholar]
- 67.Winter, D. C., E. Y. Choe, and R. Li. 1999. Genetic dissection of the budding yeast Arp2/3 complex: a comparison of the in vivo and structural roles of individual subunits. Proc. Natl. Acad. Sci. USA 96:7288-7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wolfe, D., T. Reiner, J. L. Keeley, M. Pizzini, and R. L. Keil. 1999. Ubiquitin metabolism affects cellular response to volatile anesthetics in yeast. Mol. Cell. Biol. 19:8254-8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang, S., M. J. Cope, and D. G. Drubin. 1999. Sla2p is associated with the yeast cortical actin cytoskeleton via redundant localization signals. Mol. Biol. Cell 10:2265-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yashiroda, H., T. Oguchi, Y. Yasuda, E. Toh, and Y. Kikuchi. 1996. Bul1, a new protein that binds to the Rsp5 ubiquitin ligase in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:3255-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zeng, G., and M. Cai. 1999. Regulation of the actin cytoskeleton organization in yeast by a novel serine/threonine kinase Prk1p. J. Cell Biol. 144:71-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.˙Zołądek, T., A. Tobiasz, G. Vaduva, M. Boguta, N. C. Martin, and A. K. Hopper. 1997. MDP1, a Saccharomyces cerevisiae gene involved in mitochondrial/cytoplasmic protein distribution, is identical to the ubiquitin-protein ligase gene RSP5. Genetics 145:595-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.˙Zołądek, T., G. Vaduva, L. A. Hunter, M. Boguta, B. D. Go, N. C. Martin, and A. K. Hopper. 1995. Mutations altering the mitochondrial-cytoplasmic distribution of Mod5p implicate the actin cytoskeleton and mRNA 3′ ends and/or protein synthesis in mitochondrial delivery. Mol. Cell. Biol. 15:6884-6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zuk, D., J. P. Belk, and A. Jacobson. 1999. Temperature-sensitive mutations in the Saccharomyces cerevisiae MRT4, GRC5, SLA2 and THS1 genes result in defects in mRNA turnover. Genetics 153:35-47. [DOI] [PMC free article] [PubMed] [Google Scholar]